Interferon (IFN)-stimulated gene (ISG) induction through IFN signaling is important to create an antiviral state and usually directly inhibits virus infection. The present study first demonstrated that PDCoV nsp5 can cleave mRNA-decapping enzyme 1a (DCP1A) to attenuate its antiviral activity. Furthermore, cleaving DCP1A is a common characteristic of nsp5 proteins from different coronaviruses (CoVs), which represents a common immune evasion mechanism of CoVs. Previous evidence showed that CoV nsp5 cleaves the NF-κB essential modulator and signal transducer and activator of transcription 2. Taken together, CoV nsp5 is a potent IFN antagonist because it can simultaneously target different aspects of the host IFN system, including IFN production and signaling and effector molecules.
KEYWORDS: IFN-stimulated genes, porcine deltacoronavirus, antiviral activity, mRNA-decapping enzyme 1a, DCP1A, nonstructural protein 5, nsp5
ABSTRACT
Porcine deltacoronavirus (PDCoV) is an emerging swine enteropathogenic coronavirus. The nonstructural protein nsp5, also called 3C-like protease, is responsible for processing viral polyprotein precursors in coronavirus (CoV) replication. Previous studies have shown that PDCoV nsp5 cleaves the NF-κB essential modulator and the signal transducer and activator of transcription 2 to disrupt interferon (IFN) production and signaling, respectively. Whether PDCoV nsp5 also cleaves IFN-stimulated genes (ISGs), IFN-induced antiviral effector molecules, remains unclear. In this study, we screened 14 classical ISGs and found that PDCoV nsp5 cleaved the porcine mRNA-decapping enzyme 1a (pDCP1A) through its protease activity. Similar cleavage of endogenous pDCP1A was also observed in PDCoV-infected cells. PDCoV nsp5 cleaved pDCP1A at glutamine 343 (Q343), and the cleaved pDCP1A fragments, pDCP1A1–343 and pDCP1A344–580, were unable to inhibit PDCoV infection. Mutant pDCP1A-Q343A, which resists nsp5-mediated cleavage, exhibited a stronger ability to inhibit PDCoV infection than wild-type pDCP1A. Interestingly, the Q343 cleavage site is highly conserved in DCP1A homologs from other mammalian species. Further analyses demonstrated that nsp5 encoded by seven tested CoVs that can infect human or pig also cleaved pDCP1A and human DCP1A, suggesting that DCP1A may be the common target for cleavage by nsp5 of mammalian CoVs.
IMPORTANCE Interferon (IFN)-stimulated gene (ISG) induction through IFN signaling is important to create an antiviral state and usually directly inhibits virus infection. The present study first demonstrated that PDCoV nsp5 can cleave mRNA-decapping enzyme 1a (DCP1A) to attenuate its antiviral activity. Furthermore, cleaving DCP1A is a common characteristic of nsp5 proteins from different coronaviruses (CoVs), which represents a common immune evasion mechanism of CoVs. Previous evidence showed that CoV nsp5 cleaves the NF-κB essential modulator and signal transducer and activator of transcription 2. Taken together, CoV nsp5 is a potent IFN antagonist because it can simultaneously target different aspects of the host IFN system, including IFN production and signaling and effector molecules.
INTRODUCTION
Coronaviruses (CoVs), which belong in the family Coronaviridae of the order Nidovirales, are enveloped positive single-strand RNA viruses that cause respiratory, enteric, hepatic, or neurological diseases in both human and animals (1–3). CoVs are classified into four genera, including Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus (4). As a member of the Deltacoronavirus genus, porcine deltacoronavirus (PDCoV), a newly identified swine enteropathogenic coronavirus, was first reported in Hong Kong during a molecular epidemiology study in 2012 (4). Since 2014, at least 20 states of the United States have reported PDCoV outbreaks, which caused watery diarrhea, vomiting, and mortality in piglets (5–9). Thereafter, PDCoV was detected in many countries worldwide, such as Canada (10), South Korea (11, 12), Japan (13), China (14, 15), Thailand (16), and Vietnam (17, 18), and has become a serious public health and economic issue with enough attention (7, 19–21).
As a first line to defend against invading viruses, interferon (IFN) and IFN-induced Janus kinase-signal transducer and activator of transcription (JAK-STAT) signaling pathway can enhance the transcription of hundreds of IFN-stimulated genes (ISGs), which help to control viral infection (22–25). Many ISGs can target viral life cycles (22, 26, 27), such as transcription and translation processes, and can target the viral RNA directly. For example, the IFN-induced protein with tetratricopeptide repeats (IFIT) family members IFIT1, IFIT2, IFIT3, and IFIT5 act as sensors to bind viral single-stranded RNA bearing a 5′-triphosphate group (28–30). IFN-induced 2′,5′-oligoadenylate synthetases synthesize 2′,5′-oligoadenylate (2-5A), eventually activating viral RNA cleavage by RNase L (31–33). mRNA-decapping enzyme 1a (DCP1A), located at processing bodies, helps to remove the 5′ N7-methylguanosine cap of mRNA and induce subsequent mRNA degradation (34–37). Due to their important antiviral function, viruses also develop different strategies to counteract the function of ISGs (38–41). For example, murine CoV (MHV) ns2 protein, the virus-encoded phosphodiesterase, cleaves 2-5A to impair RNase L activation (42). Additionally, Middle East respiratory syndrome coronavirus (MERS-CoV) NS4b mediates RNase L inhibition through the enzymatic degradation of 2-5A (43).
As an emerging enteropathogenic CoV, mechanisms utilized by PDCoV to antagonize the host IFN system remain largely unknown (1). Our previous studies showed that PDCoV infection inhibited Sendai virus-mediated IFN production and that the viral accessory protein NS6 interfered with RIG-I/MDA5 binding to double-stranded RNA (44, 45). We also found that PDCoV nonstructural protein 5 (nsp5), the 3C-like protease that mediates the cleavage of viral polyprotein precursors, is able to cleave the NF-κB essential modulator (NEMO) and STAT2 to impair IFN production and the IFN-induced JAK-STAT pathway, respectively, and these cleavages depend on the protease activity of nsp5 (46, 47). However, whether PDCoV or its encoded proteins counteract the function of ISGs has not yet been reported. In consideration of the cleavage activity of PDCoV nsp5, we investigated whether PDCoV nsp5 can cleave ISGs to antagonize the antiviral effector of ISGs. Here, we cloned 14 classical ISGs and screened the possible cleavage of these ISGs mediated by PDCoV nsp5. Porcine DCP1A (pDCP1A) was identified as the target of PDCoV nsp5 for cleavage, which impaired the antiviral activity of pDCP1A. Moreover, we found that nsp5 encoded by other CoVs also cleaved DCP1A, revealing a possible common immune evasion mechanism of CoVs.
RESULTS
PDCoV nsp5 cleaves pDCP1A via its protease activity.
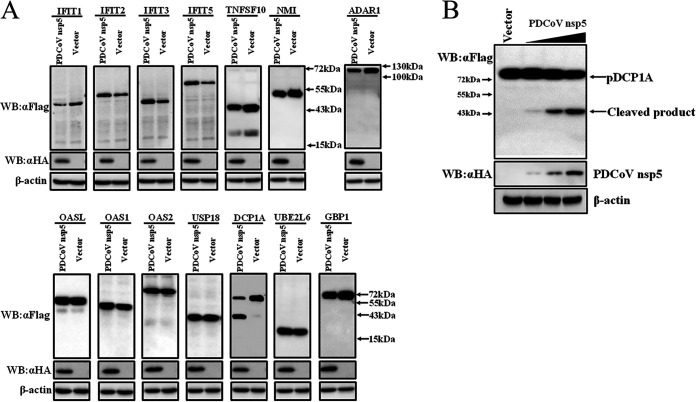
To investigate whether PDCoV nsp5 can cleave ISGs, 14 classical ISGs, including IFIT1, IFIT2, IFIT3, IFIT5, OASL, OAS1, OAS2, DCP1A, tumor necrosis factor superfamily member 10 (TNFSF10), N-myc and STAT interactor (NMI), RNA-specific adenosine deaminase I (ADAR1), ubiquitin-specific peptidase 18 (USP18), ubiquitin-conjugating enzyme E2 L6 (UBE2L6), and guanylate-binding protein 1 (GBP1), were cloned from PK-15 cells (a porcine kidney cell line) into the pCAGGS-Flag vector. These ISGs have been reported to restrict infections of some viruses by interacting with viral components or altering cellular physiology (30, 48–53). Expression constructs harboring these ISGs were cotransfected with PDCoV nsp5 eukaryotic expression plasmid into HEK-293T cells, followed by Western blotting with anti-Flag antibody. As shown in Fig. 1A, one faster-migrating protein band, representing the possible cleavage production, was observed in samples cotransfected with pCAGGS-Flag-pDCP1A and pCAGGS-nsp5-HA. No cleavage product was detected after cotransfection with the expression plasmids of other ISGs, while IFIT1, TNFSF10, and ADAR1 expression levels slightly decreased under the expression of PDCoV nsp5 (Fig. 1A). These results imply that pDCP1A is a possible target cleaved by PDCoV nsp5. To further confirm the possible cleavage mediated by PDCoV nsp5, pCAGGS-Flag-pDCP1A was cotransfected with different doses of pCAGGS-nsp5-HA into HEK-293T cells. Western blotting showed that pDCP1A cleavage gradually increased in a PDCoV nsp5-dose-dependent manner (Fig. 1B).
