Host miRNAs have been reported to regulate JEV-induced inflammation in the CNS. We found that JEV infection could reduce expression of host miRNA. The helicase region of the NS3 protein bound specifically to miRNA precursors and could lead to incorrect unwinding of miRNA precursors, thereby reducing the expression of mature miRNAs. This observation led to two major findings. First, our results suggested that JEV NS3 protein induced miR-466d-3p degradation, which promoted IL-1β expression and JEV replication. Second, arginine molecules on NS3 were the main miRNA-binding sites, because we demonstrated that miRNA degradation was abolished if arginines at R226 and R202 were mutated. Our study provides new insights into the molecular mechanism of JEV and reveals several amino acid sites that could be mutated for a JEV vaccine.
KEYWORDS: Japanese encephalitis virus, microRNA, nonstructural protein 3, RNA-protein interactions
ABSTRACT
Japanese encephalitis virus (JEV) infection alters microRNA (miRNA) expression in the central nervous system (CNS). However, the mechanism contributing to miRNA regulation in the CNS is not known. We discovered global degradation of mature miRNA in mouse brains and neuroblastoma (NA) cells after JEV infection. Integrative analysis of miRNAs and mRNAs suggested that several significantly downregulated miRNAs and their targeted mRNAs were clustered into an inflammation pathway. Transfection with miRNA 466d-3p (miR-466d-3p) decreased interleukin-1β (IL-1β) expression and inhibited JEV replication in NA cells. However, miR-466d-3p expression increased after JEV infection in the presence of cycloheximide, indicating that viral protein expression reduced miR-466d-3p expression. We generated all the JEV coding proteins and demonstrated NS3 helicase protein to be a potent miRNA suppressor. The NS3 proteins of Zika virus, West Nile virus, and dengue virus serotype 1 (DENV-1) and DENV-2 also decreased miR-466d-3p expression. Results from helicase-blocking assays and in vitro unwinding assays demonstrated that NS3 could unwind pre-miR-466d and induce miRNA dysfunction. Computational models and an RNA immunoprecipitation assay revealed arginine-rich domains of NS3 to be crucial for pre-miRNA binding and degradation of host miRNAs. Importantly, site-directed mutagenesis of conserved residues in NS3 revealed that R226G and R202W reduced the binding affinity and degradation of pre-miR-466d. These results expand the function of flavivirus helicases beyond unwinding duplex RNA to degrade pre-miRNAs. Hence, we revealed a new mechanism for NS3 in regulating miRNA pathways and promoting neuroinflammation.
IMPORTANCE Host miRNAs have been reported to regulate JEV-induced inflammation in the CNS. We found that JEV infection could reduce expression of host miRNA. The helicase region of the NS3 protein bound specifically to miRNA precursors and could lead to incorrect unwinding of miRNA precursors, thereby reducing the expression of mature miRNAs. This observation led to two major findings. First, our results suggested that JEV NS3 protein induced miR-466d-3p degradation, which promoted IL-1β expression and JEV replication. Second, arginine molecules on NS3 were the main miRNA-binding sites, because we demonstrated that miRNA degradation was abolished if arginines at R226 and R202 were mutated. Our study provides new insights into the molecular mechanism of JEV and reveals several amino acid sites that could be mutated for a JEV vaccine.
INTRODUCTION
Japanese encephalitis virus (JEV) is a single-strand, positive-sense RNA virus belonging to the genus Flavivirus of the family Flaviviridae. Its genome encodes three structural proteins in the order envelope (E), capsid (C), and premembrane (prM) and seven nonstructural (NS) proteins: NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5. During the JEV life cycle in cells, the RNA genome replicase complex (of which NS3 and NS5 are major components required for genome replication and capping) initiates replication (1). The JEV polyprotein NS3 belongs to the superfamily 2 helicases, which have protease, nucleoside triphosphatase, and ATP-dependent RNA helicase activities and unwind double-stranded genomes during viral replication (2).
JEV is a neurotropic virus causing neuroinflammation and neuronal damage that results in Japanese encephalitis (JE) (3). Even though vaccines are available to control JEV, ∼67,900 cases of JE are reported worldwide each year, half of which occur in mainland China (4). Due to a lack of efficacious antiviral drugs against JEV, the fatality rate of JE is about 25 to 32%, and 50% of survivors suffer from neuropsychiatric sequelae (5). JEV causes neuroinflammation and neuronal damage in mammalian hosts by modulating cytokine/chemokine production (6) and causing the activation and migration of neuroglial cells (7, 8). JEV infection increases the production of inflammatory cytokines and microglial activity, which facilitates viral pathogenesis and dissemination of the virus into the central nervous system (CNS) (9, 10). Most studies on the innate immune response in the CNS have focused on the molecular components of JEV recognition and signaling regulators. Recent studies have revealed that Toll-like receptor 7 (TLR7) in neurons and TLR3 in microglial cells recognize JEV (11, 12). Following JEV recognition, the adapter proteins of TLRs activate myeloid differentiation primary response 88. Furthermore, JEV infection triggers protein kinases, such as kinase B, phosphoinositide 3-kinase, p38 mitogen-activated protein kinase, and signal-regulated kinase (13). However, the involvement of JEV components in modulating the inflammatory response is not known.
Several recent studies have shown that during infection with Zika virus, NS5 of Zika virus facilitates inflammasomes to induce interleukin-1β (IL-1β) secretion (14). NS3 of JEV interacts with the Src protein tyrosine kinase to promote secretion of inflammatory cytokines (15). NS1 of dengue virus (DENV) has been reported to interact with signal transducer and activator of transcription 3 to enhance the secretion of tumor necrosis factor alpha (TNF-α) and IL-6 (13). Hence, the NS proteins of the Flaviviridae play an important part in the inflammatory response. However, the specific signaling pathway and target sites of the NS proteins of JEV are not known.
MicroRNAs (miRNAs) are small noncoding RNAs containing ∼22 nucleotides (nt) that control posttranscriptional gene regulation by targeting mRNAs. In animal cells, following transcription by RNA polymerase II (RNA Pol II), nuclear RNA polymerase III (RNA Pol III) Drosha processes the primary transcripts of miRNAs (pri-miRNAs) into precursors 60 to 100 nt in size (pre-miRNAs). Then, the pre-miRNAs are shuttled into the cytoplasm and processed further by RNA Pol II Dicer into double-stranded RNA (dsRNA) products ∼22 nt in size containing the mature miRNA “guide” strand and the “passenger” miRNA strand. The mature miRNA is loaded onto the RNA-induced silencing complex (RISC) and binds 3′ untranslated regions (UTRs) of the target mRNA, which leads to translation repression and/or mRNA degradation.
Through the repression of a target, miRNA modulates a broad range of gene expression programs during development, immunoreactions, and pathogen infection. Accumulating evidence also suggests that an abundance of cellular miRNAs are involved in multiple stages of the JEV replication cycle. For instance, JEV infection induces miR-155, miR-15b, miR-19b, and miR-29b and activates the innate immune response (16–19). JEV infection also reduces expression of miR-33a and miR-432 to facilitate virus replication (20, 21). Therefore, regulation of miRNA expression is important for cellular biological processes and mRNA homeostasis during JEV infection.
Unlike the biosynthesis of miRNA, under physiological or pathological conditions, some miRNA decays rapidly. Most studies on miRNA degradation have demonstrated the mechanisms involved to be sequence organization, RNase activity, transcription rates, and energy metabolism. For example, several 3′ → 5′ or 5′ → 3′ miRNA-degrading ribonucleases degrade different miRNAs in various cell types (22, 23). A sequence target-dependent mechanism has also been identified, in which cleavage is mediated by miRNA and high complementary interaction, such as viral noncoding transcripts (24, 25), miRNAs, and miRNA hybrids (26), or the extent of sequence complementarity (27). Although miRNA function during viral replication is well defined, the turnover dynamics of miRNA and the mechanisms involved are poorly defined.
Numerous studies have shown that viruses regulate host miRNA to control cellular protein expression, suggesting a critical role for host miRNAs in immune evasion and the viral replication cycle. DNA and RNA viruses have developed several mechanisms to degrade or promote cellular miRNA to benefit viral infection and replication. Specifically, herpesvirus saimiri (HVS) and murine cytomegalovirus (MCMV) degrade host miR-27 directly using a virus-encoded noncoding RNA (ncRNA) containing a miR-27 sequence-specific binding site (24, 28). Similarly, human cytomegalovirus (HCMV) decreases expression of mature miR-17 and miR-20a using a 15-kb miRNA decay element in the UL/b′ region of the viral genome (29). Conversely, vaccinia virus (VACV) degrades host miRNA by adding tails using viral poly(A) polymerase (30). Kaposi’s sarcoma-associated herpesvirus represses MCPIP1 (monocyte chemotactic protein-induced protein 1) expression to facilitate expression of viral miRNA (31).
The direct interaction between host miRNAs and viral components is poorly understood. There are two approaches to predicting RNA-protein interactions (RPIs). One cost-effective approach is a computational method that could predict RPIs and protein-RNA-binding sites. Another method, RNA-protein immunoprecipitation, provides a specific and accurate predication of RPIs. A combination of these two approaches is especially valuable for mapping miRNA-protein binding sites and characterizing the biologic function of viral components in viral replication.
In the present study, we demonstrated that JEV NS3 disrupts cellular pre-miRNA directly and reduces levels of mature miRNA. Through computational and RNA immunoprecipitation (RIP) analyses, we showed that binding and turnover of miRNA by NS3 were dependent upon the arginine-rich domains of NS3: R226G and R202W. Also, NS3-mediated miRNA degradation was critical in the inflammation-related pathway, which may promote an irreversible inflammatory response leading to neuron death. The present study provides insights into the role of flavivirus helicase in the regulation of host miRNA turnover.
RESULTS
JEV mediates downregulation of global miRNA in the mouse nervous system.
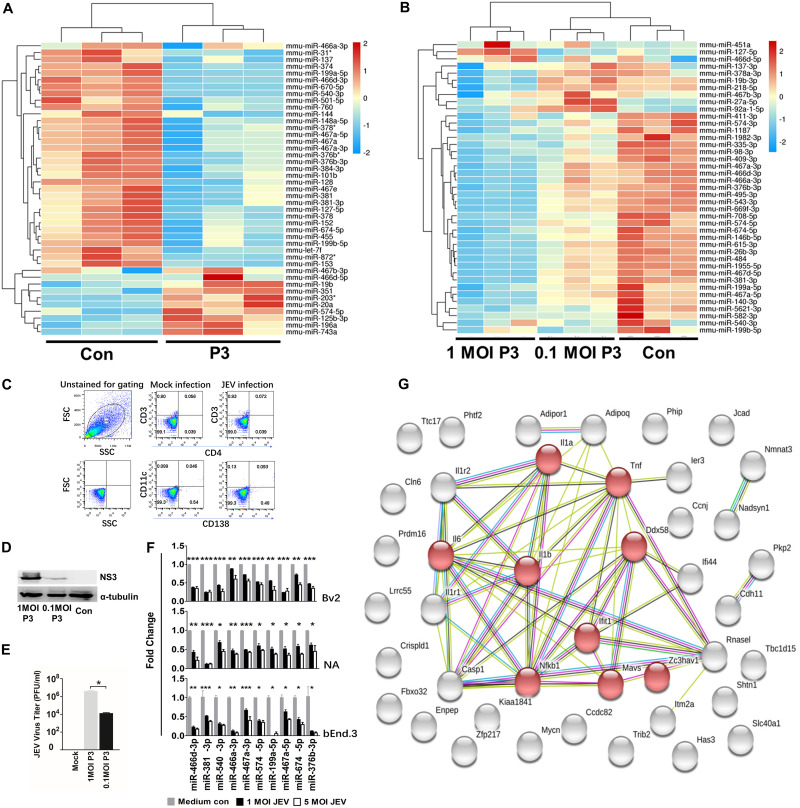
We used miRNA microarrays to assess host miRNA expression profiles in mouse brains infected by pathogenic JEV (strain P3). The miRNA expression profiles showed that JEV infection altered an abundance of host miRNAs (Fig. 1A). Notably, expression of 33 out of the 41 miRNAs (P < 0.05; change > 2.0-fold) in mouse brains was downregulated during JEV infection. We also used small-RNA deep-sequencing analysis on miRNA expression of P3-infected neuroblastoma (NA) cells at multiplicities of infection (MOI) of 1 and 0.1 for 48 h (Fig. 1B). The results confirmed that JEV infection led to a global decrease in host miRNA. We used flow cytometry to determine the change in the number of dendritic cells (DCs) (cluster of differentiation 11c [CD11c]), T cells (CD3 and CD4) and plasma cells (CD138) in the brain after JEV infection but found no significant change in the proportion of DCs, T cells, or plasma cells (Fig. 1C). Other cell types made up a minority of the total cell population. Furthermore, the virus titer and NS3 expression of cells infected with P3 at an MOI of 1 was higher than that for cells infected with P3 at an MOI of 0.1; infection at an MOI of 1 decreased miRNA expression significantly more than infection at an MOI of 0.1, suggesting that the JEV-induced global miRNA decrease was related to JEV replication (Fig. 1D and E). Quantitative reverse transcription-PCR (qRT-PCR) in JEV-infected NA, BV2, and bEnd.3 cells confirmed the miRNA profile data (Fig. 1F). To predict precisely the target mRNA, we also measured mRNA expression in mouse brains after JEV infection (see “Data availability” below). Thirty-three miRNAs with downregulated expression and 580 mRNAs with upregulated expression (P < 0.05; change > 2.0-fold) from miRNA and mRNA microarray data sets were used to predict the major targets of miRNAs with downregulated expression using integrative analysis. Of these, 42 significant miRNA-mRNA pairs were identified, consisting of 32 miRNAs and 42 mRNAs. Gene ontology (GO) analysis revealed that most of the target genes were enriched in terms of inflammation-based signaling processes. Forty-two interacting proteins with 177 interactions were retrieved from the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database, and eight proteins were segregated to one subgroup, which was related to the immune process and the inflammation process. Of these eight proteins, IL-1β expression was the most significantly upregulated and was in the middle of the protein interaction network (Fig. 1G). Furthermore, IL-1β was a target of miR-466d-3p, which was also significantly downregulated according to analysis by miRNA microarray and RNA deep sequencing.
