Epidemics of equine-origin H3N8 and avian-origin H3N2 influenza viruses in canine populations are examples of successful cross-species transmission of influenza A viruses. Genetic analysis showed that the PA-X genes of equine H3N8 or avian H3N2 influenza viruses were full-length, with X-ORFs encoding 61 amino acids; however, those of equine-origin H3N8 or avian-origin H3N2 CIVs were truncated, suggesting that PA-X truncation occurred after transmission to dogs. In this study, we extended the PA-X genes of H3N8 and H3N2 CIVs and compared the biological characteristics of CIVs bearing different lengths of PA-X. We demonstrated that for both H3N8 and H3N2 viruses, truncation of PA-X increased virus yields in MDCK cells and enhanced viral replication, pathogenicity, and transmission in dogs. These results might reflect enhanced suppression of host gene expression and upregulation of genes related to inflammatory responses. Collectively, our data partially explain the conservation of truncated PA-X in CIVs.
KEYWORDS: PA-X, truncation, X-ORF, canine influenza viruses, inflammatory responses, suppression of host gene expression, transmission, virulence
ABSTRACT
Equine-origin H3N8 and avian-origin H3N2 canine influenza viruses (CIVs) prevalent in dogs are thought to pose a public health threat arising from intimate contact between dogs and humans. However, our understanding of CIV virulence is still limited. Influenza A virus PA-X is a fusion protein encoded in part by a +1 frameshifted open reading frame (X-ORF) in segment 3. The X-ORF can be translated in full-length (61-amino-acid) or truncated (41-amino-acid) form. Genetic analysis indicated that the X-ORFs of equine H3N8 and avian H3N2 influenza viruses encoded 61 amino acids but were truncated after introduction into dogs. To determine the effect of PA-X truncation on the biological characteristics of CIVs, we constructed four recombinant viruses on H3N8 and H3N2 CIV backgrounds bearing truncated or full-length PA-Xs. We observed that truncation of PA-X increased growth of both H3N8 and H3N2 CIVs in MDCK cells and suppressed expression from cotransfected plasmids in MDCK cells. Furthermore, truncation of PA-X enhanced viral pathogenicity in dogs, as shown by aggravated clinical symptoms and histopathological changes, increased viral replication in the respiratory system, and prolonged virus shedding. Additionally, CIVs with truncated PA-Xs were transmitted more efficiently in dogs. Global gene expression profiling of the lungs of infected dogs revealed that differentially expressed genes were mainly associated with inflammatory responses, which might contribute to the pathogenicity of PA-X-truncated CIVs. Our findings revealed that truncation of PA-X might be important for the adaptation of influenza viruses to dogs.
IMPORTANCE Epidemics of equine-origin H3N8 and avian-origin H3N2 influenza viruses in canine populations are examples of successful cross-species transmission of influenza A viruses. Genetic analysis showed that the PA-X genes of equine H3N8 or avian H3N2 influenza viruses were full-length, with X-ORFs encoding 61 amino acids; however, those of equine-origin H3N8 or avian-origin H3N2 CIVs were truncated, suggesting that PA-X truncation occurred after transmission to dogs. In this study, we extended the PA-X genes of H3N8 and H3N2 CIVs and compared the biological characteristics of CIVs bearing different lengths of PA-X. We demonstrated that for both H3N8 and H3N2 viruses, truncation of PA-X increased virus yields in MDCK cells and enhanced viral replication, pathogenicity, and transmission in dogs. These results might reflect enhanced suppression of host gene expression and upregulation of genes related to inflammatory responses. Collectively, our data partially explain the conservation of truncated PA-X in CIVs.
INTRODUCTION
Canine influenza can be caused by a variety of influenza A viruses. Influenza viruses from avian (H3N2, H5N1, and H9N2), equine (H3N8), or human (H1N1/2009 and H3N2) species can be transmitted to dogs by crossing host species barriers (1–5). Furthermore, both avian-type α2,3- and human-type α2,6-sialic acid-linked receptors were detected in the endothelial cells of the respiratory tracts and other organs of dogs (6), indicating that dogs might represent “mixing vessels” for influenza viruses. Reassortment between H3N2 canine influenza viruses (CIVs) and other influenza viruses, including H1N1/2009 and H5N1 viruses, has been repeatedly reported (7–9). CIVs usually cause mild respiratory symptoms, and thus, animal owners and veterinarians often neglect treating these infections. This creates an opportunity for CIVs to circulate and further adapt in dogs, increasing the risk for zoonotic infection.
Cross-species transmission of viruses from one host species to another is a crucial feature of the ecology and epidemiology of influenza A viruses (10, 11). These host-transferred viruses are usually inefficiently replicated or transmitted and disappear after causing a small number of infections in the new host. In some cases, however, a novel virus gains the ability to spread efficiently and consequently can cause major epidemics or pandemics in immunologically naive populations. In 2004, an equine-origin H3N8 influenza virus caused an extensive epizootic of respiratory disease in dogs in Florida (2). Subsequently, transmission of an H3N2 avian influenza virus (AIV) to dogs was reported in South Korea in 2007 (4). Equine-origin H3N8 and avian-origin H3N2 influenza viruses are capable of adaptation to canine populations and have established stable lineages in America, Europe, and Asia (4, 12–16). H3N8 CIVs have predominantly circulated in the United States since 2004 (2, 17), and H3N2 CIVs are responsible for the bulk of infections in China and South Korea (4, 18). Recently, it was determined that gene segment 3 of influenza A viruses encodes not only the polymerase acidic (PA) protein but also a novel protein, PA-X, that is translated as a +1 frameshifted open reading frame (X-ORF) extension of the growing PA polypeptide (19, 20). Comprehensive evolutionary analysis showed that the PA-X gene was conserved and that the X-ORF could be divided into two types according to protein length that were associated with particular host species (21). The PA-Xs of avian and equine influenza viruses were both full-length, while those of H3N8 and H3N2 CIVs transmitted from avian and equine populations were all truncated (21). This finding implied that the PA-Xs of these CIVs were truncated after circulating in dogs. Therefore, we hypothesized that CIVs bearing truncated PA-X proteins may have a selective advantage in dogs.
In the present study, we constructed H3N8 and H3N2 CIVs bearing full-length or truncated PA-Xs. We found that elongation of PA-X decreased growth of H3N8 and H3N2 CIVs in vitro and in vivo. More importantly, elongation of PA-X attenuated the pathogenicity and transmission of CIVs in dogs. Using microarray analysis, we found that CIVs expressing truncated PA-Xs enhanced host responses in the lungs, as shown by high activation of numerous genes associated with inflammatory responses. Overall, our findings reveal the important role of PA-X truncation in the adaptation of CIVs to dogs.
RESULTS
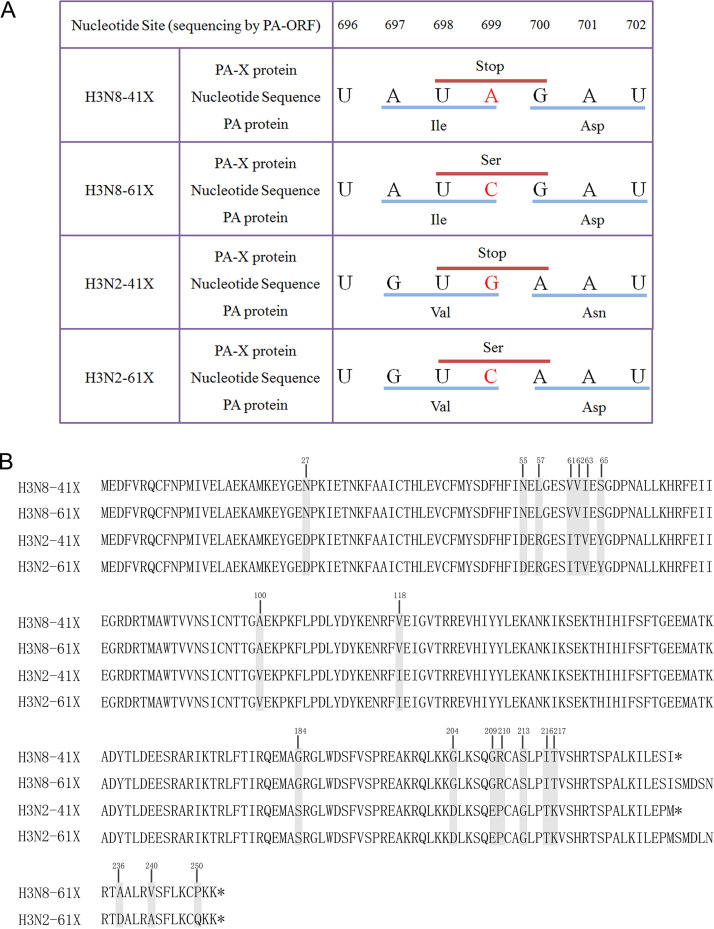
To investigate the effect of PA-X length on the virulence of CIVs, a reverse genetics system was used to generate H3N8 CIV A/canine/Colorado/6723-8/2008 (H3N8-41X) and H3N2 CIV A/canine/Beijing/362/2009 (H3N2-41X), both expressing truncated PA-X proteins of 41 amino acids in the X domain. We then introduced a G/A699C mutation in the PA ORF to generate H3N8-61X or H3N2-61X viruses, bearing full-length PA-X proteins of 61 amino acids in the X domain, without altering the PA ORF (Fig. 1A).