FIG 1.
PDCoV nsp5 targets pDCP1A for cleavage. (A) HEK-293T cells cultured in 6-well plates were cotransfected with 0.2 μg of PDCoV nsp5 expression plasmid (pCAGGS-nsp5-HA) or vector along with 2.5 μg of Flag-tagged IFIT1, IFIT2, IFIT3, IFIT5, OASL, OAS1, OAS2, TNFSF10, NMI, ADAR1, USP18, DCP1A, UBE2L6, or GBP1 expression plasmid. After 28 h, cells were lysed and detected by Western blotting with anti-Flag antibody. (B) HEK-293T cells were cotransfected with an N-terminal Flag-tagged pDCP1A and various doses of pCAGGS-nsp5-HA. After 28 h, cells were lysed for Western blotting.
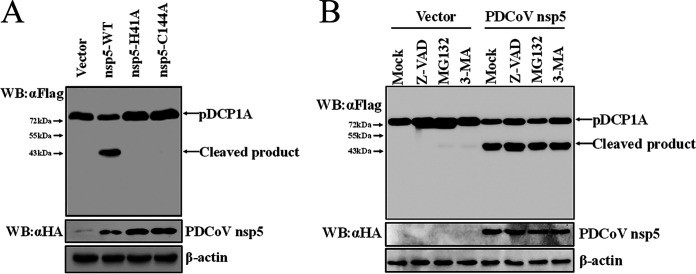
To further investigate whether PDCoV nsp5-mediated pDCP1A cleavage is associated with its protease activity, two nsp5 mutants, expression constructs encoding nsp5 H41A and C144A that do not show protease activity (46), were tested by cotransfection with pCAGGS-Flag-pDCP1A. As shown in Fig. 2A, the protease activity mutants nsp5 H41A and C144A failed to cleave pDCP1A (Fig. 2A), suggesting that PDCoV nsp5 cleaves pDCP1A via its protease activity.
FIG 2.
PDCoV nsp5 cleaves pDCP1A depending on its protease activity. (A) HEK-293T cells were cotransfected with expression constructs encoding wild-type PDCoV nsp5 or its protease-defective mutants (H41A or C144A) and pDCP1A expression plasmid. After 28 h, cells were lysed for Western blotting. (B) HEK-293T cells were cotransfected with pCAGGS-nsp5-HA and pCAGGS-Flag-pDCP1A. After 24 h, cells were treated with MG132 (final concentration of 20 μM), Z-VAD-FMK (final concentration of 20 μM), or 3-MA (final concentration of 5 mM) for another 8 h. Cells were then lysed for Western blotting. WT, wild type.
Previous studies suggested that some viral 3C or 3C-like proteases, such as Seneca Valley virus (SVV) 3C protease, not only mediate the cleavage of host and viral proteins but also degrade some host proteins (54–56). The ubiquitin proteasome system, autophagy, and apoptosis are three major intracellular protein degradation pathways in eukaryotic cells. Therefore, we evaluated PDCoV nsp5-mediated pDCP1A cleavage following treatment with the proteasome inhibitor MG132, the caspase inhibitor Z-VAD-FMK, or the autophagy inhibitor 3-MA. PDCoV nsp5-mediated pDCP1A cleavage was not affected by these inhibitors, confirming this cleavage occurred independently of cellular caspases or the proteasome (Fig. 2B).
PDCoV infection cleaves endogenous pDCP1A.
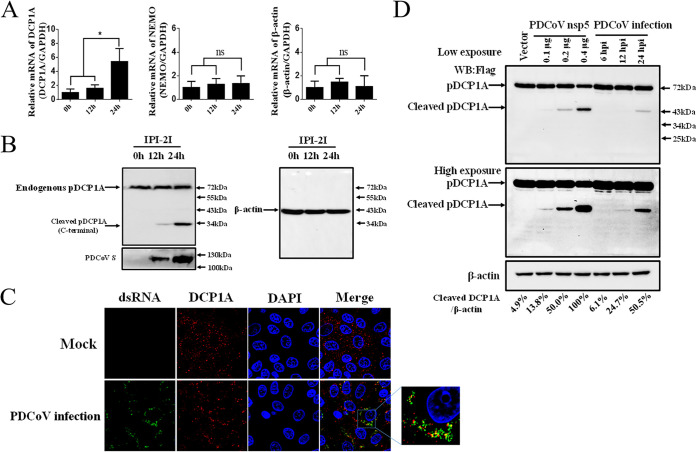
To further confirm whether pDCP1A cleavage under ectopic expression is relevant to PDCoV biology, endogenous pDCP1A was evaluated in PDCoV-infected cells. Quantitative real-time PCR (RT-qPCR) showed that pDCP1A mRNA transcription was slightly induced in PDCoV-infected IPI-2I cells (a porcine ileum epithelial cell line) at 24 h postinfection (hpi), while NEMO or β-actin mRNA had no obvious change and the fluorescence signal of each gene were detected in the control samples without reverse transcription. (Fig. 3A). These results suggested that PDCoV infection upregulated pDCP1A mRNA expression. We further evaluated pDCP1A protein expression in PDCoV-infected cells. Because an antibody against pDCP1A is not currently available, we used a rabbit monoclonal antibody against human DCP1A (hDCP1A) to detect endogenous pDCP1A protein in PDCoV-infected IPI-2I cells. As shown in Fig. 3B, an evident cleavage fragment was observed at both 12 hpi and 24 hpi; however, the protein levels of pDCP1A (including the cleaved fragments) were not affected, even at the late time point during infection. In addition, because the antibody used targets the C-terminal domain of hDCP1A and the cleaved product is approximately 34 kDa, we speculated that the cleaved site may be at the middle or C-terminal domain of pDCP1A. Due to lack of the antibody against PDCoV nsp5, it is difficult to investigate the colocalization of nsp5 and pDCP1A in the context of PDCoV infection. Thus, we examined the colocalization of endogenous pDCP1A and double-stranded RNA (dsRNA) during PDCoV infection by using a monoclonal antibody against dsRNA. As shown in Fig. 3C, endogenous pDCP1A was presented as a punctate distribution and partly colocalized with dsRNA in PDCoV-infected cells.
FIG 3.
PDCoV infection cleaves endogenous pDCP1A. (A) PDCoV strain CHN-HN-2014 was used to infect IPI-2I cells (MOI = 0.5) and then harvested to detect pDCP1A mRNA by RT-qPCR at 0, 12, or 24 h postinfection (hpi). (B) IPI-2I cells were infected with PDCoV as described for panel A. Endogenous pDCP1A was detected by Western blotting with antibody against human DCP1A (hDCP1A) at 0, 12, and 24 hpi. (C) IPI-2I cells were infected with PDCoV (MOI = 0.5). At 24 hpi, cells were fixed and then stained with mouse monoclonal antibody specific for dsRNA and rabbit monoclonal antibody against DCP1A, followed by incubation with Alexa Fluor 488-conjugated donkey anti-mouse IgG antibody (green) and 594-conjugated donkey anti-rabbit IgG antibody (red). Nuclei were stained with DAPI (blue). (D) IPI-2I cells were cotransfected with pCAGGS-Flag-pDCP1A and various doses of PDCoV nsp5 expression plasmid. In parallel, IPI-2I cells were transfected with 2.5 μg of pCAGGS-Flag-pDCP1A and then infected with PDCoV (MOI = 0.5). The cotransfected cells were collected at 28 h posttransfection, and the infected cells were collected at different time points (6, 12, 24 hpi) for Western blotting. Quantification of the Western blots was performed by grayscale value analysis with ImageJ software, and the relative cleaved DCP1A data were normalized to those for β-actin. *, P < 0.01; ns, not signficant.
In order to evaluate the difference of 3CLpro activity between nsp5 overexpression and PDCoV-infected systems, we compared the cleavage of pDCP1A in PDCoV-infected and plasmid-transfected IPI-2I cells. As shown in Fig. 3D, pDCP1A was cleaved by PDCoV nsp5 in a dose-dependent manner under the overexpression of nsp5, and the cleavage of pDCP1A was also detected at 12 hpi and 24 hpi. Quantification of the Western blots showed that the cleavage of pDCP1A at 24 hpi was consistent with that of transfection with 0.2 μg of pCAGGS-nsp5-HA. These results suggested that the expression level of nsp5 in PDCoV-infected cells should be enough to cleave pDCP1A.
PDCoV nsp5 cleaves pDCP1A at residue Q343.