FIG 1.
JEV infection downregulates global miRNA expression in the mouse nervous system. (A) Changes in miRNA expression upon JEV infection of mouse brains. Four-week-old BALB/c mice were infected with 103 PFU of P3 or medium control (Con). After 9 days of infection, miRNA expression of JEV-infected mice was compared with that from the noninfected control. The color scale is based on log2 changes in expression. (B) Changes in miRNA expression upon JEV infection of NA cells. NA cells were infected with P3 or medium control at an MOI of 0.1 and an MOI of 0.01. After 48 h of infection, miRNA expression of JEV-infected cells was compared with that for the noninfected control. The color scale is based on log2 changes in expression. (C) Fluorescence-activated cell sorter (FACS) analysis of the cell population in mouse brains. Four-week-old mice were infected with 103 FFU of JEV or mock infected for 5 days intracerebrally. Brain cells were analyzed by FACS analysis with the indicated antibodies. FSC, forward scatter; SSC, side scatter. (D) Western blot of NA cells infected with P3 or medium control at an MOI of 1 and an MOI of 0.1 after 48 h. (E) Virus titers of NA cells infected with P3 at an MOI of 1 and an MOI of 0.1 after 48 h. (F) qRT-PCR analyses of mature miRNA levels in BV2, NA, and bEnd.3 cells after 48 h of JEV infection (at an MOI of 1 and an MOI of 5). The results are shown as means and standard deviations (SD). (G) A protein interaction network was constructed by STRING based on the candidate genes (42 genes) of common mRNA from the miRNA and mRNA expression profile. The light-blue lines indicate interactions from curated databases. The purple lines denote experimentally validated interactions. The green lines show predicted interactions from gene neighborhoods. The red lines indicate predicted interactions from gene fusions. The dark-blue lines show predicted interactions from gene cooccurrence. The yellow lines represent interactions from text mining. The light-purple lines denote interactions from protein homology. The black lines show interactions from coexpression. The network is divided into many groups according to the biological process (GO). The red nodes in the network represent the genes that had a role in the immune and inflammatory process. The average number of connections per node is 2.81 (protein-protein interaction enrichment; P < 1.015). Significance was assessed using Student's t test; *, P ≤ 0.05.
JEV infection induces incorrect processing of host miRNAs.
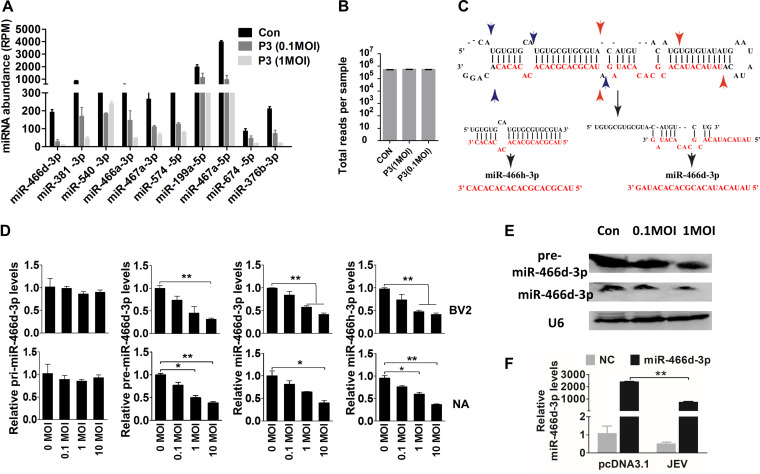
Sequence-specific reads were determined by deep sequencing of the nt 18 to 24 fraction and analyzed by the miRDeep2 module using reads per million (RPM) (Fig. 2A and B) (32). The number of reads for each miRNA was decreased significantly, but the total numbers for each sample were similar, indicating that the numbers of reads obtained per sample decreased as the number of samples in a multiplex increased. Furthermore, expression of some miRNAs from the same precursor were all downregulated. For example, expression of miR-466h-3p and miR-466d-3p generated from the miR-466d-3p precursor (Fig. 2C) was downregulated significantly in NA cells (Fig. 2D). Notably, expression of pre-miR-466d-3p and mature miR-466d-3p was reduced in a dose-dependent manner, but expression of pri-miR-466d-3p appeared unmodified. Expression of pre-miR-466d-3p and mature miR-466d-3p was also confirmed by Northern blotting using digoxigenin (DIG)-labeled probes (Fig. 2E). JEV significantly decreased the abundance of miR-466d-3p derived from an exogenously constructed miRNA mimic compared with the medium control in NA cells (Fig. 2F). Therefore, these experiments indicated that JEV infection decreased expression of pre-miRNA to downregulate miR-466d-3p expression.
FIG 2.
JEV infection induces incorrect processing of host miRNAs. (A and B) Sequencing of multiplexed miRNA after JEV infection. (A) Read numbers of miRNAs in NA cells infected with JEV at an MOI of 0.1 or 1 or mock infected. (B) Total numbers of reads per sample. (C) Schematic of pre-miR-466d-3p processing and types of mature miRNAs (miR-466d-3p and miR-466h-3p). The black arrows indicate the processing direction of miRNA, the blue arrowheads represent miR-466h-3p located in pre-miR-466d, and the red arrowheads represent miR-466d-3p located in pre-miR-466d. (D) Expression of mature miR-466d-3p, miR-466h-3p, pre-miR-466d-3p, or pri-miR-466d-3p using qRT-PCR in NA or BV2 cells infected with P3 at the indicated MOI after 48 h of infection. (E) Detection of miRNA by Northern blotting. Total RNA was isolated with RNAiso Plus and detected with a DIG-labeled probe. (F) Quantification of degradation of exogenous miR-466d-3p mimic in NA cells by qRT-PCR. NA cells were infected with P3 at an MOI of 0.1 and transfected with miR-466d-3p at 36 hpi. After 48 h of infection, the total RNA from NA cells was quantified. For all graphs, results are shown as means and SD. Significance was assessed using Student's t test. *, P ≤ 0.05; **, P ≤ 0.01.
JEV NS3 mediates miR-466d-3p degradation in neurons.
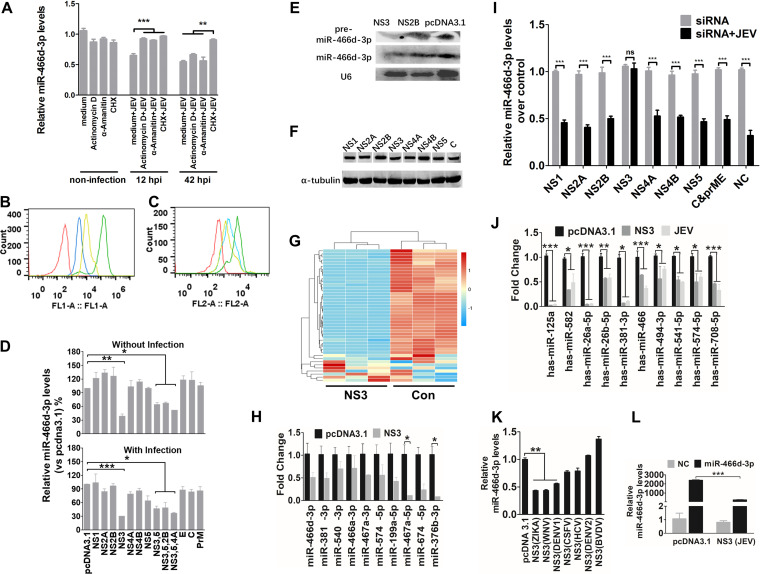
Expression of the NS protein or noncoding subgenomic RNA of a virus is required for viral pathogenicity, which affects miRNA processing in the host (33). JEV infection of neurons results in the release of viral genomic RNA, followed by translation of viral protein. The transcription inhibitor actinomycin D, α-amanitin, or the translation inhibitor cycloheximide (CHX) was used to identify the viral components induced by miR-466d-3p degradation (34, 35). All early treatments with α-amanitin, actinomycin D, and CHX 12 h postinfection (hpi) severely blocked JEV replication and inhibited miR-466d-3p degradation in NA cells. In contrast, late treatment with CHX, but not α-amanitin or actinomycin D, at 42 hpi blocked miR-466d-3p degradation (Fig. 3A). Total RNA biosynthesis in cells treated with α-amanitin and actinomycin D was significantly lower than that in nontreated cells (Fig. 3B and C). Total protein synthesis in cells treated with CHX was significantly lower than that in nontreated cells. These data suggested that the posttranscriptional modification or inappropriate processing of host miRNAs was dependent upon production of viral proteins or viral proteins with host miRNA interaction in mouse neurons. JEV transcription did not suppress host miRNA, so to determine which viral protein was sufficient to confer host miRNA degradation, 10 pcDNA 3.1 vectors encoding each structural protein (E, C, and PrM) and each NS protein (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) of JEV were constructed. miR-466d-3p expression was measured 48 hpi. Only NS3 induced a significant decrease in mature miR-466d-3p expression (40% reduction compared with the pcDNA 3.1 control) (Fig. 3D). Several other NS proteins of JEV associate with NS3 to facilitate RNA assembly and viral replication (36, 37). However, cotransfection of NS2B, NS4A, and NS5 with NS3 did not significantly change miR-466d-3p expression compared with that in the NS3-only transfection group. The effect of NS3 was confirmed further by Northern blotting (Fig. 3E). Expression levels of NS1, NS2A, NS2B, NS3, NS4A, NS4B, NS5, and C protein were similar to that of α-tubulin (Fig. 3F). Moreover, NS3 expression in NA cells induced global downregulation of expression of endogenous miRNAs (Fig. 3G) and the same miRNAs shown in Fig. 1E (as confirmed by qRT-PCR) (Fig. 3H). We further assessed the miR-466d-3p degradation induced by NS3 with small interfering RNA (siRNA). JEV infection and NS3 transfection induced miR-466d-3p degradation, which could be blocked significantly by siRNA targeting NS3 but not siRNA targeting the other proteins of JEV (Fig. 3I). Similarly, JEV infection and NS3 transfection also reduced miRNA expression in SK-N-SH cells (Fig. 3J). JEV NS3 played a critical role in the turnover of host miRNA. Hence, we constructed several pcDNA 3.1 vectors carrying NS3 plasmids of Zika virus, West Nile virus (WNV), DENV serotype 1 (DENV-1), DENV-2, classical swine fever virus (CSFV), bovine viral diarrhea virus (BVDV), and hepatitis C virus (HCV) to identify whether helicases from other flaviviruses could degrade host miRNAs. NS3 from Zika virus, WNV, and DENV-1 decreased miR-466d-3p expression (Fig. 3K). Furthermore, an exogenously constructed miRNA mimic that contained a mature miRNA guide strand and a passage (miRNA*) strand was synthesized. Transfection of the miRNA mimic increased miR-466d-3p expression in NA cells significantly, and NS3 could severely block the miR-466d-3p derived from the miRNA mimic (Fig. 3L).
FIG 3.