FIG 1.
Sequences of CIV PA-X genes and proteins. (A) Mutations (A→C or G→C at position 699 of the PA ORF) were introduced into the PA genes of H3N8-41X or H3N2-41X to extend the X-ORF, respectively, yielding H3N8-61X or H3N2-61X expressing full-length PA-Xs. (B) Amino acid sequence alignment of truncated or full-length PA-X from H3N8 or H3N2 CIVs. Gray indicates the different amino acids between H3N2 and H3N8 CIV PA-Xs, and the positions are marked. Asterisks represent stop codons.
Truncated PA-X increased CIV replication and enhanced host shutoff in vitro.
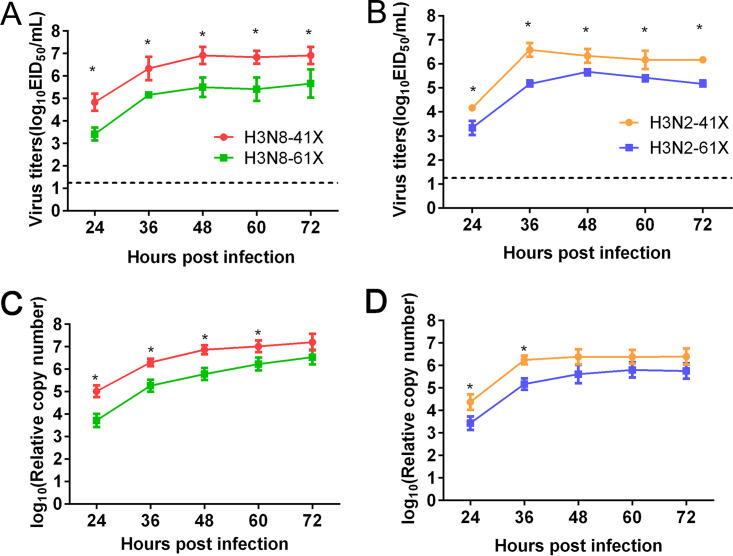
To evaluate the effect of PA-X length on virus replication, H3N8 and H3N2 CIVs bearing truncated or full-length PA-X were used to infect Madin-Darby canine kidney (MDCK) cells at a multiplicity of infection (MOI) of 0.1. Viral titers in supernatants were determined by 50% egg infectious dose (EID50) assay at different time points. As shown in Fig. 2A and B, titers of H3N8-41X were significantly higher than those of H3N8-61X in MDCK cells at 24, 36, 48, 60, and 72 h postinfection (hpi) (mean titers, 1.41-, 1.17-, 1.42-, 1.41-, and 1.26-fold higher, respectively) (P < 0.05). Similarly, titers of H3N2-41X were significantly higher than those of H3N2-61X at 24, 36, 48, 60, and 72 hpi (mean titers, 0.83-, 1.42-, 0.67-, 0.75-, and 1.00-fold higher, respectively) (P < 0.05). Because some mutations in the PA-X protein might influence viral growth in embryonated eggs (22), we also assessed viral growth kinetics using real-time reverse transcription (RT)-PCR analysis (Fig. 2C and D). The results also showed that PA-X truncation improved growth of CIVs in vitro.
FIG 2.
Growth curves of CIVs in MDCK cells. MDCK cells were infected with H3N8-41X and H3N8-61X viruses (A and C) and H3N2-41X and H3N2-61X viruses (B and D) at an MOI of 0.1. Viral titers were assessed by EID50 assay (A and B) or real-time RT-PCR (C and D) at the indicated time points. The results are shown as the means ± standard deviations (n = 3 biological replicates and n = 3 technical replicates). *, P < 0.05.
To identify factors contributing to the improved replication of CIVs with truncated PA-X proteins, we evaluated the polymerase activity of ribonucleoproteins (RNPs) from H3N8-41X, H3N8-61X, H3N2-41X, and H3N2-61X CIVs using a minigenome replication assay. No significant differences in polymerase activity were observed among these CIVs (Fig. 3).
FIG 3.
RNP polymerase activity of CIVs bearing truncated or full-length PA-Xs. MDCK cells were transfected with plasmids encoding PB2, PB1, NP, and PA from H3N8 (A) or H3N2 (B) CIVs, together with pYHNS1-Luci and pRL-TK. RNP polymerase activity was determined using a minigenome replication assay. Reference H3N8-41X or H3N2-41X CIVs were set as 100%. The results are shown as the means ± standard deviations (n = 3 biological replicates and n = 3 technical replicates).
The PA-X protein plays a major role in suppression of host protein synthesis in infected cells (19, 23, 24). To study the impact of PA-X truncation on the suppression of nonviral protein synthesis, MDCK cells were cotransfected with a green fluorescent protein (GFP) expression plasmid, pcDNA-GFP, and pRK5-Flag-PA-X vectors encoding truncated or full-length PA-X from H3N8 or H3N2 CIVs. GFP expression was significantly higher following cotransfection of cells with a vector encoding full-length PA-X than with one encoding truncated PA-X, regardless of subtype (Fig. 4A). To further assess host shutoff activity, MDCK cells were cotransfected with a pRL-SV40 plasmid expressing Renilla luciferase (Rluc) and pRK5-Flag-PA-X vectors encoding truncated or full-length PA-X from H3N8 or H3N2 CIVs. Rluc expression was assessed using a dual-luciferase reporter assay. Consistently, both full-length and truncated PA-X suppressed pRL-SV40 expression, but Rluc expression was significantly higher in cells expressing the full-length PAX-X than in those expressing truncated PA-X, regardless of CIV subtype (P < 0.05) (Fig. 4B). All PA-X proteins were expressed at similar levels (Fig. 4C). These results indicated that truncated PA-X enhanced suppression of synthesis proteins from cotransfected expression vectors. Interestingly, the host shutoff activity of H3N8 CIV PA-X was stronger than that of the corresponding H3N2 PA-X. Amino acids potentially contributing to differential inhibition of host gene expression by H3N8 and H3N2 CIV PA-Xs were identified by sequence alignment (Fig. 1B, gray).
FIG 4.
Host shutoff activity of PA-Xs with different X-ORF lengths. MDCK cells were transfected with plasmids encoding either full-length or truncated Flag-tagged PA-X from H3N8 or H3N2 CIVs, together with GFP (A) or Rluc (B) expression vectors. (A) At 24 h posttransfection, cells expressing GFP were observed using an inverted fluorescence microscope under identical exposure conditions. The experiments were repeated three times, with similar results. (B) Cells transfected with pRL-SV40 were lysed for Rluc expression analysis using the dual-luciferase reporter assay system. Values represent means and standard deviations (n = 3 biological replicates and n = 3 technical replicates) *, P < 0.05. (C) PA-X and cellular β-actin protein expression levels were analyzed by Western blotting. Molecular mass markers are indicated on the left. The experiments were repeated three times, with similar results.
Truncated PA-X improved CIV virulence in dogs.