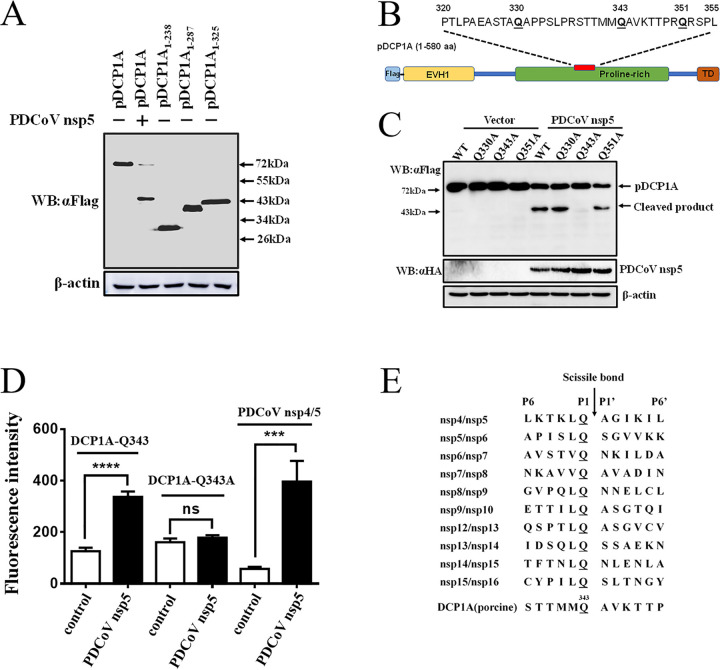
To identify the recognition site of pDCP1A in PDCoV nsp5-mediated cleavage, we generated three truncated pDCP1A mutants, pDCP1A1–238, pDCP1A1–287, and pDCP1A1–325. Western blotting showed that the size of cleaved N-terminally Flag-tagged pDCP1A product was near 43 kDa, which seemed slightly longer than that for the pDCP1A1–325 mutant, indicating the cleavage site was located near residue 325 of pDCP1A (Fig. 4A). Previous studies showed that CoVs nsp5 only recognizes the glutamine (Q) residue at the P1 position for substrate cleavage (57, 58). The possible Q residue at the P1 position was analyzed, and three Q residues exist among the peptide of pDCP1A320–355 (Fig. 4B). Thus, these three Q residues (Q330, Q343, and Q351) were mutated to alanine (A). Cotransfection of pCAGGS-nsp5-HA and expression plasmids of mutant pDCP1A-Q330A, -Q343A, or -Q351A in HEK-293T cells showed that mutant pDCP1A-Q343A resisted PDCoV nsp5-mediated cleavage, while cleavage of mutant pDCP1A-Q330A or -Q351A was not changed (Fig. 4C). These results indicated that Q343 may be the nsp5 cleavage site per se or that mutation at this site may affect potential cleavage in pDCP1A. Considering the conserved Q residue at the P1 position and the size of cleaved product, the cleavage P1 position must be the Q residue which is near the C-terminal pDCP1A 325 residue. Due to the successful cleavage of pDCP1A-Q330 and Q351 mutants, Q343 is a unique site which resisted the cleavage. The cleavage of Q343 was further detected in vitro by fluorescence resonance energy transfer (FRET) assays. To this end, three fluorogenic peptide substrates derived from pDCP1A [STTMMQ(343)AVKTTP; P6 to P6′], pDCP1A-Q343A [TTMMA(343)AVK], and nsp4/nsp5 in PDCoV replicase polyprotein (LKTKLQ/AGIKIL) were incubated with purified PDCoV nsp5. FRET assays showed that PDCoV nsp5 was able to cleave the substrates containing the Q343 site of pDCP1A and nsp4/nsp5 in PDCoV replicase polyprotein but not the substrate derived from pDCP1A-Q343A (Fig. 4D). A comparation between the P6 to P6′ sequence from the DCP1A cleavage site and substrates corresponding to cleavage sites of the polyproteins 1a and 1ab was conducted, which exhibited a high conserved Q residue at the P1 position (Fig. 4E). The P1′ positions of substrates based on the cleavage sites of DCP1A and the polyproteins 1a and 1ab were mostly occupied by a relatively conserved A residue (Fig. 4E). These results further supported the standpoint that PDCoV nsp5 targets DCP1A at Q343.
FIG 4.
PDCoV nsp5 mediates pDCP1A cleavage at residue Q343. (A) The estimated pDCP1A cleavage product by PDCoV nsp5. HEK-293T cells were transfected with expression constructs encoding pDCP1A1–238, pDCP1A1–287, or pDCP1A1–325 truncated mutants and collected after 28 h for Western blotting. (B) Schematic representation of pDCP1A and its mutant sites. (C) HEK-293T cells were cotransfected with pCAGGS-nsp5-HA along with expression constructs encoding wild-type pDCP1A or pDCP1A mutant, including pDCP1A-Q330A, pDCP1A-Q343A, and pDCP1A-Q351A. Cells were then lysed after 28 h and evaluated by Western blotting. (D) Cleavage of pDCP1A-derived substrate. The purified PDCoV nsp5 was incubated with three fluorogenic peptide substrates (Dabcyl-STTMMQ↓AVKTTP-E-Edans, Dabcyl-TTMMA↓AVK-E-Edans, and Dabcyl-LKTKLQ↓AGIKIL-E-Edans) containing the Q343 site of pDCP1A, A343 site of pDCP1A-Q343A, and N-terminal autocleavage site of PDCoV nsp5 (nsp4/nsp5), respectively. The fluorescence was monitored at 485 nm, with excitation at 340 nm, using a fluorescence spectrophotometer. (E) Comparation between 12 amino acids sequences from P6 to P6′ corresponding to conservation within pp1a/1ab and substrate from the DCP1A cleavage site. ns, not significant; ***, P < 0.001; ****, P < 0.0001.
PDCoV nsp5-mediated cleavage impairs pDCP1A’s antiviral activity.
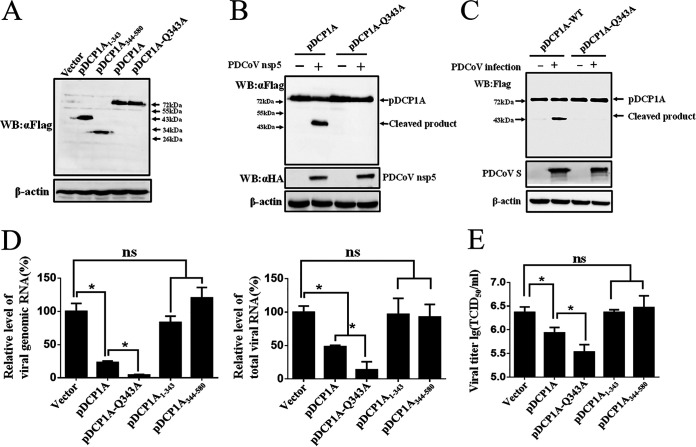
To investigate whether PDCoV nsp5-induced cleavage affects the antiviral activity of pDCP1A, two truncated mutants, pDCP1A1-343 and pDCP1A344–580, were generated. The expression of pDCP1A1–343, pDCP1A344–580, pDCP1A-Q343A, and wild-type pDCP1A was detected in IPI-2I cells (Fig. 5A). Similar to observations in HEK-293T cells, PDCoV nsp5 successfully cleaved pDCP1A in IPI-2I cells but failed to cleave the pDCP1A-Q343A mutant (Fig. 5B). The cleavage of pDCP1A or pDCP1A-Q343A was further evaluated in the context of PDCoV infection. As shown in Fig. 5C, the cleaved pDCP1A product was also detected in a sample of pDCP1A overexpression and PDCoV infection, demonstrating that pDCP1A can be cleaved in PDCoV-infected cells.
FIG 5.
PDCoV nsp5-induced cleavage of pDCP1A impairs its antiviral activity. (A) Expression of wild-type pDCP1A and pDCP1A mutants in IPI-2I cells. Wild-type pDCP1A, pDCP1A-Q343A, pDCP1A1–343, or pDCP1A344–580 was transfected into IPI-2I cells and analyzed by Western blotting after 24 h. (B) IPI-2I cells were cotransfected with PDCoV nsp5 along with wild-type pDCP1A or pDCP1A-Q343A and collected after 28 h for Western blotting. (C) Flag-tagged pDCP1A or pDCP1A-Q343A mutant expression plasmids were transfected into IPI-2I cells. After 12 h, cells were infected with PDCoV (MOI = 0.5) for 24 h. The cells were harvested and lysed for Western blot analysis. (D and E) IPI-2I cells were transfected with expression plasmid encoding wild-type pDCP1A, pDCP1A-Q343A, pDCP1A1–343, or pDCP1A344–580. After 24 h, cells were infected with PDCoV (MOI = 0.5). PDCoV genomic RNA and total RNA or viral titer was detected by RT-qPCR (D) or TCID50 assay (E), respectively, at 24 hpi. ns, not significant; *, P < 0.01.
Antiviral activities of wild-type pDCP1A or its mutants were examined in IPI-2I cells. Expression constructs encoding wild-type pDCP1A, pDCP1A-Q343A, pDCP1A1–343, pDCP1A344–580, or empty vector were transfected into IPI-2I cells. At 24 h after transfection, cells were infected with PDCoV (multiplicity of infection [MOI] = 0.5) for another 24 h. RT-qPCR with primers targeting the PDCoV nsp16-coding sequence or N gene was performed to analyze viral genomic RNA and total RNA. As shown in Fig. 5D, no statistical differences were observed among pDCP1A1–343, pDCP1A344–580, and empty vector, indicating that the antiviral activities of the two cleavage fragments, pDCP1A1–343 and pDCP1A344–580, were nearly abolished (Fig. 5D). Compared with that of the wild-type pDCP1A, pDCP1A-Q343A, which resists nsp5-mediated cleavage, exhibited a stronger antiviral effect (Fig. 5D). These observations were further confirmed by a 50% tissue culture infective dose (TCID50) assay (Fig. 5E). Overall, these findings indicate that PDCoV nsp5-induced cleavage of pDCP1A impairs its antiviral activity, exemplifying the importance of pDCP1A cleavage for PDCoV infection.
pDCP1A and hDCP1A are common targets of different coronavirus nsp5s.