NS3 of JEV mediates miR-466d degradation in neurons. (A) Transcription and translation inhibition assay. NA cells were mock treated with medium or infected with P3 at an MOI of 5, and actinomycin D (10 mΜ), α-amanitin (10 mΜ), or CHX (100 nM) was added to the NA cells at 12 hpi or 42 hpi. After 48 h of infection, total RNA from the NA cells was used to quantify the expression of miR-466d-3p. (B and C) FACS analysis of RNA and protein synthesis. (B) NA cells (106 cells/ml) were treated with vehicle or CHX (100 nM) for 30 min at 37°C. Subsequently, the cells were incubated for an additional 30 min with fresh aliquots of medium containing protein label or protein label and CHX. FACS analysis of the negative-control (red line), background (azide only; blue line), positive-control (protein label; green line), and CHX-treated (yellow line) cell populations was performed. (C) NA cells (106 cells/ml) were pretreated with vehicle, α-amanitin (10 mΜ), or actinomycin D (10 mΜ) for 4 h at 37°C. Subsequently, the cells were incubated with EZClick RNA label for 1 h. FACS analysis of negative-control (red line), positive-control (RNA label), α-amanitin-treated (blue line), and actinomycin D-treated (yellow line) cells was done according to the protocol. (D) Relative analyses of miR-466d-3p expression in NA cells transfected with the indicated expression plasmids with and without JEV infection. After 48 h of transfection/infection, the total RNA from NA cells was used to quantify relative expression of miR-466d-3p (versus pcDNA 3.1 control) by qRT-PCR. (E) Detection of miRNA by Northern blotting. Total RNA was isolated with RNAiso Plus and detected with a DIG-labeled probe. (F) Western blotting of expression of NS1, NS2A, NS2B, NS3, NS4A, NS4B, NS5, and C protein. (G) Changes in miRNA expression upon transfection of NS3 in NA cells. NA cells were transfected with NS3 or pcDNA 3.1 control (Con). After 48 h of transfection, miRNA expression of NS3-transfected cells was measured by RNA deep sequencing and compared with that for the pcDNA 3.1-transfected control. The color scale is based on log2 changes in expression. (H) qRT-PCR of levels of mature miRNA in NA cells after 48 h of transfection of NS3. (I) qRT-PCR of miR-466d-3p in NA cells after 48 h of infection with P3 at an MOI of 0.1 and transfection with the indicated siRNAs. (J) qRT-PCR of human miRNA in NA cells after 48 h of transfection with NS3 or infection with P3 at an MOI of 0.1. (K) qRT-PCR of miR-466d-3p in NA cells after 48 h of transfection with NS3 proteins of ZIKA virus, WNV, DENV-1, DENV-2, CSFV, BVDV, and HCV. (L) Quantification of degradation of exogenous miR-466d-3p mimic in NA cells by qRT-PCR. NA cells were transfected with NS3, and after 48 h, the NA cells were transfected with miR-466d-3p mimic. After 54 h of transfection of NS3, the total RNA from the NA cells was used for quantification. For all graphs, results are shown as means and SD. Significance was assessed using Student's t test. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ns, not significant (P > 0.05).
miR-466d-3p blockade enhances IL-1β secretion and promotes JEV replication.
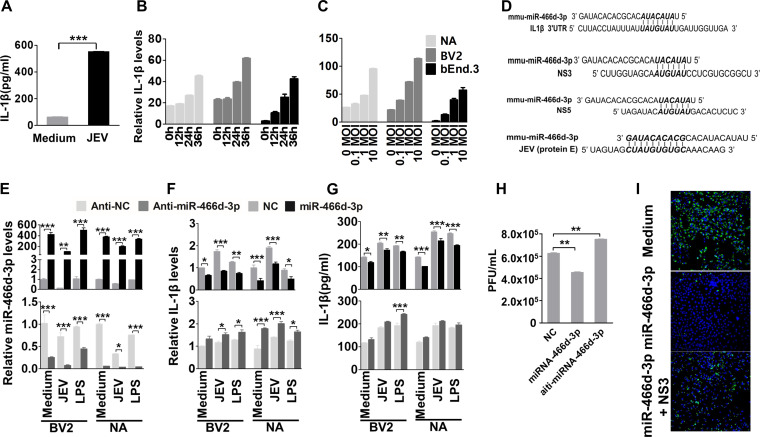
We verified the in silico analysis of miRNA-mRNA interactions using enzyme-linked immunosorbent assays (ELISAs) to measure IL-1β expression in JEV-infected mouse brains and cells. IL-1β expression in mouse brains was enhanced significantly at 9 days postinfection (dpi) (Fig. 4A). IL-1β expression was increased significantly in a dose-dependent and time-dependent manner in NA, BV2, and bEnd.3 cells (Fig. 4B and C).
FIG 4.
miR-466d blockade enhances IL-1β secretion and promotes JEV replication (A) Homogenized mouse brains were collected at 9 dpi, and IL-1β expression in the mouse brains was determined by ELISA. (B and C) Cell supernatants from NA cells were collected at the indicated infection time points and infection doses, and IL-1β expression was determined by qRT-PCR and ELISA. (D) Introduction of miR-466d-3p binding sites in IL-1β, NS3, NS5, and E. miR-466d-3p sequences complementary to the coding sequences of IL-1β, NS3, NS5, and E are indicated in boldface italic. (E to G) The synthetic mimic of miR-466d-3p reduced IL-1β expression and blocked JEV replication, and the inhibitor of miR-466d-3p had the opposite effect. NA or BV2 cells were infected/treated with JEV at an MOI of 0.01, LPS (100 ng/ml), or medium control. After 48 h of infection/treatment, the cells were transfected with a mimic of miR-466d-3p or inhibitor of miR-466d-3p. (E) After 6 h of transfection, total RNA from NA cells was used to quantify the relative expression of miR-466d-3p (versus the negative control) by qRT-PCR. (F) After 6 h of transfection, total RNA from NA cells was used to quantify the relative expression of IL-1β (versus the negative control) by qRT-PCR. (G) After 6 h of transfection, cell supernatants were collected to quantify IL-1β expression by ELISA. (H) After 24 h of transfection, cell supernatants were collected to determine the JEV titer. (I) miR-466d target site-fused GFP was cotransfected with NS3 and/or miR-466d-3p mimic, and after 48 h of transfection, the cells were stained with DAPI (4′,6-diamidino-2-phenylindole). Fluorescence was observed under a fluorescence microscope. For all graphs, results are shown as means and SD. Significance was assessed using Student's t test. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
The sequences of miR-466d-3p and its target site on the 3′ UTR of IL-1β were aligned using TargetScan (www.targetscan.org/mmu_72/) (Fig. 4D). To determine if miR-466-3p targeted IL-1β mRNA, expression of IL-1β was measured in BV2 and NA cells after transfection with miR-466d-3p mimics or miR-466d-3p inhibitors. Overexpression of miR-466d-3p suppressed expression of IL-1β mRNA and protein significantly in BV2 and NA cells, and similar results were observed in BV2 and NA cells after JEV infection or treatment with lipopolysaccharide (LPS) (Fig. 4E to G, top). In contrast, the miR-466d-3p inhibitor significantly increased expression of IL-1β mRNA and protein secretion in BV2 and NA cells, respectively (Fig. 4E to G, bottom). Moreover, miR-466d-3p expression in cells treated with LPS or poly(I·C) was similar to that in medium-treated cells and significantly higher than that in JEV-infected cells. However, the miR-466d-3p mimic could continue to inhibit LPS- or poly(I·C)-induced IL-1β overtranscription and overexpression, indicating that miR-466d-3p was a common regulator of IL-1β but not specific to JEV infection. Interestingly, comparison of miRNA and JEV genes indicated that miR-466d-3p also targeted the coding sequences of the E, NS3, and NS5 genomes of JEV (Fig. 4D). There was a substantial reduction of the viral titer in NA cells transfected with miR-466d-3p. Moreover, inhibition of miR-466d-3p expression led to a small increase in JEV replication (Fig. 4H). To determine whether miR-466d-3p targeted the seed sequence of JEV directly, we constructed a miR-466d-3p vector containing two miR-466-3p target seed sequences of JEV NS3 and E fused to green fluorescent protein (GFP). As expected, transfection by the miR-466d-3p mimic resulted in complete loss of fluorescence in miR-466d-3p target site-fused GFP-expressing cells, whereas cotransfection of NS3 recovered some expression of GFP, suggesting that NS3 cleaved miR-466d-3p to block the miRNA function (Fig. 4I). Our data indicated that IL-1β was a possible target of miR-466d-3p, which was also a negative regulator of JEV replication.
NS3 helicase activity blocks the silencing function of miR-466d-3p.
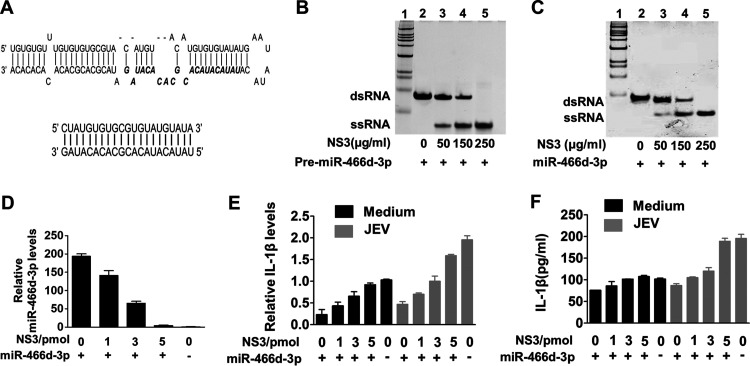
JEV NS3 has been identified as an RNA helicase that “unwinds” dsRNA (2). Transactivating response RNA-binding protein and Argonaute 2 contain several dsRNA-binding domains, which help dsRNA fragments go into the RISC to target mRNA (38, 39). We hypothesized that host pre-miRNAs might also be unwound into a single-strand RNA by NS3, in which the RISC could not recognize the single-strand fragment or target mRNA. In vitro unwinding assays revealed that a synthesized pre-miR-466d-3p containing a hairpin structure (Fig. 5A, top) was unwound into a single-strand RNA in a dose-dependent manner (Fig. 5B, lane 2 to lane 5). We used an in vitro degradation assay to determine whether NS3 could unwind the miR-466d-3p mimic directly, like endogenous host miRNAs. NS3 unwound the synthetic double-stranded miR-466d-3p mimic (Fig. 5A, bottom) into a single strand in a dose-dependent manner (Fig. 5C, lane 2 to lane 5).
FIG 5.
NS3 helicase activity blocks the silencing function of miR-466d-3p. (A) Schematic of the synthetic double-stranded pre-miR-466d-3p mimic and double-stranded miR-466d-3p mimic. The sequence of miR-466d-3p in pre-miR-466d-3p is highlighted in boldface italics. (B and C) The synthetic double-stranded pre-miR-466d-3p and double-stranded mimic of miR-466d-3p were unwound by purified NS3 generated from NA cells. The FLAG-tagged NS3 was expressed in NA cells and purified with an affinity gel. The indicated concentrations of NS3 (0, 50, 150, and 250 μg/ml) were incubated with 20 pmol of pre-miR-466d-3p mimic (B) or miR-466d-3p mimic (C). After 2 h of incubation at 37°C, the pre-miR-466d-3p mimic or miR-466d-3p mimic was determined by 10% native polyacrylamide gel electrophoresis. (D) The miR-466d-3p mimic and pre-miR-466d-3p mimic were degraded after unwinding with NS3. After 24 h of transfection, total RNA of NA cells was used to quantify the relative expression of miR-466d-3p (versus the negative control) by qRT-PCR. (E and F) Unwinding of the miR-466d-3p mimic by NS3 reduced IL-1β expression. (E) Total RNA of NA cells was used to quantify the relative expression of IL-1β (versus the negative control) by qRT-PCR. (F) Supernatants of NA cells were used to determine IL-1β expression by ELISA. For all graphs, results are shown as means and SD.
We wondered if unwound miRNA could continue to be transported into the RISC and execute the normal function of RNA silencing with host mRNA. Fragments of unwound miR-466d-3p mimic and pre-miR-466d-3p were recycled with TRIzol reagent after the in vitro unwinding assay. After transfection with the unwound miR-466d-3p mimic, expression of mature miR-466d-3p was similar to that in the negative-control group, indicating that the unwound miR-466d-3p mimic was unstable in host cells (Fig. 5D). mRNA expression and protein secretion of IL-1β did not decrease after transfection of the unwound miR-466d-3p mimic, with or without JEV infection (Fig. 5E and F). Our data demonstrated that NS3 of JEV contributed to miR-466d-3p dysfunction in host cells.
NS3 has a specific binding affinity with pre-miRNA.
