Chinook salmon are a keystone fish species of great ecological and commercial significance in their native northern Pacific range and in regions to which they have been introduced. Threats to salmon populations include habitat degradation, climate change, and infectious agents, including viruses. While the first isolation of a flavivirus from wild migrating salmon may indicate an emerging disease threat, characterization of the genome provides insights into the ecology and long evolutionary history of this important group of viruses affecting humans and other animals and into an expanding group of recently discovered aquatic flaviviruses.
KEYWORDS: aquatic, flavivirus, novel, salmon
ABSTRACT
The first isolation of a flavivirus from fish was made from moribund Chinook salmon (Oncorhynchus tshawytscha) from the Eel River, California, USA. Following the observation of cytopathic effect in a striped-snakehead fish cell line, 35-nm virions with flaviviral morphology were visualized using electron microcopy. Next-generation sequencing and rapid amplification of cDNA ends obtained the complete genome. Reverse transcriptase quantitative PCR (RT-qPCR) confirmed the presence of viral RNA in formalin-fixed tissues from the wild salmon. For the first time, in vivo replication of an aquatic flavivirus was demonstrated following intracoelomic injection in a Chinook salmon model of infection. RT-qPCR demonstrated viral replication in salmon brains up to 15 days postinjection. Infectious virus was then reisolated in culture, fulfilling Rivers’ postulates. Only limited replication occurred in the kidneys of Chinook salmon or in tissues of rainbow trout (Oncorhynchus mykiss). The proposed salmon flavivirus (SFV) has a 10.3-kb genome that encodes a rare dual open reading frame, a feature uncharacteristic of classical flaviviruses. Phylogenetic analysis places SFV in a basal position among a new subgroup of recently recognized aquatic and bat flaviviruses distinct from the established mosquito-borne, tick-borne, insect-only, and unknown-vector flavivirus groups. While the pathogenic potential of the virus remains to be fully elucidated, its basal phylogeny and the in vivo infection model will allow SFV to serve as a prototype for aquatic flaviviruses. Ongoing field and laboratory studies will facilitate better understanding of the potential impacts of SFV infection on ecologically and economically important salmonid species.
IMPORTANCE Chinook salmon are a keystone fish species of great ecological and commercial significance in their native northern Pacific range and in regions to which they have been introduced. Threats to salmon populations include habitat degradation, climate change, and infectious agents, including viruses. While the first isolation of a flavivirus from wild migrating salmon may indicate an emerging disease threat, characterization of the genome provides insights into the ecology and long evolutionary history of this important group of viruses affecting humans and other animals and into an expanding group of recently discovered aquatic flaviviruses.
INTRODUCTION
The Chinook salmon (Oncorhynchus tshawytscha) (CS) is a keystone fish species in the Pacific Northwest of North America, important for its ecological roles, cultural significance to indigenous peoples, and high economic value. Multiple population segments exist in the Pacific Northwest and face a host of environmental and disease threats. The Sacramento River winter-run and Upper Columbia River spring-run Chinook salmon populations are considered endangered under the Endangered Species Act. Eel River fall-run Chinook salmon and six other populations are listed as threatened (1).
Adults of this anadromous species migrate from salt water into rivers to reproduce. Juvenile stages remain in freshwater for as long as a year and then migrate to the ocean to grow and mature before returning to freshwater to spawn. Native Chinook salmon populations have declined dramatically since European settlement in California in the mid-1800s. Increased water temperatures, poor water quality, and increased anthropogenic pressures resulting in altered water flows, habitat degradation, and obstructed access to spawning grounds caused by dams and culverts are factors impacting survival, migration, and spawning success (2–4). Emerging and reemerging pathogens, including viruses, also contribute to the decline of wild salmonid populations.
Flavivirus is a genus in the family Flaviviridae. The small virions, 37 to 60 nm in diameter, are spherical when mature, are enveloped, and have 10- to 11-kb genomes composed of single-stranded, positive-sense RNA. The Flavivirus genus can be further categorized into four well-established subgroups: the mosquito-borne, tick-borne, insect-only, and unknown-vector subgroups. The mosquito-borne and tick-borne subgroups include many human pathogens with complex transmission cycles that involve wildlife reservoirs as well as mosquito or tick vectors. Some flaviviruses in the unknown-vector subgroup, such as Modoc virus and Rio Bravo virus, produce no disease or only rare disease in humans or other animals. Flaviviruses in the insect-only subgroup are restricted entirely to insects (5). Despite a common genomic organization, the high degree of divergence among members of the Flaviviridae suggests a long evolutionary history (6, 7).
For decades, the Tamana bat virus (TABV), isolated in 1973 from the insectivorous bat Pteronotus parnellii, was considered an “unclassified” flavivirus that did not fall within one of the four established subgroups. The virus produced mortality and was reisolated from a mouse inoculation model of infection (8). Recent discoveries in marine aquatic animals have further expanded the known diversity of unassigned flaviviruses. The first fish flavivirus, Cyclopterus lumpus virus (CLuV), was detected in lumpfish (Cyclopterus lumpus) by next-generation sequencing (NGS). The virus was anecdotally associated with mortalities of >50% (9). The Wenzhou shark flavivirus was described in metagenomic sequences of the Pacific spadenose shark (Scoliodon macrorhynchos) (10) and was later rediscovered in the gazami crab (Portunus trituberculatus), raising the hypothesis of horizontal transmission between the two distantly related hosts in the ocean ecosystem (11). Additional aquatic flaviviruses found through the analysis of public sequence databases include three crustacean viruses (Gammarus chevreuxi flavivirus, Gammarus pulex flavivirus, and Crangon crangon flavivirus) and two cephalopod flaviviruses (southern pygmy squid flavivirus and firefly squid flavivirus) (11). The full host ranges and tissue tropisms of these aquatic flaviviruses remain elusive, since none have been isolated in culture or studied in in vivo models of infection.
During a disease investigation among migrating adult Chinook salmon in California, a highly divergent aquatic flavivirus was successfully isolated from the brain in cell culture. Following the characterization of its genome by NGS and phylogenetic analysis, the virion morphology was described using electron microscopy, and three in vivo challenges were conducted to investigate the disease potential, replication, and tissue tropism of the virus in two salmonid fish species.