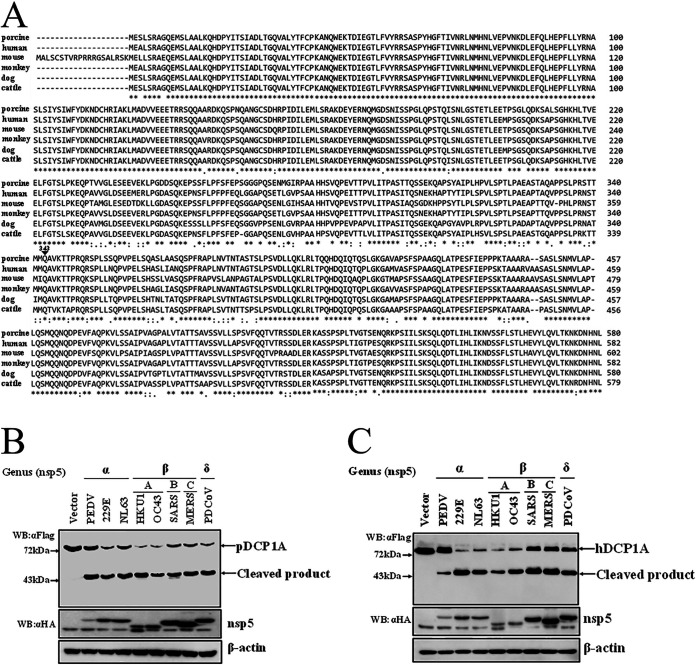
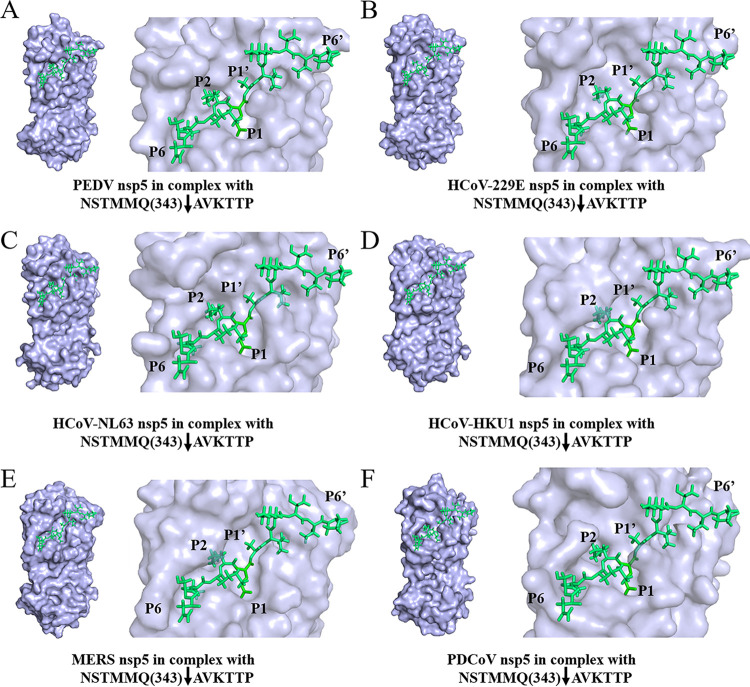
To investigate whether DCP1A cleavage is species-restricted, we analyzed the diversity of DCP1A homologs among different mammalian species. Multiple-sequence alignment showed that the amino acid sequence of pDCP1A is more than 85% identical to that of human, monkey, mouse, dog, and cattle DCP1A (Fig. 6A). Notably, the Q343 residue recognized by PDCoV nsp5 is strictly conserved among different mammalian species of DCP1A (Fig. 6A), suggesting that DCP1A cleavage may not be species dependent. In support of this view, we tested whether nsp5 encoded by other mammalian CoVs also cleaves DCP1A. Several alphacoronaviruses, including porcine epidemic diarrhea virus (PEDV), human coronavirus (HCoV)-229E, HCoV-NL63, and betacoronaviruses, including HCoV-OC43, HCoV-HKU1, severe acute respiratory syndrome coronavirus (SARS-CoV), and MERS-CoV, were chosen because these CoVs can infect humans or pigs. nsp5s of these CoVs were cloned or synthesized. Intriguingly, after cotransfection with pDCP1A, we found that all nsp5s from different CoVs were able to cleave pDCP1A (Fig. 6B). Furthermore, we also found that all nsp5s were able to cleave hDCP1A (Fig. 6C). These results indicate that DCP1A cleavage is a conserved mechanism among different CoVs. We further analyzed the complex of peptides NSTMMQ(343)↓AVKTTP of hDCP1A (P6 to P6′; downward arrow indicates cleavage site) and nsp5 of different CoVs through homology modeling. Normally P1, P2, and P1′ residues mainly determine the substrate recognition of CoV nsp5 (59, 60). As shown in Fig. 7, these residues of the hDCP1A peptide both fit comfortably in the pockets of nsp5s of different CoVs, implying possible strong interactions.
FIG 6.
DCP1A is a common target of nsp5 of different coronaviruses. (A) Alignment of the amino acid sequence of pDCP1A (GenBank accession number NP_001231287.1) with other DCP1A homologs from human (GenBank accession number NP_060873.4), cattle (GenBank accession number NP_001095800.1), mouse (GenBank accession number NP_598522.3), monkey (GenBank accession number XP_001083504.2), and dog (GenBank accession number XP_849483.2). Asterisks represent identical amino acid residues, and dots indicate similar amino acid residues. (B) HEK-293T cells cultured in 6-well plates were cotransfected with Flag-tagged pDCP1A expression plasmid and nsp5 encoded by PEDV, HCoV-229E, HCoV-NL63, HCoV-HKU1, HCoV-OC43, SARS, MERS, or PDCoV. After 28 h, cells were lysed and detected by Western blotting. (C) hDCP1A expression plasmid was transfected into HEK-293T cells with nsp5 of PEDV, HCoV-229E, HCoV-NL63, HCoV-HKU1, HCoV-OC43, SARS-CoV, MERS-CoV, or PDCoV. After 28 h, cells were lysed and detected by Western blotting.
FIG 7.
Homology modeling of nsp5 of different CoVs with the cleaved hDCP1A peptide substrate. Based on the complex structure of PEDV nsp5 mutant (C144A) with a peptide substrate (PDB accession number 4ZUH), the model of the cleaved peptide substrate NSTMMQ(343)↓AVKTTP PDCoV nsp5 was generated by replacing the original peptide substrate. The molding structure of PEDV nsp5 (PDB accession number 4XFQ) (A), HCoV-229E nsp5 (PDB accession number 2ZU2) (B), HCoV-NL63 nsp5 (PDB accession number 3TLO) (C), HCoV-HKU1 nsp5 (PDB accession number 3D23) (D), MERS nsp5 (PDB accession number 4RSP) (E), and PDCoV nsp5 (model based on the structure of PEDV nsp5) (F) combined with the cleaved peptide substrate NSTMMQ(343)↓AVKTTP (P6 to P6′ of hDCP1A; downward arrows indicate cleavage sites) were analyzed using PyMOL software (https://pymol.org/2/). The positions of P6 to P6′ of the peptide substrate are labeled.
DISCUSSION
IFNs generate an antiviral state through ISG induction as a defense mechanism against viral infection. Hundreds of ISGs have been identified, the majority of which have been reported to inhibit viral infection via diverse mechanisms (22). To combat these antiviral effects of ISGs, many viruses, including CoVs, have evolved elaborate mechanisms, such as altering subcellular localization or inducing ISG degradation, to antagonize their antiviral functions (39, 61). To our knowledge, some proteins encoded by CoVs, such as PEDV N protein, PEDV nsp1, PDCoV nsp5, PDCoV ns6, MHV nsp15, and SARS 8b, have been demonstrated to hijack IFN signaling to reduce ISG production indirectly (45, 46, 62–65). In the present study, we established that PDCoV nsp5 directly targets pDCP1A, a classical ISG, for cleavage. Notably, endogenous pDCP1A was also cleaved in PDCoV-infected cells. Furthermore, the cleaved fragments of pDCP1A1–343 and pDCP1A344–580 lost anti-PDCoV effects, while mutant pDCP1A-Q343A, which resists nsp5-mediated cleavage, exhibited stronger inhibition of PDCoV infection than the wild-type pDCP1A. These findings verified the biological effect of PDCoV nsp5-mediated pDCP1A cleavage during PDCoV infection.
Generally, CoV nsp5, the 3C-like protease, is responsible for cleavage of polyprotein precursors to produce mature nonstructural proteins from nsp4 to nsp16 (66, 67). Previously, our group demonstrated that PDCoV nsp5 cleaves NEMO and STAT2 to disrupt IFN production or signaling depending on its protease activity (46, 47). In our present study, we showed that PDCoV nsp5-mediated pDCP1A cleavage also relies on its protease activity but not on the ubiquitin proteasome, apoptotic, or autophagy-lysosome pathways. Taken together, PDCoV nsp5 is an effective antagonist against the IFN system through multiple strategies. During screening of ISGs cleaved by PDCoV nsp5, we also found that the expressions of IFIT1, TNFSF10, and ADAR1 were significantly decreased after coexpression with PDCoV nsp5, even though no cleavage products were detected. A previous study demonstrated that SVV 3C protease degrades IRF3 and IRF7 through an unidentified mechanism (54). Whether PDCoV nsp5 can mediate the degradation of these ISGs requires further study. In addition, only a limited number of well-characterized classical ISGs were tested in our screening. Besides pDCP1A, PDCoV nsp5 may cleave other ISGs. Systematic screening of host ISGs will be helpful to clarify the relationship between ISGs and PDCoV infection.