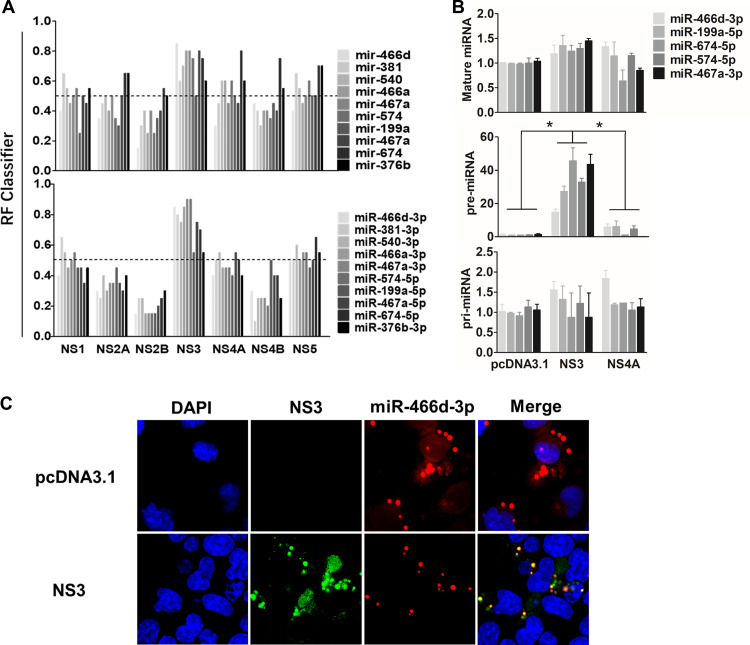
Many viral suppressors of RNA silencing (VSRs) have been reported to block RNA interference (RNAi) by binding siRNA or miRNAs (40, 41). Several ribonucleoproteins create posttranscriptional regulatory networks that are mediated by RPIs, and some computational models help identify RPIs and predict the RNA- and protein binding sites (42–45). We assessed whether the binding ability of miRNAs was required for NS3-induced miRNA degradation using the RPISeq Web server (http://pridb.gdcb.iastate.edu/RPISeq/) (43). RPISeq calculated a random forest (RF) for the indicated miRNAs between the NS proteins of JEV to evaluate the RPI. The probability threshold of RF used for positive RPIs was >0.5, and the values of pre-miRNAs or mature miRNAs that interacted with NS3 were >0.5. The mean RFs of NS3 with pre-miRNA and mature miRNA were 0.675 and 0.69, respectively, significantly higher than those of all other JEV proteins (Fig. 6A).
FIG 6.
NS3 binds specifically with pre-miRNA. (A) The binding affinities of pre-miRNA and mature miRNA of the indicated NS proteins of JEV were predicated with a Web server named RPISeq (http://pridb.gdcb.iastate.edu/RPISeq/), and the RF calculated by RPISeq was used to evaluate the RPI. The probability threshold of RF used for positive RPIs was >0.5. (B) Expression of pri-, pre-, and mature miRNAs of miR-466d-3p, miR-199a-5p, miR-674-5p, miR-574-5p, and miR-467a-3p was selected to determine the binding affinity between NS3 and miRNA by RIP. NA cells were transfected with the plasmid encoding FLAG-tagged NS3, NS4A, or pcDNA 3.1, and the FLAG-tagged proteins were purified on an affinity gel after 48 h of transfection. After purification, total RNA on the FLAG-tagged protein was used to quantify the relative binding affinity of the indicated pri-, pre-, and mature miRNAs (versus the pcDNA 3.1 control) by qRT-PCR. The fold change of each RIP reaction from qRT-PCR data was calculated as follows: fold enrichment = 2(−ΔΔCT [experimental/pcDNA 3.1]), where ΔΔCT [experimental/pcDNA 3.1] = ΔCT [experimental] − ΔCT [pcDNA 3.1] and ΔCT = CT [RIP] − [CT [input] − log2(input/RIP dilution factor)]. (C) The pre-miR-466d-3p mimic and NS3 were colocalized in NA cells. After 48 h of transfection, the CY3-labeled miR-466d-3p mimic and NS3 stained with fluorescein isothiocyanate (FITC) in NA cells were detected by fluorescence microscopy. The images are representative of the results of three independent experiments. For all graphs, results are shown as means and SD. Significance was assessed using Student's t test. *, P ≤ 0.05.
We confirmed the in silico analysis using an RIP assay with a FLAG-fused protein to identify the binding affinity of NS3 and miRNA. The fold enrichment of NS protein over pcDNA 3.1 was used to determine the RIP value (see Materials and Methods for details). NS3 had a significantly higher pre-miRNA (i.e., miR-466d-3p, miR-199a-5p, miR-674-5p, miR-574-5p, and miR-467a-3p) binding affinity than pcDNA 3.1 (Fig. 6B). In contrast to NS3, the miRNA-binding ability of NS4A was similar to that of pcDNA 3.1. However, contrary to these in silico analyses, the RIP assay revealed that the binding ability of pre-miR-466d-3p on NS3 was 9-fold more than that for miR-466d-3p, and there was no significant difference in binding affinity between NS3, pcDNA 3.1, and NS4A (Fig. 6B). Furthermore, a CY3-labeled miR-466d-3p mimic colocalized with NS3 expression in the cytoplasm of NA cells (Fig. 6C). However, the miR-466d-3p mimic was evenly distributed in NA cells without NS3 expression. These results indicated that NS3 was bound specifically to pre-miRNA and that its binding ability may be essential for miRNA degradation.
NS3-mediated miRNA degradation is dependent on arginine-rich motifs (ARMs).
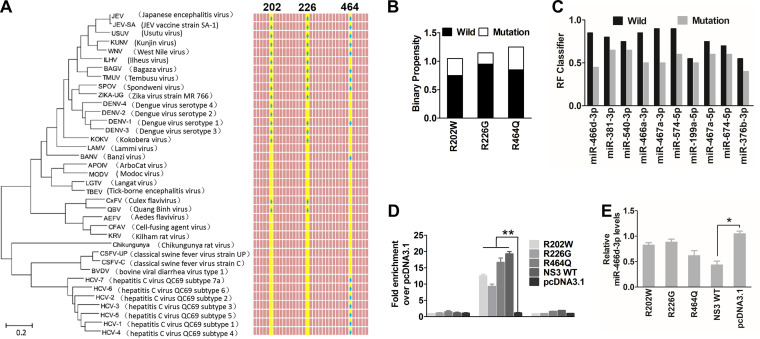
Computational methods have been used to predict the amino acid binding sites of PRIs. However, sequence-based predictors usually have high sensitivity but low specificity; conversely, structure-based predictors tend to have high specificity but low sensitivity (44). To combine the advantages of the two methods, we used four types of software (RPISeq, aaRNA, Pprint, and PRIdictor) to predict the RPI binding sites of NS3 (analysis available online) (42–45). The RNA-binding sites on the NS3 protein of JEV were analyzed using a Protein Data Bank (PDB) format of NS3 (PDB entry 2Z83) with aaRNA (http://sysimm.ifrec.osaka-u.ac.jp/aarna/). Four arginines (R202, R226, R388, and R464) on NS3 had higher scores (>0.5) than all other amino acid sites (data not shown). Furthermore, Pprint (http://www.imtech.res.in/raghava/pprint/) and PRIdictor (http://bclab.inha.ac.kr/pridictor) predicted that R202, R226, or R464 was located in the helicase region of NS3, which has high miRNA-binding ability. Similarly, previous studies have reported that arginine is a stronger RNA-binding amino acid than other amino acids (46). Interestingly, almost all amino acid (aa) 202, 226, or 464 sites are arginine on the NS3 protein of Flavivirus, but not on that of Hepacivirus or Pestivirus (Fig. 7A). Sequence alignment (Fig. 3G) confirmed that only Flavivirus species had reduced miRNA expression. When these arginine sites were mutated into other amino acids, the binary propensity (data analyses by aaRNA) or RF value (data analyses by RPISeq) of R202W, R226G, and R464Q decreased sharply (Fig. 7B and C). We constructed three mutant substitution vectors of NS3 named R202W, R226G, and R464Q to confirm the in silico analyses. The mutants or parent plasmids were used to investigate the role of miRNA-binding affinity in inducing miRNA turnover in NA cells by RIP analyses and miRNA expression. Compared with pcDNA 3.1, the mutants of NS3 were also bound specifically to pre-miR-466d-3p (Fig. 7C), and the pre-miR-466d-3p binding abilities of R226G and R202W were significantly lower than that of wild-type NS3 (Fig. 7D). Furthermore, R226G and R202W, but not R464Q, reduced miR-466d-3p degradation significantly (Fig. 7E). Thus, the arginine sites at positions 202 and 226 of NS3 contributed to pre-miRNA binding and promoted miRNA degradation.
FIG 7.
NS3-mediated miRNA degradation is dependent on arginine-rich motifs. (A) Phylogenetic tree and alignment based on the full-length NS3 sequences of 37 Flaviviridae strains. The phylogenetic tree was deduced from a full-length alignment of NS3 sequences from the indicated viruses using the neighbor-joining method as implemented in MEGA7 (left). Phylogenetic analysis of 37 NS3 gene nucleotide sequences from Flaviviridae included the JEV vaccine strain SA-14-14-2 (GenBank accession number M55506.1), a lineage II WNV strain (GenBank accession number M12294.2), Dengue virus serotype 1 isolate TM100 (GenBank accession number KU666942.1), Dengue virus serotype 2 isolate TM26 (GenBank accession number KU666944.1), Zika virus strain MR 766 (GenBank accession number AY632535.2), classical swine fever virus strain C (GenBank accession number Z46258.1), hepatitis C virus QC69 subtype 7a (GenBank accession number EF108306.2), and bovine viral diarrhea virus type 1 isolate MA_101_05 (GenBank accession number LT968777.1). (Right) Alignment of amino acid sequences of NS3 proteins of 37 Flaviviridae. The three conserved amino acids are highlighted in yellow, and arginine is indicated with a blue spot. (B and C) RNA-binding sites on NS3 were predicted with aaRNA and PRIdictor. (B) The sequences of these predicted RNA-binding sites were changed to another amino acid as indicated, and we evaluated the RNA-binding affinity by binary propensity. (C) After changing all three amino acid sites (R202W, R226G, and R464Q), the probability threshold of RF was evaluated. The probability threshold of RF used for positive RPIs was >0.5. (D) The pri-, pre-, and mature miR-466d-3p binding affinities of the substitution mutant of NS3 were determined by the RIP assay. Three substitution mutant vectors of NS3 named R202W, R226G, and R464Q were constructed by overlap extension PCR. After 48 h of transfection, FLAG-tagged NS3 was purified with affinity gel, and total RNA on NS3 was used to quantify the relative binding affinity of pre-miR-466d-3p (versus the pcDNA 3.1 control) by qRT-PCR. The fold change of each RIP reaction from qRT-PCR data was calculated as follows: fold enrichment = 2(−ΔΔCT [experimental/pcDNA 3.1]). (E) Relative analyses of miR-466d-3p expression in NA cells transfected with the indicated expression plasmids. After 48 h of transfection, total RNA from NA cells was used to quantify the relative expression of miR-466d-3p (versus the pcDNA 3.1 control) by qRT-PCR. For all graphs, results are shown as means and SD. Significance was assessed using Student's t test. *, P ≤ 0.05; **, P ≤ 0.01.
Arginine mutations at NS3 positions 202 and 226 of P3 reduce viral replication and pathogenicity.
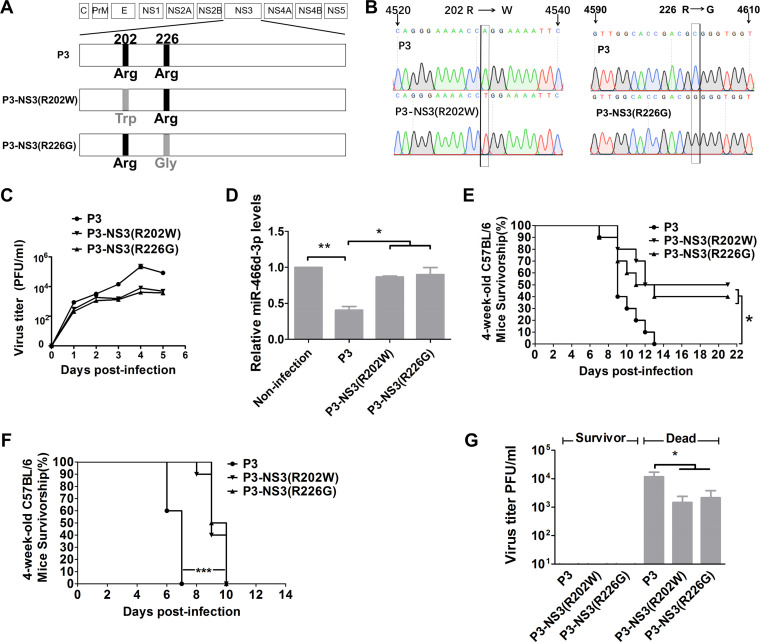
To test whether the Arg202 → Trp202 or Arg226 → Gly226 mutation alone was responsible for the effect on viral replication, miRNA expression, or viral pathogenicity, we introduced mutations in JEV strain P3 using site-directed mutagenesis. The mutant viruses were named P3-NS3(R202W) and P3-NS3(R226G) (Fig. 8A). T replaced A at nt 4531 in the P3-NS3(R202W) genome. G replaced C at nt 4,603 in P3-NS3(R226G) (Fig. 8B). The growth curves of P3-NS3(R202W) and P3-NS3(R226G) were lower than those of the parental JEV strain P3 in NA cells (Fig. 8C). miR-466d-3p expression was measured at 48 hpi in NA cells at an MOI of 0.1, and P3 showed a decrease in miR-466d-3p expression compared with that of P3-NS3(R202W) or P3-NS3(R226G) (Fig. 8D). These results indicated that Arg202 and Arg226 in NS3 were essential for suppression of JEV-mediated miRNA expression and viral replication.
