Porcine reproductive and respiratory syndrome virus (PRRSV) is a major respiratory disease agent in pigs that causes enormous economic losses to the global swine industry. PRRSV helicase nsp10 is a multifunctional protein with translocation and unwinding activities and plays a vital role in viral RNA synthesis. Here, we report the first structure of full-length nsp10 from the arterivirus PRRSV at 3.0-Å resolution. Our results show that the 1B domain of PRRSV nsp10 adopts a novel open state and has a unique C-terminal domain structure, which plays a crucial role in nsp10 helicase activity. Furthermore, mutagenesis and structural analysis revealed conservation of the helicase catalytic domain across the order Nidovirales (families Arteriviridae and Coronaviridae). Importantly, our results will provide a structural basis for further understanding the function of helicases in the order Nidovirales.
KEYWORDS: helicase, nonstructural protein 10, porcine reproductive and respiratory syndrome virus, structure
ABSTRACT
Currently, an effective therapeutic treatment for porcine reproductive and respiratory syndrome virus (PRRSV) remains elusive. PRRSV helicase nsp10 is an important component of the replication transcription complex that plays a crucial role in viral replication, making nsp10 an important target for drug development. Here, we report the first crystal structure of full-length nsp10 from the arterivirus PRRSV, which has multiple domains: an N-terminal zinc-binding domain (ZBD), a 1B domain, and helicase core domains 1A and 2A. Importantly, our structural analyses indicate that the conformation of the 1B domain from arterivirus nsp10 undergoes a dynamic transition. The polynucleotide substrate channel formed by domains 1A and 1B adopts an open state, which may create enough space to accommodate and bind double-stranded RNA (dsRNA) during unwinding. Moreover, we report a unique C-terminal domain structure that participates in stabilizing the overall helicase structure. Our biochemical experiments also showed that deletion of the 1B domain and C-terminal domain significantly reduced the helicase activity of nsp10, indicating that the four domains must cooperate to contribute to helicase function. In addition, our results indicate that nidoviruses contain a conserved helicase core domain and key amino acid sites affecting helicase function, which share a common mechanism of helicase translocation and unwinding activity. These findings will help to further our understanding of the mechanism of helicase function and provide new targets for the development of antiviral drugs.
IMPORTANCE Porcine reproductive and respiratory syndrome virus (PRRSV) is a major respiratory disease agent in pigs that causes enormous economic losses to the global swine industry. PRRSV helicase nsp10 is a multifunctional protein with translocation and unwinding activities and plays a vital role in viral RNA synthesis. Here, we report the first structure of full-length nsp10 from the arterivirus PRRSV at 3.0-Å resolution. Our results show that the 1B domain of PRRSV nsp10 adopts a novel open state and has a unique C-terminal domain structure, which plays a crucial role in nsp10 helicase activity. Furthermore, mutagenesis and structural analysis revealed conservation of the helicase catalytic domain across the order Nidovirales (families Arteriviridae and Coronaviridae). Importantly, our results will provide a structural basis for further understanding the function of helicases in the order Nidovirales.
INTRODUCTION
Porcine reproductive and respiratory syndrome virus (PRRSV) is a positive single-stranded RNA virus belonging to the family Arteriviridae (a family that also includes equine arteritis virus [EAV], lactate dehydrogenase-elevating virus, and simian hemorrhagic fever virus) in the order Nidovirales (1, 2). A worldwide outbreak of PRRSV in 2006 posed a threat to swine and resulted in decreased reproductive performance and increased respiratory problems in pigs (3–5). Unfortunately, there is still no effective treatment for PRRSV infection (6, 7). Therefore, it is important to explore the process and mechanisms of PRRSV genome replication to aid the development of new strategies for PRRSV prevention and therapy.
Similar to that of other nidoviruses, the genomic RNA of PRRSV is approximately 15.6 kb in length (8). Arterivirus genomes encode an exceptionally large number of nonstructural protein domains, which mediate the key functions required for genomic RNA synthesis (1, 8). The PRRSV genome encodes six structural proteins (the GP2 to GP5, M, and N proteins) (9, 10) and at least 16 distinct nonstructural proteins (nsp1α, nsp1β, nsp2 to nsp6, nsp7α, nsp7β, nsp8, nsp9, nsp10, nsp11, and nsp12) via ribosomal frameshift sites and four proteinases: the papain-like cysteine proteinases 1α (PLP1α; nsp1α), PLP1β (nsp1β), and PLP2 (nsp2) and the main serine proteinase (SP; nsp4) (1, 8, 11, 12). The structural proteins are the components of viral particle assembly, and the nonstructural proteins carry out viral replication. Among nidoviruses, arteriviruses and coronaviruses use a similar discontinuous transcription mechanism to produce a nested set of subgenomic mRNAs for structural gene expression (1, 8, 13). RNA-dependent RNA polymerases (arterivirus nsp9 and coronavirus nsp12) and helicases (arterivirus nsp10 and coronavirus nsp13) are particularly important components of the Nidovirus RNA synthetic machinery and have therefore attracted much attention (1, 8, 14–16).
Helicases are nucleoside triphosphate (NTP)-dependent motor proteins and are classified into six superfamilies (SF1 to SF6) (17). SF1 helicases can be divided into two classes based on mechanism: SF1A helicases, which translocate along their nucleic acid substrates with 3′–5′ polarity, and SF1B helicases, which translocate in a 5′–3′ direction (17). Nidovirus helicases (arterivirus nsp10 and coronavirus nsp13) belong to the SF1B helicase family (14–16). To date, many crystal structures of SF1 helicases have been reported, such as those of the Escherichia coli Rep helicase (18), the eukaryotic RNA helicase Upf1 (19, 20), and the helicases of the RNA viruses tomato mosaic virus (21), chikungunya virus (22), Zika virus (23), EAV (16), severe acute respiratory syndrome coronavirus (SARS-CoV) (14), and Middle East respiratory syndrome coronavirus (MERS-CoV) (15). Structural data obtained for Upf1 in complexes with phosphate, ADP, and ADPNP (hydrolysable ATP analog) reveal that a conformational change accompanies binding of ATP (19). EAV nsp10 has an N-terminal zinc-binding domain (ZBD), a 1B domain, and an SF1 helicase core (containing 1A and 2A domains), but the C-terminal structure has not been determined (16). The 1B domain of EAV nsp10 undergoes a large conformational change upon substrate binding, and the 1B and 1A domains of the helicase core together form a channel that accommodates single-stranded nucleic acids (16). The nsp13 of coronaviruses (SARS-CoV and MERS-CoV) similarly consists of multiple domains: a ZBD, a stalk domain, a 1B domain, and 1A and 2A domains, which have been shown to coordinate in conducting the final unwinding process (14, 15). Furthermore, structural analyses have shown that the domain organizations of EAV nsp10 and coronavirus nsp13 are closely related to the equivalent domains of the eukaryotic Upf1 helicase (14–16).
Previous research has shown that UPF1 exhibits RNA-dependent ATPase activity essential for the complete degradation of NMD substrates (24). Upf1 from human and yeast has RNA-dependent ATPase and 5′ to 3′ helicase activities (25, 26), and human Upf1 is an RNA-binding protein, the RNA-binding activity of which is modulated by ATP (26). In the order Nidovirales, EAV nsp10 has ATPase activity that is strongly stimulated by poly(dT), poly(U), and poly(dA) (27). Coronavirus nsp13 can hydrolyze nucleoside triphosphates and deoxynucleoside triphosphates, can unwind DNA and RNA duplexes with a 5′–3′ directionality, and has RNA 5′-triphosphatase activity (28, 29). In SARS-CoV nsp13, the active sites of NTP hydrolysis are located in the cleft at the base between the 1A and 2A domains (14). Moreover, all the domains (ZBD, 1B, 1A, and 2A) in SARS-CoV are involved in the double-stranded DNA (dsDNA) unwinding process, and the β19-β20 loop on the 1A domain is directly involved in the unwinding process (14). Moreover, the interaction between nsp12 (RdRp) and nsp13 (helicase) is critical for the unwinding process in SARS-CoV (14). Previous research also indicated that the Nsp9- and Nsp10-coding regions of highly pathogenic PRRSV (HP-PRRSV) together contribute to replication efficiency in vitro and in vivo and are related to the increased pathogenicity and fatal virulence of HP-PRRSV in piglets (30). However, the specific position of the key active site in the structure of PRRSV nsp10 and how the helicase functions are coordinated between domains remain unclear. Hence, analysis of the structure of full-length nsp10 provides not only a structural basis for studying the molecular mechanism of PRRSV replication but also a new target for drug design.