Our previous study showed that porcine reproductive and respiratory syndrome virus (PRRSV) nsp4 cleaves pDCP1A but not the human or monkey DCP1A homolog (68). Contrary to the species-specific cleavage mediated by PRRSV nsp4 at residue E238 of pDCP1A, residue Q343 and the peptide nearby are quite conserved among different mammalian species of DCP1A, providing a natural substrate for CoV nsp5. As expected, both pDCP1A and hDCP1A were readily cleaved by nsp5 of alphacoronaviruses (PEDV, HCoV-229E, and HCoV-NL63), betacoronaviruses (HCoV-OC43, HCoV-HKU1, SARS, and MERS), and deltacoronavirus (PDCoV). Interestingly, the expressions of both pDCP1A and hDCP1A were significantly affected by nsp5 of HCoV-NL63, HCoV-229E, HCoV-OC43, and HCoV-HKU1, which can induce severe infections in infants, children, and elderly individuals (69). Thus, whether nsp5-mediated hDCP1A cleavage affects HCoV replication requires further investigation.
In addition, as shown by Western blotting, these cleaved N-terminal fragments seem to be located at the same position near 43 kDa, implying a common P1 recognition residue. At present, more evidence in sequence databases exhibited that all six human coronaviruses, including HCoV-NL63, HCoV-229E, HCoV-OC43, HCoV-HKU1, SARS-CoV, and MERS-CoV, have animal origins (69, 70). A previous study demonstrated that inhibitors targeting the conserved substrate-binding pocket (S2-S1′) exhibited broad anticoronaviral activity (71). Here, DCP1A cleavage further demonstrated that nsp5 of porcine and human CoVs is able to target the same substrate [STTMMQ(343)↓AVKTTP]. Thus, we speculated that a peptide including the DCP1A cleavage site may serve as a new broad anti-CoV polypeptide drug. Further detailed affinity analysis and determination of the complete crystal structures of nsp5 and the cleaved peptide of DCP1A will provide more evidence to understand the substrate recognition mechanism, guiding the design of broad anti-CoV drugs targeting nsp5.
In addition to CoVs, many other virus-encoded proteases are reported to target host proteins. For example, vaccinia virus-encoded I7 protease cleaves Dicer protein to suppress microRNA (miRNA) processing (72). The leader protease of foot-and-mouth disease virus (FMDV) cleaves RNA helicase LGP2 to mediate immune evasion (73). SVV 3C protease targets both MAVS, TRIF, and TANK in IFN induction for cleavage (56). This evidence implies that virus-encoded proteases exhibit distinct immune regulation functions, except for their role in processing viral polyprotein precursors. Based on the protease activity, whether CoV nsp5 could target more unknown host proteins to expand its function is interesting. Our present work only identified the common target DCP1A. Further detailed study focused on CoV nsp5 will help better understand its function and substrate recognition mechanism.
MATERIALS AND METHODS
Cells and viruses.
IPI-2I and HEK-293T cells were both obtained from the China Center for Type Culture Collection (Wuhan, China). LLC-PK1 cells were purchased from the ATCC (ATCC CL-101). All cells were cultured in Dulbecco’s modified Eagle’s medium (Invitrogen, Madison, WI, USA) supplemented with 10% fetal bovine serum at 37°C in 5% CO2. PDCoV strain CHN-HN-2014 (GenBank accession number KT336560) used in this study was isolated from a piglet with severe diarrhea in China in 2014 (14).
RNA extraction and RT-qPCR.
To determine DCP1A transcription in PDCoV-infected cells, IPI-2I cells cultured in 24-well plates were infected with PDCoV (MOI = 0.5). Cells were collected with TRIzol reagent (Invitrogen) at 0, 12, and 24 hpi. Total RNA extracted from infected cells was then reverse transcribed into cDNA with avian myeloblastosis virus reverse transcriptase (TaKaRa, Shiga, Japan). The no-reverse-transcription experiments for each sample RNA were also performed without adding avian myeloblastosis virus reverse transcriptase. RT-qPCR experiments were performed in triplicates and evaluated the nsp16-coding sequence using the Applied Biosystems ViiA 7 RT-qPCR system (Life Technologies). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), β-actin, and NF-κB essential modulator (NEMO) were detected as controls. The mRNA level of the target gene was normalized to that of GAPDH. RT-qPCR primers are listed in Table 1.
TABLE 1.
PCR primers used in the study
| Primera | Sequences (5′→3′) |
|---|---|
| pIFIT1-F | TTTGGTACCATGAGTAATAATGCTGATGAAGA |
| pIFIT1-R | TTTAGATCTTTAGGGATCAAGTCCCACAGATT |
| pIFIT2-F | TTTGGTACCATGAGTGAGACCACAAAGAAC |
| pIFIT2-R | GTTCTCGAGTTATTCCTCAGCTAAAGATAC |
| pIFIT3-F | TATGGTACCATGAGTGAGGTCAACAAGAATTCT |
| pIFIT3-R | TTTAGATCTTCAGCCACTATTCCAGGCGCCCTG |
| pIFIT5-F | TTTGGTACCATGAGTGAAATTCCTAAGGACTCCT |
| pIFIT5-R | TTAAGATCTTTAAATGGAAAGTCGAAGCTCACA |
| pTNFSF10-F | TTTGGTACCATGGCGGTGATGCAGACTC |
| pTNFSF10-R | GTGCTCGAGTTAGCCAATTAAAAAGG |
| pNMI-F | GGAGAATTCATGGCAACGGATGAAAAGG |
| pNMI-R | GAGGGTACCCTATTCTTCAAAGTATACT |
| pADAR1-F | TTTGAATTCATGGACCCGAGGCAGGGGTG |
| pADAR1-R | GTTCTCGAGTTACACTGGGCAGAGGT |
| pOASL-F | GGTGAATTCATGGAGCTATTTTACACC |
| pOASL-R | TTTAGATCTTCAGTCACAGCCTTTGGCT |
| pOAS1-F | TTTCTCGAGATGGATACCCCTGTTAGGGACCT |
| pOAS1-R | TTAAGATCTCTAGATATCTTCCTCCTGTGGAG |
| pOAS2-F | TTTGAATTCATGGGAAACTGGGGGTCCCAT |
| pOAS2-R | TTTGGTACCTCAGTCCATGAATCTCCAACGT |
| pUSP18-F | TTTGAATTCATGGGCCCGGTCGGTTTGCACAA |
| pUSP18-R | ATTAGATCTTCAGGACTCAGCCATCATGTAAA |
| pDCP1A-F | TTTGAATTCATGGAGTCGCTGAGTCGAGCT |
| pDCP1A-R | GTTGGTACCTCATAGGTTGTGGTTGTCTT |
| pUBE2L6-F | TTTGAATTCATGACGGCGAGCAAGCGGGT |
| pUBE2L6-R | TTTAGATCTTTAGGAGGGCCGGTCCACT |
| pGBP1-F | TTTCTCGAGATGGCCTCAAAGGTGCACATG |
| pGBP1-R | ATTAGATCTTTAGCTCAGGAAACATTCTTTC |
| hDCP1A-F | ATAGAATTCATGGAGGCGCTGAGTCGAGCT |
| hDCP1A-R | GCGCTCGAGTCATAGGTTGTGGTTGTCTTTG |
| pDCP1Aq-F | AGCAGAGCCAAGGATGAGTATG |
| pDCP1Aq-R | CAGTGCTTCCCAGATTGGAGAT |
| pNEMOq-F | TACCACCAGCTTTTCCAGGA |
| pNEMOq-R | CTCCTCCTTCAGCTTGTCGA |
| PDCoV-nsp16q-F | GCCCTCGGTGGTTCTATCTT |
| PDCoV-nsp16q-R | TCCTTAGCTTGCCCCAAATA |
| PDCoV-Nq-F | AGCTGCTACCTCTCCGATTC |
| PDCoV-Nq-R | ACATTGGCACCAGTACGAGA |
| pβ-ACTINq-F | GCAAATGCTTCTAGGCGGAC |
| pβ-ACTINq-R | GCGTCCATCACAGCTTCTCA |
| pGAPDHq-F | ACATGGCCTCCAAGGAGTAAGA |
| pGAPDHq-R | GATCGAGTTGGGGCTGTGACT |
F, forward; R, reverse.
Plasmids.