RESULTS AND DISCUSSION
During an investigation of unexplained mortalities in returning Chinook salmon (CS) in California’s lower Eel River in November 2015, approximately 150 diseased fish were observed exhibiting signs of lethargy, congregating at the riverbanks, decreased avoidance of humans, and lenticular opacity. Gross necropsy of affected salmon revealed petechial hemorrhages in the optic lobes, cerebellums, and spinal cords of 3/3 fish (Fig. 1). Trematode parasites of the genus Diplostomum were associated with lenticular opacities (see Fig. S1 in the supplemental material). More-detailed gross and histologic findings are presented in the supplemental material. To investigate the potential involvement of a viral etiology, pooled organ homogenates were used to inoculate three fish cell lines. In the striped-snakehead (SSN-1) fish cell line only, cytopathic effect (CPE) consisting of vacuolation was detected 4 days postinoculation (dpi), followed by cell rounding, lysis, and lifting of the monolayer by 10 dpi (Fig. 2A). A filtrate of this lysed SSN-1 cell suspension was serially diluted (100 to 10−7) and inoculated further into Chinook salmon embryo-214 (CHSE-214), epithelioma papulosum cyprini (EPC), fathead minnow (FHM), koi fin (KF), SSN-1, and bluegill fin (BF-2) cell lines. CPE occurred in both SSN-1 (dilutions, 100 to 10−5) and CHSE-214 (dilutions, 100 to 10−2) cells but began earlier in the SSN-1 cell line. Complete lysis occurred in both by day 10. The concentrations of virus for SSN-1 and CHSE-214 cells were 9.6 × 107 and 1.7 × 104 50% tissue culture infective doses (TCID50)/ml, respectively. No CPE was observed in the EPC, FHM, KF, or BF-2 cell cultures.
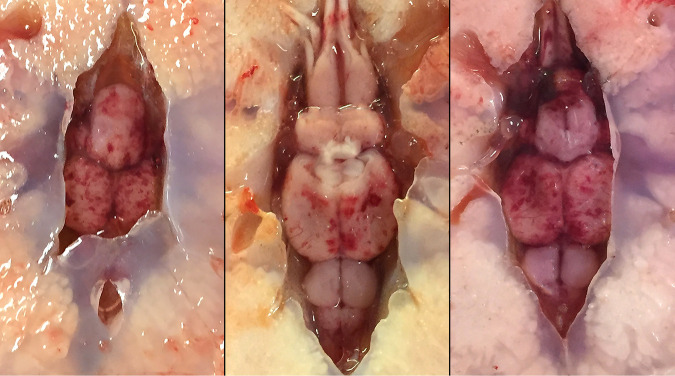
FIG 1.
Examination of moribund Chinook salmon (Oncorhynchus tshawytscha) observed congregating along the banks of the lower Eel River, California, revealed petechial hemorrhages on the surfaces of the forebrain, optic lobes, cerebellum, and brain stem. The gross findings prompted investigation of a potential viral etiology as a cause of the clinical signs observed.
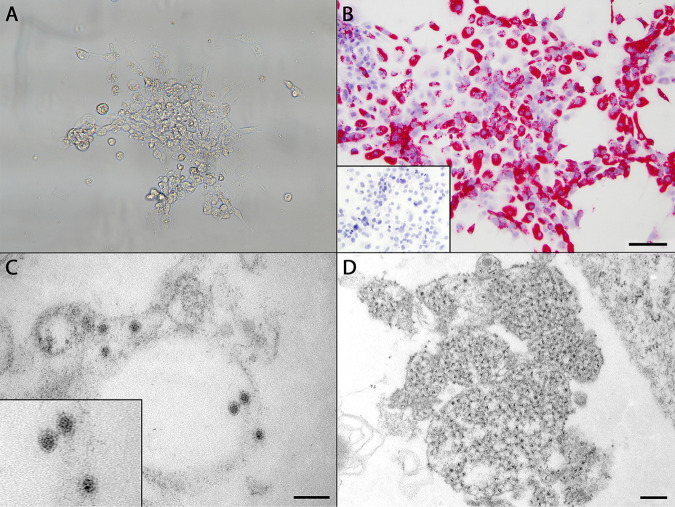
FIG 2.
(A) Cytopathic effect in SFV-infected striped-snakehead (SSN-1) cells incubated at 20°C, beginning with mild vacuolation and rounding. By day 10 postinoculation, all cells had lifted from the flask or lysed. (B) RNAscope in situ hybridization assay demonstrating strong positive cytoplasmic staining of SFV-infected SSN-1 cells by Vulcan Fast Red. Bar, 100 μm. (Inset) Lack of staining in uninoculated control cells. (C) Transmission electron microscopic image of SFV prepared from infected SSN-1 cells. Viral particles were roughly round, with an average diameter of 35.17 nm, and possessed an electron-dense core surrounded by an outer membrane. Particles were present primarily within the cisternae of the endoplasmic reticulum and in vesicles. Bar, 100 nm. (Inset) Enlarged image of three viral particles with typical flaviviral morphology. (D) Cytoplasmic replication complex of SFV within infected SSN-1 cells filled by ill-defined, uncondensed material believed to represent early RNA synthesis. Bar, 200 nm.
Virus particles with typical flavivirus morphology (12) were observed during electron microscopic examination of SSN-1 cells exhibiting CPE following inoculation with Chinook salmon tissue homogenates. Virions averaged 35.17 ± 2.49 nm in diameter (n = 50) and were roughly round, with an electron-dense core surrounded by an envelope of reduced electron density (Fig. 2C). Particles were found primarily within cytoplasmic cisternae, presumably of the endoplasmic reticulum, and vesicles. Also present were large cytoplasmic replication complexes filled with ill-defined, uncondensed material possibly associated with early RNA synthesis (Fig. 2D) (6, 13, 14).
Next-generation sequencing of the salmon viral isolate revealed a divergent virus related to members of the Flaviviridae. The salmon flavivirus (SFV) genome is composed of 10,322 nucleotides (nt) (GenBank accession no. MT075326), and its coding region shares <30% identity with those of various flaviviruses in a translated protein homology search (blastx). At least 400× NGS coverage was obtained in the coding region. We further extended and confirmed the 5′ noncoding region (NCR) sequence by rapid amplification of cDNA ends (RACE).
Typical members of the Flaviviridae, including species of the genus Flavivirus, contain a single open reading frame (ORF) flanked by NCRs. The ORF encodes a polyprotein that is cleaved into at least 10 nonstructural (NS) and structural proteins, the latter group including the capsid (C), membrane (M), and envelope (E) (6). The SFV genome has a distinctive organization with two split ORFs, containing an 1,108-amino-acid (aa) ORF1 and a 2,229-aa ORF2, and a unique ORF1/2 junction located between NS1 and NS2A, which share a 1-nt overlap (Fig. 3). The junction and adjacent sequences were manually inspected for consistency using the >400× NGS sequence coverage. A programmed −1 ribosomal frameshifting signal, conforming to the heptanucleotide “slippery sequence” (UUUAAAC), was detected at the 3’ end of ORF1. Recent reports of CLuV from lumpfish and the three cephalopod flaviviruses have also identified this dual ORF organization (9, 11). The SFV genome codes for 10 proteins in typical flavivirus gene order (Fig. 3). The cleavage map of SFV was derived by alignment with flavivirus reference sequences.