At present, the structure of PRRSV nsp10 has not been determined, and structural information regarding PRRSV nsp10 remains lacking, especially in the C-terminal domain of the arteriviruses nsp10. In this study, we performed structural and functional analyses of nsp10 to elucidate the mechanism underlying the function of the PRRSV helicase. We report the first crystal structure of the PRRSV helicase nsp10 and demonstrate that the fold of the helicase active sites is widely conserved among members of the order Nidovirales (families Arteriviridae and Coronaviridae). Importantly, we found that the 1B domain may undergo dynamic conformational changes during the execution of helicase function and a unique C-terminal domain structure that participates in stabilizing overall helicase structure. Here, we elaborate on the structural mechanism underlying this finding.
RESULTS AND DISCUSSION
Structure determination.
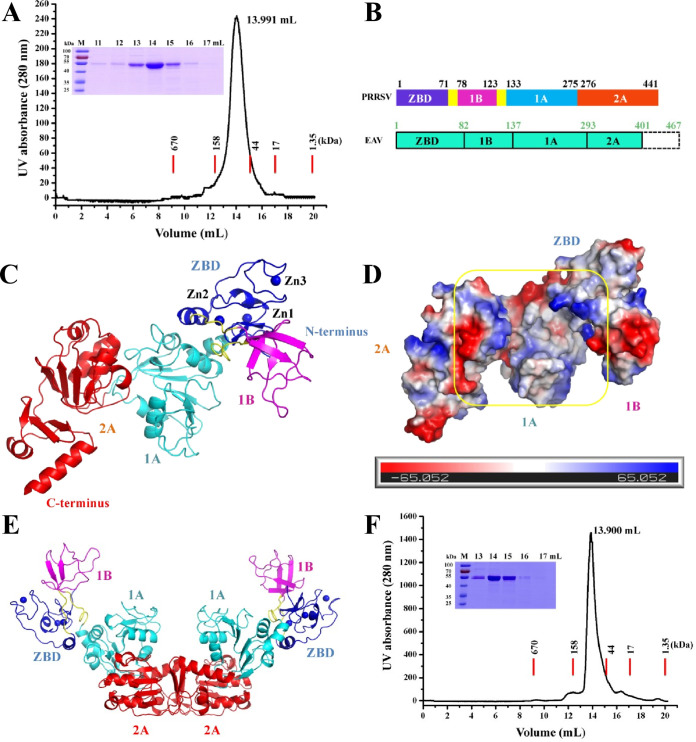
To investigate the structure of nsp10, recombinant full-length PRRSV nsp10 was expressed and purified (∼55 kDa, Fig. 1A). nsp10 was crystallized in the P41212 space group and diffracted to a resolution of 3.0 Å, and the electron density was of sufficient quality to trace the entire chain (Ser4 to Glu441, excluding Gly1-Lys3 and the N- and C-terminal His6 tags) (Fig. 1B and C). Structural data analysis showed two molecules in the asymmetric unit with the 1A and 2A domains providing the interaction interface (Fig. 1E). However, the results of gel filtration chromatography showed that nsp10 (alone or with dsDNA) was a monomer (Fig. 1A and F). Moreover, in the dimer structure, the interaction between the two molecules through the 1A and 2A domains may affect the binding of nucleic acid substrates (Fig. 1E), indicating that the arrangement of the two nsp10 molecules may be attributed to crystal packing. Similarly, other nidovirus helicases (SARS-CoV nsp13 and MERS-CoV nsp13) also perform unwinding functions in a monomer form (14, 15). Data collection and refinement statistics are summarized in Table 1.
FIG 1.
Overall structure of PRRSV nsp10. (A) Oligomerization state of nsp10. Nsp10 was eluted from a Superdex 75 10/300 GL column precalibrated with gel filtration standards (thyroglobulin, 670 kDa; γ-globulin, 158 kDa; ovalbumin, 44 kDa; myoglobin, 17 kDa; vitamin B12, 1.35 kDa). The results of SDS-PAGE analysis are also shown. (B) Chart showing the domain organization of the arterivirus (PRRSV and EAV) helicases. (C) Cartoon model of PRRSV nsp10 containing the ZBD (blue) and 1B (magenta), 1A (cyan), and 2A (red) domains. Zinc ions are shown as blue balls. (D) Nsp10 is shown as a molecular surface model colored according to electrostatic potential (red for negatively charged regions and blue for positively charged regions). The putative nucleic acid binding groove is highlighted with a yellow square. (E) Crystal packing arrangement of two nsp10 molecules. The ZBD and 1B, 1A, and 2A domains are shown as in panel C. (F) Oligomerization state of the nsp10 incubated with dsDNA. The details are as described in panel A.
TABLE 1.
Statistics of data collection and refinement
| Parameter | Valuea
|
|
|---|---|---|
| nsp10 | Se-Met nsp10 | |
| Data collection statistics | ||
| Space group | P 41212 | P 41212 |
| Cell parameter | ||
| a, b, c (Å) | 93.62, 93.62, 357.41 | 117.55, 117.55, 365.86 |
| α, β, γ (°) | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 |
| Wavelength (Å) | 0.97918 | 0.97918 |
| Resolution (Å) (range) | 48.57–3.00 | 47.76−3.50 |
| Completeness (%) | 99.4 (100.0) | 97.4 (100.0) |
| Rmergeb (%) | 8.60 (66.10) | 21.10 (76.50) |
| I/δ (last shell) | 51.29 (3.15) | 29.78 (2.17) |
| Redundancy (last shell) | 24.80 (23.70) | 24.90 (25.30) |
| Refinement statistics | ||
| Resolution range (Å) | 46.81–3.00 | |
| No. of reflections | 32,988 | |
| Rwork/Rfreec (%) | 22.52/28.07 | |
| No. of atoms | ||
| Protein | 6,877 | |
| Solvent | 110 | |
| RMSD | ||
| Bond length (Å) | 0.01 | |
| Bond angle (°) | 1.23 | |
| Avg B factor (Å2) | 56.96 | |
| Ramachandran plot (%) | ||
| Core | 91.34 | |
| Allowed | 6.83 | |
| Disallowed | 1.82 | |
The highest-resolution values are indicated in parentheses.
Rmerge = ΣΣ|Ii –〈I〉|/ΣΣIi, where is Ii the intensity measurement of reflection h and 〈I〉is the average intensity from multiple observations.
Rwork = ΣǁFo| – |Fcǁ/Σ|Fo|, where Fo and Fc are the observed and calculated structure factors, respectively, and Rfree is equivalent to Rwork, but 5% of the measured reflections have been excluded from the refinement and set aside for cross-validation.
The crystal structure of nsp10 can be divided into three major parts: the N-terminal ZBD (Ser4-Val71), the intermediate 1B domain (1B, Thr78-Thr123), and the C-terminal helicase core domains 1A (Asn133-Thr275) and 2A (Ile276-Glu441) (Fig. 1B and C). The 1B domain is connected to the ZBD and the catalytic domain (1A and 2A) through a linker loop (Pro72 to Arg77 and Ala124 to Ile132, respectively). The two domains 1B and 1A form a cavity to be positively charged, which is associated with binding to the nucleic acid substrate (16) (Fig. 1D). Although the functions of nidovirus helicases are conserved, their structures are diverse; alignment of the Cα atoms of PRRSV nsp10 with those of EAV nsp10 (16), SARS-CoV Nsp13 (14), and MERS-CoV Nsp13 (15) yielded RMSDs of 2.8, 5.2, and 5.3 Å, respectively (Table 2). Unexpectedly, compared to the nidovirus helicase structure, domain 1B in our structure adopted a novel open conformation (1B domain away from 1A and 2A domains) (Fig. 1C). Moreover, previous research indicated that the C-terminal 65 amino acids of EAV nsp10 may regulate nsp10 helicase-mediated functions in vivo and communicate with the nsp10 active site (16). However, the only available helicase structure among the arteriviruses family is truncated at its C-terminal end (EAV nsp10) (16); therefore, our structure is the first to reveal the C-terminal conformation (Asp382-Glu441) (Fig. 1B and C).