Full-length cDNA of porcine IFIT1 (GenBank accession number NM_001244363.1), IFIT2 (GenBank accession number NM_001315658.1), IFIT3 (GenBank accession number NM_001204395.2), IFIT5 (GenBank accession number NM_001315572.1), TNFSF10 (GenBank accession number NM_001024696.1), NMI (GenBank accession number XM_003359414.4), ADAR1 (GenBank accession number NM_001111.5), OASL (GenBank accession number NM_001031790.1), OAS1 (GenBank accession number NM_214303.2), OAS2 (GenBank accession number NM_001031796.1), USP18 (GenBank accession number NM_213826.1), DCP1A (GenBank accession number NM_001244358.1), UBE2L6 (GenBank accession number NM_001246215.1), and GBP1 (GenBank accession number NM_001128473.1) were amplified from PK-15 cells and then cloned into pCAGGS-Flag vector containing an N-terminal Flag tag. The human DCP1A gene (GenBank accession number NM_018403.7) was amplified from cDNA of HEK-293T cells and cloned into the pCAGGS-Flag vector. pDCP1A substitution mutants, including pDCP1A-Q330A, -Q343A, and -Q351A, and truncated mutants, including pDCP1A1–238, pDCP1A1–287, pDCP1A1–325, pDCP1A1–343, and pDCP1A344–580, were also cloned into pCAGGS-Flag vector. Nsp5-coding sequences of PDCoV strain CHN-HN-2014 and mutants were both amplified and cloned into pCAGGS-HA-C with a hemagglutinin (HA) tag in the C terminus. Prokaryotic expression plasmid of PDCoV nsp5 was generated by cloning the cDNA sequence of nsp5 into the pGEX-6p-1 vector with an N-terminal glutathione S-transferase (GST) tag. All primers used for PCR are listed in Table 1. The constructed plasmids were confirmed by sequencing.
Western blotting.
Experiments for DCP1A cleavage were conducted in 6-well plates. We cotransfected 2.5 μg of DCP1A or mutants with 0.2 μg of nsp5 or empty vector. Cells were also treated with 20 μM broad caspase inhibitor (Z-VAD-FMK; Beyotime, Shanghai, China), 20 μM proteasome inhibitor (MG132; Beyotime), or 5 mM autophagy inhibitor (3-MA; Sigma-Aldrich, Saint Louis, MO, USA) for 8 h. Cells were then collected in lysis buffer (Beyotime), added to sample loading buffer (Beyotime), and boiled for 10 min. Subsequently, the same samples were separated in parallel on the different gels and transferred to polyvinylidene difluoride membranes (Millipore, Burlington, MA, USA) separately. Anti-Flag antibody (Macgene, Beijing, China), anti-HA antibody (MBL, Nagoya, Japan), and anti-β-actin antibody (Antgene, Wuhan, China) were used to detect respective proteins. Rabbit DCP1A polyclonal antibody (Abcam, Cambridge, UK) was used to detect endogenous DCP1A protein in PDCoV infection.
Antiviral analysis of pDCP1A.
IPI-2I cells cultured in 24-well plates were transfected with pDCP1A, pDCP1A-Q343A, pDCP1A1–343, pDCP1A344–580, or empty vector (0.8 μg for each plasmid). After 24 h, cells were infected with PDCoV (MOI = 0.5). At 24 hpi, cells were collected in TRIzol reagent (Invitrogen), and PDCoV genomic RNA copies or viral total RNA was detected by primers targeting the nsp16-coding sequence or N gene, respectively, through RT-qPCR. In addition, infected cells were directly collected and subjected to three freeze-thaw cycles. Viral titers in LLC-PK1 cells were determined by the 50% tissue culture infective dose (TCID50) assay as described previously (14).
Indirect immunofluorescence assay.
IPI-2I cells were seeded on glass coverslips (NEST) in 24-well plates and infected with PDCoV (MOI = 0.5). At 24 hpi, cells were washed with phosphate-buffered saline (PBS) three times and separately fixed with 4% paraformaldehyde and methanol for 15 min. After that, bovine serum albumin (5%) in PBS was applied to block cells for 1 h. The monoclonal antibody against dsRNA or rabbit monoclonal antibody against DCP1A was used to incubate cells for 1 h. After three washes with PBS, the cells were incubated with Alexa Fluor 488-conjugated donkey anti-mouse IgG antibody and 594-conjugated donkey anti-rabbit IgG antibody for another 1 h. Nuclei were stained with DAPI (4′,6-diamidino-2-phenylindole) and then washed with PBS three times. Fluorescence images were examined by confocal laser scanning microscopy (Olympus IX73 inverted microscope; Olympus).
Protein expression and purification.
First, the cDNA containing the PDCoV nsp5-encoding sequence was inserted into vector pGEX-6p-1, and the recombinant plasmid was transformed into Escherichia coli BL21 cells. Isopropyl-β-d-thiogalactopyranoside (IPTG) (0.8 mM) was added to the culture medium for 18 h at 18°C. The protein purification was conducted as described previously (74). The GST region of fused PDCoV nsp5 was removed by cleavage using GST-3C rhinovirus protease (homemade). Eventually, 6.1 mg/ml of the purified protein was obtained.
FRET-based assays for enzymatic characteristics.
The fluorogenic peptide substrates (Nanjing GenScript Company) Dabcyl-STTMMQ↓AVKTTP-E-Edans, Dabcyl-TTMMA↓AVK-E-Edans, and Dabcyl-LKTKLQ↓AGIKIL-E-Edans were designed based on the cleaved peptides derived from pDCP1A [STTMMQ(343)AVKTTP; P6 to P6′], pDCP1A-Q343A [TTMMA(343)AVK], and the N-terminal autocleavage site of PDCoV nsp5. As described in previous studies, the protease activity was detected during the incubation with the fluorogenic peptide substrate based on intramolecular FRET (75–77). The purified PDCoV nsp5 (final concentration of 600 nM) and fluorogenic peptide substrate (final concentration of 10 μM) were incubated in solutions containing 20 mM Tris-HCl (pH 7.4), 100 mM NaCl, and 5 mM dithiothreitol (DTT) at 37°C for 8 h. The fluorescence was monitored at 485 nm, with excitation at 340 nm, using a fluorescence spectrophotometer (77).
Sequence alignment.
Amino acid sequences from different species, including pDCP1A (GenBank accession number NP_001231287.1), hDCP1A (GenBank accession number NP_060873.4), cattle DCP1A (GenBank accession number NP_001095800.1), mouse DCP1A (GenBank accession number NP_598522.3), monkey DCP1A (GenBank accession number XP_001083504.2), and dog DCP1A (GenBank accession number XP_849483.2), were collected. Multiple-sequence alignment was conducted using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/).
Statistical analysis.
All experiments were conducted in triplicate. Data are presented as the means ± standard deviations (SDs). Significant differences were measured through Student's t test. P values of <0.05 were considered statistically significant.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (31730095), the National Key R&D Plan of China (2016YFD0500103), and the Major S&T Project of Hubei Province (2017ABA138).
REFERENCES
- 1.Zhang Q, Yoo D. 2016. Immune evasion of porcine enteric coronaviruses and viral modulation of antiviral innate signaling. Virus Res 226:128–141. doi: 10.1016/j.virusres.2016.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paul Masters S, Perlman S, 2013. Coronaviridae, p 825–858. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, vol I Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 3.Weiss SR, Navas-Martin S. 2005. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol Mol Biol Rev 69:635–664. doi: 10.1128/MMBR.69.4.635-664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. 2012. Virus taxonomy: 9th report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, Inc, San Diego, CA. [Google Scholar]
- 5.Wang L, Byrum B, Zhang Y. 2014. Detection and genetic characterization of deltacoronavirus in pigs, Ohio, USA, 2014. Emerg Infect Dis 20:1227–1230. doi: 10.3201/eid2007.140296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang J. 2016. Porcine deltacoronavirus: overview of infection dynamics, diagnostic methods, prevalence and genetic evolution. Virus Res 226:71–84. doi: 10.1016/j.virusres.2016.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jung K, Hu H, Saif LJ. 2016. Porcine deltacoronavirus infection: etiology, cell culture for virus isolation and propagation, molecular epidemiology and pathogenesis. Virus Res 226:50–59. doi: 10.1016/j.virusres.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang L, Byrum B, Zhang Y. 2014. Porcine coronavirus HKU15 detected in 9 US states, 2014. Emerg Infect Dis 20:1594–1595. doi: 10.3201/eid2009.140756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Homwong N, Jarvis MC, Lam HC, Diaz A, Rovira A, Nelson M, Marthaler D. 2016. Characterization and evolution of porcine deltacoronavirus in the United States. Prev Vet Med 123:168–174. doi: 10.1016/j.prevetmed.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ajayi T, Dara R, Misener M, Pasma T, Moser L, Poljak Z. 2018. Herd-level prevalence and incidence of porcine epidemic diarrhoea virus (PEDV) and porcine deltacoronavirus (PDCoV) in swine herds in Ontario, Canada. Transbound Emerg Dis 65:1197–1207. doi: 10.1111/tbed.12858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee JH, Chung HC, Nguyen VG, Moon HJ, Kim HK, Park SJ, Lee CH, Lee GE, Park BK. 2016. Detection and phylogenetic analysis of porcine deltacoronavirus in Korean swine farms, 2015. Transbound Emerg Dis 63:248–252. doi: 10.1111/tbed.12490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jang G, Kim SH, Lee YJ, Kim S, Lee DS, Lee KK, Lee C. 2018. Isolation and characterization of Korean porcine deltacoronavirus strain KNU16-07. J Vet Sci 19:577–581. doi: 10.4142/jvs.2018.19.