FIG 3.
Genome characteristics of SFV, sequencing coverage, and comparisons of the SFV genome and genes to those of representatives of the Flavivirus genus. (A) Genome characteristics of SFV, which is composed of typical flaviviral proteins organized in two open reading frames, unlike classical flaviviruses. (B) Sliding-window analysis showing pairwise distances of SFV amino acids from those of other flaviviruses. Protein sequences from the two SFV ORFs are concatenated for analysis. HCV, hepatitis C virus; YFV, yellow fever virus; DV, dengue virus. Intergenus divergence (SFV-HCV) was compared with intragenus/interspecies divergence (DV-YFV, SFV-YFV, and SFV-DV) and intergenotype divergence (between DV genotypes). Pairwise distance is shown along the y axis. (C) Continuous high sequence coverage used to construct the SFV genome, with the 5′ and 3′ NCR further confirmed by RACE and Sanger sequencing. (D) Pairwise identities (expressed as percentages) of SFV proteins with those of representatives of the Flavivirus genus.
Host signalase motifs (typically Ala-X-Ala preceding the cleavage site) are moderately conserved and were detected in cleavage sites preceding C, NS1, NS2A, and NS2B. Flaviviruses typically utilize a host signal peptidase cleavage between C and prM, between prM and E, between E and NS1, and between 2K and NS4B, while a virus-encoded serine protease NS2B-3 process cleaves at NS2A–NS2B, NS2B–NS3, NS3–NS4A, NS4A–2K, and NS4B–NS5 (6). The signal peptidase cleavage sites for the SFV are most conserved at prM–E and E–NS1 but not at C–prM and 2K–NS4B. The serine protease cleavage sites for SFV are moderately conserved at NS2B–NS3, NS3–NS4A, NS4A–2K, and NS4B–NS5 but not at NS2A–NS2B. The deletion of approximately 30 amino acids at the NS2A–NS2B junction relative to other flaviviruses prevents accurate prediction of the cleavage site.
Intraspecies, intragenus, and intergenus sequence divergence between SFV and different members of the Flaviviridae was determined using sliding-window analysis (Fig. 3). Sequence divergence between SFV and members of the Flavivirus genus is slightly higher than that among different flavivirus species, such as dengue virus and yellow fever virus, throughout the lengths of their genomes. However, it is still lower than the intergenus range. These findings indicate that SFV is a divergent species of Flavivirus and support its placement in a basal phylogenetic position. The most conserved protein among these flaviviruses is NS5, while the least conserved proteins are NS2A and NS2B.
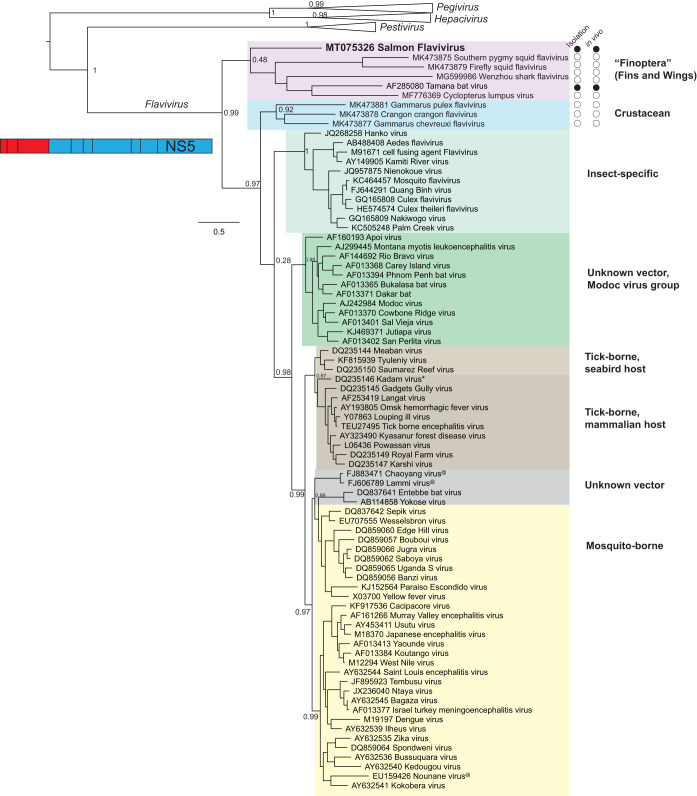
Phylogenetic analysis of the NS5 protein (Fig. 4) demonstrated that SFV belongs in a new subgroup that includes “unclassified” flaviviruses from bats (TABV), lumpfish (CLuV), sharks/gazami crabs (Wenzhou shark flavivirus), and squid (10, 11). For ease of discussion, we tentatively refer to this clade as the “finoptera” (fins and wings) subgroup, considering the aquatic-animal and bat origins of its member viruses. The fact that TABV is a recognized Flavivirus species supports the placement of SFV and other members of this subclade within the Flavivirus genus.
FIG 4.
NS5 maximum likelihood phylogeny of SFV in relationship to representatives of the Flavivirus genus. Classical Flavivirus subgroups are labeled according to the ICTV classification (as described in Materials and Methods); two recent subgroups containing aquatic flaviviruses are tentatively marked “Finoptera” and “Crustacean,” with purple and blue shading, respectively. The current status of viral isolation and in vivo infection, indicating the confirmation of host range and tropism, is shown for the two subgroups (filled circles, successful; open circles, not attempted or not successful). A few classical flaviviruses whose hosts were not experimentally confirmed are marked with asterisks; those whose hosts were not yet known are marked with “@.” Representatives of the genera Pegivirus, Hepacivirus, and Pestivirus within the family Flaviviridae were included in the analysis, but their taxa collapsed in final graphics. The location of NS5 in relation to the flaviviral genome is shown as a schematic.
Furthermore, phylogenetic analysis of SFV provides new insights into the evolution of flaviviruses. We found strong phylogenetic support for placing the finoptera subgroup in a basal position ancestral to the classical flaviviruses and the crustacean group (Fig. 4). Of note, the phylogenetic position of the crustacean flaviviruses is slightly different from that presented in a recent genuswide Flavivirus tree (11). We speculate that our inclusion of the divergent SFV together with outgroups from Flaviviridae family representatives provides better resolution. However, both studies were concordant with the ancestral position of the finoptera subgroup. SFV is currently one of the most divergent flaviviruses described in the genus Flavivirus.
