MERS-CoV is an important emerging pathogen and causes severe respiratory infection in humans. MERS-CoV strains from early epidemic clade A and contemporary epidemic clade B have not been phenotypically characterized to compare their abilities to infect cells and mice. In this study, we showed that a clade B virus ChinaGD01 strain caused more severe disease in mice, with delayed viral clearance, increased inflammatory cytokines, and decreased antiviral T cell responses, than the early clade A virus EMC/2012. Given the differences in pathogenicity of different clades of MERS-CoV, periodic assessment of currently circulating MERS-CoV is needed to monitor potential severity of zoonotic disease.
KEYWORDS: MERS-CoV, T cell response, pathogenicity, phylogenetics, virulence
ABSTRACT
Middle East respiratory syndrome coronavirus (MERS-CoV) causes severe acute respiratory disease in humans. MERS-CoV strains from early epidemic clade A and contemporary epidemic clade B have not been phenotypically characterized to compare their abilities to infect cells and mice. We isolated the clade B MERS-CoV ChinaGD01 strain from a patient infected during the South Korean MERS outbreak in 2015 and compared the phylogenetics and pathogenicity of MERS-CoV EMC/2012 (clade A) and ChinaGD01 (clade B) in vitro and in vivo. Genome alignment analysis showed that most clade-specific mutations occurred in the orf1ab gene, including mutations that were predicted to be potential glycosylation sites. Minor differences in viral growth but no significant differences in plaque size or sensitivity to beta interferon (IFN-β) were detected between these two viruses in vitro. ChinaGD01 virus infection induced more weight loss and inflammatory cytokine production in human DPP4-transduced mice. Viral titers were higher in the lungs of ChinaGD01-infected mice than with EMC/2012 infection. Decreased virus-specific CD4+ and CD8+ T cell numbers were detected in the lungs of ChinaGD01-infected mice. In conclusion, MERS-CoV evolution induced changes to reshape its pathogenicity and virulence in vitro and in vivo and to evade adaptive immune response to hinder viral clearance.
IMPORTANCE MERS-CoV is an important emerging pathogen and causes severe respiratory infection in humans. MERS-CoV strains from early epidemic clade A and contemporary epidemic clade B have not been phenotypically characterized to compare their abilities to infect cells and mice. In this study, we showed that a clade B virus ChinaGD01 strain caused more severe disease in mice, with delayed viral clearance, increased inflammatory cytokines, and decreased antiviral T cell responses, than the early clade A virus EMC/2012. Given the differences in pathogenicity of different clades of MERS-CoV, periodic assessment of currently circulating MERS-CoV is needed to monitor potential severity of zoonotic disease.
INTRODUCTION
Middle East respiratory syndrome coronavirus (MERS-CoV) is a newly emerging pathogen that causing zoonotic disease on the Arabian Peninsula and which may be transmitted by travelers to other parts of the world (1). MERS-CoV infection results in severe pneumonia and a high mortality rate in humans. At the end of January 2020, there were 2,519 cases with 866 deaths (34.3% mortality) reported to the World Health Organization (WHO). Epidemiological studies indicated that dromedary camels are the source of human infection (2, 3), although bats could be a natural reservoir (4, 5). While clusters of human-to-human transmission have occurred within health care facilities, MERS-CoV does not appear to transmit efficiently between humans in the community (6). Human dipeptidyl peptidase 4 (hDPP4; also known as CD26) was identified as the functional receptor for MERS-CoV (7, 8) and is primarily expressed in the lower respiratory tract in humans, which could be a possible reason for limited human-to-human transmission (9, 10). So far there are no specific treatments or vaccines available for MERS (11, 12).
Despite intensive studies over the past several years, the pathogenesis of MERS-CoV infection is not well understood. Recent autopsy and histopathological studies revealed that there was diffuse alveolar damage, necrotizing pneumonia, and acute renal injury in MERS-CoV-infected patients and that viral particles were localized in the pneumocytes, epithelial syncytial cells, and pulmonary macrophages in the lung and renal proximal tubular epithelial cells in the kidney (13, 14). In 2015, a traveler returning from the Middle East initiated a MERS-CoV outbreak in South Korea which resulted in 186 confirmed cases and 36 deaths (15). One of the patients who got infected in South Korea travelled to China and was cared for in China (16). Another new case of MERS was imported into South Korea in September 2018, indicating that long-term and continuous preparedness was required for MERS prevention and control worldwide (17).