TABLE 2.
Pairwise comparison of the isolated ZBD/CH, 1B, and helicase core (1A and 2A) domains of PRRSV nsp10, EAV nsp10, SARS-CoV nsp13, and MERS-CoV nsp13a
| Domain | Comparison | Dali Z-score | RMSD (Å) |
|---|---|---|---|
| ZBD/CH | PRRSV vs EAV | 6.3 | 2.7 |
| PRRSV vs SARS-CoV | 3.3 | 2.4 | |
| PRRSV vs MERS-CoV | 3.2 | 3.0 | |
| 1B | PRRSV vs EAV | 7.0 | 1.4 |
| PRRSV vs SARS-CoV | <2.0 | ||
| PRRSV vs MERS-CoV | 2.0 | 2.6 | |
| 1A/RecA1 | PRRSV vs EAV | 20.7 | 1.7 |
| PRRSV vs SARS-CoV | 12.9 | 2.4 | |
| PRRSV vs MERS-CoV | 13.0 | 2.4 | |
| 2A/RecA2 | PRRSV vs EAV | 19.0 | 1.3 |
| PRRSV vs SARS-CoV | 9.6 | 2.6 | |
| PRRSV vs MERS-CoV | 11.2 | 3.0 | |
| Helicase core domains (1A and 2A) | PRRSV vs EAV | 28.7 | 2.0 |
| PRRSV vs SARS-CoV | 19.3 | 2.9 | |
| PRRSV vs MERS-CoV | 20.9 | 3.2 | |
| Overall structure | PRRSV vs EAV | 32.7 | 2.8 |
| PRRSV vs SARS-CoV | 17.6 | 5.2 | |
| PRRSV vs MERS-CoV | 14.3 | 5.3 |
Structural comparison of PRRSV nsp10 with other nidovirus helicases.
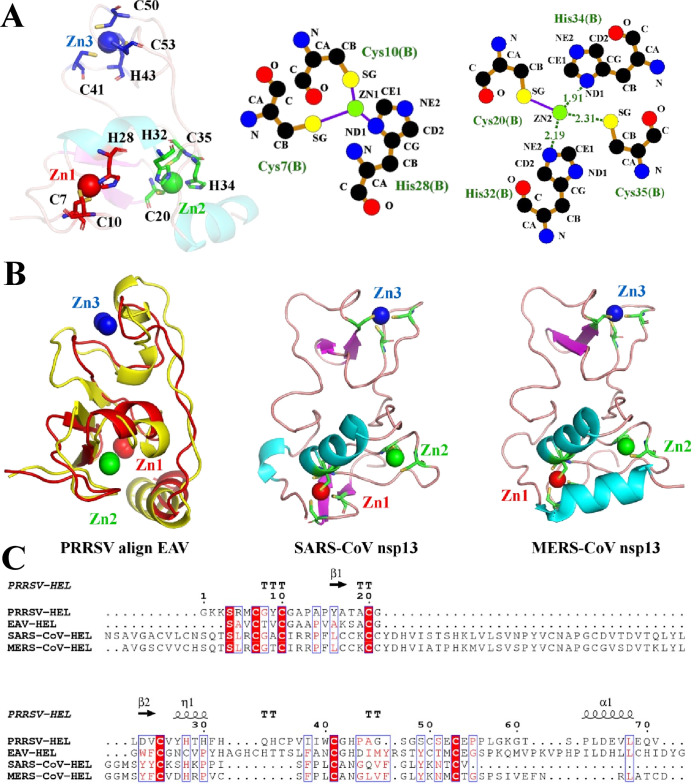
The N-terminal ZBD, which is composed of two α-helices and two β-sheets, binds three zinc ions (Zn1 to Zn3). The ZBD can be subdivided into three molecules: ring-like molecules containing Zn1 and Zn2, a third zinc finger containing Zn3, and a linker loop connected to the 1B domain. As shown in Fig. 2A, Zn1 binds the first zinc finger via Cys7, Cys10, and His28; Zn2 binds the second zinc finger via Cys20, His32, His34, and Cys35; and Zn3 binds the third zinc finger via Cys41, His43, Cys50, and Cys53. The structure of the ZBD is relatively conserved, with a root mean square deviation (RMSD) between the Cα atoms of monomeric PRRSV Nsp10 and EAV Nsp10 of 2.7 Å (Fig. 2B; Table 2). However, the amino acid sequence of the arterivirus nsp10 ZBD is shorter than that of the coronavirus nsp13 ZBD (Fig. 2C), and their structures are significantly different (Fig. 2B; Table 2). In addition, Zn1 and Zn2 are located at the interface between the ZBD and 1A domains, and Zn3 is oriented away and does not interact with other regions of the protein. Previous studies have shown that the ZBD is critical for the ATP-hydrolysis and unwinding functions of nidovirus helicases (14, 16, 31). Indeed, site mutations (H32A) of PRRSV nsp10 impaired both ssDNA- and dsDNA-binding and dsDNA-unwinding activities but no obvious influence on ATPase activity (31). In our structure, His32 stabilizes the binding of Zn2 through hydrogen bonding (Fig. 2A).
FIG 2.
Structural characterization of the PRRSV ZBD. (A) PRRSV nsp10 ZBD. Zn1, Zn2, and Zn3 are colored red, green, and blue, respectively. The intermolecular interactions between Zn1, Zn2, and surrounding amino acid sites were analyzed. (B) Structural comparison of the ZBDs from PRRSV, EAV, SARS-CoV, and MERS-CoV helicases. (C) Sequence alignment of arterivirus and coronavirus helicase ZBDs. Secondary structure elements of PRRSV nsp10 are marked on the top of the alignment (helices with squiggles, β-strands with arrows, and turns with the letters TT). The sequences were aligned using ClustalW2, and the alignment was drawn with ESPript 3.0.
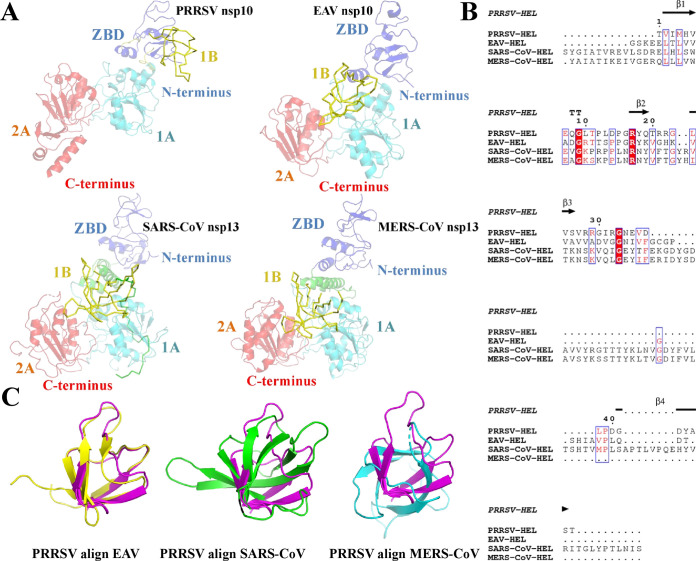
The 1B domain consists of four β-sheets, which are connected with the ZBD and helicase core structure domain (1A and 2A) by a long loop. Structure comparison showed that the overall structure of PRRSV nsp10 1B is conserved relative to that of EAV nsp10 (with an RMSD of 1.4 Å) but significantly different from those of SARS-CoV nsp13 and MERS-CoV nsp13 (Fig. 3C; Table 2). Furthermore, amino acid sequence alignment revealed significant differences in the nidovirus 1B domain sequences, and coronaviruses had a longer 1B amino acid sequence than arteriviruses (Fig. 3B). Importantly, analysis of the overall structure of the nidovirus helicases (EAV nsp10, SARS-CoV nsp13, and MERS-CoV nsp13) showed that the 1B domain adopts a “down” conformation (14–16), while the PRRSV helicase 1B domain adopts an “up” conformation (Fig. 3A and 4A).