The 5′ NCRs of flaviviruses are important promoters of viral RNA replication (15). Unlike the 5′ NCRs of sister genera, such as Hepacivirus, which contain a long internal ribosome entry site (IRES) that is required for the initiation of translation (16), the flaviviral 5′ NCR is short, since the translation of flaviviral RNA is initiated not by an IRES but by ribosome scanning of a 5′ cap structure (17). Resembling other flaviviruses, SFV contains a 121-nt 5′ NCR and a 188-nt 3′ NCR, confirmed both by unbiased deep sequencing and by targeted 5′ and 3′ RACE. Flavivirus 5′ NCR sequences are poorly conserved between species, although secondary structural elements, such as a bifurcating 5′ stem-loop, have been identified. The SFV 5′ NCR does not exhibit any nucleotide homology with other flaviviruses, as shown by a BLASTn search. The 121-nt 5′ NCR of SFV is similar to the typical 89- to 132-nt 5′ NCRs of other flaviviruses (18).
To investigate the pathogenic potential of SFV for fish, three laboratory-controlled challenge trials were performed in fingerling rainbow trout (RBT) and CS by immersion and by intracoelomic injection (IC) routes. In the immersion challenge, 60 fish of each species were immersed in a viral inoculum or a control medium for 1 h. At the end of the 30-day trial, 1/60, 2/60, and 1/60 RBT died when exposed to final concentrations of 7.4 × 106 TCID50/ml, 7.4 × 104 TCID50/ml, and 7.4 × 102 TCID50/ml of SFV inoculum, respectively. When challenged with corresponding levels of the virus, 3/60, 1/60, and 0/60 CS died. Low background levels of mortality also occurred in control RBT (3/60) and CS (1/60). For both the test and control groups, no CPE occurred in cell cultures from fresh dead or surviving fish after 4 weeks of incubation. These results suggest either that fish were briefly infected, with rapid clearance of the virus, or that the immersion route may not be a suitable model for inducing infection.
In the initial in vivo challenge, 10 fish of each species received intracoelomic injections of 2.1 × 107 TCID50/fish. Ten controls received injections of the SSN-1 cell flask medium only. During the 14-day trial, 1/10 RBT died. No mortalities occurred in CS or negative controls of either species. Upon completion of the trial, organ tissue pools were cultured on SSN-1 cells and were tested for SFV RNA using reverse transcriptase quantitative PCR (RT-qPCR). Cultures from 3/5 RBT exhibited compatible CPE, and 3/5 were PCR positive, while 5/5 CS samples were both culture and PCR positive. No control fish cultures produced CPE, and all were PCR negative. Higher viral RNA copy numbers were detected in CS tissues than in RBT tissues by a t test (degrees of freedom [df] = 4.8; P = 0.05) (data not shown).
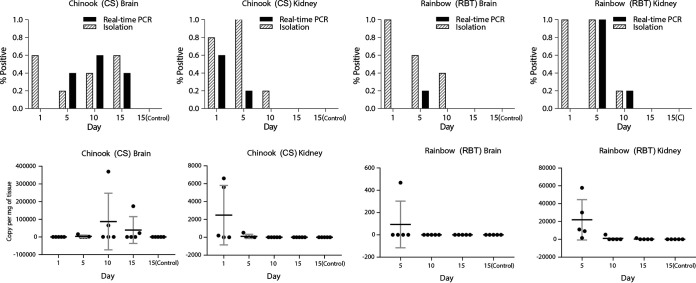
Based on the observed 14-day persistence of SFV in fish tissues from the first injection trial, a second in vivo injection challenge was conducted so as to better elucidate tissue tropisms and infection characteristics. Following intracoelomic injection, five RBT and five CS were sampled at 5-day intervals over a 15-day trial and were evaluated by culture and RT-qPCR (Fig. 5). SFV replication levels were greatest in the brains of CS, where 3/5 tested positive by culture and 2/5 by RT-qPCR on day 15. Importantly, the numbers of SFV genome copies in the brains of CS sampled on day 10 were higher than those on day 5, suggesting viral replication, and remained high on day 15. In comparison, the brains of RBT and the kidney samples of RBT and CS were negative on day 15, following decreases by day 10, when the virus was detected in the brains of RBT (2/5) and the kidneys of RBT and CS (1/5 each) by one or both detection methods. Although RNA degradation precluded RT-qPCR testing of samples from day 1, the number of viral genome copies in the brains of RBT decreased from day 5 to day 10, when they were nondetectable. Greater replication of SFV in the brains of CS than in those of RBT is not unexpected, since the virus was originally isolated from CS and is likely better adapted to CS, suggesting that SFV has a narrow host range. All control fish samples were negative by culture and RT-qPCR throughout the trial. The complete findings are summarized in Fig. 5.
FIG 5.
Detection of salmon flavivirus in Chinook salmon (Oncorhynchus tshawytscha) (CS) and rainbow trout (Oncorhynchus mykiss) (RBT) fingerlings exposed experimentally by intracoelomic injection. Brains and kidneys of fingerlings (n, 5 per species) were collected at different time points postinoculation, and virus was detected using RT-qPCR or viral isolation. The upper panels show the frequency of detection by each method, and the lower panels show the numbers of viral genome copies by RT-qPCR. Results for control animals at day 15 [15(Control)] are also shown. In the two graphs on the lower right, viral copies for RBT on day 1 of the experiment are not shown due to RNA degradation. Note that the scales of viral copy numbers are different in all four graphs.
Evaluation of fish tissues for histopathologic lesions indicative of SFV-induced damage and the application of an SFV-specific in situ hybridization (ISH) assay developed using RNAscope Technology took place in conjunction with the second injection challenge. Despite supporting culture and RT-qPCR results, at no time point were any abnormal changes observed in tissues, including central nervous system tissue, in challenged fingerlings of either fish species. While the absence of microscopic changes may indicate that SFV is nonpathogenic to CS and RBT, it is possible that >10 days are required for the development of lesions. Alternatively, the virus may require an adult rather than a juvenile host or additional stressors, such as increased water temperatures, to induce pathological changes. However, gross and microscopic changes in the original adult wild salmon were limited to nonspecific petechial hemorrhages in their brains and parasitic cataracts.
ISH probes targeting 1,476 bp of the viral envelope gene produced strong positive staining of infected SSN-1 cell culture monolayers and no staining of negative-control cells, validating their utility as controls in the tissue studies (Fig. 2B). In concordance with the lack of histologic findings, the ISH probe did not demonstrate the presence of SFV within any tissues of CS fingerlings collected on day 10 of the challenge, when viral RNA copy numbers were highest, or in the eyes and brains of the adult salmon. The negative staining results are presumably due to the lower detection limits of the ISH assay, since these tissues tested positive by the more-sensitive method RT-qPCR.