To determine whether PA-X truncation affected the pathogenicity of CIVs in dogs, beagles were inoculated intranasally with 107 EID50s of H3N8-41X, H3N8-61X, H3N2-41X, or H3N2-61X. Dogs inoculated with H3N8-41X CIV showed clinical symptoms starting from 2 days postinoculation (dpi). The symptoms worsened and overall clinical scores peaked at 2.43 at 5 dpi. Symptoms included moderately rough hair, persistent anorexia, and depression, as well as copious mucus production and persistent cough. In contrast, dogs inoculated with H3N8-61X had mild clinical symptoms, with mildly rough hair, anorexia, and copious mucus production. The overall clinical scores of H3N8-61X-infected dogs were significantly lower than those of H3N8-41X-infected dogs from 5 dpi to 8 dpi (P < 0.05) (Fig. 5A). The clinical symptoms of dogs inoculated with the H3N2-41X virus were more serious and the clinical scores were significantly higher from 3 dpi to 8 dpi than for dogs inoculated with H3N2-61X (P < 0.05) (Fig. 5C). Assessment of rectal body temperature showed that dogs inoculated with H3N8-41X had fevers (>39°C) starting at 2 dpi that lasted for 5 days. Dogs inoculated with H3N8-61X experienced fever starting at 3 dpi, 1 day later than for H3N8-41X (Fig. 5B). Similarly, dogs inoculated with H3N2-41X experienced fever starting at 2 dpi and lasting for 6 days, 1 day earlier and 1 day longer than for H3N2-61X (Fig. 5D).
FIG 5.
Clinical scores and body temperatures of dogs infected with CIVs. Dogs were intranasally inoculated with 107 EID50s of H3N8 or H3N2 CIVs bearing truncated or full-length PA-Xs. Clinical symptoms (A and C) and body temperatures (B and D) were scored and recorded for dogs infected with H3N8 (A and B) and H3N2 (C and D) CIVs. The results are shown as the means ± standard deviations (n = 3). *, P < 0.05.
To further examine the effects of PA-X truncation on viral pathology, histopathological examination of the lungs was performed at 3 and 5 dpi. Dogs inoculated with H3N8-41X or H3N2-41X viruses showed severe pneumonia with extensive infiltration of mesenchymal and inflammatory cells, including lymphocytes and plasma cells into the lungs at 5 dpi. In contrast, mild interstitial pneumonia was observed in the lungs of dogs inoculated with H3N8-61X or H3N2-61X virus (Fig. 6A to H and M). For both H3N8 and H3N2 CIVs, dogs infected by viruses with truncated PA-X proteins showed increased CIV-positive signals in the lungs compared with those in dogs infected by viruses with full-length PA-X (Fig. 6I to L). The immunohistochemistry (IHC) was quantified by assessing integrated optical densities (IOD), and the average IOD in lungs infected with H3N8-41X and H3N2-41X CIVs were significantly higher than those of the corresponding 61X groups (P < 0.05) (Fig. 6N). Thus, truncation of PA-X aggravated the pathogenicity of CIVs in dogs.
FIG 6.
Histopathological changes and IHC staining in the lungs of dogs infected with CIVs. Hematoxylin and eosin staining of lung sections from dogs infected with H3N8-41X (A and E), H3N8-61X (B and F), H3N2-41X (C and G), or H3N2-61X (D and H) at 3 dpi (A, B, C, and D) or 5 dpi (E, F, G, and H). Microscopic lesions were counted (M). IHC staining of lungs of dogs infected with H3N8-41X (I), H3N8-61X (J), H3N2-41X (K), or H3N2-61X (L) was performed using an anti-NP antibody, and the results are presented as IOD values from five random fields (N). Values represent means and standard deviations. *, P < 0.05.
Truncated PA-X enhanced virus replication and transmissibility of CIVs in dogs.
To assess virus shedding and transmissibility of CIVs with different lengths of PA-X, three inoculated dogs from each group were removed to a separate room containing three naive dogs at 1 dpi. Virus shedding was assessed by daily collection and titration of nasal washes. Titers of H3N8-41X in nasal swabs were significantly higher than titers of H3N8-61X at 2 and 3 dpi (0.9-fold and 0.8-fold higher, respectively) (P < 0.05) (Fig. 7A and B). Furthermore, dogs inoculated with H3N8-41X shed virus for 1 day longer than dogs inoculated with H3N8-61X. At 7 dpi, H3N8-41X was still detectable in two of three inoculated dogs, but no H3N8-61X was detectable in any animal. Consistently, viral shedding was detectable in dogs cohoused with H3N8-41X-infected dogs starting at 4 dpi, while viral shedding was delayed in dogs in contact with H3N8-61X-infected dogs. Similarly, in the background of H3N2 CIV, titers of H3N2-41X in nasal swabs were significantly higher than titers of H3N2-61X at 3 and 4 dpi (1.2-fold and 1-fold higher, respectively) (P < 0.05) (Fig. 7C and D). Viral shedding was detectable in all three dogs in contact with H3N2-41X-infected dogs since 5 dpi, while shedding was not detected in dogs cohoused with H3N2-61X-infected dogs. Overall, truncation of PA-X improved virus shedding and transmissibility of both H3N8 and H3N2 CIVs.
FIG 7.
Virus shedding in inoculated and contact dogs. Dogs were intranasally inoculated with 107 EID50s of the indicated viruses. At 1 dpi, three contact dogs were introduced to assess transmission. Nasal swabs were collected from inoculated and contact dogs exposed to H3N8-41X (A), H3N8-61X (B), H3N2-41X (C), or H3N2-61X (D) CIV every day. Viruses were titrated by EID50 assay. Each color bar represents the viral titers from an individual animal. The dashed black horizontal lines indicate the lower limits of detection.
To evaluate the replication of different subtypes of CIVs bearing truncated or full-length PA-Xs in dogs, tissues, including nasal turbinates, tracheas, tonsils, and lungs from the three dogs infected with each virus, were collected and titrated at 3 and 5 dpi. Viral loads of H3N8-41X were significantly higher than those of H3N8-61X at 3 dpi in the nasal turbinates, tonsils, tracheas, and lungs (4.15-, 1.45-, 2.45-, and 2.25-fold higher, respectively) (P < 0.05) (Fig. 8A). Similarly, viral loads of H3N8-41X were significantly higher than those of H3N8-61X at 5 dpi in the nasal turbinates, tracheas, and lungs (2.4-, 4.3-, and 1.4-fold higher, respectively), suggesting that truncated PA-X increased the growth of H3N8 CIVs in dogs (P < 0.05) (Fig. 8B). In the background of H3N2 CIVs, truncation of PA-X significantly improved virus replication in turbinates, tonsils, tracheas, and lungs (1.25-, 0.75-, 0.75-, and 1.25-fold higher at 3 dpi and 1.05-, 0.75-, 0.85-, and 0.75-fold higher at 5 dpi) (P < 0.05) (Fig. 8C and D). These results indicated that truncation of PA-X improved virus replication in dogs regardless of CIV subtype.
FIG 8.
Replication of CIVs in vivo. The nasal turbinates, tracheas, tonsils, and lungs were collected from dogs infected with H3N8 (A and B) or H3N2 (C and D) CIVs at 3 dpi (A and C) or 5 dpi (B and D) and processed for virus titration. Values represent means and standard deviations (n = 3). The dashed black horizontal lines indicate the lower limits of detection. *, P < 0.05.
PA-X truncation regulated expression of genes associated with inflammatory responses.
To understand the mechanism underlying the increased virulence of CIVs with truncated PA-Xs in dogs, we used microarray analysis to compare global transcriptional responses in the lungs of dogs infected with H3N8-41X and H3N8-61X. Heat map profiles of sequences showing ≥2-fold changes in expression showed that H3N8-41X CIV induced a host transcriptional response different from that of H3N8-61X CIV, particularly at earlier time points (Fig. 9A). Significantly differentially expressed (SDE) genes at 3 dpi were subjected to integrity pathway analysis. The lungs of dogs infected with H3N8-41X and H3N8-61X differed greatly in the induction of genes related to the infectious disease, inflammatory response, cell death and survival, cell movement, and hematological system development and function (Fig. 9B). Interestingly, infectious disease, cell movement, and hematological system development and function are direct consequences of the inflammatory response associated with antiviral activity. Canonical-pathway analysis further demonstrated that SDE genes were involved in interferon and interferon regulatory factor activation related to the inflammatory response biofunction (Fig. 9C).