FIG 8.
The changes at NS3 positions 202 and 226 of P3 affect viral replication and pathogenicity. (A) Schematic of construction of pMW219-P3, P3-NS3(R202W), and P3-NS3(R226G). (B) Viral genome RNAs isolated from NA cells infected with P3-NS3(R202W) and P3-NS3(R226G) were used as templates to carry out RT-PCR and sequencing. (C) Multistep growth curves of pMW219-P3, P3-NS3(R202W), and P3-NS3(R226G) in NA cells. NA cells were infected with the indicated viruses at an MOI of 0.1, and the virus titers in infected cell supernatants were determined at 1, 2, 3, 4, and 5 dpi. (D) Relative analyses of miR-466d-3p expression in NA cells transfected with the indicated expression plasmids with or without JEV infection. After 48 h of transfection/infection, total RNA from NA cells was used to quantify the relative expression of miR-466d-3p (versus the pcDNA 3.1 control) by qRT-PCR. (E and F) Four-week-old C57BL/6 mice were inoculated with 103 FFU of the indicated virus (i.m.) (E), and 1-day-old C57BL/6 mice were inoculated with 102 FFU of the indicated virus intracerebrally (F). Survival was observed for 21 days; the asterisks indicate that the differences between survival curves was significant using the log rank (Mantel-Cox) test (***, P < 0.001; *, 0.01 < P < 0.05). (G) Virus titers in the brains of dead and surviving mice inoculated with the indicated virus by the intramuscular or intracerebral route. For all graphs except survivorship analysis, results are shown as means and SD. Significance was assessed using Student's t test. *, P ≤ 0.05.
The pathogenicity of P3-NS3(R202W) and P3-NS3(R226G) was evaluated in C57BL/6 mice via different inoculation routes. We injected (intramuscularly [i.m.]) 4-week-old mice with 103 focus-forming units (FFU) of the indicated JEV. For 21 days, we observed the development of clinical symptoms of JEV infection (limb paralysis and encephalitis). Survival was evaluated using a log-rank (Mantel-Cox) test. Significantly more mice infected with P3-NS3(R202W) (50%) and P3-NS3(R226G) (40%) survived the challenge than mice infected with parental P3 (0%) (Fig. 8E). Intracerebral inoculation with 102 FFU of mutant or P3 JEV induced death in all the mice, but the average day of death for mice infected with P3-NS3(R202W) and P3-NS3(R226G) was significantly later than that for parental P3 (Fig. 8F). In mice that succumbed to JEV, viral titers reached ≥103 PFU/ml [P3, 104 PFU/ml; P3-NS3(R202W), 103 PFU/ml; P3-NS3(R226G), 103.25 PFU/ml], and JEV was not found in the brains of survivors (Fig. 8G). These results supported the importance of Arg202 and Arg226 in NS3 for viral pathogenicity.
DISCUSSION
Increasing numbers of studies have shown that miRNAs play an important part in the replication and propagation of viruses, including defense against pathogenic viral infections and promotion of viral replication through complex regulatory pathways (47). In the early steps of viral infection, innate virus detection sensors of host cells, such as pathogen recognition receptors, recognize a wide spectrum of pathogens and activate downstream antiviral pathways that include miRNAs (48). However, the defense function of host miRNAs can be counteracted by VSRs that inhibit host antiviral responses by interacting with the critical components of cellular RNA-silencing machinery or participating in host RNA degradation (47, 49). Recent studies have suggested that JEV infection can regulate functional miRNAs (16–21). Several scholars have examined the host biogenesis regulating miRNA during viral infection, but relatively little is known about miRNA regulation by JEV. Given the importance of miRNA in establishing JEV infection, we explored the interplay between JEV and host miRNAs.
An intriguing finding of our study was that JEV globally decreased expression of host miRNA independently of Dicer or the RISC. We demonstrated that JEV NS3 could unwind pre-miR-466d and induce miR-466d-3p dysfunction. We also observed that miR-466d-3p degradation could enhance JEV replication. Taken together, these results suggest a role for miRNA degradation in enhancement of JEV replication. Similarly, poly(A) polymerase of VACV and the 3′ UTR of MCMV degrade host miRNA via different mechanisms (28, 30). Although this is the first report of a link between degradation of host miRNA and JEV infection, several studies have reported a role of miRNA machinery in the replication of flaviviruses, especially DENV and Kunjin virus (33, 50). Some analogous systems in the degradomes of bacteria and exosomes of eukaryotes have revealed that RNA helicases are involved in RNA degradation (51, 52). Similarly, JEV helicase seems to be involved in degrading host miRNA.
How did JEV evolve an miRNA degradation process during viral infection in the CNS? There are two possible reasons why the JEV infection-induced global degradation of host miRNA might promote viral infection and replication. First, the host cell miRNA binds to miRNA-binding sites of viral genomes to influence viral function (53, 54). Consistent with this phenomenon is our observation that restoration of miR-466d-3p expression during JEV infection reduces viral replication. Conversely, the virus-induced degradation of host miRNA blocks the cleavage of the viral genome and viral proteins. Second, changes in expression of host miRNA could also lead to enhanced antiviral effectors, resulting in reduced viral replication (16, 55). However, some viral infections mediate the degradation of host miRNAs, leading to downstream changes in the host transcriptome that can benefit virus replication and pathogenicity (30). Our hypotheses were also confirmed with NS3 proteins of other flaviviruses, including Zika virus, which could reduce miR-466d-3p expression, resulting in enhanced inflammation.
Eubacteria, archaebacteria, and eukaryotes have developed dedicated pathways and complexes to check the accuracy of RNAs and “feed” undesired RNA into exoribonucleases to degrade the RNA. Furthermore, these complexes usually contain adaptor proteins, such as RraA (a regulator of RNase E and DEAD box helicases), RhlB (a DEAD box RNA helicase) (51), Ski2-like RNA helicases (52, 56), and Suv3 RNA helicase (57). In addition to RNA degradation, Up-frameshift protein 1 (UPF1) helicase can dissociate miRNAs from their mRNA targets and promote miRNA degradation by Tudor staphylococcal/micrococcal-like nuclease (58). P68 helicase promotes unwinding of the human let-7 microRNA precursor duplex, which helps let-7 microRNA load into the silencing complex. p72 (DDX17) interacts with the Drosha-containing microprocessor complex and facilitates processing of a subset of pri-miRNAs in the nucleus and miRNA-guided mRNA cleavage (59). Although the host helicase may initially seem to benefit only the processing and immune systems of a cell, multiple studies have shown that viral helicase promotes viral replication and disrupts the immune system. The NS3 proteins of flaviviruses can unwind double-stranded DNA or RNA, which were not produced by flaviviruses themselves (2, 60). Similarly, we observed that NS3 of JEV could reduce exogenously constructed ds-miRNA mimics and disable an miRNA mimic.
Some single mutations of amino acids in arginines R538, R225, and R268 of DENV affect DENV replication (61, 62). Another study reported that the Asp285 → Ala substitution of the JEV NS3 protein abolished ATPase and RNA helicase activities (2). Furthermore, the helicase domains of NS3 induce neuron apoptosis (63, 64). These findings suggested that NS3, particularly the helicase domain, contributes to the efficient replication of flaviviruses. We used miRNA deep sequencing to investigate the details of NS3 involvement in miRNA degradation. Subtype sequence analysis of miR-466d-3p during JEV infection and NS3 overexpression showed that the percentage of incorrect splicing products of mature miRNA was higher than normal. Furthermore, in vitro unwinding assays demonstrated that NS3 could unwind pre-miR-466d-3p and induce miRNA dysfunction. However, the molecular mechanism of miRNA degradation after unwinding by NS3 has yet to be determined.
We showed that the arginine molecules of NS3 were critical for host pre-miRNA binding and promoted global miRNA turnover in the host. We constructed three arginine variants of NS3 that had single amino acid substitutions on R202W, R226G, and R464Q which reduced the pre-miRNA-binding affinity of NS3 and abrogated miRNA degradation. Interestingly, these sites were located in the helicase region of NS3, spanning the protein sequence of NS3 from positions 163 to 619. Similar to our findings, several proteins containing ARMs are known to bind RNA and are involved in regulating RNA processing in viruses and cells. For example, the ARM of lambdoid bacteriophage N protein and the human immunodeficiency virus type 1 (HIV-1) Rev protein bind RNAs and regulate RNA transport and splicing (65, 66). Four single-amino-acid substitutions of arginine on the HIV-1 Rev protein strongly decreased RNA-binding ability. Our results extend the function of flavivirus helicases beyond that of unwinding duplex RNA to regulate miRNA pathways by decay of pre-miRNAs—a new mechanism for helicases. We suggest that the helicases of flaviviruses may be able to regulate various cellular miRNAs as part of a general viral response to overcome host defense mechanisms. Furthermore, we consider investigations of JEV helicase in miRNA degradation during viral propagation to be important for the development of efficacious vaccines and antiviral drugs.
MATERIALS AND METHODS
Cells, viruses, and reagents.
The mouse microglial cell line BV2, the mouse neuroblastoma cell line NA, and the mouse brain endothelial cell line bEnd.3 were gifts from Huazhong Agricultural University (Wuhan, China). The cell lines were maintained in RPMI 1640 medium (Thermo Fisher, Boston, MA, USA) supplemented with 10% fetal bovine serum (FBS) (Gibco, Carlsbad, CA, USA) at 37°C in an atmosphere of 5% CO2. The Syrian baby hamster kidney cell line BHK-21 was a gift from Huazhong Agricultural University. The cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (high glucose; Thermo Fisher) containing 10% FBS. The human neuroblastoma cell line SK-N-SH was a gift from Huazhong Agricultural University. The cells were cultured in minimum essential medium (MEM) (Thermo Fisher) containing 10% FBS.
The P3 strain of JEV was propagated in suckling BALB/c mice (purchased from Vital River Laboratories, Beijing, China) as described previously (37). Briefly, 1-day-old suckling mice were inoculated with 10 μl of viral inoculum via the intracerebral route. When moribund, mice were euthanized, and brains were removed. A 10% (wt/vol) suspension was prepared by homogenizing the brains in DMEM and centrifugation at 10,000 × g for 5 min to remove cellular debris. The brain suspension was passed through sterile filters (pore size, 0.22 μm; Millipore, Bedford, MA, USA) and subpackaged at −80°C until use. DRB [5,6-dichloro-1-(β-d-ribofuranosyl) benzimidazole] and ivermectin were purchased from Apexbio (Houston, TX, USA).
miRNA and mRNA microarray analyses.
Total RNA was isolated from mouse brains with TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and quantified using a 2100 Bioanalyzer (Agilent Technologies, Wilmington, DE, USA). miRNA and mRNA microarray hybridization was carried out by the Shanghai Bio Analysis System (Shanghai, China) with 8,000-by-15,000 Agilent mouse microRNA microarrays (Agilent Technologies) and 4,000-by-44,000 Agilent whole-mouse genome oligonucleotide microarrays (Agilent Technologies), respectively. Data analyses were undertaken using GeneSpring GX (Agilent Technologies). miRNA was designated as being expressed significantly if expression in JEV infection was ≥1.5-fold compared with that in the control group. mRNAs that caused significant changes (≥2.0-fold; P < 0.05) by JEV infection in mouse brains were clustered using GO and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment tools (Shanghai Bio Analysis System).
Usually, there were hundreds of target genes for each miRNA. We used three miRNA target prediction databases: TargetScan (67), miRDB (68), and microRNA (69). The integrative analysis of miRNAs and mRNAs allowed us to predict the major target of miRNAs with decreasing expression. To accurately elucidate the correlation between the pattern of mRNA expression and miRNA-targeted regulation, miRNA expression files were measured with mRNA microarrays to predict the miRNAs with major decreasing expression in mouse brains. First, 41 miRNAs (with a threshold of ≥2.0-fold change; P < 0.01) were identified from the miRNA expression files, and these miRNA-targeted mRNAs were compared to the mRNA expression profiles to find the common mRNAs. Using the common genes as “seeds,” a protein-protein interaction network was constructed to discover the core gene of the miRNA target. Forty-two interacting proteins with 177 interactions (enrichment of protein-protein interaction; P < 1.015) were retrieved from the STRING database (70). The k-means clustering algorithm was applied to segregate the network of interacting proteins into different subgroups.
RNA sequencing.