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suzuki T, Shibahara T, Imai N, Yamamoto T, Ohashi S. 2018. Genetic characterization and pathogenicity of Japanese porcine deltacoronavirus. Infect Genet Evol 61:176–182. doi: 10.1016/j.meegid.2018.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong N, Fang L, Yang H, Liu H, Du T, Fang P, Wang D, Chen H, Xiao S. 2016. Isolation, genomic characterization, and pathogenicity of a Chinese porcine deltacoronavirus strain CHN-HN-2014. Vet Microbiol 196:98–106. doi: 10.1016/j.vetmic.2016.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang YW, Yue H, Fang W, Huang YW. 2015. Complete genome sequence of porcine deltacoronavirus strain CH/Sichuan/S27/2012 from Mainland China. Genome Announc 3:e00945-15. doi: 10.1128/genomeA.00945-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saeng-Chuto K, Stott CJ, Wegner M, Senasuthum R, Tantituvanont A, Nilubol D. 2017. Retrospective investigation and evolutionary analysis of a novel porcine deltacoronavirus strain detected in Thailand from 2008 to 2015. Arch Virol 162:2103–2108. doi: 10.1007/s00705-017-3331-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saeng-Chuto K, Lorsirigool A, Temeeyasen G, Vui DT, Stott CJ, Madapong A, Tripipat T, Wegner M, Intrakamhaeng M, Chongcharoen W, Tantituvanont A, Kaewprommal P, Piriyapongsa J, Nilubol D. 2017. Different lineage of porcine deltacoronavirus in Thailand, Vietnam and Lao PDR in 2015. Transbound Emerg Dis 64:3–10. doi: 10.1111/tbed.12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le VP, Song S, An BH, Park GN, Pham NT, Le DQ, Nguyen VT, Vu TTH, Kim KS, Choe S, An DJ. 2018. A novel strain of porcine deltacoronavirus in Vietnam. Arch Virol 163:203–207. doi: 10.1007/s00705-017-3594-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma Y, Zhang Y, Liang X, Lou F, Oglesbee M, Krakowka S, Li J. 2015. Origin, evolution, and virulence of porcine deltacoronaviruses in the United States. mBio 6:e00064-15. doi: 10.1128/mBio.00064-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jung K, Hu H, Saif LJ. 2017. Calves are susceptible to infection with the newly emerged porcine deltacoronavirus, but not with the swine enteric alphacoronavirus, porcine epidemic diarrhea virus. Arch Virol 162:2357–2362. doi: 10.1007/s00705-017-3351-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Q, Vlasova AN, Kenney SP, Saif LJ. 2019. Emerging and re-emerging coronaviruses in pigs. Curr Opin Virol 34:39–49. doi: 10.1016/j.coviro.2018.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schneider WM, Chevillotte MD, Rice CM. 2014. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol 32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levy D, Larner A, Chaudhuri A, Babiss LE, Darnell JE Jr. 1986. Interferon-stimulated transcription: isolation of an inducible gene and identification of its regulatory region. Proc Natl Acad Sci U S A 83:8929–8933. doi: 10.1073/pnas.83.23.8929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schindler C, Fu XY, Improta T, Aebersold R, Darnell JE Jr. 1992. Proteins of transcription factor ISGF-3: one gene encodes the 91-and 84-kDa ISGF-3 proteins that are activated by interferon alpha. Proc Natl Acad Sci U S A 89:7836–7839. doi: 10.1073/pnas.89.16.7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sadler AJ, Williams BR. 2008. Interferon-inducible antiviral effectors. Nat Rev Immunol 8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goujon C, Moncorge O, Bauby H, Doyle T, Ward CC, Schaller T, Hue S, Barclay WS, Schulz R, Malim MH. 2013. Human MX2 is an interferon-induced post-entry inhibitor of HIV-1 infection. Nature 502:559–562. doi: 10.1038/nature12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu SY, Aliyari R, Chikere K, Li G, Marsden MD, Smith JK, Pernet O, Guo H, Nusbaum R, Zack JA, Freiberg AN, Su L, Lee B, Cheng G. 2013. Interferon-inducible cholesterol-25-hydroxylase broadly inhibits viral entry by production of 25-hydroxycholesterol. Immunity 38:92–105. doi: 10.1016/j.immuni.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diamond MS. 2014. IFIT1: a dual sensor and effector molecule that detects non-2′-O methylated viral RNA and inhibits its translation. Cytokine Growth Factor Rev 25:543–550. doi: 10.1016/j.cytogfr.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abbas YM, Pichlmair A, Gorna MW, Superti-Furga G, Nagar B. 2013. Structural basis for viral 5′-PPP-RNA recognition by human IFIT proteins. Nature 494:60–64. doi: 10.1038/nature11783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar P, Sweeney TR, Skabkin MA, Skabkina OV, Hellen CU, Pestova TV. 2014. Inhibition of translation by IFIT family members is determined by their ability to interact selectively with the 5′-terminal regions of cap0-, cap1- and 5′ppp- mRNAs. Nucleic Acids Res 42:3228–3245. doi: 10.1093/nar/gkt1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Justesen J, Hartmann R, Kjeldgaard NO. 2000. Gene structure and function of the 2′-5′-oligoadenylate synthetase family. Cell Mol Life Sci 57:1593–1612. doi: 10.1007/pl00000644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han Y, Whitney G, Donovan J, Korennykh A. 2012. Innate immune messenger 2-5A tethers human RNase L into active high-order complexes. Cell Rep 2:902–913. doi: 10.1016/j.celrep.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 33.Hovanessian AG, Justesen J. 2007. The human 2′-5′oligoadenylate synthetase family: unique interferon-inducible enzymes catalyzing 2′-5′ instead of 3′-5′ phosphodiester bond formation. Biochimie 89:779–788. doi: 10.1016/j.biochi.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 34.Cougot N, Babajko S, Seraphin B. 2004. Cytoplasmic foci are sites of mRNA decay in human cells. J Cell Biol 165:31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Franks TM, Lykke-Andersen J. 2008. The control of mRNA decapping and P-body formation. Mol Cell 32:605–615. doi: 10.1016/j.molcel.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.She M, Decker CJ, Sundramurthy K, Liu Y, Chen N, Parker R, Song H. 2004. Crystal structure of Dcp1p and its functional implications in mRNA decapping. Nat Struct Mol Biol 11:249–256. doi: 10.1038/nsmb730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tritschler F, Braun JE, Motz C, Igreja C, Haas G, Truffault V, Izaurralde E, Weichenrieder O. 2009. DCP1 forms asymmetric trimers to assemble into active mRNA decapping complexes in metazoa. Proc Natl Acad Sci U S A 106:21591–21596. doi: 10.1073/pnas.0909871106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sorgeloos F, Jha BK, Silverman RH, Michiels T. 2013. Evasion of antiviral innate immunity by Theiler’s virus L* protein through direct inhibition of RNase L. PLoS Pathog 9:e1003474. doi: 10.1371/journal.ppat.1003474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kueck T, Neil SJ. 2012. A cytoplasmic tail determinant in HIV-1 Vpu mediates targeting of tetherin for endosomal degradation and counteracts interferon-induced restriction. PLoS Pathog 8:e1002609. doi: 10.1371/journal.ppat.1002609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, Yu XF. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302:1056–1060. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- 41.Silverman RH, Weiss SR. 2014. Viral phosphodiesterases that antagonize double-stranded RNA signaling to RNase L by degrading 2-5A. J Interferon Cytokine Res 34:455–463. doi: 10.1089/jir.2014.0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao L, Jha BK, Wu A, Elliott R, Ziebuhr J, Gorbalenya AE, Silverman RH, Weiss SR. 2012. Antagonism of the interferon-induced OAS-RNase L pathway by murine coronavirus ns2 protein is required for virus replication and liver pathology. Cell Host Microbe 11:607–616. doi: 10.1016/j.chom.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thornbrough JM, Jha BK, Yount B, Goldstein SA, Li Y, Elliott R, Sims AC, Baric RS, Silverman RH, Weiss SR. 2016. Middle East respiratory syndrome coronavirus NS4b protein inhibits host RNase L activation. mBio 7:e00258-16. doi: 10.1128/mBio.00258-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luo J, Fang L, Dong N, Fang P, Ding Z, Wang D, Chen H, Xiao S. 2016. Porcine deltacoronavirus (PDCoV) infection suppresses RIG-I-mediated interferon-beta production. Virology 495:10–17. doi: 10.1016/j.virol.2016.