FIG 3.
Structural characterization of the PRRSV 1B domain. (A) Conformational states of the 1B domains from PRRSV, EAV (PDB ID 4N0N), SARS-CoV (PDB ID 6JYT), and MERS-CoV helicases (PDB ID 5WWP). The 1B domain is shown as a yellow ribbon. (B) Sequence alignment of arterivirus and coronavirus helicase 1B domains. The sequence alignment shown is marked as in Fig. 2C. (C) Structural alignment of 1B domains from PRRSV nsp10 (magenta), EAV nsp10 (yellow), SARS-CoV nsp13 (green), and MERS-CoV Nsp13 (cyan). The PDB ID is consistent with panel A.
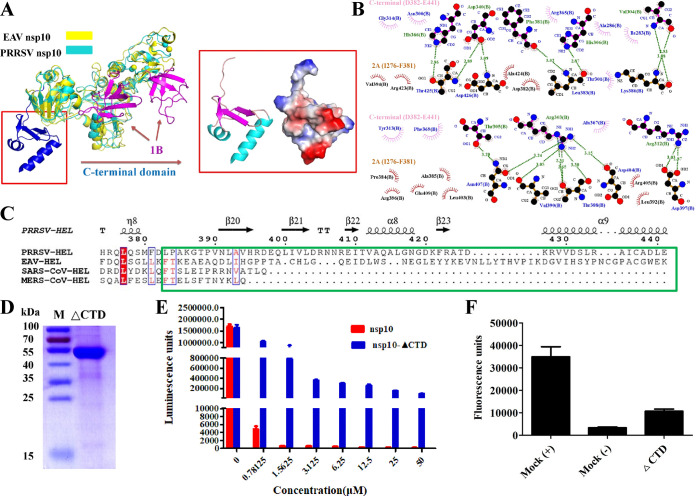
FIG 4.
Structural characterization of the PRRSV nsp10 C-terminal domain. (A) Structural alignment of PRRSV nsp10 and EAV nsp10 (PDB ID 4N0N). The structures of PRRSV nsp10 and EAV nsp10 are shown as cyan and yellow, respectively, and the 1B domains are shown as magenta. The electrostatic potential surface of the C-terminal domain is also shown as in Fig. 1D. (B) Analysis of the intermolecular forces between the C-terminal and 2A domains. All of the residues in the C-terminal (D382-E441) and 2A (ILe276-Phe381) domains that interact were determined using LIGPLOT. Carbon, nitrogen, and oxygen atoms are shown as black, blue, and red spheres, respectively. Hydrogen bond interactions are shown as blue dashed lines between the respective donor and acceptor atoms along with the bond distance. Hydrophobic interactions are indicated by arcs with rods radiating toward the atoms they contact. (C) Sequence alignment of arterivirus and coronavirus helicase C-terminal domains. The sequence alignment shown is marked as in Fig. 2C. (D) SDS-PAGE analysis of the nsp10 ΔCTD. (E) Analysis of the ATPase hydrolysis activities of nsp10 and nsp10 ΔCTD. Each protein was assayed at different concentrations (0, 0.78125, 1.5625, 3.125, 6.25, 12.5, 25, and 50 μM). (F) Analysis of the helicase activity of nsp10 ΔCTD. The reaction with nsp10 WT or without protein are indicated as as “Mock (+)” or “Mock (–),” respectively.
The C-terminal domain is poorly conserved among nidovirus helicases (Fig. 1B and 4C). Sequence alignment of amino acids showed that arterivirus helicases are approximately 50 amino acids longer than coronavirus helicases (Fig. 4C). The C-terminal structure presents a novel composition: three β-folds and two vertical α-helices connected to the upstream structure by a long loop (Fig. 4A). However, our analysis revealed that the C-terminal is far from the active core region of helicase. Hence, the C-terminal may be not directly involved in the helicase’s function. To further study its role, the segment was analyzed in terms of its structural interactions with other domains, as shown in Fig. 4B. The C-terminal domain and the 2A domain interact via many hydrogen bonds and hydrophobic interactions (Fig. 4B), showing that the C-terminal domain is very important for maintaining the stability of the helicase structure. To investigate the effect of the C-terminal domain on helicase function, we expressed mutant nsp10 protein in which the C-terminal domain had been deleted (nsp10-ΔCTD) (Fig. 4D). As shown in Fig. 4E and F, the wild-type (WT) PRRSV nsp10 protein had very high ATPase activity and helicase activity. However, nsp10-ΔCTD exhibited significantly reduced function, suggesting that the C-terminal domain is crucial for the PRRSV helicase function. The results of our structural analysis and biochemical experiments are consistent and indicate that the C-terminal domain can promote the hydrolysis and unwinding activity of nsp10 through interaction with the 2A domain.
Movements of the 1B domain between PRRSV nsp10 and other nidovirus helicases.
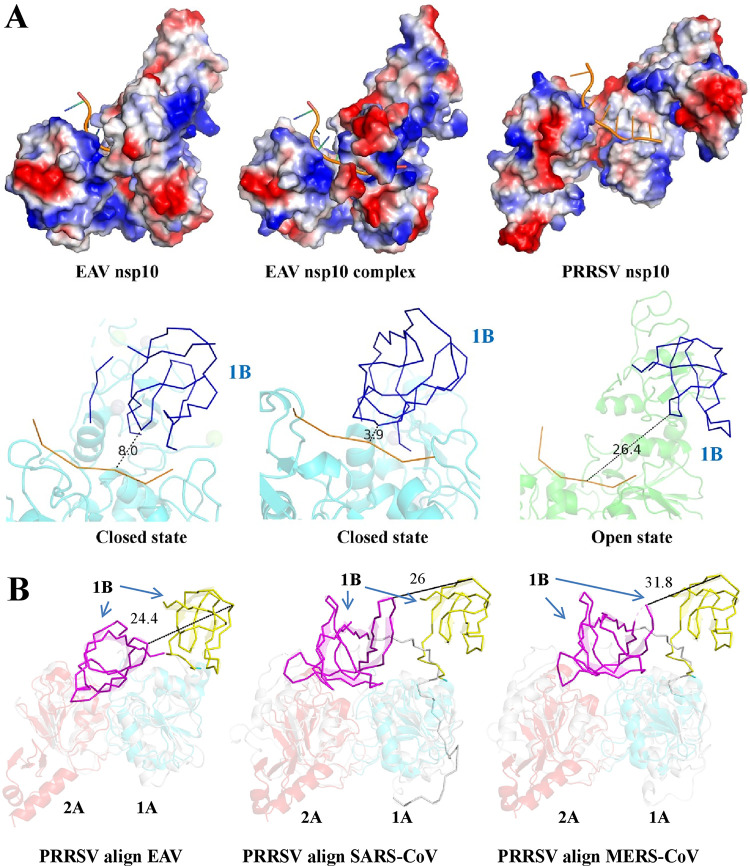
To analyze the effect of 1B conformation on helicase function, we used an EAV nsp10 complex structure to simulate the conformations of ssDNA-bound EAV and PRRSV (Fig. 5A). The results indicated that when EAV nsp10 does not bind ssDNA, the binding cavity formed by 1B and 1A domains is small (with a distance between ssDNA and the 1B domain of ∼8.0 Å). In contrast, when EAV nsp10 binds ssDNA, the binding cavity formed by the 1B and 1A domains becomes smaller (with a distance between ssDNA and the 1B domain of ∼3.9 Å). However, PRRSV nsp10 is in an open state, and the binding cavity formed by domains 1B and 1A is large (with a distance between ssDNA and the 1B domain of ∼26.4 Å). Moreover, we observed that the helicase core domains (1A and 2A) of PRRSV match well with EAV, SARS-CoV, and MERS-CoV (Fig. 5B and Table 2). However, the 1B domain of PRRSV deviated significantly from EAV, SARS-CoV, and MERS-CoV by approximately 24.4, 26, and 31.8 Å, respectively (Fig. 5B). Previous research has also shown that conformational changes in the structure of the EAV DNA-helicase complex occurs between the 1B domain and helicase structures, which are rotated relative to one another by 28.7 Å (16). Because the 1B domain connects with the ZBD and 1A domain through two long loops, the 1B domain may undergo large rotational movements. In addition, when the helicase binds dsDNA, it requires a large binding cavity to accommodate the nucleic acid substrate. Hence, we speculate that during the process of unwinding, the 1B domain binds the substrate mainly by conformational changes. The open state of the 1B domain may be an intermediate state in the process of substrate binding.