Total RNA was extracted from cultured NA cells using a total-RNA extractor kit (B511311; Sangon Biotech, Shanghai, China) according to the manufacturer’s protocols. RNA (2 μg) from each sample was used for library preparation according to the instructions in the VAHTS mRNA-seq V2 library prep kit for Illumina (San Diego, CA, USA). The 3′ adapter (TGGAATTCTCGGGTGCCAAGGAACTC) and 5′ adapter (GUUCAGAGUUCUACAGUCCGACGAUC) were ligated to each end of total-RNA samples. Then, the adapter-ligated products were reverse transcribed with an RNA RT primer (5′-GCCTTGGCACCCGAGAATTCCA) and library amplified by PCR using standard Illumina primers (5′-phosphate/AATGATACGGCGACCACCGAGATCTACACGTTCAGAGTTCTACAGTCCGACGATC-3′ phosphate/) and a primer containing barcode sequences. The amplified cDNA constructs were separated on 12% polyacrylamide gels, and 140- to 150-bp bands were excised. The library fragments were purified with an AMPure XP system (Beckman Coulter, Beverly, MA, USA). PCR products were purified (AMPure XP system), and library quality was assessed on a Bioanalyzer 2100 (Agilent Technologies). Then, the libraries were quantified and pooled. The paired-end sequencing of the library was done on HiSeq XTen sequencers (Illumina). FastQC v0.11.2 (Babraham Institute, Cambridge, United Kingdom) was used for evaluating the quality of the sequenced data. Raw reads were filtered with Trimmomatic v0.36 according to protocols. The remaining “clean” data were used for further analyses. Gene expression of the transcripts was computed with StringTie v1.3.3b. Principal-component analysis (PCA) and principal-coordinates analysis (PCoA) were carried out to reflect the distance and difference between samples. Transcripts per million (TPM) eliminates the influence of gene lengths and sequencing discrepancies to enable direct comparison of gene expression between samples. DESeq2 v1.12.4 was used to determine differentially expressed genes (DEGs) between two samples.
Analyses of mature miRNA and its isomiRs with miRDeep2.
Expression profiling of miRNAs was done by means of miRDeep2 (www.mdc-berlin.de/rajewsky/miRDeep) as described previously (32). The miRDeep2 software package identifies miRNAs in small-RNA deep-sequencing data. miRDeep2 can convert raw sequencing reads to FASTA files and can make the data useful for downstream analysis. miRDeep2 consists of a series of scripts that make use of other software, such as the short-read aligner Bowtie and the prediction tool for RNA secondary structure RNA fold from the Vienna RNA package. Before mapping to the reference genome, we used a mapper module to remove the adapter (TGGAATTCTCGGGTGCCAAGGAACTC) and redundant sequences. All reads were aligned to the mouse miRNA miRBase to quantify the differential expression of all known miRNAs. The results of miRDeep2 show the number of reads that map to natural sequences, the precursor-miRNA secondary structure, and a density plot of reads mapped to the precursor. Based on the reads mapped to the precursor, we analyzed the abundance of miRNA isoforms (isomiRs). Finally, we calculated the RPM expression value for one (71). MicroRNA counts within each sample were normalized to RPM values by adding a pseudocount of one to each microRNA, dividing by the total number of reads that aligned to all microRNA loci within that sample, multiplying by 106, and then applying a log2 transformation. The RPM data for all samples are available upon request.
Plaque assay.
JEV was titrated on the BHK-21 cell line by a viral plaque formation assay as described previously (20). A monolayer of BHK cells was cocultured with JEV (a serial 10-fold dilution prepared in DMEM without FBS) at 37°C. After 2 h of incubation, DMEM containing 3% FBS and 4% sodium carboxymethyl cellulose (Sigma-Aldrich, Saint Louis, MO, USA) was added to the cells, and the cells were cultured for 5 days until the appearance of visible plaques. The cells were fixed with 10% formaldehyde overnight, followed by staining with crystal violet for 2 h. Visible plaques were counted, and viral titers were calculated. The data were expressed as means of triplicate samples.
siRNA, miRNA mimic, or miRNA inhibitor transfection.
Mouse miR-466d-3p mimics, inhibitors, mimic controls, and negative controls were purchased from GenePharma (Beijing, China). The sequences of the mimics, inhibitors, mimic controls, and control oligonucleotides were as follows: miR-466d-3p mimics, 5′-UAUACAUACACGCACACAUAG-3′ (forward) and 5′-AUGUGUGCGUGUAUGUAUAUU-3′ (reverse); mimic negative controls, 5′-UUCUCCGAACGUGUCACGUTT-3′ (forward) and 5′-ACGUGACACGUUCGGAGAATT-3′ (reverse); miR-466d-3p inhibitor, 3′-CUAUGUGUGCGUGUAUGUAUA-5′; inhibitor negative control, 3′-CAGUACUUUUGUGUAGUACAA-0.5′. All miRNA mimics (1.5 μmol/well), inhibitors (1.5 μmol/well), and controls (3 μmol/well) were transfected into BV2 cells or NA cells (106 cells/well) in 12-well plates using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s instructions. The JEV protein siRNA and nontargeting control (NC) siRNA were purchased from GenePharma (Beijing, China), and the sequences of the siRNAs are available upon request. All the siRNAs (0.5 μmol/well) were transfected into NA cells (106 cells/well) in 12-well plates using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s instructions.
Viral infection.
BV2 cells, NA cells, or bEnd.3 cells were seeded in 12-well plates (106 cells/well). Until growth to 80% confluence, the cells were incubated with serum-free medium or JEV at an MOI of 0.1 at 37°C for 2 h. After discarding unbound virus and medium, the cells were cultured in RPMI 1640 with 10% FBS, 1% penicillin-streptomycin liquid at 37°C in an atmosphere of 5% CO2 for 48 h. The cells and supernatants were collected 48 hpi.
RNA isolation and qRT-PCR.
Total RNA of cells was isolated with TRIzol reagent (Thermo Fisher) according to the manufacturer’s recommendations and used for qRT-PCR in a 7500 real-time PCR system (Applied Biosystems, Foster City, CA, USA) as described previously. For mRNA evaluation and to obtain cDNA, total RNA (1 μg) was reverse transcribed using a PrimeScript RT reagent kit with gDNA Eraser (TaKaRa Biotechnology, Shiga, Japan). IL-1β mRNA expression in NA cells was quantified with a SYBR green quantitative-PCR (qPCR) kit (TaKaRa Biotechnology) using gene-specific primers according to the manufacturer’s instructions. Amplification was undertaken at 95°C for 30 s, 95°C for 5 s, and 60°C for 34 s, followed by 40 cycles of 95°C for 15 s, 60°C for 1 min, 95°C for 30 s, and 60°C for 15 s. IL-1β expression was normalized to that of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) using the 2−△△CT method as described previously (72). For miRNA expression, a specific stem-loop structure was added to the 5′ ends of the reverse-transcribed primers of all miRNAs. Pri-miRNA, pre-miRNA, and mature miRNA expression of cells was quantified with the SYBR green qPCR kit using miRNA sequence-specific primers. Amplification was done at 95°C for 30 s, 95°C for 5 s, and 60°C for 34 s, followed by 40 cycles of 95°C for 15 s, 60°C for 1 min, 95°C for 30 s, and 60°C for 15 s. Relative expression of microRNAs was normalized to that of U6, determined by the 2−△△CT method as described previously (17). PCR primers for amplification of DNA inserts were ordered from Huada (Beijing, China), and the sequences are available upon request.
ELISA.
The culture supernatants of the experimental group and control group cells or brain tissue lysates were collected and stored at −80°C. Protein expression of IL-1β in cell cultures or mouse brain tissue lysates was determined using ELISA kits (eBioscience, San Diego, CA, USA) according to the manufacturer’s instructions.
Western blotting.
Cells were lysed with RIPA lysis and extraction buffer (Thermo Scientific). Protein expression was measured with an enhanced bicinchoninic acid (BCA) protein assay kit (Sigma-Aldrich). Extracts that contained 25 μg of total protein were subjected to SDS-PAGE using 10% gels. Protein blots were transferred to nitrocellulose membranes after electrophoresis. The nitrocellulose membranes were washed with Tris-buffered saline and Tween 20 (TBST) and blocked in 5% skim milk at 4°C overnight. Primary antibodies were prepared at a dilution of 1:1,000 in 1% bovine serum albumin (BSA) in 1× phosphate-buffered saline with Tween 20 (PBST), followed by mixing with horseradish peroxidase-conjugated secondary antibodies (Sigma-Aldrich) for 1 h at room temperature. The blots were detected with enhanced chemiluminescence reagent (Thermo Scientific) and developed by exposure in the Tanon 5200 imaging system (Tanon Science and Technology, Beijing, China) using Tanon MP software. α-Tubulin was the internal control.
RIP.
RIP was carried out as described previously (73) with slight modification. Briefly, NA cells were transfected with FLAG-tagged NS3, FLAG-tagged NS2B, or FLAG-tagged pcDNA 3.1 plasmid for 48 h. Cells (107) expressing FLAG fusion protein were harvested with 2.5% trypsin and resuspended in 5 ml of phosphate-buffered saline (PBS). Then, 143 μl of 37% formaldehyde was added to the resuspension to cross-link for 10 min on a shaker, and 685 μl of 2 M glycine was used to block the formaldehyde. The cells were washed twice with 5 ml of ice-cold PBS and centrifuged for 2 min at 400 × g to collect the cell pellet. Then, 1 ml of CelLytic lysis buffer (Sigma-Aldrich) containing 20 μl of 0.1 M phenylmethylsulfonyl fluoride, 20 μl of a protease inhibitor cocktail (Sigma-Aldrich), and 5 μl of 40 U/μl RNase inhibitor (Invitrogen) was added to the cells. The cells were kept on ice and sonicated for 2 min (on for 10 s, off for 10 s; amplitude, 15 μm) until the lysate was clear. The lysate was centrifuged for 3 min at 14,000 × g to collect the supernatant. Each 1 ml of cell lysate was added to 40 μl of anti-FLAG M2 affinity gel (Sigma-Aldrich) at 4°C for 4 h on a shaker at 10 rpm. The next step was centrifugation of the resin for 30 s at 8,200 × g and supernatant removal. After the resin had been washed thrice with 0.5 ml of TBS, total RNA was extracted with TRIzol reagent and analyzed by RT-qPCR.
The fold change of each RIP reaction from RT-qPCR data was calculated by the 2−△△CT method as described previously with minor modification (74) (the formulas are shown below). The cycle threshold (CT) value of each specimen (FLAG-tagged NS3, FLAG-tagged NS2B, or non-FLAG-fused blank pcDNA 3.1) was normalized to that of the input RNA to eliminate possible differences in preparation of RNA samples (ΔCT). To obtain ΔΔCT, the normalized experimental RIP fraction (ΔCT) was normalized to the nonspecific background as an internal control (ΔCT normalized to the pcDNA 3.1 sample). Finally, ΔΔCT [experimental/pcDNA 3.1] was undertaken with linear conversion to calculate the fold enrichment. The formulas are as follows: ΔCT = CT [RIP] − [ΔCT [input] − log2 (input/RIP dilution factor)]; ΔΔCT [experimental/pcDNA 3.1] = ΔCT [experimental] − ΔCT [pcDNA 3.1]; fold enrichment = 2−ΔΔCT [experimental/pcDNA 3.1].
Construction of mutant NS3 plasmids.
The plasmid encoding FLAG-tagged E, C, PrM, NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5 cDNA clones in pcDNA 3.1(+) eukaryotic expression plasmid (Addgene catalog number V790-20) was flanked by KpnI ribozyme and XbaI ribozyme sequences as described previously (75). The amino acid residues at R202W, R226G, and R464Q of NS3 in the SA strain were swapped individually or together by overlap extension PCR as described previously. PCR primers for amplification of the DNA inserts were ordered from Huada, and the sequences are available upon request. Briefly, in a PCR, 1 μl of full-length NS3 gene cDNA was mutated and amplified with 20 μM each 202, 226, or 464 site mutation forward and reverse primers and 20 μM each JEV-NS3 forward and reverse primers using PrimeStar GXL DNA polymerase (TaKaRa Biotechnology) according to the manufacturer’s instructions. The size of the PCR products was 1.86 kb, and the products were purified with a gel purification kit. The PCR mixture was heated at 94°C for 2 min, followed by 35 cycles of amplification at 98°C for 10 s, 55°C for 30 s, and 68°C for 1 min 45 s and a final extension at 68°C for 10 min. All NS3 fragments with FLAG tags that included amino acid mutations and NS3 cDNA vectors were digested with the enzyme set KpnI and XbaI (Thermo Scientific). Following digestion, the NS3 fragments with FLAG tags and NS3 cDNA vectors were ligated together at an approximate molar ratio of 1:3 using a DNA ligation kit (TaKaRa Biotechnology) according to the manufacturer’s instructions.
Northern blotting.