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fang P, Fang L, Ren J, Hong Y, Liu X, Zhao Y, Wang D, Peng G, Xiao S. 2018. Porcine deltacoronavirus accessory protein NS6 antagonizes interferon beta production by interfering with the binding of RIG-I/MDA5 to double-stranded RNA. J Virol 92:e00712-18. doi: 10.1128/JVI.00712-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu X, Wang D, Zhou J, Pan T, Chen J, Yang Y, Lv M, Ye X, Peng G, Fang L, Xiao S. 2017. Porcine deltacoronavirus nsp5 antagonizes type I Interferon signaling by cleaving STAT2. J Virol 91:e00003-17. doi: 10.1128/JVI.00003-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu X, Fang L, Wang D, Yang Y, Chen J, Ye X, Foda MF, Xiao S. 2017. Porcine deltacoronavirus nsp5 inhibits interferon-beta production through the cleavage of NEMO. Virology 502:33–38. doi: 10.1016/j.virol.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Habjan M, Hubel P, Lacerda L, Benda C, Holze C, Eberl CH, Mann A, Kindler E, Gil-Cruz C, Ziebuhr J, Thiel V, Pichlmair A. 2013. Sequestration by IFIT1 impairs translation of 2′O-unmethylated capped RNA. PLoS Pathog 9:e1003663. doi: 10.1371/journal.ppat.1003663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ibsen MS, Gad HH, Andersen LL, Hornung V, Julkunen I, Sarkar SN, Hartmann R. 2015. Structural and functional analysis reveals that human OASL binds dsRNA to enhance RIG-I signaling. Nucleic Acids Res 43:5236–5248. doi: 10.1093/nar/gkv389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu J, Zhang Y, Ghosh A, Cuevas RA, Forero A, Dhar J, Ibsen MS, Schmid-Burgk JL, Schmidt T, Ganapathiraju MK, Fujita T, Hartmann R, Barik S, Hornung V, Coyne CB, Sarkar SN. 2014. Antiviral activity of human OASL protein is mediated by enhancing signaling of the RIG-I RNA sensor. Immunity 40:936–948. doi: 10.1016/j.immuni.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dougherty JD, Reineke LC, Lloyd RE. 2014. mRNA decapping enzyme 1a (Dcp1a)-induced translational arrest through protein kinase R (PKR) activation requires the N-terminal enabled vasodilator-stimulated protein homology 1 (EVH1) domain. J Biol Chem 289:3936–3949. doi: 10.1074/jbc.M113.518191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pujantell M, Franco S, Galván-Femenía I, Badia R, Castellví M, Garcia-Vidal E, Clotet B, de Cid R, Tural C, Martínez MA, Riveira-Muñoz E, Esté JA, Ballana E. 2018. ADAR1 affects HCV infection by modulating innate immune response. Antiviral Res 156:116–127. doi: 10.1016/j.antiviral.2018.05.012. [DOI] [PubMed] [Google Scholar]
- 53.Li LF, Yu J, Li Y, Wang J, Li S, Zhang L, Xia SL, Yang Q, Wang X, Yu S, Luo Y, Sun Y, Zhu Y, Munir M, Qiu HJ. 2016. Guanylate-binding protein 1, an interferon-induced GTPase, exerts an antiviral activity against classical swine fever virus depending on its GTPase activity. J Virol 90:4412–4426. doi: 10.1128/JVI.02718-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xue Q, Liu H, Zhu Z, Yang F, Ma L, Cai X, Xue Q, Zheng H. 2018. Seneca Valley virus 3C(pro) abrogates the IRF3- and IRF7-mediated innate immune response by degrading IRF3 and IRF7. Virology 518:1–7. doi: 10.1016/j.virol.2018.01.028. [DOI] [PubMed] [Google Scholar]
- 55.Xue Q, Liu H, Zhu Z, Yang F, Xue Q, Cai X, Liu X, Zheng H. 2018. Seneca Valley virus 3C protease negatively regulates the type I interferon pathway by acting as a viral deubiquitinase. Antiviral Res 160:183–189. doi: 10.1016/j.antiviral.2018.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qian S, Fan W, Liu T, Wu M, Zhang H, Cui X, Zhou Y, Hu J, Wei S, Chen H, Li X, Qian P. 2017. Seneca Valley virus suppresses host type I interferon production by targeting adaptor proteins MAVS, TRIF, and TANK for cleavage. J Virol 91:e00823-17. doi: 10.1128/JVI.00823-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ziebuhr J, Siddell SG. 1999. Processing of the human coronavirus 229E replicase polyproteins by the virus-encoded 3C-like proteinase: identification of proteolytic products and cleavage sites common to pp1a and pp1ab. J Virol 73:177–185. doi: 10.1128/JVI.73.1.177-185.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chuck CP, Chow HF, Wan DC, Wong KB. 2011. Profiling of substrate specificities of 3C-like proteases from group 1, 2a, 2b, and 3 coronaviruses. PLoS One 6:e27228. doi: 10.1371/journal.pone.0027228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hegyi A, Ziebuhr J. 2002. Conservation of substrate specificities among coronavirus main proteases. J Gen Virol 83:595–599. doi: 10.1099/0022-1317-83-3-595. [DOI] [PubMed] [Google Scholar]
- 60.Chuck CP, Chong LT, Chen C, Chow HF, Wan DC, Wong KB. 2010. Profiling of substrate specificity of SARS-CoV 3CL. PLoS One 5:e13197. doi: 10.1371/journal.pone.0013197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hrecka K, Hao C, Gierszewska M, Swanson SK, Kesik-Brodacka M, Srivastava S, Florens L, Washburn MP, Skowronski J. 2011. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474:658–661. doi: 10.1038/nature10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ding Z, Fang L, Jing H, Zeng S, Wang D, Liu L, Zhang H, Luo R, Chen H, Xiao S. 2014. Porcine epidemic diarrhea virus nucleocapsid protein antagonizes beta interferon production by sequestering the interaction between IRF3 and TBK1. J Virol 88:8936–8945. doi: 10.1128/JVI.00700-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang Q, Shi K, Yoo D. 2016. Suppression of type I interferon production by porcine epidemic diarrhea virus and degradation of CREB-binding protein by nsp1. Virology 489:252–268. doi: 10.1016/j.virol.2015.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Athmer J, Fehr AR, Grunewald ME, Qu W, Wheeler DL, Graepel KW, Channappanavar R, Sekine A, Aldabeeb DS, Gale M Jr, Denison MR, Perlman S. 2018. Selective packaging in murine coronavirus promotes virulence by limiting type I interferon responses. mBio 9:e00272-18. doi: 10.1128/mBio.00272-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wong HH, Fung TS, Fang S, Huang M, Le MT, Liu DX. 2018. Accessory proteins 8b and 8ab of severe acute respiratory syndrome coronavirus suppress the interferon signaling pathway by mediating ubiquitin-dependent rapid degradation of interferon regulatory factor 3. Virology 515:165–175. doi: 10.1016/j.virol.2017.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ziebuhr J, Snijder EJ, Gorbalenya AE. 2000. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J Gen Virol 81:853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
- 67.Thiel V, Ivanov KA, Putics Á, Hertzig T, Schelle B, Bayer S, Weißbrich B, Snijder EJ, Rabenau H, Doerr HW, Gorbalenya AE, Ziebuhr J. 2003. Mechanisms and enzymes involved in SARS coronavirus genome expression. J Gen Virol 84:2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- 68.Tao R, Fang L, Bai D, Ke W, Zhou Y, Wang D, Xiao S. 2018. Porcine reproductive and respiratory syndrome virus nonstructural protein 4 cleaves porcine DCP1a to attenuate its antiviral activity. J Immunol 201:2345–2353. doi: 10.4049/jimmunol.1701773. [DOI] [PubMed] [Google Scholar]
- 69.Su S, Wong G, Shi W, Liu J, Lai ACK, Zhou J, Liu W, Bi Y, Gao GF. 2016. Epidemiology, Genetic Recombination, and pathogenesis of coronaviruses. Trends Microbiol 24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Forni D, Cagliani R, Clerici M, Sironi M. 2017. Molecular evolution of human coronavirus genomes. Trends Microbiol 25:35–48. doi: 10.1016/j.tim.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang H, Xie W, Xue X, Yang K, Ma J, Liang W, Zhao Q, Zhou Z, Pei D, Ziebuhr J, Hilgenfeld R, Yuen KY, Wong L, Gao G, Chen S, Chen Z, Ma D, Bartlam M, Rao Z. 2005. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol 3:e324. doi: 10.1371/journal.pbio.0030324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen JS, Li HC, Lin SI, Yang CH, Chien WY, Syu CL, Lo SY. 2015. Cleavage of Dicer protein by I7 protease during vaccinia virus infection. PLoS One 10:e0120390. doi: 10.1371/journal.pone.0120390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rodríguez Pulido M, Sánchez-Aparicio MT, Martínez-Salas E, García-Sastre A, Sobrino F, Sáiz M. 2018. Innate immune sensor LGP2 is cleaved by the Leader protease of foot-and-mouth disease virus. PLoS Pathog 14:e1007135. doi: 10.1371/journal.ppat.1007135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang D, Chen J, Yu C, Zhu X, Xu S, Fang L, Xiao S. 2019. Porcine reproductive and respiratory syndrome virus nsp11 antagonizes type I interferon signaling by targeting IRF9. J Virol 93:e00623-19. doi: 10.1128/JVI.00623-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen J, Wang D, Sun Z, Gao L, Zhu X, Guo J, Xu S, Fang L, Li K, Xiao S. 2019. Arterivirus nsp4 antagonizes interferon beta production by proteolytically cleaving NEMO at multiple sites. J Virol 93:e00385-19. doi: 10.1128/JVI.00385-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Matayoshi E, Wang G, Krafft G, Erickson J. 1990. Novel fluorogenic substrates for assaying retroviral proteases by resonance energy transfer. Science 247:954–958. doi: 10.1126/science.2106161. [DOI] [PubMed] [Google Scholar]
- 77.Ye G, Deng F, Shen Z, Luo R, Zhao L, Xiao S, Fu ZF, Peng G. 2016. Structural basis for the dimerization and substrate recognition specificity of porcine epidemic diarrhea virus 3C-like protease. Virology 494:225–235. doi: 10.1016/j.virol.2016.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]