FIG 5.
Dynamic conformational states of the 1B domain in arterivirus helicases. (A) The structures of PRRSV nsp10 and EAV nsp10 (PDB ID 4N0N and 4N0O) are shown and are colored according to electrostatic potential as described in Fig. 1D. The ssDNA from the EAV nsp10 complex (PDB ID 4N0O) was docked into PRRSV nsp10 and EAV nsp10 (PDB ID 4N0N). The distance between the ssDNA substrate and the 1B domain was measured by using PyMOL software. (B) Structural alignment between PRRSV and other nidovirus helicases (EAV nsp10, SARS-CoV nsp13, and MERS-CoV nsp13). The PDB ID is consistent with Fig. 3A.
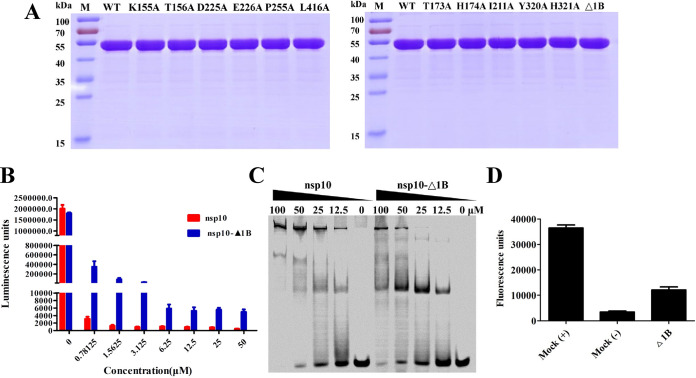
To further investigate the effect of 1B on helicase function, we expressed mutant nsp10 protein in which the 1B domain had been deleted (nsp10-Δ1B) (Fig. 6A). As shown in Fig. 6B, the WT PRRSV nsp10 protein had a very high ATPase activity and a fast reaction rate and could completely hydrolyze ATP. However, nsp10-Δ1B exhibited obviously reduced hydrolysis, suggesting that the 1B domain of the PRRSV helicase is crucial for the nsp10’s ATPase activity. Electrophoretic mobility shift assay (EMSA) confirmed that deletion of the 1B domain decreased nucleic acid-binding capacity (Fig. 6C). Furthermore, nsp10-Δ1B exhibited obviously reduced helicase activity (Fig. 6D). Our experimental results confirmed that the 1B domain is important for the function of nsp10. Previous research has shown that two nonenzymatic domains (ZBD and 1B) may regulate helicase function (14). In the EAV nsp10-DNA complex, the polynucleotide substrate channel formed by domains 1A and 1B is capable of holding and binding ssDNA but not dsDNA (16). However, nidovirus helicases can bind dsDNA/dsRNA, although how they do so is unclear. In contrast to the structure of other nidovirus helicases (14–16), our structure of PRRSV nsp10 reveals an alternative 1B conformation that would potentially allow this domain to create enough space to accommodate and bind dsDNA/dsRNA during unwinding. Since the dsRNA has a diameter ∼20 Å, in the process of nsp10 unwinding dsRNA, the distance between 1B and 1A may be a changing process. Hence, we speculate that when nsp10 binds dsRNA, the distance between the 1B and 1A domain will become smaller in order to stably bind dsRNA.
FIG 6.
Deletion of the 1B domain attenuates nsp10 helicase activity. (A) SDS-PAGE analysis of WT and mutants (K155A, T156A, D225A, E226A, P255A, L416A; T173A, H174A, I211A, Y320A, H321A, and Δ1B) nsp10. Molecular weight markers are shown. (B) Analysis of the ATPase hydrolysis activities of nsp10 and nsp10 Δ1B. Each protein was assayed at different concentrations (0, 0.78125, 1.5625, 3.125, 6.25, 12.5, 25, and 50 μM). (C) The dsDNA binding abilities of nsp10 and nsp10-Δ1B were determined by EMSA. Each protein was assayed at different concentrations (0, 12.5, 25, and 50 μM) with 1 μM Cy5-labeled dsDNA. (D) Analysis of the helicase activity of nsp10 Δ1B. The reaction with nsp10 WT or without protein was used are indicated as “Mock (+)” or “Mock (–),” respectively.
In addition, domain movements are also observed in the complexes structure of the E. coli Rep helicase bound to ssDNA or ssDNA, and the ADP and 2B domain undergoes very large movements in the Rep monomer (18). Previous research indicated that the relative movements would facilitate the translocation of the Rep dimer (18, 32). Therefore, during the unwinding process of nidovirus helicases, the overall closed and open conformational state of the 1B domain may also exist in other nidovirus helicases. Furthermore, we analyzed the causes of the novel conformation of 1B. It is possible that we obtained only one of many conformations of 1B by crystallographic methods. Therefore, it is necessary to determine whether there is a dynamic change process during the functioning of 1B in helicase. A second possibility is that this conformational state of 1B exists during the helicase unwinding process and that we happened to capture this conformational state. Due to the prokaryotic expression system we use, whether the folding and processing of the protein after production is completely consistent with the protein produced during virus replication requires experimental determination.
ATP activity and helicase activity.
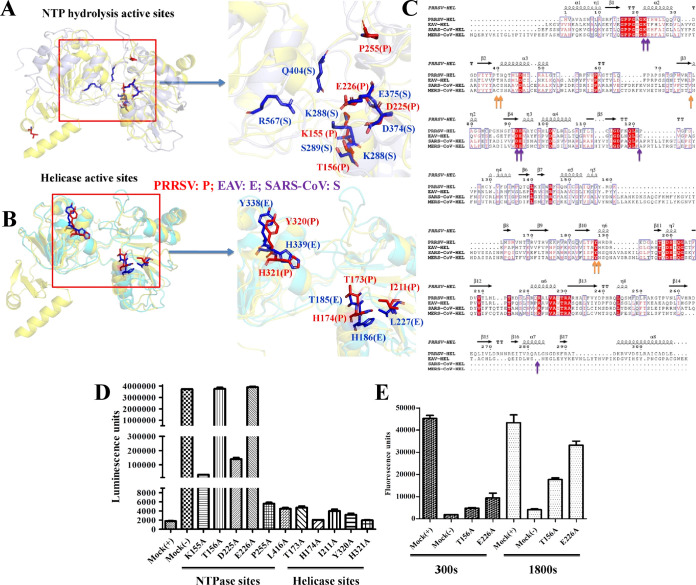
Although the amino acid sequences of the nidovirus helicases exhibit low homology, the composition and orientation of the core (1A and 2A) domains are well conserved (Fig. 7A to C). The structures of the helicase catalytic (1A and 2A) domains can be nearly perfectly superimposed (the RSMDs following alignment of PRRSV nsp10 with EAV nsp10, SARS-CoV nsp13, and MERS-CoV nsp13 are 2.0, 2.9, and 3.2 Å, respectively), especially in the ATP hydrolysis and helicase active sites (Fig. 7A and B; Table 2). To identify the key amino acids that affect helicase function, we identified potential ATP hydrolysis and helicase active sites by structural comparison and amino acid sequence alignment (Fig. 7A to C). Structural comparison demonstrated that residues in the potential ATP hydrolysis active sites from nsp10 (Lys155, Thr156, Asp225, and Glu226) superimposed well with the corresponding residues of SARS nsp13 (Fig. 7A). In addition, the potential helicase active-site residues Thr173, His174, Ile211, Tyr320, and His321 superimposed well with the corresponding residues in EAV nsp10 (Fig. 7B). These nine residues are clustered together in the cleft at the base between the 1A and 2A domains in Fig. 7A and B. Furthermore, our biochemical experiments showed that the ATPase activity level was significantly lower in mutant (T156A and E226A) nsp10 than in WT nsp10 (Fig. 7D), indicating that these residues are important ATPase active sites. However, the mutation of residues in the helicase active sites did not appear to affect ATP hydrolytic activity (Fig. 7D). The helicase activities of the two mutants (T156A and E226A) were consistent with their ATPase activities, indicating that the unwinding activity of PRRSV nsp10 is dependent on ATP hydrolysis. Our results show that active-site residues are highly conserved between arteriviruses and coronaviruses and that similar substrate recognition and helicase mechanisms are shared among nidoviruses within the families Arteriviridae and Coronaviridae.