FIG 9.
Microarray analysis of lungs of dogs infected with H3N8-41X or H3N8-61X CIV. Lungs were collected from dogs infected with H3N8-41X or H3N8-61X CIV at 3 and 5 dpi. (A) Expression profile heat maps showing genes with ≥2-fold expression differences between H3N8-41X and H3N8-61X infection at 3 and 5 dpi. Each column represents data from an individual experiment, while rows represent unique sequences. (B and C) Biofunction and pathway analysis of genes whose expression levels differed significantly (P < 0.05; ≥2-fold expression difference) between H3N8-41X- and H3N8-61X-infected lung tissues at 3 dpi. (B) Top biofunctions of SDE genes. (C) Top canonical pathways of SDE genes. (D) Expression of genes of interest was confirmed by real-time RT-PCR. The expression level of each cytokine was normalized to that of glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Data are presented as the fold change in expression in H3N8-61X-infected dogs compared with H3N8-61X-infected dogs. Values represent means and standard deviations (n = 3). *, P < 0.05.
To confirm the results of microarray analysis, we determined mRNA levels of a set of representative cytokine genes. The cellular transcript levels mirrored the genome microarray findings (Fig. 9D). Expression of ISG15, MX1, OAS1, OASL, CCL8, and CXCL10 was upregulated to a greater extent by H3N8-41X than by H3N8-61X viruses.
Collectively, these results indicated that truncation of PA-X altered host gene expression, especially of genes involved in inflammatory responses, which may contribute to the increased virulence of CIVs in dogs.
DISCUSSION
Influenza A viruses can cross species barriers through mutation or reassortment to acquire virulence factors required for adaptation in the new host, and they are thus major potential threats to public health (25–27). Unfortunately, current understanding of how influenza viruses jump species barriers is limited. Epidemics of equine-origin H3N8 and avian-origin H3N2 influenza viruses in canine populations are typical examples of successful cross-species transmission (2, 4). Genetic analysis revealed that equine H3N8 and avian H3N2 influenza viruses possessed full-length PA-X, while their PA-Xs all became truncated because of a stop codon at position 42 in the X-ORF after introduction into dogs (21). To confirm whether truncation of PA-X contributed to the adaptation of influenza viruses to dogs, we constructed H3N8 and H3N2 CIVs bearing truncated or full-length PA-Xs. We found that H3N8 and H3N2 CIVs bearing truncated PA-Xs showed increased replication in MDCK cells and enhanced pathogenicity, replication, and transmissibility in dogs compared with CIVs bearing full-length PA-Xs. The host shutoff activity of truncated PA-X was higher than that of full-length PA-X, and CIVs bearing truncated PA-Xs triggered significantly increased expression of genes related to inflammatory responses.
PA-X is a virulence factor (19, 22, 23, 28–32), and previous studies showed that the H1N1/2009, H5N1, and H9N2 influenza viruses bearing full-length PA-Xs showed higher replication levels in A549 cells than those bearing truncated PA-Xs. Furthermore, full-length PA-X enhanced viral replication and pathogenicity in mice (23, 32). In contrast, truncation of PA-X increased the virulence and transmissibility of H1N2 swine influenza virus (SIV) in pigs (33). Coincidentally, all human influenza viruses possess full-length PA-X in nature, except for the 2009 H1N1 virus, whose PA gene is of swine origin. SIVs possessed full-length PA-Xs prior to 1985, but since that time SIVs with truncated PA-Xs have gradually increased in frequency and become dominant (21). These data, along with the results of the present study, suggested a strong species-specific association in the length of PA-X.
PA-X plays an important role in host shutoff (19, 28, 30, 34–38), which inhibits cellular gene expression to hinder the induction of antiviral responses (39) and to divert ribosomes toward translation of viral mRNAs (40). Feng et al. showed that the gene suppression activity of truncated H3N8 PA-X was higher than of the elongated form in human embryonic kidney 293T (HEK293T) cells (41). Similarly, we observed that truncation of PA-X enhanced host shutoff activity along with increased viral replication for both H3N8 and H3N2 CIVs in MDCK cells. Meanwhile, the clinical symptoms of dogs infected by H3N8 and H3N2 CIVs bearing truncated PA-Xs were more obvious than those of dogs infected with the corresponding full-length PA-X viruses. Symptoms included moderately rough hair, persistent anorexia, and depression as well as copious mucus production and persistent cough. Analysis of global gene expression in the lungs of dogs infected with CIVs bearing truncated or full-length PA-Xs demonstrated that SDE genes were associated with inflammatory responses. Thus, enhanced pathogenicity may have been associated with heightened inflammation. Histopathological examination showed that PA-X truncation enhanced virus pathogenicity in dogs, with extensive infiltration of mesenchymal and inflammatory cells into the lungs of dogs infected with H3N8 or H3N2 CIVs. Because the increased host shutoff activity was expected to weaken host antiviral responses, the stronger inflammatory response might result from improvements in viral replication.
Inhibition of host protein expression by PA-X and NS1 is determined by a strict balance between these two proteins that can affect viral pathogenesis and fitness (42–44). Nogales et al. found that H3N8 CIV NS1 was unable to block gene expression in cells, while H3N2 CIV NS1 possessed host shutoff activity (45). Interestingly, we found that the host shutoff activity of H3N8 PA-X was stronger than that of the corresponding H3N2 PA-X, which might partially compensate for loss of H3N8 NS1 inhibition of gene expression.
Successful replication and transmission in a new host are among the key requirements for cross-species transmission of influenza viruses and have been demonstrated to depend on multiple genetic variants (46). The receptor-binding specificity determined by the hemagglutinin (HA) protein is the first barrier that must be crossed. Human influenza viruses preferentially bind to sialic acid linked to galactose through an α2,6-linkage (SAα2,6Gal). This preference is matched by the SAα2,6Gal present on epithelial cells in the human trachea. In contrast, AIVs preferentially recognize SAα2,3Gal, matched by the SAα2,3Gal present on epithelial cells in the intestinal tracts of waterfowl (the main replication site of AIVs) (47). Adaptation of the viral polymerase to host factors plays an essential role in interspecies transmission. The available evidence indicates that most adaptive mutations are in the PB2 subunit of the polymerase. Position 627 in PB2 is a particularly remarkable host-associated genetic signature. In different highly pathogenic AIVs, the presence of PB2-627K leads to high levels of virulence in experimentally infected mice, guinea pigs, and ferrets (48–51) and has led to fatal outcomes of several zoonotic infections in humans (52, 53). However, this substitution is not present in H1N1/2009 influenza viruses or their mouse-adapted strains (54). Introduction of PB2 E627K into a H1N1/2009 virus did not result in enhanced virulence or transmission (55). Thus, the effect of this mutation appears to be strain specific.
Both the molecular determinants associated with CIVs and their preference for canine hosts are rare. Dogs possess both avian-type α2,3- and human-type α2,6-sialic acid-linked receptors in the endothelial cells of the respiratory tract, indicating that dogs might be “mixing vessels” for influenza viruses (6). Influenza viruses circulating in human, avian, and equine species have all been detected in dogs (1–3). As one of the most common companion animals, dogs are in close contact with humans and may act as an intermediate host for transfer of animal influenza viruses to humans. Thus, understanding the molecular mechanisms responsible for canine specificity will help prevention of occurrence of novel influenza viruses, which might emerge at the canine-human interface. Limited data have indicated that the neuraminidase (NA), nucleoprotein (NP), and M genes of CIVs were critical for the adaptation of avian H3N2 viruses in dogs by promoting viral growth (56). A 2-amino-acid insertion in the NA stalk acquired by an avian-origin H3N2 CIV enhanced viral replication in MDCK cells (57). A W222L mutation in the hemagglutinin receptor binding site could facilitate adaptation of equine influenza A (H3N8) virus to dogs (58). Introducing the K186E substitution into the H3N8 CIV NS1 protein restored its ability to inhibit host gene expression; H3N2 CIV harbored this substitution, suggesting that it might be important for the adaptation of avian-origin influenza viruses to mammals (45). In the present study, we found that a stop codon (TAG) occurred at position 42 of the X-ORF, resulting in truncation of the PA-X proteins of equine-origin H3N8 and avian-origin H3N2 CIVs compared with their ancestors. The PA-X truncation enhanced the virulence and transmissibility of both H3N8 and H3N2 CIVs in dogs. These results indicated that the effect of this substitution is not dependent on the viral gene background but is related to canine tropism.