While the significance of SFV to the health of CS remains uncertain, ongoing sampling in 2019 revealed the continued presence of the virus in California wild adult salmon. SFV was isolated from the ovarian fluids of returning females, and RT-qPCR detected SFV RNA in the brains of adult male and female CS (data not shown). Retrospectively, RT-qPCR also confirmed the presence of SFV in at least one formalin-fixed ocular, renal, or gastric tissue sample from each of the three original case CS in 2015 (data not shown). As a result, the remaining questions concerning SFV pathogenicity, routes of transmission, and potential vectors warrant further study.
Many arthropod-borne flaviviruses are neurotropic as well as neuropathogenic and are responsible for several diseases of growing global concern in humans and domestic animals (19). Disease pathogenesis is complex and has been fully elucidated for only a few viruses. Clinical presentations can range from subclinical to fulminating (19, 20). In contrast, the bat and rodent flaviviruses with no known vectors, such as TABV and Modoc virus, do not appear to regularly cause overt disease in their reservoir hosts (5, 7). However, cases of disease, including meningitis and encephalitis, have followed natural or laboratory exposure of humans, nonhuman primates, and rodents to various flaviviruses in the no-known-vector group (5). Although the findings demonstrate that SFV can replicate in the nervous systems of CS, no compelling evidence of pathogenic potential for the virus was established within the current scope of immersion or injection challenges. It is possible that SFV causes disease only within specific temperature ranges, as is seen with many established viral fish pathogens (21, 22), or that changing environmental conditions create stressful conditions that promote opportunistic disease. Environmental factors, such as increasing water temperatures, are expected to alter disease emergence, severity, and existing patterns in the future (23, 24).
Any role for vectors in the transmission of aquatic flaviviruses is largely unknown. Salmon share their aquatic environment with other fish species and parasites that can potentially serve as carriers or vectors of disease (25, 26). While the findings indicate that CS are a better host for the replication of SFV than the related species RBT, it is not known whether additional fish species serve as potential reservoirs of SFV or harbor additional flaviviruses related to SFV. The possible role of vector-borne transmission also remains to be investigated. The presence of Wenzhou shark flavivirus RNA in shark and crab species suggests that horizontal transmission may occur between the unrelated species (11), although this has not been demonstrated experimentally. Transmission of SFV by crustacean parasites or via the natural food chain is also possible. Alternatively, flaviviruses such as Zika virus can be transmitted vertically in utero and at birth, as well as in sexual fluids (27). Despite the differing reproductive strategies of fish and humans, detection of SFV in the ovarian fluids of prespawning CS make it reasonable to speculate that vertical transmission of the virus may occur.
In this study, we characterize a novel flavivirus, the first to be isolated in culture from a piscine source. The proposed salmon flavivirus assumes a basal phylogenic position within an expanding subclade of unclassified aquatic flaviviruses identified in vertebrate and invertebrate species. Despite its isolation from moribund CS returning upriver to spawn, no clear relationship of SFV to clinical signs could be established. Similarly, no specific pathogenic effects were demonstrated in challenge trials involving CS and RBT. However, the tissue tropism of SFV and its replication in the brains of CS were established using RT-qPCR. The application of this assay and others to future disease surveillance and experimental trials is expected to further elucidate the pathogenic potential and transmission cycle of SFV in ecologically and economically important salmonid species.
MATERIALS AND METHODS
Case history.
In November 2015, Eel River Recovery Project (ERRP), National Oceanic and Atmospheric Administration (NOAA), and California Department of Fish and Wildlife (CDFW) biologists observed diseased prespawning adult Chinook salmon (CS) in the lower Eel River, upstream of Fernbridge, CA. Approximately 10% of the salmon in an estimated group of 1,500 fish localized to a shallow pool were clinically affected. Two affected adult females, 87 and 95 cm long, and one male salmon, 65 cm long, were collected for diagnostic evaluation. The fish were euthanized by shocking in ice water, individually bagged, and iced for approximately 6 h prior to examination. No similar disease signs were observed in salmon that had completed the upriver migration.
Necropsy and histological, microbiological, and parasitological analyses.
Necropsies were performed, and samples of gill, eye, brain, heart, liver, spleen, posterior kidney, intestine, and skeletal muscle were variously processed for histological, microbiological, and molecular evaluation. Tissues fixed in 10% neutral buffered formalin were processed routinely and were stained with hematoxylin and eosin (H&E) for histological examination. Formalin-fixed tissues were also used later for detection and flavivirus quantification by reverse transcriptase quantitative PCR (RT-qPCR).
Samples of brain, posterior kidney, and a retrobulbar swab were inoculated onto plates containing tryptic soy agar with 5% sheep blood and modified Thayer Martin agar; then the samples were incubated at 20°C for 2 weeks. For virus isolation, pooled samples of brain, spleen, kidney, and gonad were homogenized, centrifuged, inoculated into cells of the Chinook salmon embryo-214 (CHSE-214), epithelioma papulosum cyprini (EPC), and striped-snakehead (SSN-1) lines, and incubated at 15°C (CHSE-214 and EPC cells) or 20°C (SSN-1 cells) for 2 weeks. To compare growth in different cell lines, the medium collected from a flask exhibiting complete CPE was centrifuged and frozen at –80°C for stock. A vial of frozen stock was thawed and diluted 1:20 with MEM-2 plus HEPES before 10-fold dilutions were made in MEM-2 plus HEPES. Five 50-μl replicates of each dilution were added to individual wells of a 96-well plate, and 50 μl of a suspension of either SSN-1, CHSE-214, EPC, fathead minnow (FHM), koi fin (KF), or bluegill fin (BF-2) cells was added to each well before incubation at either 15°C or 20°C for 3 weeks. The methods used in the molecular identification of the ocular trematode can be found in the supplemental material and in references 28, to ,32.
TEM.
The medium was removed from an SSN-1 cell culture exhibiting cytopathic effect (CPE) and the flask flooded with 2% paraformaldehyde and 2.5% glutaraldehyde in 0.1 M sodium cacodylate (pH 7.4). Cells were removed from the flask using a cell scraper, transferred to a 1.5-ml microcentrifuge tube, pelleted, and enrobed in agar prior to postfixation in osmium tetroxide. The cells were then dehydrated in a series of ethanol (30% to 100%), acetone, and propylene oxide steps before the infiltration of Epon-Araldite (Electron Microscopy Sciences, Hatfield, PA) and overnight polymerization at 80°C. Ultrathin sections were cut on a Reichert Ultracut S ultramicrotome (Leica, Inc., Deerfield, IL), stained with lead citrate, and examined on a JEM-1210 transmission electron microscope (TEM) (JEOL USA, Inc., Peabody, MA).
Viral metagenomic, bioinformatic, and genomic analyses.