FIG 7.
The structures of the 1A and 2A domains reveal the conservation of the nidovirus helicases. (A) Structural alignment of the 1A and 2A domains from PRRSV nsp10 and SARS-CoV nsp13. The potential ATPase hydrolysis sites in PRRSV nsp10 are labeled and shown as sticks (red), and those in SARS-CoV nsp13 are labeled and shown as sticks (blue). (B) Structural alignment of the 1A and 2A domains in PRRSV nsp10 and EAV nsp10. The potential helicase sites in PRRSV nsp10 and EAV nsp10 are labeled as in panel A. (C) Relationships between the sequences of arterivirus and coronavirus helicase domains. The key potential residues at the ATPase hydrolysis and helicase active sites of PRRSV nsp10 are marked with blue and orange arrows at the bottom. (D and E) Mutagenesis studies of PRRSV nsp10 ATPase hydrolysis and helicase activities.
Implications for PRRSV replication.
During arterivirus replication, the viral genome can be directly used to complete viral RNA synthesis in the first round. Then, the dsRNA intermediate product must be untwisted by nsp10 to complete the synthesis of viral RNA. Undoubtedly, nsp10 plays a crucial role in arterivirus replication. Moreover, nsp10 participates in the formation of replication transcription complexes (RTCs) as a helicase. The 1B and C-terminal domains play an important role in the function of nsp10. Meanwhile, in RTCs, they may interact with other nonstructural proteins and play a key role in the arterivirus replication. Therefore, the mechanisms by which the 1B and C-terminal domains affect the function of nsp10 in PRRSV replication need further study.
Conclusions.
In summary, we provide here the first structural information regarding the multiple functions of the PRRSV helicase nsp10, which contains a 1B domain with a novel conformation that distinguishes it from EAV nsp10 and coronavirus nsp13. The 1B domain of PRRSV nap10 adopts an open state, and the cavity in which the nucleic acid substrate binds is large and sufficient to hold dsDNA/dsRNA for unwinding. Hence, the open state of the 1B domain probably represents an intermediate state during binding to a nucleic acid substrate. Moreover, our results showed that the unique structure of the C-terminal domain helps stabilize the helicase structure. In addition, our analyses demonstrated that the 1A and 2A domains of PRRSV nsp10 are conserved relative to those of EAV nsp10, SARS-CoV nsp13, and MERS-CoV nsp13, especially with regard to the key amino acids in the active sites. These results enhance our understanding of nsp10 helicase activity and provide a novel structure that may aid the structure-based design of drugs against PRRSV in the future.
MATERIALS AND METHODS
Plasmid construction.
The sequence encoding the 441-residue nsp10 gene corresponds to nucleotides 10851 to 11520 in the genome of the PRRSV WUH3 strain (GenBank accession no. HM853673) (33). Wild-type (WT) nsp10 flanked by an N-terminal His6 tag and S tag and a C-terminal His6 tag was cloned into pET-30a(+) between the BamHI and XhoI restriction sites. The point mutant Δ1B (missing Pro75-Thr128), ΔCTD (missing Val390-Glu441), and point mutations in the predicted ATPase hydrolysis sites (K155A, T156A, D225A, E226A, P255A, and L416A) and predicted helicase sites (T173A, H174A, I211A, Y320A, and H321A) were engineered using overlap-extension PCR, and the fragments were cloned into pET-30a(+) via the same method. All constructs were validated by DNA sequencing.
Protein expression and purification.
To express WT and mutant nsp10, the recombinant plasmids were transformed into E. coli strain BL21(DE3) (Beijing TransGen Biotech Co., Ltd.). Transformed cells were cultured at 37°C in Luria-Bertani medium containing 50 μg/ml kanamycin. Protein expression was induced with 0.8 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) when the culture density reached an optical density at 600 nm of 0.8, and cell growth continued for an additional 16 h at 18°C. To solve the phase problem, selenomethionine (SeMet)-labeled nsp10 was expressed in BL21(DE3) cells according to our previously reported procedure (34).
Protein purification was performed as described previously (34). Briefly, the cell supernatant was filtered with a 0.45-μm filter and loaded onto a nickel-charged HisTrap HP column (GE Healthcare). The proteins were eluted with elution buffer (20 mM Tris-HCl, 500 mM NaCl, 500 mM imidazole [pH 7.4]). The harvested protein was then concentrated to approximately 2.0 ml and filtered using a Superdex 200 gel filtration column (GE Healthcare) equilibrated with buffer (20 mM Tris-HCl and 200 mM NaCl [pH 7.4]). For crystallization, the purified protein was concentrated to approximately 7 mg/ml, flash-frozen with liquid nitrogen, and stored at –80°C. The concentration of purified PRRSV nsp10 was determined based on the absorbance at 280 nm (A280) using a NanoDrop 2000c UV-Vis spectrophotometer (Thermo Fisher Scientific).
Oligomerization of the WT nsp10 protein (alone or with dsDNA) was analyzed using a Superdex 75 10/300 GL column (GE Healthcare) with a buffer containing 20 mM Tris-HCl (pH 7.4) and 200 mM NaCl at a flow rate of 0.5 ml/min (4°C). The dsDNA was formed by chains (5′-TTTTTTTTTTTTTTTCGAGCACCGCTGCGGCTGCACC-3′ and 5′-GGTGCAGCCGCAGCGGTGCTCG-3′). The eluted nsp10 proteins were then analyzed by SDS-PAGE. Equal volumes (200 μl) of Bio-Rad size exclusion standards (catalog no. 151-1901; 1 ml/vial) were analyzed under the same buffer conditions. The results were analyzed using Origin 8.0 software.
Crystallization, data collection, and structure determination.
Crystallization screening, data collection, and structure determination were performed as described previously (34). The best crystallization conditions for both WT and SeMet-labeled nsp10 were crystallization by vapor diffusion in hanging drops consisting of 3 μl of reservoir solution (0.1 M HEPES [pH 7.5]; 2.0 M NaCl) and 3 μl of concentrated protein solution (7 mg/ml protein in 20 mM Tris-HCl and 200 mM NaCl [pH 7.4]), followed by incubation at 20°C for 7 days. Then, the crystals were flash-cooled in liquid nitrogen in a cryoprotectant solution containing 30% ethylene glycol and 70% reservoir solution (0.1 M HEPES [pH 7.5]; 2.4 M NaCl). Data collection was performed at the Shanghai Synchrotron Radiation Facility (SSRF) with beam line BL17U1 (wavelength, 0.97918 Å; temperature, 100 K). Reflections were integrated, merged, and scaled using HKL-3000 (35), and the resulting statistics are listed in Table 1. The structure of nsp10 was solved from a SeMet derivative of nsp10 by the single-wavelength anomalous dispersion method. All nine potential selenium atoms in the nsp10 monomer were located, and the initial phases were calculated using the program AutoSol from the PHENIX software suite (36). Manual model rebuilding and refinement were performed using COOT (37) and the PHENIX software suite.