MATERIALS AND METHODS
Ethics statement.
All animal experiments were approved by the Beijing Association for Science and Technology (approval identifier [ID] SYXK [Beijing] 2007-0023) and performed in compliance with the Beijing Laboratory Animal Welfare and Ethics guidelines as issued by the Beijing Administration Committee of Laboratory Animals. Animal protocols were in accordance with the China Agricultural University (CAU) Institutional Animal Care and Use Committee guidelines (ID SKLAB-B-2012-005) approved by the Animal Welfare Committee of CAU.
Cells.
Human embryonic HEK293T and MDCK cells were maintained in Dulbecco’s modified Eagle’s medium (Life Technologies, Rockville, MD) supplemented with 10% fetal bovine serum (Life Technologies), 100 units/ml of penicillin, and 100 μg/ml of streptomycin and incubated at 37°C under a humidified atmosphere containing 5% CO2.
Generation of recombinant viruses by reverse genetics.
The genome sequence of an H3N8 subtype CIV, A/canine/Colorado/6723-8/2008 (H3N8-41X) (accession no. CY067547 to CY064554), was downloaded from the NCBI GenBank database and synthesized by Sangon Biotech Company (Shanghai, China). The H3N2 subtype CIV A/canine/Beijing/362/2009 (H3N2-41X) (GenBank accession no. JX101382 to JX101389) was isolated and kept in our laboratory. All eight gene segments were amplified by RT-PCR and cloned into the dual-promoter plasmid pHW2000. Reverse-genetics systems for H3N8-41X or H3N2-41X were then established as previously described (59). The wild-type H3N8-41X and H3N2-41X viruses expressed truncated-length PA-Xs encoding 41 amino acids in the X domain. We extended the length of the X domain of PA-X to 61 amino acids by introducing an A→C or G→C substitution at position 699 in the PA of H3N8-41X or H3N2-41X, respectively, using site-directed mutagenesis. The H3N8-61X and H3N2-61X viruses expressing full-length PA-X were reconstructed by reverse genetics. Viral RNA was extracted and analyzed by RT-PCR, and each viral segment was sequenced.
Host shutoff assay.
The GFP expression plasmid pcDNA-GFP was constructed by inserting the GFP ORF into the pcDNA3.1+ plasmid between the NheI and XbaI sites. Truncated or full-length PA-Xs from H3N8 or H3N2 CIVs were amplified by overlap PCR and inserted into pRK5-Flag (Addgene; catalog number 42332) by In-Fusion cloning using the Seamless Assembly Cloning kit (CloneSmarter, China). The resulting vectors encoded PA-X proteins bearing N-terminal Flag tags. MDCK cells (6-well plate format, in triplicate) were transiently cotransfected with (i) 50 ng/well of pRK5-Flag plasmids encoding either truncated or full-length PA-X proteins from H3N8 or H3N2 CIVs or empty plasmid as an internal control and (ii) 1000 ng/well of pcDNA-GFP or pRL-SV40 (Rluc) using Lipofectamine 3000 (Thermo Fisher Scientific, Waltham, MA). After 24 h, cells were evaluated for GFP expression under a fluorescence microscope and Rluc activity was determined using a dual-luciferase reporter assay system (Promega, Madison, WI) and a GloMax 96 microplate luminometer (28). Three independent experiments were performed.
Western blotting.
Total cell lysates were extracted from MDCK cells with radioimmunoprecipitation assay buffer. Total protein concentration was determined using a bicinchoninic acid protein assay kit (Beyotime, China). Cell lysates were heated at 100°C for 10 min and separated using 12% SDS-PAGE. Proteins were transferred to a polyvinylidene difluoride membrane (Bio-Rad, Hercules, CA) and subsequently incubated with anti-β-actin (Beyotime, China) or anti-Flag (Sigma-Aldrich, St. Louis, MO) primary antibodies. The secondary antibodies were horseradish peroxidase-conjugated anti-mouse antibodies (Beyotime, China). Blots were visualized using a Western Lightning chemiluminescence kit (Amersham Pharmacia, Germany) following the manufacturer’s protocols.
Virus titration and replication kinetics.
The EID50 was determined in 10-day-old embryonated chicken eggs with 10-fold serially diluted virus by infection at 35°C for 48 h. EID50 values were calculated using the method of Reed and Muench (60). Multistep replication kinetics were determined by inoculating MDCK cells at an MOI of 0.1. Supernatants were sampled at 24, 36, 48, 60, and 72 hpi. Viral titers were determined by EID50 assay. Three independent experiments were performed.
Polymerase activity assay.
A dual-luciferase reporter assay system (Promega) was used to compare the polymerase activities of viral RNP complexes. The PB2, PB1, NP, and PA genes (the lattermost encoding truncated or full-length PA-X from H3N8 or H3N2 CIVs) were cloned into the pcDNA3.1+ expression plasmid. MDCK cells were transfected with PB2, PB1, PA, and NP expression plasmids (125 ng of each plasmid) from each CIV, together with a pYHNS1-Luci plasmid expressing a firefly luciferase reporter under the control of the canine RNA polymerase I promoter (10 ng) and an internal control plasmid (pRL-TK) encoding Rluc (2.5 ng). Cell lysates were analyzed 24 h posttransfection to measure firefly and Renilla luciferase activities using a GloMax 96 microplate luminometer. Three independent experiments were performed.
Dog studies.
Eight-week-old beagles (Vital River Laboratory, Beijing, China) were serologically tested using a hemagglutination inhibition assay against 1% chicken red blood cells. The animals were negative for H3N8 and H3N2 CIVs, human influenza viruses (H1N1, H3N2, and influenza virus B), and avian viruses (H5N1 and H9N2). Dogs in each experimental group (n = 9) were anesthetized with 80 mg/kg (of body weight) of tiletamine-zolazepam (Zoletil; Virbac, France) and inoculated intranasally with 2 ml (107 EID50s) of virus diluted in phosphate-buffered saline. Three inoculated dogs from each group were removed to a separate room containing another three naive dogs at 1 day postinoculation (dpi). The three inoculated dogs and contact dogs in each group were monitored daily for clinical symptoms and body temperature at fixed times on each day throughout the duration of the study (14 days). Scoring of clinical symptoms was based on activity and respiratory signs as previously described (61). Dog body temperatures above 39°C were considered fever. Nasal swabs were obtained from the three inoculated and contact dogs daily after inoculation. At 3 and 5 dpi, three dogs from each inoculation group were anesthetized and their nasal turbinates, tracheas, tonsils, and lungs were collected for virus titration (0.5 g per organ).
Microscopic pathological and IHC examinations of the lungs of each dog were performed as previously described (62). Portions of lung collected at 3 and 5 dpi were preserved in 10% phosphate-buffered formalin, then processed for paraffin embedding, and cut into 5-μm-thick sections. One section from each tissue sample was stained with hematoxylin and eosin. Lung microscopic lesions were blindly evaluated from 0 to 4 in five random fields to account for the distribution and severity of interstitial pneumonia. Another section was stained with a monoclonal antibody specific for NP protein (Abcam, United Kingdom) at a 1:1,000 dilution for IHC examination of CIV antigen. Staining intensity was quantified using integrated optical densities (IOD) in five random fields using Image-Pro Plus 6.0 software (Media Cybernetics, Rockville, MD).
Microarray analysis.