A sensitive, nonradioactive Northern blotting method to detect miRNAs was employed as described previously (76). Briefly, total RNA of cells was extracted using RNAiso Plus (TaKaRa Biotechnology) according to the manufacturer’s instructions, eluted in RNase-free water, and stored at −80°C. Total RNA (20 μg) was separated on a denaturing 10% polyacrylamide-8 M urea gel. Then, the RNA was transferred to a nylon membrane (Solarbio Life Sciences, Beijing, China) in a semidry transfer cell (Bio-Rad Laboratories, Hercules, CA, USA) for 30 min at 250 mA. The nylon membrane was blocked and hybridized to DIG-labeled miR-466d-3p and pre-miR-466d-3p probes synthesized by Sangon Biotech (77) and washed. The miR-466d-3p probe sequence was 5′-CTATGTGTGCGTGTATGTATA-DIG-3′. The pre-miR-466d-3p probe sequence was 5′-DIG-CCTGTGTGTGTGTGTGCGTGCGTATCTATGTGTGCGTGTATGTATATGTATATTCATATACACACATGTACATGTACGCACACACAAACACACATG-3′. CDP-Star chemiluminescent substrate (Sigma-Aldrich) was used for luminescence detection. The nylon membrane was exposed to the Tanon 5200 imaging system (Tanon) using Tanon MP software at room temperature and analyzed using Image-Pro plus 6 (Media Cybernetics, Rockville, MD, USA).
Construction of recombinant virus cDNA clones and virus rescue.
The full-length cDNA clone pMW219-P3 (pMW219 is a vector derived from pBR322) was kindly provided by Shengbo Cao (State Key Laboratory of Agricultural Microbiology, Huazhong Agricultural University). The full-length cDNA clone pMW219-P3 was flanked by a T7 promoter at the 5′ end and poly(A) at the 3′ end. The amino acid residues at R202W and R262G of NS3 in the P3 strain were swapped individually with a Fast Mutagenesis System kit (TransGen Biotech, Beijing, China). Briefly, in a PCR, 10 ng of full-length cDNA was mutated and amplified with 200 μM each 202 or 226 site mutation forward and reverse primers (P3-R202W and P3-R226G) according to the manufacturer’s instructions. PCR primers for amplification of DNA inserts were ordered from Huada, and the primer sequences were as follows: P3-R202W(+), 5′-GGG AAA ACC TGG AAA ATT CTG CCA C-3′, and P3-R202W(−), 5′-CAG AAT TTT CCA GGT TTT CCC TGA A-3′; P3-R226G(+), 5′-GTG TTG GCA CCG ACG GGG GTG GTA G-3′, and P3-R226G(−), 5′-GCT ACC CCC GTC GGT GCC AAC A-3′. The PCR mixture was heated at 94°C for 2 min, followed by 35 cycles of amplification at 98°C for 10 s, 55°C for 30 s, and 68°C for 5 min and a final extension at 68°C for 10 min. The size of the PCR products was 15,231 bp, and the products were purified with a GeneJET gel extraction kit (Thermo Fisher Scientific). Rescue of the infectious virus was carried out as described previously (78). Parental pMW219-P3 and mutant full-length cDNA were cotransfected with T7 RNA polymerase into VERO E6 cells using Lipofectamine 3000 (Life Technologies). Five days posttransfection, the supernatants were collected and used to infect BHK cells. The recombinant viruses were passed twice, and virus stocks were titrated by plaque assays.
Ethics statement.
Experimental infection studies were carried out in strict accordance with the Guide for the Care and Use of Laboratory Animals published by the Monitoring Committee of Hubei Province, China. The study protocol was approved by the Scientific Ethics Committee of Huazhong Agricultural University (Hzaumo-2015-018). All efforts were made to minimize animal suffering.
Statistical analyses.
Experiments were carried out at least thrice and elicited similar results. The data generated were analyzed using Prism 5 (GraphPad, San Diego, CA, USA). For all tests, a P value of <0.05 was considered significant.
Data availability.
Transcriptome-sequencing (RNA-seq) data have been deposited in the Sequence Read Archive (SRA) under accession number PRJNA616441. The data for mRNA microarrays and miRNA microarrays have been deposited in the Gene Expression Omnibus (GEO) with accession number GSE148031.
ACKNOWLEDGMENTS
This work was supported by grants from the National Key Research and Development Program of China (grant 2016YFD0500406 to Y.Y.), the National Natural Science Foundation of China (grant 31602071 to Y.Y.), and the Inner Mongolia Autonomous Region Basic Research Project (grant 201503001 to G.L.).
We declare that no competing interests exist.
Y.Y. and G.L. conceived the study; H.J., L.B., L.J., Z.B., J.S., T.Q., Y.Y., and J.Y. performed all experiments; Y.Y. and H.J. wrote the manuscript; and X.W., M.C., S.C., G.W., V.B., and G.L. edited the manuscript. We all critically reviewed and approved the final manuscript.
REFERENCES
- 1.Chen CJ, Kuo MD, Chien LJ, Hsu SL, Wang YM, Lin JH. 1997. RNA-protein interactions: involvement of NS3, NS5, and 3’ noncoding regions of Japanese encephalitis virus genomic RNA. J Virol 71:3466–3473. doi: 10.1128/JVI.71.5.3466-3473.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Utama A, Shimizu H, Morikawa S, Hasebe F, Morita K, Igarashi A, Hatsu M, Takamizawa K, Miyamura T. 2000. Identification and characterization of the RNA helicase activity of Japanese encephalitis virus NS3 protein. FEBS Lett 465:74–78. doi: 10.1016/S0014-5793(99)01705-6. [DOI] [PubMed] [Google Scholar]
- 3.Morita K, Nabeshima T, Buerano CC. 2015. Japanese encephalitis. Rev Sci Tech 34:441–452. doi: 10.20506/rst.34.2.2370. [DOI] [PubMed] [Google Scholar]
- 4.Campbell GL, Hills SL, Fischer M, Jacobson JA, Hoke CH, Hombach JM, Marfin AA, Solomon T, Tsai TF, Tsu VD, Ginsburg AS. 2011. Estimated global incidence of Japanese encephalitis: a systematic review. Bull World Health Organ 89:766–774, 774A-774E. doi: 10.2471/BLT.10.085233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Solomon T, Dung NM, Kneen R, Gainsborough M, Vaughn DW, Khanh VT. 2000. Japanese encephalitis. J Neurol Neurosurg Psychiatry 68:405–415. doi: 10.1136/jnnp.68.4.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen CJ, Chen JH, Chen SY, Liao SL, Raung SL. 2004. Upregulation of RANTES gene expression in neuroglia by Japanese encephalitis virus infection. J Virol 78:12107–12119. doi: 10.1128/JVI.78.22.12107-12119.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen CJ, Ou YC, Lin SY, Raung SL, Liao SL, Lai CY, Chen SY, Chen JH. 2010. Glial activation involvement in neuronal death by Japanese encephalitis virus infection. J Gen Virol 91:1028–1037. doi: 10.1099/vir.0.013565-0. [DOI] [PubMed] [Google Scholar]
- 8.Das S, Mishra MK, Ghosh J, Basu A. 2008. Japanese encephalitis virus infection induces IL-18 and IL-1beta in microglia and astrocytes: correlation with in vitro cytokine responsiveness of glial cells and subsequent neuronal death. J Neuroimmunol 195:60–72. doi: 10.1016/j.jneuroim.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 9.Ghoshal A, Das S, Ghosh S, Mishra MK, Sharma V, Koli P, Sen E, Basu A. 2007. Proinflammatory mediators released by activated microglia induces neuronal death in Japanese encephalitis. Glia 55:483–496. doi: 10.1002/glia.20474. [DOI] [PubMed] [Google Scholar]
- 10.Khanna N, Mathur A, Chaturvedi UC. 1994. Regulation of vascular permeability by macrophage-derived chemotactic factor produced in Japanese encephalitis. Immunol Cell Biol 72:200–204. doi: 10.1038/icb.1994.30. [DOI] [PubMed] [Google Scholar]
- 11.Han YW, Choi JY, Uyangaa E, Kim SB, Kim JH, Kim BS, Kim K, Eo SK. 2014. Distinct dictation of Japanese encephalitis virus-induced neuroinflammation and lethality via triggering TLR3 and TLR4 signal pathways. PLoS Pathog 10:e1004319. doi: 10.1371/journal.ppat.1004319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nazmi A, Mukherjee S, Kundu K, Dutta K, Mahadevan A, Shankar SK, Basu A. 2014. TLR7 is a key regulator of innate immunity against Japanese encephalitis virus infection. Neurobiol Dis 69:235–247. doi: 10.1016/j.nbd.2014.05.036. [DOI] [PubMed] [Google Scholar]
- 13.Chua JJ, Bhuvanakantham R, Chow VT, Ng ML. 2005. Recombinant non-structural 1 (NS1) protein of dengue-2 virus interacts with human STAT3beta protein. Virus Res 112:85–94. doi: 10.1016/j.virusres.2005.03.025. [DOI] [PubMed] [Google Scholar]
- 14.Wang W, Li G, De W, Luo Z, Pan P, Tian M, Wang Y, Xiao F, Li A, Wu K, Liu X, Rao L, Liu F, Liu Y, Wu J. 2018. Zika virus infection induces host inflammatory responses by facilitating NLRP3 inflammasome assembly and interleukin-1beta secretion. Nat Commun 9:106. doi: 10.1038/s41467-017-02645-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raung SL, Chen SY, Liao SL, Chen JH, Chen CJ. 2007. Japanese encephalitis virus infection stimulates Src tyrosine kinase in neuron/glia. Neurosci Lett 419:263–268. doi: 10.1016/j.neulet.2007.04.036. [DOI] [PubMed] [Google Scholar]
- 16.Thounaojam MC, Kundu K, Kaushik DK, Swaroop S, Mahadevan A, Shankar SK, Basu A. 2014. MicroRNA 155 regulates Japanese encephalitis virus-induced inflammatory response by targeting Src homology 2-containing inositol phosphatase 1. J Virol 88:4798–4810. doi: 10.1128/JVI.02979-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu B, Ye J, Nie Y, Ashraf U, Zohaib A, Duan X, Fu ZF, Song Y, Chen H, Cao S. 2015. MicroRNA-15b modulates Japanese encephalitis virus-mediated inflammation via targeting RNF125. J Immunol 195:2251–2262. doi: 10.4049/jimmunol.1500370. [DOI] [PubMed] [Google Scholar]
- 18.Ashraf U, Zhu B, Ye J, Wan S, Nie Y, Chen Z, Cui M, Wang C, Duan X, Zhang H, Chen H, Cao S. 2016. MicroRNA-19b-3p modulates Japanese encephalitis virus-mediated inflammation via targeting RNF11. J Virol 90:4780–4795. doi: 10.1128/JVI.02586-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thounaojam MC, Kaushik DK, Kundu K, Basu A. 2014. MicroRNA-29b modulates Japanese encephalitis virus-induced microglia activation by targeting tumor necrosis factor alpha-induced protein 3. J Neurochem 129:143–154. doi: 10.1111/jnc.12609. [DOI] [PubMed] [Google Scholar]
- 20.Chen Z, Ye J, Ashraf U, Li Y, Wei S, Wan S, Zohaib A, Song Y, Chen H, Cao S. 2016. MicroRNA-33a-5p modulates Japanese encephalitis virus replication by targeting eukaryotic translation elongation factor 1A1. J Virol 90:3722–3734. doi: 10.1128/JVI.03242-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma N, Kumawat KL, Rastogi M, Basu A, Singh SK. 2016. Japanese encephalitis virus exploits the microRNA-432 to regulate the expression of Suppressor of Cytokine Signaling (SOCS) 5. Sci Rep 6:27685. doi: 10.1038/srep27685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ibrahim F, Rymarquis LA, Kim EJ, Becker J, Balassa E, Green PJ, Cerutti H. 2010. Uridylation of mature miRNAs and siRNAs by the MUT68 nucleotidyltransferase promotes their degradation in Chlamydomonas. Proc Natl Acad Sci U S A 107:3906–3911. doi: 10.1073/pnas.0912632107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chatterjee S, Grosshans H. 2009. Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature 461:546–549. doi: 10.1038/nature08349. [DOI] [PubMed] [Google Scholar]