Viral metagenomic analysis was carried out using previously described protocols (33–35). Briefly, the SSN-1 culture supernatant was first pushed through a 0.2-μm filter to enrich for viral particles and then depleted of host nucleic acid by using nucleases. Viral nucleic acid was extracted using the QIAquick viral RNA column purification system (Qiagen), which was amplified by an unbiased sequence-independent, single-primer amplification (SISPA) protocol. A sequencing library was prepared using the Nextera XT DNA sample preparation kit (Illumina, San Diego, CA) with dual-indexed barcoding. Sequencing was performed using an Illumina MiSeq system with 2 × 250 bp paired-end sequencing reagents (Illumina MiSeq reagents, v2; 500 cycles). Virome analysis of the NGS data using de novo assembly and blastx protein homology searches was performed as described previously (36).
For genomic analysis, sequences from all ICTV-recognized members of the family Flaviviridae were retrieved from GenBank and the ICTV Flaviviridae Study Group wiki (https://talk.ictvonline.org/ictv_wikis/flaviviridae/w/sg_flavi). The protein-coding sequences, including that of the salmon flavivirus, were aligned using MUSCLE (37), where flaviviral genes were inferred by comparing the cleavage sites of related species. Maximum likelihood phylogenetic trees were constructed on the NS5 protein using PhyML (38), where the best nucleotide substitution model was analyzed by Smart Model Selection based on the Bayesian information criterion (39). Tree annotation and midpoint rooting were performed using FigTree (http://tree.bio.ed.ac.uk/software/figtree/). Sliding-window analysis was performed for representative sequences with SSE (40) using a window size of 18 residues with an increment of 3. Pairwise nucleotide identity between sequences was calculated using the Sequence Demarcation Tool using the MUSCLE function (41).
Reverse transcriptase quantitative PCR.
RNA was isolated from supernatants of SSN-1 viral cultures using the TRIzol reagent (Life Technologies), followed by DNase treatment (AMP-D1; Sigma) and cDNA synthesis using SuperScript II reverse transcriptase (Life Technologies). Conventional PCR was performed to amplify a 270-bp portion of the major capsid protein (MCP) gene using the primer set comprising Salmon ENV_AF (5′-AGATTGATTTCGGCAACAGC-3′) and Salmon ENV_AR (5′-GTTGGAAGACGTTGGACGTT-3′). The amplified fragment was cloned into a pGEM-T Easy vector (Promega). Three plasmid clones carrying the MCP gene were obtained; they were sequenced by the UC Davis DNA Sequencing Facility (http://dnaseq.ucdavis.edu/) and confirmed to be identical to the reference sequence obtained by metagenomic analysis. A series of plasmid DNA standards (1:10 serial dilutions; 10 to 1 × 1010 copies per 5 μl) was prepared. The primers and probe for the flavivirus qPCR were designed by Primer Express, v3.0 (Applied Biosystems), against the MCP gene (Table 1). The qPCRs were performed with Maxima Probe/ROX qPCR master mix (Thermo Fisher) using an ABI 7900HT Fast system at the UC Davis Real-Time PCR Research and Diagnostics Core Facility (https://pcrlab.vetmed.ucdavis.edu/) to generate a standard curve (Fig. S2). The ABI standard cycling reaction conditions were used for the reactions: 50°C for 2 min, 95°C for 10 min, 40 cycles of 95°C for 15 s, and 60°C for 60 s.
TABLE 1.
Primer and probe sequences for the flavivirus qPCR assay developed in this study
| Primer or probe | Sequence (5′–3′) |
|---|---|
| SalmonENV_98Fw | AGCCCTCGCAGGTTCTTTC |
| SalmonENV_135Rv | GTTGCCCCATTGCACCAT |
| Salmon_ENV_118P (FAM-MGB)a | CCCGCAACCTTGAC |
FAM, 6-carboxyfluorescein.
For the detection of the virus and determination of the viral copy number in formalin-fixed, paraffin-embedded (FFPE) tissues of the three Chinook salmon, total RNA was isolated with the WaxFree RNA extraction kit (TrimGen), followed by DNase treatment and cDNA synthesis as described above. Reactions were run in triplicate. The flavivirus genome possesses only one copy of the MCP gene, so the MCP gene copy numbers obtained from RT-qPCR were reported as viral copy numbers. The viral copy number was standardized by the wet weight of fish tissue used for RNA isolation.
Infectious challenges.
All challenges were conducted under protocols approved by the University of California, Davis School of Veterinary Medicine Institutional Animal Care and Use Committee.
To test Rivers’ postulates and determine the distribution of the flavivirus in tissues, experimental challenges were performed by immersion and intracoelomic injection using rainbow trout (RBT) and CS fingerlings obtained from a source with no history of compatible disease signs. For all experiments, challenge and control fish were maintained in separate 15-liter tanks receiving aerated freshwater at 18 ± 1°C at a flow rate of 1.5 liters/min. During all trials, mortality observations were made twice daily, and at the conclusion of the trials, fish were euthanized by overdose with 250 mg/liter buffered tricaine methanesulfonate (MS-222; Sigma-Aldrich).
For the immersion challenge, RBT and CS (average weights, 2.2 and 1.2 g, respectively) were maintained at 10 fish per tank. Six replicate RBT and CS tanks were used for each challenge dose and as negative controls. The water flow was stopped, and fish were challenged by immersion in 7.4 × 106 TCID50/ml, 7.4 × 104 TCID50/ml, or 7.4 × 102 TCID50/ml for 1 h before the flow was restored. Control fish were treated similarly with media from uninoculated flasks of SSN-1 cells diluted 1:100 in tank water. After 30 days, surviving fish were euthanized, and the heads and viscera of 10 fish per treatment were pooled for virus isolation.
In an initial intracoelomic injection challenge, RBT and CS (average weights, 2.6 and 3.1 g, respectively) were housed 10 per tank. Fish were anesthetized with 50 mg/liter of buffered MS-222 and were injected with 2.1 × 107 TCID50/fish or, for controls, SSN-1 cell flask medium. The fish were returned to their original tanks and were monitored for 14 days, at which time five fish from each tank were euthanized. The brain, posterior kidney, heart, and gills from each fish were pooled and processed for virus isolation, and a non-pooled subsample was used for molecular diagnosis. Pooled tissues used for molecular diagnosis were frozen at –80°C, and total RNA was extracted using the TRIzol reagent (Thermo Fisher) for quantification of the flavivirus by RT-qPCR.