Gene expression analysis was performed using RNA isolated from the lungs of inoculated dogs (n = 3) at 3 and 5 dpi. Total RNA was extracted using TRIzol-chloroform and then purified using a NucleoSpin miRNA kit (catalog no. 740971.250; Macherey-Nagel). Target preparation for microarray processing was carried out using the GeneChip 3′ IVT PLUS reagent kit. A total of 500 ng of RNA was used for two rounds of cDNA synthesis. After fragmentation of the second-cycle single-stranded cDNA, samples were labeled with biotin using terminal deoxynucleotidyltransferase. Samples were hybridized to the Affymetrix GeneChip Canine 2.0 array (catalog no. 900727) for 16 to 18 h at 45°C. Following hybridization, the microarrays were washed and stained with phycoerythrin-conjugated streptavidin on the Affymetrix Fluidics Station 450. Microarrays were scanned using Affymetrix GeneChip Command Console installed on a GeneChip Scanner 3000 7G.
Data were analyzed with the robust multichip analysis algorithm using default analysis settings and global scaling as the normalization method. Values presented represent log2 robust microarray analysis signal intensity. Normalized data were further analyzed using moderated t tests to identify differentially expressed genes. We defined genes with fold changes of more than 2 or less than −2 and P values of <0.05 as SDE genes. These were selected for further analysis. The Database for Annotation, Visualization and Integrated Discovery (DAVID) was used to determine processes and pathways of major biological significance and importance based on integrity pathway analysis (IPA) and Gene Ontology (GO) annotation.
Real-time RT-PCR.
RNA was extracted from allantoic fluid or lung tissue using the QIAamp viral RNA minikit (Qiagen, Germany). An 8-μl aliquot of RNA was treated with RNase-free DNase I (New England BioLabs, Ipswich, MA) in a total reaction volume of 10 μl according to the manufacturer’s instructions. DNase-treated RNA (4 μl) was used as the template for amplification of target sequences using specific primers (Table 1) with the SuperScript III Platinum One-Step quantitative RT-PCR (qRT-PCR) kit (Invitrogen, Carlsbad, CA). The RT-PCR consisted of 1 cycle of 50°C for 5 min and 95°C for 2 min and then 40 cycles of 95°C for 3 s and 60°C for 30 s. RT-PCR experiments were conducted using the LightCycler 96 system (Roche, Switzerland) and LightCycler 96 system software version 1.1. The threshold was set automatically, and threshold cycle (CT) was determined. For CIV detection, RNA standards were prepared from pGEMT-M plasmids encoding CIV Matrix 1 protein using the Riboprobe in vitro transcription system (Promega). For all runs, samples (assayed in triplicate), RNA standards, and positive and negative (no-template) controls were included.
TABLE 1.
Primers for amplification in real-time RT-PCR
| Primer name | Sequence (5′–3′) |
|---|---|
| CIV M-F | TAAGGGGATTTTAGGGTTTGTGTTC |
| CIV M-R | ATTTAACTGCCCTGTCCATGTTGTT |
| ISG15-F | CTGGAGAGAGGAGCATTCCC |
| ISG15-R | GCTTGCAGCTACTCTCCTGT |
| MX1-F | TGTGTTGGAGGCTGCATTGA |
| MX1-R | ATGCATGGGCGTACCTTCTC |
| OAS1-F | AACCCAGGCCTGTGATTCTG |
| OAS1-R | GCAGGTTTTACCAGCACGTC |
| OASL-F | GCTGGAATGTGAAGAAGGCAC |
| OASL-R | CGGCTTTGCCACATCTTCTC |
| CCL8-F | TCAGCCAGATTCAGTTTCCATC |
| CCL8-R | CTCCCTGGATGCTTTGGTCTT |
| CXCL10-F | TTCCTGCAAGTCCATCGTGT |
| CXCL10-R | AGATCTTCTAGACCTTTTCTTGCT |
| GAPDH-F | CTGAACGGGAAGCTCACTGG |
| GAPDH-R | TCCGATGCCTGCTTCACTAC |
Statistical analysis.
All statistical analyses were performed using GraphPad Prism software version 6.01 (GraphPad Software Inc., San Diego, CA). The two treatments were compared using the two-tailed Student t test, and multiple comparisons were carried out with two-way analysis of variance considering time and virus as factors. P values of <0.05 were considered statistically significant.
Accession number(s).
The sequences determined in this study for the H3N8-61X and H3N2-61X viruses expressing full-length PA-X have been deposited in GenBank under accession numbers MT378379 and MT378380.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (31672573, 31761133003, and 31961130381), the National Natural Science Fund for Outstanding Young Scholars (31522058), Newton Advanced Fellowships from the Royal Society (NAF\R1\191166), and grants from the Chang Jiang Scholars Program.
REFERENCES
- 1.Sun Y, Shen Y, Zhang X, Wang Q, Liu L, Han X, Jiang B, Wang R, Sun H, Pu J, Lin D, Xia Z, Liu J. 2014. A serological survey of canine H3N2, pandemic H1N1/09 and human seasonal H3N2 influenza viruses in dogs in China. Vet Microbiol 168:193–196. doi: 10.1016/j.vetmic.2013.10.012. [DOI] [PubMed] [Google Scholar]
- 2.Crawford PC, Dubovi EJ, Castleman WL, Stephenson I, Gibbs EP, Chen L, Smith C, Hill RC, Ferro P, Pompey J, Bright RA, Medina M-J, Johnson CM, Olsen CW, Cox NJ, Klimov AI, Katz JM, Donis RO. 2005. Transmission of equine influenza virus to dogs. Science 310:482–485. doi: 10.1126/science.1117950. [DOI] [PubMed] [Google Scholar]
- 3.Songserm T, Amonsin A, Jam-On R, Sae-Heng N, Pariyothorn N, Payungporn S, Theamboonlers A, Chutinimitkul S, Thanawongnuwech R, Poovorawan Y. 2006. Fatal avian influenza A H5N1 in a dog. Emerg Infect Dis 12:1744–1747. doi: 10.3201/eid1211.060542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song D, Kang B, Lee C, Jung K, Ha G, Kang D, Park S, Park B, Oh J. 2008. Transmission of avian influenza virus (H3N2) to dogs. Emerg Infect Dis 14:741–746. doi: 10.3201/eid1405.071471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Su S, Zhou P, Fu X, Wang L, Hong M, Lu G, Sun L, Qi W, Ning Z, Jia K, Yuan Z, Wang H, Ke C, Wu J, Zhang G, Gray GC, Li S. 2014. Virological and epidemiological evidence of avian influenza virus infections among feral dogs in live poultry markets, China: a threat to human health? Clin Infect Dis 58:1644–1646. doi: 10.1093/cid/ciu154. [DOI] [PubMed] [Google Scholar]
- 6.Ning ZY, Wu XT, Cheng YF, Qi WB, An YF, Wang H, Zhang GH, Li SJ. 2012. Tissue distribution of sialic acid-linked influenza virus receptors in beagle dogs. J Vet Sci 13:219–222. doi: 10.4142/jvs.2012.13.3.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moon H, Hong M, Kim JK, Seon B, Na W, Park SJ, An DJ, Jeoung HY, Kim DJ, Kim JM, Kim SH, Webby RJ, Webster RG, Kang BK, Song D. 2015. H3N2 canine influenza virus with the matrix gene from the pandemic A/H1N1 virus: infection dynamics in dogs and ferrets. Epidemiol Infect 143:772–780. doi: 10.1017/S0950268814001617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Su S, Cao N, Chen J, Zhao F, Li H, Zhao M, Wang Y, Huang Z, Yuan L, Wang H, Zhang G, Li S. 2012. Complete genome sequence of an avian-origin H3N2 canine influenza A virus isolated in farmed dogs in southern China. J Virol 86:10238. doi: 10.1128/JVI.01601-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song D, Moon HJ, An DJ, Jeoung HY, Kim H, Yeom MJ, Hong M, Nam JH, Park SJ, Park BK, Oh JS, Song M, Webster RG, Kim JK, Kang BK. 2012. A novel reassortant canine H3N1 influenza virus between pandemic H1N1 and canine H3N2 influenza viruses in Korea. J Gen Virol 93:551–554. doi: 10.1099/vir.0.037739-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taubenberger JK, Kash JC. 2010. Influenza virus evolution, host adaptation, and pandemic formation. Cell Host Microbe 7:440–451. doi: 10.1016/j.chom.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuiken T, Holmes EC, McCauley J, Rimmelzwaan GF, Williams CS, Grenfell BT. 2006. Host species barriers to influenza virus infections. Science 312:394–397. doi: 10.1126/science.1122818. [DOI] [PubMed] [Google Scholar]