- 24.Cazalla D, Yario T, Steitz JA, Steitz J. 2010. Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science 328:1563–1566. doi: 10.1126/science.1187197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marcinowski L, Tanguy M, Krmpotic A, Rädle B, Lisnić VJ, Tuddenham L, Chane-Woon-Ming B, Ruzsics Z, Erhard F, Benkartek C, Babic M, Zimmer R, Trgovcich J, Koszinowski UH, Jonjic S, Pfeffer S, Dölken L. 2012. Degradation of cellular mir-27 by a novel, highly abundant viral transcript is important for efficient virus replication in vivo. PLoS Pathog 8:e1002510. doi: 10.1371/journal.ppat.1002510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen PS, Su JL, Cha ST, Tarn WY, Wang MY, Hsu HC, Lin MT, Chu CY, Hua KT, Chen CN, Kuo TC, Chang KJ, Hsiao M, Chang YW, Chen JS, Yang PC, Kuo ML. 2011. miR-107 promotes tumor progression by targeting the let-7 microRNA in mice and humans. J Clin Invest 121:3442–3455. doi: 10.1172/JCI45390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ameres SL, Horwich MD, Hung JH, Xu J, Ghildiyal M, Weng Z, Zamore PD. 2010. Target RNA-directed trimming and tailing of small silencing RNAs. Science 328:1534–1539. doi: 10.1126/science.1187058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Libri V, Helwak A, Miesen P, Santhakumar D, Borger JG, Kudla G, Grey F, Tollervey D, Buck AH. 2012. Murine cytomegalovirus encodes a miR-27 inhibitor disguised as a target. Proc Natl Acad Sci U S A 109:279–284. doi: 10.1073/pnas.1114204109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee S, Song J, Kim S, Kim J, Hong Y, Kim Y, Kim D, Baek D, Ahn K. 2013. Selective degradation of host microRNAs by an intergenic HCMV noncoding RNA accelerates virus production. Cell Host Microbe 13:678–690. doi: 10.1016/j.chom.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 30.Backes S, Shapiro JS, Sabin LR, Pham AM, Reyes I, Moss B, Cherry S, tenOever BR. 2012. Degradation of host microRNAs by poxvirus poly(A) polymerase reveals terminal RNA methylation as a protective antiviral mechanism. Cell Host Microbe 12:200–210. doi: 10.1016/j.chom.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Happel C, Ramalingam D, Ziegelbauer JM. 2016. Virus-mediated alterations in miRNA factors and degradation of viral miRNAs by MCPIP1. PLoS Biol 14:e2000998. doi: 10.1371/journal.pbio.2000998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Friedlander MR, Mackowiak SD, Li N, Chen W, Rajewsky N. 2012. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res 40:37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kakumani PK, Ponia SS, S RK, Sood V, Chinnappan M, Banerjea AC, Medigeshi GR, Malhotra P, Mukherjee SK, Bhatnagar RK. 2013. Role of RNA interference (RNAi) in dengue virus replication and identification of NS4B as an RNAi suppressor. J Virol 87:8870–8883. doi: 10.1128/JVI.02774-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schneider-Poetsch T, Ju J, Eyler DE, Dang Y, Bhat S, Merrick WC, Green R, Shen B, Liu JO. 2010. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat Chem Biol 6:209–217. doi: 10.1038/nchembio.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Casse C, Giannoni F, Nguyen VT, Dubois MF, Bensaude O. 1999. The transcriptional inhibitors, actinomycin D and alpha-amanitin, activate the HIV-1 promoter and favor phosphorylation of the RNA polymerase II C-terminal domain. J Biol Chem 274:16097–16106. doi: 10.1074/jbc.274.23.16097. [DOI] [PubMed] [Google Scholar]
- 36.Murray CL, Jones CT, Rice CM. 2008. Architects of assembly: roles of Flaviviridae non-structural proteins in virion morphogenesis. Nat Rev Microbiol 6:699–708. doi: 10.1038/nrmicro1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lindenbach BD, Thiel HJ, Rice CM. 2007. Flaviviridae: the viruses and their replication. Lippincott-Raven Publishers, Philadelphia, PA. [Google Scholar]
- 38.van Rij RP, Saleh MC, Berry B, Foo C, Houk A, Antoniewski C, Andino R. 2006. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev 20:2985–2995. doi: 10.1101/gad.1482006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. 2005. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schnettler E, de Vries W, Hemmes H, Haasnoot J, Kormelink R, Goldbach R, Berkhout B. 2009. The NS3 protein of rice hoja blanca virus complements the RNAi suppressor function of HIV-1 Tat. EMBO Rep 10:258–263. doi: 10.1038/embor.2009.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haasnoot J, de Vries W, Geutjes EJ, Prins M, de Haan P, Berkhout B. 2007. The Ebola virus VP35 protein is a suppressor of RNA silencing. PLoS Pathog 3:e86. doi: 10.1371/journal.ppat.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumar M, Gromiha MM, Raghava GP. 2008. Prediction of RNA binding sites in a protein using SVM and PSSM profile. Proteins 71:189–194. doi: 10.1002/prot.21677. [DOI] [PubMed] [Google Scholar]
- 43.Muppirala UK, Honavar VG, Dobbs D. 2011. Predicting RNA-protein interactions using only sequence information. BMC Bioinformatics 12:489. doi: 10.1186/1471-2105-12-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li S, Yamashita K, Amada KM, Standley DM. 2014. Quantifying sequence and structural features of protein-RNA interactions. Nucleic Acids Res 42:10086–10098. doi: 10.1093/nar/gku681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tuvshinjargal N, Lee W, Park B, Han K. 2016. PRIdictor: protein-RNA interaction predictor. Biosystems 139:17–22. doi: 10.1016/j.biosystems.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 46.Janas T, Widmann JJ, Knight R, Yarus M. 2010. Simple, recurring RNA binding sites for L-arginine. RNA 16:805–816. doi: 10.1261/rna.1979410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo YE, Steitz JA. 2014. Virus meets host microRNA: the destroyer, the booster, the hijacker. Mol Cell Biol 34:3780–3787. doi: 10.1128/MCB.00871-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang P, Hou J, Lin L, Wang C, Liu X, Li D, Ma F, Wang Z, Cao X. 2010. Inducible microRNA-155 feedback promotes type I IFN signaling in antiviral innate immunity by targeting suppressor of cytokine signaling 1. J Immunol 185:6226–6233. doi: 10.4049/jimmunol.1000491. [DOI] [PubMed] [Google Scholar]
- 49.Burgyan J, Havelda Z. 2011. Viral suppressors of RNA silencing. Trends Plant Sci 16:265–272. doi: 10.1016/j.tplants.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 50.Moon SL, Dodd BJ, Brackney DE, Wilusz CJ, Ebel GD, Wilusz J. 2015. Flavivirus sfRNA suppresses antiviral RNA interference in cultured cells and mosquitoes and directly interacts with the RNAi machinery. Virology 485:322–329. doi: 10.1016/j.virol.2015.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Py B, Higgins CF, Krisch HM, Carpousis AJ. 1996. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature 381:169–172. doi: 10.1038/381169a0. [DOI] [PubMed] [Google Scholar]
- 52.Evguenieva-Hackenberg E, Walter P, Hochleitner E, Lottspeich F, Klug G. 2003. An exosome-like complex in Sulfolobus solfataricus. EMBO Rep 4:889–893. doi: 10.1038/sj.embor.embor929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Song L, Liu H, Gao S, Jiang W, Huang W. 2010. Cellular microRNAs inhibit replication of the H1N1 influenza A virus in infected cells. J Virol 84:8849–8860. doi: 10.1128/JVI.00456-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng Z, Ke X, Wang M, He S, Li Q, Zheng C, Zhang Z, Liu Y, Wang H. 2013. Human microRNA hsa-miR-296-5p suppresses enterovirus 71 replication by targeting the viral genome. J Virol 87:5645–5656. doi: 10.1128/JVI.02655-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Slonchak A, Shannon RP, Pali G, Khromykh AA. 2015. Human microRNA miR-532-5p exhibits antiviral activity against West Nile virus via suppression of host genes SESTD1 and TAB3 required for virus replication. J Virol 90:2388–2402. doi: 10.1128/JVI.02608-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jia H, Wang X, Anderson JT, Jankowsky E. 2012. RNA unwinding by the Trf4/Air2/Mtr4 polyadenylation (TRAMP) complex. Proc Natl Acad Sci U S A 109:7292–7297. doi: 10.1073/pnas.1201085109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dziembowski A, Piwowarski J, Hoser R, Minczuk M, Dmochowska A, Siep M, van der Spek H, Grivell L, Stepien PP. 2003. The yeast mitochondrial degradosome. Its composition, interplay between RNA helicase and RNase activities and the role in mitochondrial RNA metabolism. J Biol Chem 278:1603–1611. doi: 10.1074/jbc.M208287200. [DOI] [PubMed] [Google Scholar]
- 58.Elbarbary RA, Miyoshi K, Hedaya O, Myers JR, Maquat LE. 2017. UPF1 helicase promotes TSN-mediated miRNA decay. Genes Dev 31:1483–1493. doi: 10.1101/gad.303537.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Salzman DW, Shubert-Coleman J, Furneaux H. 2007. P68 RNA helicase unwinds the human let-7 microRNA precursor duplex and is required for let-7-directed silencing of gene expression. J Biol Chem 282:32773–32779. doi: 10.1074/jbc.M705054200. [DOI] [PubMed] [Google Scholar]
- 60.Chernov AV, Shiryaev SA, Aleshin AE, Ratnikov BI, Smith JW, Liddington RC, Strongin AY. 2008. The two-component NS2B-NS3 proteinase represses DNA unwinding activity of the West Nile virus NS3 helicase. J Biol Chem 283:17270–17278. doi: 10.1074/jbc.M801719200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Swarbrick CMD, Basavannacharya C, Chan KWK, Chan SA, Singh D, Wei N, Phoo WW, Luo D, Lescar J, Vasudevan SG. 2017. NS3 helicase from dengue virus specifically recognizes viral RNA sequence to ensure optimal replication. Nucleic Acids Res 45:12904–12920. doi: 10.1093/nar/gkx1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chiang PY, Wu HN. 2016. The role of surface basic amino acids of dengue virus NS3 helicase in viral RNA replication and enzyme activities. FEBS Lett 590:2307–2320. doi: 10.1002/1873-3468.12232. [DOI] [PubMed] [Google Scholar]
- 63.Yiang GT, Chen YH, Chou PL, Chang WJ, Wei CW, Yu YL. 2013. The NS3 protease and helicase domains of Japanese encephalitis virus trigger cell death via caspase dependent and independent pathways. Mol Med Rep 7:826–830. doi: 10.3892/mmr.2013.1261. [DOI] [PubMed] [Google Scholar]
- 64.Yang TC, Shiu SL, Chuang PH, Lin YJ, Wan L, Lan YC, Lin CW. 2009. Japanese encephalitis virus NS2B-NS3 protease induces caspase 3 activation and mitochondria-mediated apoptosis in human medulloblastoma cells. Virus Res 143:77–85. doi: 10.1016/j.virusres.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 65.Su L, Radek JT, Hallenga K, Hermanto P, Chan G, Labeots LA, Weiss MA. 1997. RNA recognition by a bent alpha-helix regulates transcriptional antitermination in phage lambda. Biochemistry 36:12722–12732. doi: 10.1021/bi971408k. [DOI] [PubMed] [Google Scholar]
- 66.Frankel AD, Young JA. 1998. HIV-1: fifteen proteins and an RNA. Annu Rev Biochem 67:1–25. doi: 10.1146/annurev.biochem.67.1.1. [DOI] [PubMed] [Google Scholar]
- 67.Agarwal V, Bell GW, Nam JW, Bartel DP. 2015. Predicting effective microRNA target sites in mammalian mRNAs. Elife 4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wong N, Wang X. 2015. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res 43:D146–152. doi: 10.1093/nar/gku1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Betel D, Wilson M, Gabow A, Marks DS, Sander C. 2008. The microRNA.org resource: targets and expression. Nucleic Acids Res 36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, Mering CV. 2019. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 72.Yang Y, Ye J, Yang X, Jiang R, Chen H, Cao S. 2011. Japanese encephalitis virus infection induces changes of mRNA profile of mouse spleen and brain. Virol J 8:80. doi: 10.1186/1743-422X-8-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Peritz T, Zeng F, Kannanayakal TJ, Kilk K, Eiriksdottir E, Langel U, Eberwine J. 2006. Immunoprecipitation of mRNA-protein complexes. Nat Protoc 1:577–580. doi: 10.1038/nprot.2006.82. [DOI] [PubMed] [Google Scholar]
- 74.Marmisolle FE, Garcia ML, Reyes CA. 2018. RNA-binding protein immunoprecipitation as a tool to investigate plant miRNA processing interference by regulatory proteins of diverse origin. Plant Methods 14:9. doi: 10.1186/s13007-018-0276-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ye J, Chen Z, Zhang B, Miao H, Zohaib A, Xu Q, Chen H, Cao S. 2013. Heat shock protein 70 is associated with replicase complex of Japanese encephalitis virus and positively regulates viral genome replication. PLoS One 8:e75188. doi: 10.1371/journal.pone.0075188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kim SW, Li Z, Moore PS, Monaghan AP, Chang Y, Nichols M, John B. 2010. A sensitive non-radioactive Northern blot method to detect small RNAs. Nucleic Acids Res 38:e98. doi: 10.1093/nar/gkp1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wu W, Gong P, Li J, Yang J, Zhang G, Li H, Yang Z, Zhang X. 2014. Simple and nonradioactive detection of microRNAs using digoxigenin (DIG)-labeled probes with high sensitivity. RNA 20:580–584. doi: 10.1261/rna.042150.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.de Wispelaere M, Frenkiel M-P, Desprès P. 2015. A Japanese encephalitis virus genotype 5 molecular clone is highly neuropathogenic in a mouse model: impact of the structural protein region on virulence. J Virol 89:5862–5875. doi: 10.1128/JVI.00358-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Transcriptome-sequencing (RNA-seq) data have been deposited in the Sequence Read Archive (SRA) under accession number PRJNA616441. The data for mRNA microarrays and miRNA microarrays have been deposited in the Gene Expression Omnibus (GEO) with accession number GSE148031.