Structural analysis and sequence alignment.
The RMSD between arterivirus nsp10 and coronavirus nsp13 was determined using Dali server (http://ekhidna2.biocenter.helsinki.fi/dali/) (38), and structure figures were generated using PyMOL (Schrödinger). Detailed molecular interactions were determined using LIGPLOT (39). In addition, the amino acid sequences of arterivirus nsp10 and coronaviruses nsp13 were aligned using ClustalW2 (40) and visualized with the ESPript 3 server (http://espript.ibcp.fr) (41). The GenBank accession numbers of the sequences used are as follows: EAV nsp10 (NP_705591.1), SARS-CoV nsp13 (AY291315), and MERS-CoV nsp13 (YP_009047202).
ATPase activity assay.
The ATPase activity of PRRSV nsp10 was detected with a Kinase-Glo luminescent kinase assay platform kit according to a previously reported procedure (31). Briefly, Kinase-Glo buffer was added to the Kinase-Glo substrate bottle, mixed well, and stored at –20°C. The following were added to a 96-well plate and protected from light: 15 μl of 0.9 mM ATP, 3.9 μl of nsp10 (1 mg/ml), and 26.1 μl of buffer C solution (20 mM Tris-HCl [pH 8.0], 50 mM NaCl, 5% glycerinum, 10 mM MgCl2). The reaction was then mixed by shaking and incubated at 37°C for 30 min; the same volume of kinase reagent was added and mixed by shaking, and the chemiluminescence was measured with a multifunction microplate reader. Four replicates of each experimental group were used, and three data replicates were selected for statistical analysis using GraphPad Prism 5 software.
Helicase activity assay.
Fluorescence resonance energy transfer was used as described previously (31) to detect nsp10 helicase activity. The following fluorescently labeled single-stranded DNA fragments were chemically synthesized by Sangon Biotech (Shanghai): chain F, 5′-TTTTTTTTTTTTTTTCGAGCACCGCTGCGGCTGCACC-(Cy5)-3′; and chain Q, 5′-(BHQ-2)-GGTGCAGCCGCAGCGGTGCTCG-3′. Chains F and Q were mixed at a ratio of 1:1.5 and then slowly annealed by PCR (95°C, 20 min; 60°C, 10 min; 37°C, 10 min; 20°C, 10 min) to form a dsDNA substrate. Meanwhile, to prevent the formation of double-stranded chains F and Q, capture chain C (5′-CGAGCACCGCTGCGGCTGCACC-3′) was used to immediately capture the Q chain, avoiding detection of the Cy5 fluorophore. Initial fluorescence in the reaction system in a 96-well plate was detected with a multifunction microplate reader at an excitation wavelength of 625 nm (excitation) and an emission wavelength of 670 nm (emission). The unwinding reaction was carried out at 37°C. Four replicates from each experimental group were assessed, and three data replicates were selected for statistical analysis using GraphPad Prism 5 software.
EMSA.
An electrophoretic mobility shift assay (EMSA) was performed as described previously (42). Briefly, protein (at a final concentration of 50, 25, 12.5, or 0 μM) and 5′-Cy5-labeled dsDNA (formed by chains F and Q [without 5′-BHQ-2], 1 μM) were added to a PCR tube, mixed by gentle pipetting, and incubated at room temperature in the dark for 45 min. Samples were then run on 6.5% nondenaturing Tris-borate/EDTA polyacrylamide gels for 37 min at a voltage of 180 V, and the results were determined with an FLA-2000 fluorescent image analyzer (Fuji, Stamford, CT).
Data availability.
Coordinates and structure factors for PRRSV nsp10 were deposited in the RCSB Protein Data Bank under accession number 6LKX.
ACKNOWLEDGMENTS
We thank the staff at SSRF beamline BL17U for their help with crystallographic data collection.
This study was supported by National Natural Science Foundation of China grants 31722056 and 31702249, National Key R&D Plan of China grant 2018YFD0500100, China Postdoctoral Science Foundation grant 2019M662674, and the Huazhong Agricultural University Scientific and Technological Self-innovation Foundation (program 2662017PY028).
We declare there are no competing interests.
REFERENCES
- 1.Gorbalenya AE, Enjuanes L, Ziebuhr J, Snijder EJ. 2006. Nidovirales: evolving the largest RNA virus genome. Virus Res 117:17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Snijder EJ, Kikkert M, Fang Y. 2013. Arterivirus molecular biology and pathogenesis. J Gen Virol 94:2141–2163. doi: 10.1099/vir.0.056341-0. [DOI] [PubMed] [Google Scholar]
- 3.Tian K, Yu X, Zhao T, Feng Y, Cao Z, Wang C, Hu Y, Chen X, Hu D, Tian X, Liu D, Zhang S, Deng X, Ding Y, Yang L, Zhang Y, Xiao H, Qiao M, Wang B, Hou L, Wang X, Yang X, Kang L, Sun M, Jin P, Wang S, Kitamura Y, Yan J, Gao GF. 2007. Emergence of fatal PRRSV variants: unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS One 2:e526. doi: 10.1371/journal.pone.0000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lunney JK, Fang Y, Ladinig A, Chen N, Li Y, Rowland B, Renukaradhya GJ. 2016. Porcine reproductive and respiratory syndrome virus (PRRSV): pathogenesis and interaction with the immune system. Annu Rev Anim Biosci 4:129–154. doi: 10.1146/annurev-animal-022114-111025. [DOI] [PubMed] [Google Scholar]
- 5.Han J, Zhou L, Ge X, Guo X, Yang H. 2017. Pathogenesis and control of the Chinese highly pathogenic porcine reproductive and respiratory syndrome virus. Vet Microbiol 209:30–47. doi: 10.1016/j.vetmic.2017.02.020. [DOI] [PubMed] [Google Scholar]
- 6.Du L, Pang F, Yu Z, Xu X, Fan B, Huang K, He K, Li B. 2017. Assessment of the efficacy of two novel DNA vaccine formulations against highly pathogenic porcine reproductive and respiratory syndrome virus. Sci Rep 7:41886–41886. doi: 10.1038/srep41886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Du L, Li B, Pang F, Yu Z, Xu X, Fan B, Tan Y, He K, Huang K. 2017. Porcine GPX1 enhances GP5-based DNA vaccination against porcine reproductive and respiratory syndrome virus. Vet Immunol Immunopathol 183:31–39. doi: 10.1016/j.vetimm.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 8.Fang Y, Snijder EJ. 2010. The PRRSV replicase: exploring the multifunctionality of an intriguing set of nonstructural proteins. Virus Res 154:61–76. doi: 10.1016/j.virusres.2010.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson CR, Griggs TF, Gnanandarajah J, Murtaugh MP. 2011. Novel structural protein in porcine reproductive and respiratory syndrome virus encoded by an alternative ORF5 present in all arteriviruses. J Gen Virol 92:1107–1116. doi: 10.1099/vir.0.030213-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Firth AE, Zevenhoven-Dobbe JC, Wills NM, Go YY, Balasuriya UB, Atkins JF, Snijder EJ, Posthuma CC. 2011. Discovery of a small arterivirus gene that overlaps the GP5 coding sequence and is important for virus production. J Gen Virol 92:1097–1106. doi: 10.1099/vir.0.029264-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fang Y, Treffers EE, Li Y, Tas A, Sun Z, van der Meer Y, de Ru AH, van Veelen PA, Atkins JF, Snijder EJ, Firth AE. 2012. Efficient –2 frameshifting by mammalian ribosomes to synthesize an additional arterivirus protein. Proc Natl Acad Sci U S A 109:E2920–E2928. doi: 10.1073/pnas.1211145109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tian X, Lu G, Gao F, Peng H, Feng Y, Ma G, Bartlam M, Tian K, Yan J, Hilgenfeld R, Gao GF. 2009. Structure and cleavage specificity of the chymotrypsin-like serine protease (3CLSP/nsp4) of porcine reproductive and respiratory syndrome virus (PRRSV). J Mol Biol 392:977–993. doi: 10.1016/j.jmb.2009.