- 12.Harder TC, Vahlenkamp TW. 2010. Influenza virus infections in dogs and cats. Vet Immunol Immunopathol 134:54–60. doi: 10.1016/j.vetimm.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 13.Sun Y, Sun S, Ma J, Tan Y, Du L, Shen Y, Mu Q, Pu J, Lin D, Liu J. 2013. Identification and characterization of avian-origin H3N2 canine influenza viruses in northern China during 2009–2010. Virology 435:301–307. doi: 10.1016/j.virol.2012.09.037. [DOI] [PubMed] [Google Scholar]
- 14.Kim JK, Nam JH, Lyoo KS, Moon H, Na W, Song EJ, Yeom M, Shim SM, Jeong DG, An DJ, Kang BK, Song D. 2016. Genetic characterization of an ancestral strain of the avian-origin H3N2 canine influenza virus currently circulating in East Asia. J Microbiol Biotechnol 26:1109–1114. doi: 10.4014/jmb.1511.11047. [DOI] [PubMed] [Google Scholar]
- 15.Lee E, Kim EJ, Kim BH, Song JY, Cho IS, Shin YK. 2016. Molecular analyses of H3N2 canine influenza viruses isolated from Korea during 2013–2014. Virus Genes 52:204–217. doi: 10.1007/s11262-015-1274-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lavan R, Knesl O. 2015. Prevalence of canine infectious respiratory pathogens in asymptomatic dogs presented at US animal shelters. J Small Anim Pract 56:572–576. doi: 10.1111/jsap.12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pecoraro HL, Bennett S, Spindel ME, Landolt GA. 2014. Evolution of the hemagglutinin gene of H3N8 canine influenza virus in dogs. Virus Genes 49:393–399. doi: 10.1007/s11262-014-1102-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu H, Hughes J, Murcia PR. 2015. Origins and evolutionary dynamics of H3N2 canine influenza virus. J Virol 89:5406–5418. doi: 10.1128/JVI.03395-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jagger BW, Wise HM, Kash JC, Walters K-A, Wills NM, Xiao Y-L, Dunfee RL, Schwartzman LM, Ozinsky A, Bell GL, Dalton RM, Lo A, Efstathiou S, Atkins JF, Firth AE, Taubenberger JK, Digard P. 2012. An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science 337:199–204. doi: 10.1126/science.1222213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yewdell JW, Ince WL. 2012. Virology. Frameshifting to PA-X influenza. Science 337:164–165. doi: 10.1126/science.1225539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi M, Jagger BW, Wise HM, Digard P, Holmes EC, Taubenberger JK. 2012. Evolutionary conservation of the PA-X open reading frame in segment 3 of influenza A virus. J Virol 86:12411–12413. doi: 10.1128/JVI.01677-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hussain S, Turnbull ML, Wise HM, Jagger BW, Beard PM, Kovacikova K, Taubenberger JK, Vervelde L, Engelhardt OG, Digard P. 2019. Mutation of influenza A virus PA-X decreases pathogenicity in chicken embryos and can increase the yield of reassortant candidate vaccine viruses. J Virol 93:e01551-18. doi: 10.1128/JVI.01551-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao H, Sun Y, Hu J, Qi L, Wang J, Xiong X, Wang Y, He Q, Lin Y, Kong W, Seng L-G, Sun H, Pu J, Chang K-C, Liu X, Liu J. 2015. The contribution of PA-X to the virulence of pandemic 2009 H1N1 and highly pathogenic H5N1 avian influenza viruses. Sci Rep 5:8262. doi: 10.1038/srep08262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaucherand L, Porter BK, Levene RE, Price EL, Schmaling SK, Rycroft CH, Kevorkian Y, McCormick C, Khaperskyy DA, Gaglia MM. 2019. The influenza A virus endoribonuclease PA-X usurps host mRNA processing machinery to limit host gene expression. Cell Rep 27:776–792.e7. doi: 10.1016/j.celrep.2019.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, Xu X, Lu H, Zhu W, Gao Z, Xiang N, Shen Y, He Z, Gu Y, Zhang Z, Yang Y, Zhao X, Zhou L, Li X, Zou S, Zhang Y, Li X, Yang L, Guo J, Dong J, Li Q, Dong L, Zhu Y, Bai T, Wang S, Hao P, Yang W, Zhang Y, Han J, Yu H, Li D, Gao GF, Wu G, Wang Y, Yuan Z, Shu Y. 2013. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med 368:1888–1897. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- 26.Chen H, Yuan H, Gao R, Zhang J, Wang D, Xiong Y, Fan G, Yang F, Li X, Zhou J, Zou S, Yang L, Chen T, Dong L, Bo H, Zhao X, Zhang Y, Lan Y, Bai T, Dong J, Li Q, Wang S, Zhang Y, Li H, Gong T, Shi Y, Ni X, Li J, Zhou J, Fan J, Wu J, Zhou X, Hu M, Wan J, Yang W, Li D, Wu G, Feng Z, Gao GF, Wang Y, Jin Q, Liu M, Shu Y. 2014. Clinical and epidemiological characteristics of a fatal case of avian influenza A H10N8 virus infection: a descriptive study. Lancet 383:714–721. doi: 10.1016/S0140-6736(14)60111-2. [DOI] [PubMed] [Google Scholar]
- 27.de Jong JC, Claas EC, Osterhaus AD, Webster RG, Lim WL. 1997. A pandemic warning? Nature 389:554. doi: 10.1038/39218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Desmet EA, Bussey KA, Stone R, Takimoto T. 2013. Identification of the N-terminal domain of the influenza virus PA responsible for the suppression of host protein synthesis. J Virol 87:3108–3118. doi: 10.1128/JVI.02826-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hayashi T, MacDonald LA, Takimoto T. 2015. Influenza A virus protein PA-X contributes to viral growth and suppression of the host antiviral and immune responses. J Virol 89:6442–6452. doi: 10.1128/JVI.00319-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee J, Yu H, Li Y, Ma J, Lang Y, Duff M, Henningson J, Liu Q, Li Y, Nagy A, Bawa B, Li Z, Tong G, Richt JA, Ma W. 2017. Impacts of different expressions of PA-X protein on 2009 pandemic H1N1 virus replication, pathogenicity and host immune responses. Virology 504:25–35. doi: 10.1016/j.virol.2017.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu J, Mo Y, Wang X, Gu M, Hu Z, Zhong L, Wu Q, Hao X, Hu S, Liu W, Liu H, Liu X, Liu X. 2015. PA-X decreases the pathogenicity of highly pathogenic H5N1 influenza A virus in avian species by inhibiting virus replication and host response. J Virol 89:4126–4142. doi: 10.1128/JVI.02132-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao H, Xu G, Sun Y, Qi L, Wang J, Kong W, Sun H, Pu J, Chang KC, Liu J. 2015. PA-X is a virulence factor in avian H9N2 influenza virus. J Gen Virol 96:2587–2594. doi: 10.1099/jgv.0.000232. [DOI] [PubMed] [Google Scholar]
- 33.Xu G, Zhang X, Sun Y, Liu Q, Sun H, Xiong X, Jiang M, He Q, Wang Y, Pu J, Guo X, Yang H, Liu J. 2016. Truncation of C-terminal 20 amino acids in PA-X contributes to adaptation of swine influenza virus in pigs. Sci Rep 6:21845. doi: 10.1038/srep21845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khaperskyy DA, Emara MM, Johnston BP, Anderson P, Hatchette TF, McCormick C. 2014. Influenza A virus host shutoff disables antiviral stress-induced translation arrest. PLoS Pathog 10:e1004217. doi: 10.1371/journal.ppat.1004217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chaimayo C, Dunagan M, Hayashi T, Santoso N, Takimoto T. 2018. Specificity and functional interplay between influenza virus PA-X and NS1 shutoff activity. PLoS Pathog 14:e1007465. doi: 10.1371/journal.ppat.1007465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oishi K, Yamayoshi S, Kawaoka Y. 2018. Identification of novel amino acid residues of influenza virus PA-X that are important for PA-X shutoff activity by using yeast. Virology 516:71–75. doi: 10.1016/j.virol.