In a second intracoelomic injection challenge, RBT and CS, averaging 8.6 and 9.2 g, respectively, were maintained 15 per tank in four replicate tanks per treatment. Challenge (5.6 × 107 TCID50/fish) and control fish were treated as described above. At 1, 5, 10, and 15 days postchallenge, five fish from each treatment group were euthanized, and samples of brain and kidney were collected. Tissues were processed for virus isolation and RT-qPCR as described above. Five of the remaining challenged and control fish from each treatment were fixed whole in formalin, with their coelomic cavities incised. After decalcification in Kristensen’s solution, bodies were serially cut into 2- to 3-mm cross sections in their entirety, processed routinely, and stained with H&E for histological examination.
RNAscope in situ hybridization assay.
An RNAscope 2.5 HD RED kit from Advanced Cell Diagnostics (ACD) was used to probe for flaviviral RNA using a 1,476-bp sequence of the viral envelope gene provided to ACD for probe design using proprietary technology. The resulting probe set consisted of 20 tiled and labeled oligomers (20 ZZ’s). Application of the method to FFPE sections of experimentally infected brains and kidneys followed validation in an infected SSN-1 fish cell line. Monolayers of SSN-1 cells cultured on glass coverslip discs received either a viral inoculum or culture medium only for use as a positive or negative control, respectively. Fixation of cells in 10% buffered formalin followed 3 days of incubation at 20°C and 3 days at 15°C, when extensive CPE was observed. The prepared cells were subjected to target retrieval in kit reagents (15 min at 100°C), protease treatment (30 min at 40°C), probe hybridization to target cells (2 h at 40°C), and a series of six proprietary signal amplification steps using the reagents provided in the kit and the ACD protocol. Incubation of the slides with Vulcan Fast Red (10 min at room temperature) produced the final signal generation, followed by washing with water, counterstaining with hematoxylin, and bluing of the nuclei. Slides were dried (60°C, 15 min), covered with a coverslip using a nonaqueous mounting medium (EcoMount), and examined using bright-field microscopy. Probing of FFPE tissue sections followed the same protocol as that for the control slides with the exception of a necessary initial deparaffinization step.
Data availability.
The salmon flavivirus genome sequence is available in GenBank under accession no. MT075326.
Supplementary Material
ACKNOWLEDGMENTS
We thank the California Department of Fish and Wildlife statewide fish disease research program (2015–2018) for supporting this study.
We thank Swee Teh, Chelsea Lam, and Khiet Huynh of the Department of Anatomy, Physiology and Cell Biology, School of Veterinary Medicine, University of California, Davis, Davis, CA, for assistance.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.National Oceanic and Atmospheric Administration (NOAA). 14 January 2020, accession date. Species directory. Chinook salmon (protected) https://www.fisheries.noaa.gov/species/chinook-salmon-protected.
- 2.Cline TJ, Ohlberger J, Schindler DE. 2019. Effects of warming climate and competition in the ocean for life-histories of Pacific salmon. Nat Ecol Evol 3:935–942. doi: 10.1038/s41559-019-0901-7. [DOI] [PubMed] [Google Scholar]
- 3.Katz J, Moyle PB, Quiñones RM, Israel J, Purdy S. 2013. Impending extinction of salmon, steelhead, and trout (Salmonidae) in California. Environ Biol Fish 96:1169–1186. doi: 10.1007/s10641-012-9974-8. [DOI] [Google Scholar]
- 4.Moyle PB, Kiernan JD, Crain PK, Quinones RM. 2013. Climate change vulnerability of native and alien freshwater fishes of California: a systematic assessment approach. PLoS One 8:e63883. doi: 10.1371/journal.pone.0063883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blitvich BJ, Firth AE. 2017. A review of flaviviruses that have no known arthropod vector. Viruses 9:154. doi: 10.3390/v9060154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lindenbach BD, Murray CL, Thiel H-J, Rice CM. 2013. Flaviviridae, p 712–746. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 7.Ludwig GV, Iacono-Connors LC. 1993. Insect transmitted vertebrate viruses: Flaviviridae. In Vitro Cell Dev Biol Anim 29A:296–309. doi: 10.1007/BF02633958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Price JL. 1978. Isolation of Rio Bravo and a hitherto undescribed agent, Tamana bat virus, from insectivorous bats in Trinidad, with serological evidence of infection in bats and man. Am J Trop Med Hyg 27:153–161. doi: 10.4269/ajtmh.1978.27.153. [DOI] [PubMed] [Google Scholar]
- 9.Skoge RH, Brattespe J, Okland AL, Plarre H, Nylund A. 2018. New virus of the family Flaviviridae detected in lumpfish (Cyclopterus lumpus). Arch Virol 163:679–685. doi: 10.1007/s00705-017-3643-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi M, Lin X-D, Chen X, Tian J-H, Chen L-J, Li K, Wang W, Eden J-S, Shen J-J, Liu L, Holmes EC, Zhang Y-Z. 2018. The evolutionary history of vertebrate RNA viruses. Nature 556:197–202. doi: 10.1038/s41586-018-0012-7. [DOI] [PubMed] [Google Scholar]
- 11.Parry R, Asgari S. 2019. Discovery of novel crustacean and cephalopod flaviviruses: insights into the evolution and circulation of flaviviruses between marine invertebrate and vertebrate hosts. J Virol 93:e00432-19. doi: 10.1128/JVI.00432-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Junglen S, Kopp A, Kurth A, Pauli G, Ellerbrok H, Leendertz FH. 2009. A new flavivirus and a new vector: characterization of a novel flavivirus isolated from Uranotaenia mosquitoes from a tropical rain forest. J Virol 83:4462–4468. doi: 10.1128/JVI.00014-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doane FW, Anderson N. 1987. Electron microscopy in diagnostic virology: a practical guide and atlas, p 116–122. Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- 14.Uchil PD, Satchidanandam V. 2003. Architecture of the flaviviral replication complex. Protease, nuclease, and detergents reveal encasement within double-layered membrane compartments. J Biol Chem 278:24388–24398. doi: 10.1074/jbc.M301717200. [DOI] [PubMed] [Google Scholar]
- 15.Dong H, Zhang B, Shi PY. 2008. Terminal structures of West Nile virus genomic RNA and their interactions with viral NS5 protein. Virology 381:123–135. doi: 10.1016/j.virol.2008.07.040. [DOI] [PubMed] [Google Scholar]