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lehmann KC, Gulyaeva A, Zevenhoven-Dobbe JC, Janssen GMC, Ruben M, Overkleeft HS, van Veelen PA, Samborskiy DV, Kravchenko AA, Leontovich AM, Sidorov IA, Snijder EJ, Posthuma CC, Gorbalenya AE. 2015. Discovery of an essential nucleotidylating activity associated with a newly delineated conserved domain in the RNA polymerase-containing protein of all nidoviruses. Nucleic Acids Res 43:8416–8434. doi: 10.1093/nar/gkv838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jia Z, Yan L, Ren Z, Wu L, Wang J, Guo J, Zheng L, Ming Z, Zhang L, Lou Z, Rao Z. 2019. Delicate structural coordination of the severe acute respiratory syndrome coronavirus Nsp13 upon ATP hydrolysis. Nucleic Acids Res 47:6538–6550. doi: 10.1093/nar/gkz409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hao W, Wojdyla JA, Zhao R, Han R, Das R, Zlatev I, Manoharan M, Wang M, Cui S. 2017. Crystal structure of Middle East respiratory syndrome coronavirus helicase. PLoS Pathog 13:e1006474. doi: 10.1371/journal.ppat.1006474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng Z, Lehmann KC, Li X, Feng C, Wang G, Zhang Q, Qi X, Yu L, Zhang X, Feng W, Wu W, Gong P, Tao Y, Posthuma CC, Snijder EJ, Gorbalenya AE, Chen Z. 2014. Structural basis for the regulatory function of a complex zinc-binding domain in a replicative arterivirus helicase resembling a nonsense-mediated mRNA decay helicase. Nucleic Acids Res 42:3464–3477. doi: 10.1093/nar/gkt1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singleton MR, Dillingham MS, Wigley DB. 2007. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem 76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 18.Korolev S, Hsieh J, Gauss GH, Lohman TM, Waksman G. 1997. Major domain swiveling revealed by the crystal structures of complexes of Escherichia coli Rep helicase bound to single-stranded DNA and ADP. Cell 90:635–647. doi: 10.1016/S0092-8674(00)80525-5. [DOI] [PubMed] [Google Scholar]
- 19.Cheng Z, Muhlrad D, Lim MK, Parker R, Song H. 2007. Structural and functional insights into the human Upf1 helicase core. EMBO J 26:253–264. doi: 10.1038/sj.emboj.7601464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chakrabarti S, Jayachandran U, Bonneau F, Fiorini F, Basquin C, Domcke S, Hir HL, Conti E. 2011. Molecular mechanisms for the RNA-dependent ATPase activity of Upf1 and its regulation by Upf2. Mol Cell 41:693–703. doi: 10.1016/j.molcel.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 21.Nishikiori M, Sugiyama S, Xiang H, Niiyama M, Ishibashi K, Inoue T, Ishikawa M, Matsumura H, Katoh E. 2012. Crystal structure of the superfamily 1 helicase from Tomato mosaic virus. J Virol 86:7565–7576. doi: 10.1128/JVI.00118-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Law YS, Utt A, Tan YB, Zheng J, Wang S, Chen MW, Griffin PR, Merits A, Luo D. 2019. Structural insights into RNA recognition by the Chikungunya virus nsP2 helicase. Proc Natl Acad Sci U S A 116:9558–9567. doi: 10.1073/pnas.1900656116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu S, Ci Y, Wang L, Yang Y, Zhang L, Xu C, Qin C, Shi L. 2019. Zika virus NS3 is a canonical RNA helicase stimulated by NS5 RNA polymerase. Nucleic Acids Res 47:8693–8707. doi: 10.1093/nar/gkz650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Franks TM, Singh G, Lykke-Andersen J. 2010. Upf1 ATPase-dependent mRNP disassembly is required for completion of nonsense-mediated mRNA decay. Cell 143:938–950. doi: 10.1016/j.cell.2010.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weng Y, Czaplinski K, Peltz SW. 1996. Genetic and biochemical characterization of mutations in the ATPase and helicase regions of the Upf1 protein. Mol Cell Biol 16:5477–5490. doi: 10.1128/mcb.16.10.5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhattacharya A, Czaplinski K, Trifillis P, He F, Jacobson A, Peltz SW. 2000. Characterization of the biochemical properties of the human Upf1 gene product that is involved in nonsense-mediated mRNA decay. RNA 6:1226–1235. doi: 10.1017/s1355838200000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seybert A, van Dinten LC, Snijder EJ, Ziebuhr J. 2000. Biochemical characterization of the equine arteritis virus helicase suggests a close functional relationship between arterivirus and coronavirus helicases. J Virol 74:9586–9593. doi: 10.1128/jvi.74.20.9586-9593.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ivanov KA, Thiel V, Dobbe JC, van der Meer Y, Snijder EJ, Ziebuhr J. 2004. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J Virol 78:5619–5632. doi: 10.1128/JVI.78.11.5619-5632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adedeji AO, Lazarus H. 2016. Biochemical characterization of middle east respiratory syndrome coronavirus helicase. mSphere 1:e00235-16. doi: 10.1128/mSphere.00235-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y, Zhou L, Zhang J, Ge X, Zhou R, Zheng H, Geng G, Guo X, Yang H. 2014. Nsp9 and Nsp10 contribute to the fatal virulence of highly pathogenic porcine reproductive and respiratory syndrome virus emerging in China. PLoS Pathog 10:e1004216. doi: 10.1371/journal.ppat.1004216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Li H, Peng G, Zhang Y, Gao X, Xiao S, Cao S, Chen H, Song Y. 2015. Mutational analysis of the functional sites in porcine reproductive and respiratory syndrome virus nonstructural protein 10. J Gen Virol 96:547–552. doi: 10.1099/jgv.0.000004. [DOI] [PubMed] [Google Scholar]
- 32.Wong I, Lohman TM. 1992. Allosteric effects of nucleotide cofactors on Escherichia coli Rep helicase-DNA binding. Science 256:350–355. doi: 10.1126/science.256.5055.350. [DOI] [PubMed] [Google Scholar]
- 33.Li B, Xiao S, Wang Y, Xu S, Jiang Y, Chen H, Fang L. 2009. Immunogenicity of the highly pathogenic porcine reproductive and respiratory syndrome virus GP5 protein encoded by a synthetic ORF5 gene. Vaccine 27:1957–1963. doi: 10.1016/j.vaccine.2009.01.098. [DOI] [PubMed] [Google Scholar]
- 34.Shi Y, Li Y, Lei Y, Ye G, Shen Z, Sun L, Luo R, Wang D, Fu ZF, Xiao S, Peng G. 2016. A Dimerization-dependent mechanism drives the endoribonuclease function of porcine reproductive and respiratory syndrome virus nsp11. J Virol 90:4579–4592. doi: 10.1128/JVI.03065-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. 2006. HKL-3000: the integration of data reduction and structure solution: from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr 62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 36.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 37.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 38.Holm L. 2019. Benchmarking fold detection by DaliLite v.5. Bioinformatics 35:5326–5327. doi: 10.1093/bioinformatics/btz536. [DOI] [PubMed] [Google Scholar]
- 39.Laskowski RA, Swindells MB. 2011. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model 51:2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- 40.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 41.Robert X, Gouet P. 2014. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Min B, Collins K. 2010. Multiple mechanisms for elongation processivity within the reconstituted tetrahymena telomerase holoenzyme. J Biol Chem 285:16434–16443. doi: 10.1074/jbc.M110.119172. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Coordinates and structure factors for PRRSV nsp10 were deposited in the RCSB Protein Data Bank under accession number 6LKX.