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oishi K, Yamayoshi S, Kozuka-Hata H, Oyama M, Kawaoka Y. 2018. N-terminal acetylation by NatB Is required for the shutoff activity of influenza A virus PA-X. Cell Rep 24:851–860. doi: 10.1016/j.celrep.2018.06.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oishi K, Yamayoshi S, Kawaoka Y. 2019. Identification of amino acid residues in influenza A virus PA-X that contribute to enhanced shutoff activity. Front Microbiol 10:432. doi: 10.3389/fmicb.2019.00432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rigby RE, Wise HM, Smith N, Digard P, Rehwinkel J. 2019. PA-X antagonises MAVS-dependent accumulation of early type I interferon messenger RNAs during influenza A virus infection. Sci Rep 9:7216. doi: 10.1038/s41598-019-43632-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weber F, Haller O. 2007. Viral suppression of the interferon system. Biochimie 89:836–842. doi: 10.1016/j.biochi.2007.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feng KH, Sun M, Iketani S, Holmes EC, Parrish CR. 2016. Comparing the functions of equine and canine influenza H3N8 virus PA-X proteins: suppression of reporter gene expression and modulation of global host gene expression. Virology 496:138–146. doi: 10.1016/j.virol.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nogales A, Martinez-Sobrido L, Chiem K, Topham DJ, DeDiego ML. 2018. Functional evolution of the 2009 pandemic H1N1 influenza virus NS1 and PA in humans. J Virol 92:e01206-18. doi: 10.1128/JVI.01206-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nogales A, Rodriguez L, DeDiego ML, Topham DJ, Martinez-Sobrido L. 2017. Interplay of PA-X and NS1 proteins in replication and pathogenesis of a temperature-sensitive 2009 pandemic H1N1 influenza A virus. J Virol 91:e00720-17. doi: 10.1128/JVI.00720-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nogales A, Martinez-Sobrido L, Topham DJ, DeDiego ML. 2018. Modulation of innate immune responses by the influenza A NS1 and PA-X proteins. Viruses 10:708. doi: 10.3390/v10120708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nogales A, Chauche C, DeDiego ML, Topham DJ, Parrish CR, Murcia PR, Martinez-Sobrido L. 2017. The K186E amino acid substitution in the canine influenza virus H3N8 NS1 protein restores its ability to inhibit host gene expression. J Virol 91:e00877-17. doi: 10.1128/JVI.00877-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cauldwell AV, Long JS, Moncorge O, Barclay WS. 2014. Viral determinants of influenza A virus host range. J Gen Virol 95:1193–1210. doi: 10.1099/vir.0.062836-0. [DOI] [PubMed] [Google Scholar]
- 47.Rogers GN, Paulson JC. 1983. Receptor determinants of human and animal influenza virus isolates: differences in receptor specificity of the H3 hemagglutinin based on species of origin. Virology 127:361–373. doi: 10.1016/0042-6822(83)90150-2. [DOI] [PubMed] [Google Scholar]
- 48.Gao P, Watanabe S, Ito T, Goto H, Wells K, McGregor M, Cooley AJ, Kawaoka Y. 1999. Biological heterogeneity, including systemic replication in mice, of H5N1 influenza A virus isolates from humans in Hong Kong. J Virol 73:3184–3189. doi: 10.1128/JVI.73.4.3184-3189.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Govorkova EA, Rehg JE, Krauss S, Yen HL, Guan Y, Peiris M, Nguyen TD, Hanh TH, Puthavathana P, Long HT, Buranathai C, Lim W, Webster RG, Hoffmann E. 2005. Lethality to ferrets of H5N1 influenza viruses isolated from humans and poultry in 2004. J Virol 79:2191–2198. doi: 10.1128/JVI.79.4.2191-2198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hatta M, Gao P, Halfmann P, Kawaoka Y. 2001. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science 293:1840–1842. doi: 10.1126/science.1062882. [DOI] [PubMed] [Google Scholar]
- 51.Steel J, Lowen AC, Mubareka S, Palese P. 2009. Transmission of influenza virus in a mammalian host is increased by PB2 amino acids 627K or 627E/701N. PLoS Pathog 5:e1000252. doi: 10.1371/journal.ppat.1000252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Jong MD, Simmons CP, Thanh TT, Hien VM, Smith GJ, Chau TN, Hoang DM, Chau NV, Khanh TH, Dong VC, Qui PT, Cam BV, Ha do Q, Guan Y, Peiris JS, Chinh NT, Hien TT, Farrar J. 2006. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med 12:1203–1207. doi: 10.1038/nm1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fouchier RA, Schneeberger PM, Rozendaal FW, Broekman JM, Kemink SA, Munster V, Kuiken T, Rimmelzwaan GF, Schutten M, Van Doornum GJ, Koch G, Bosman A, Koopmans M, Osterhaus AD. 2004. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc Natl Acad Sci U S A 101:1356–1361. doi: 10.1073/pnas.0308352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ye J, Sorrell EM, Cai Y, Shao H, Xu K, Pena L, Hickman D, Song H, Angel M, Medina RA, Manicassamy B, Garcia-Sastre A, Perez DR. 2010. Variations in the hemagglutinin of the 2009 H1N1 pandemic virus: potential for strains with altered virulence phenotype? PLoS Pathog 6:e1001145. doi: 10.1371/journal.ppat.1001145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Herfst S, Chutinimitkul S, Ye J, de Wit E, Munster VJ, Schrauwen EJ, Bestebroer TM, Jonges M, Meijer A, Koopmans M, Rimmelzwaan GF, Osterhaus AD, Perez DR, Fouchier RA. 2010. Introduction of virulence markers in PB2 of pandemic swine-origin influenza virus does not result in enhanced virulence or transmission. J Virol 84:3752–3758. doi: 10.1128/JVI.02634-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee IW, Kim YI, Lim GJ, Kwon HI, Si YJ, Park SJ, Kim EH, Kim SM, Nguyen HD, Song MS, Choi YK. 2018. Comparison of the virulence and transmissibility of canine H3N2 influenza viruses and characterization of their canine adaptation factors. Emerg Microbes Infect 7:17. doi: 10.1038/s41426-017-0013-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lin Y, Xie X, Zhao Y, Kalhoro DH, Lu C, Liu Y. 2016. Enhanced replication of avian-origin H3N2 canine influenza virus in eggs, cell cultures and mice by a two-amino acid insertion in neuraminidase stalk. Vet Res 47:53. doi: 10.1186/s13567-016-0337-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wen F, Blackmon S, Olivier AK, Li L, Guan M, Sun H, Wang PG, Wan XF. 2018. Mutation W222L at the receptor binding site of hemagglutinin could facilitate viral adaption from equine influenza A(H3N8) virus to dogs. J Virol 92:e01115-18. doi: 10.1128/JVI.01115-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sun Y, Qin K, Wang J, Pu J, Tang Q, Hu Y, Bi Y, Zhao X, Yang H, Shu Y, Liu J. 2011. High genetic compatibility and increased pathogenicity of reassortants derived from avian H9N2 and pandemic H1N1/2009 influenza viruses. Proc Natl Acad Sci U S A 108:4164–4169. doi: 10.1073/pnas.1019109108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reed LJ, Muench H. 1937. A simple method of estimating fifty per cent endpoints. Am J Trop Med Hyg 27: 493–497. [Google Scholar]
- 61.Kang YM, Kim HM, Ku KB, Park EH, Yum J, Sang HS. 2013. H3N2 canine influenza virus causes severe morbidity in dogs with induction of genes related to inflammation and apoptosis. Vet Res 44:92. doi: 10.1186/1297-9716-44-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bi Y, Xie Q, Zhang S, Li Y, Xiao H, Jin T, Zheng W, Li J, Jia X, Sun L, Liu J, Qin C, Gao GF, Liu W. 2015. Assessment of the internal genes of influenza A (H7N9) virus contributing to high pathogenicity in mice. J Virol 89:2–13. doi: 10.1128/JVI.02390-14. [DOI] [PMC free article] [PubMed] [Google Scholar]