- 16.Brown EA, Zhang H, Ping LH, Lemon SM. 1992. Secondary structure of the 5′ nontranslated regions of hepatitis C virus and pestivirus genomic RNAs. Nucleic Acids Res 20:5041–5045. doi: 10.1093/nar/20.19.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruiz-Linares A, Bouloy M, Girard M, Cahour A. 1989. Modulations of the in vitro translational efficiencies of yellow fever virus mRNAs: interactions between coding and noncoding regions. Nucleic Acids Res 17:2463–2476. doi: 10.1093/nar/17.7.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Markoff L. 2003. 5′- and 3′-noncoding regions in flavivirus RNA. Adv Virus Res 59:177–228. doi: 10.1016/s0065-3527(03)59006-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maximova OA, Pletnev AG. 2018. Flaviviruses and the central nervous system: revisiting neuropathological concepts. Annu Rev Virol 5:255–272. doi: 10.1146/annurev-virology-092917-043439. [DOI] [PubMed] [Google Scholar]
- 20.Potokar M, Jorgacevski J, Zorec R. 2019. Astrocytes in Flavivirus infections. Int J Mol Sci 20:691. doi: 10.3390/ijms20030691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilad O, Yun S, Adkison MA, Way K, Willits NH, Bercovier H, Hedrick RP. 2003. Molecular comparison of isolates of an emerging fish pathogen, koi herpesvirus, and the effect of water temperature on mortality of experimentally infected koi. J Gen Virol 84:2661–2667. doi: 10.1099/vir.0.19323-0. [DOI] [PubMed] [Google Scholar]
- 22.Jung MH, Nikapitiya C, Vinay TN, Lee J, Jung SJ. 2017. Rock bream iridovirus (RBIV) replication in rock bream (Oplegnathus fasciatus) exposed for different time periods to susceptible water temperatures. Fish Shellfish Immunol 70:731–735. doi: 10.1016/j.fsi.2017.09.038. [DOI] [PubMed] [Google Scholar]
- 23.Marcos-Lopez M, Gale P, Oidtmann BC, Peeler EJ. 2010. Assessing the impact of climate change on disease emergence in freshwater fish in the United Kingdom. Transbound Emerg Dis 57:293–304. doi: 10.1111/j.1865-1682.2010.01150.x. [DOI] [PubMed] [Google Scholar]
- 24.Matanza XM, Osorio CR. 2018. Transcriptome changes in response to temperature in the fish pathogen Photobacterium damselae subsp. damselae: clues to understand the emergence of disease outbreaks at increased seawater temperatures. PLoS One 13:e0210118. doi: 10.1371/journal.pone.0210118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cusack R, Cone DK. 1986. A review of parasites as vectors of bacterial and viral diseases of fish. J Fish Dis 9:169–171. doi: 10.1111/j.1365-2761.1986.tb01000.x. [DOI] [Google Scholar]
- 26.Jakob E, Barker DE, Garver KA. 2011. Vector potential of the salmon louse Lepeophtheirus salmonis in the transmission of infectious haematopoietic necrosis virus (IHNV). Dis Aquat Organ 97:155–165. doi: 10.3354/dao02414. [DOI] [PubMed] [Google Scholar]
- 27.Gregory CJ, Oduyebo T, Brault AC, Brooks JT, Chung K-W, Hills S, Kuehnert MJ, Mead P, Meaney-Delman D, Rabe I, Staples E, Petersen LR. 2017. Modes of transmission of Zika virus. J Infect Dis 216(Suppl 10):S875–S883. doi: 10.1093/infdis/jix396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosser TG, Alberson NR, Khoo LH, Woodyard ET, Pote LM, Griffin MJ. 2016. Characterization of the life cycle of a fish eye fluke, Austrodiplostomum ostrowskiae (Digenea: Diplostomidae), with notes on two other diplostomids infecting Biomphalaria havanensis (Mollusca: Planorbidae) from catfish aquaculture ponds in Mississippi, USA. J Parasitol 102:260–274. doi: 10.1645/15-850. [DOI] [PubMed] [Google Scholar]
- 29.Morgan JA, Blair D. 1995. Nuclear rDNA ITS sequence variation in the trematode genus Echinostoma: an aid to establishing relationships within the 37-collar-spine group. Parasitol 111:609–615. doi: 10.1017/S003118200007709X. [DOI] [PubMed] [Google Scholar]
- 30.Rosser TG, Baumgartner WA, Alberson NR, Noto TW, Woodyard ET, King DT, Wise DJ, Griffin MJ. 2018. Clinostomum poteae n. sp. (Digenea: Clinostomidae), in the trachea of a double-crested cormorant Phalacrocorax auritus Lesson, 1831 and molecular data linking the life-cycle stages of Clinostomum album Rosser, Alberson, Woodyard, Cunningham, Pote & Griffin, 2017 in Mississippi, USA. Syst Parasitol 95:543–566. doi: 10.1007/s11230-018-9801-5. [DOI] [PubMed] [Google Scholar]
- 31.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 32.Labrie L, Komar C, Terhune J, Camus A, Wise D. 2004. Effect of sublethal exposure to the trematode Bolbophorus spp. on the severity of enteric septicemia of catfish in channel catfish fingerlings. J Aquat Anim Health 16:231–237. doi: 10.1577/H04-011.1. [DOI] [Google Scholar]
- 33.Ng TF, Marine R, Wang C, Simmonds P, Kapusinszky B, Bodhidatta L, Oderinde BS, Wommack KE, Delwart E. 2012. High variety of known and new RNA and DNA viruses of diverse origins in untreated sewage. J Virol 86:12161–12175. doi: 10.1128/JVI.00869-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ng TF, Kondov NO, Deng X, Van Eenennaam A, Neibergs HL, Delwart E. 2015. A metagenomics and case-control study to identify viruses associated with bovine respiratory disease. J Virol 89:5340–5349. doi: 10.1128/JVI.00064-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ng TF, Chen LF, Zhou Y, Shapiro B, Stiller M, Heintzman PD, Varsani A, Kondov NO, Wong W, Deng X, Andrews TD, Moorman BJ, Meulendyk T, MacKay G, Gilbertson RL, Delwart E. 2014. Preservation of viral genomes in 700-y-old caribou feces from a subarctic ice patch. Proc Natl Acad Sci U S A 111:16842–16847. doi: 10.1073/pnas.1410429111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dill JA, Camus AC, Leary JH, Ng T. 2018. Microscopic and molecular evidence of the first elasmobranch adomavirus, the cause of skin disease in a giant guitarfish, Rhynchobatus djiddensis. mBio 9:e00185-18. doi: 10.1128/mBio.00185-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 39.Lefort V, Longueville JE, Gascuel O. 2017. SMS: smart model selection in PhyML. Mol Biol Evol 34:2422–2424. doi: 10.1093/molbev/msx149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simmonds P. 2012. SSE: a nucleotide and amino acid sequence analysis platform. BMC Res Notes 5:50. doi: 10.1186/1756-0500-5-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muhire BM, Varsani A, Martin DP. 2014. SDT: a virus classification tool based on pairwise sequence alignment and identity calculation. PLoS One 9:e108277. doi: 10.1371/journal.pone.0108277. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The salmon flavivirus genome sequence is available in GenBank under accession no. MT075326.