Viral evolution can increase or decrease pathogen virulence depending on the environment, virus, and host (18). Thirteen MERS-CoV strains were isolated during the 2015 South Korean outbreak, and 11 of them possessed a point mutation in the receptor-binding domain (RBD) of spike (S) protein (I529T), resulting in reduced affinity to the hDPP4 molecule (19). In addition, the virus strains circulating in South Korea showed strain-specific genetic variations, having 8 novel nucleotide substitutions that were unique to the South Korea lineage and shared nucleotide substitution T258C, located in spike, with some viruses from Saudi Arabia detected earlier in 2015 (20). The occurrence of spontaneous or induced mutations in the viral genome could alter their pathogenicity and virulence (21). However, few studies were performed to address the difference in pathogenicity and virulence of different MERS-CoV strains. Clade A MERS-CoV has been replaced by clade B in both camels and humans on the Arabian Peninsula, and contemporary zoonotic MERS is mainly caused by clade B viruses (22). However, the differences in pathogenicity and virulence in vitro and in vivo between viruses from these two clades are largely unknown.
In this study, we have characterized the MERS-CoV isolate from the first imported MERS-CoV ChinaGD01 (clade B) strain in China and compared its pathogenicity and virulence with those of prototype virus EMC/2012 (clade A) in vitro and in vivo. Mice infected with the ChinaGD01 strain showed higher viral load and more inflammatory cytokine production and fewer virus-specific CD4+ and CD8+ T cells in the lungs, indicating that virus evolution changed the pathogenicity and increased virulence in vivo.
RESULTS
Isolation and identification of the MERS-CoV ChinaGD01 strain.
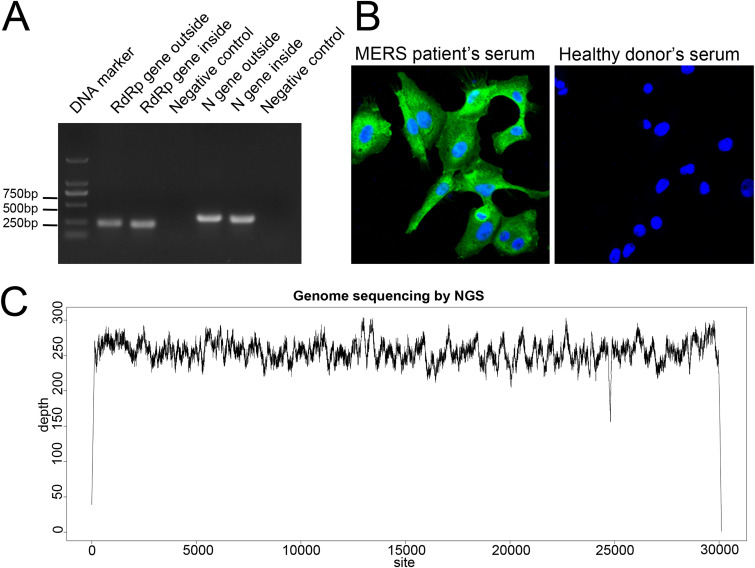
In 2015, a 43-year-old man acquired MERS in South Korean and then traveled to Guangdong, China. The patient had the first imported MERS case in China and was quarantined quickly, and nasopharyngeal swabs, sputum, and serum for diagnostic purposes were collected. The patient survived with timely treatment, as previously report (16). The specimens were subjected to virus isolation in Huh 7 cells as described in Materials and Methods. Cells inoculated were blindly passaged for three times until the cytopathic effects (CPE) were obvious. Viral replication in the cells was then confirmed by nested reverse transcription-PCR (RT-PCR) (23) and immunofluorescence assay (IFA). Nested RT-PCR for 200- to ∼300-bp fragments in the RNA-dependent RNA polymerase (RdRp) and N genes were both positive for MERS-CoV as shown in Fig. 1A. IFAs were performed using serum obtained from the same MERS patient. Cells infected with culture supernatant from the above-described procedures were strongly positive for MERS-CoV proteins by immunofluorescence with convalescent-phase serum from the patient but not with serum from a healthy donor control (Fig. 1B). The whole-genome sequence was obtained using next-generation sequencing (NGS) and showed that the isolated virus was closely related to MERS-CoV (Fig. 1C). The genome of the ChinaGD01 strain was obtained by NGS as described previously (24), and virus isolation from clinical specimens was accomplished in this study. MERS-CoV ChinaGD01 was thus successfully isolated from the patient’s respiratory tract specimens and this virus was further plaque purified twice in Vero 81 cells before being used in the experiments described below. Fewer than five passages of the ChinaGD01 and EMC/2012 strains were used in this study. The orf3, orf4a, orf4b, and orf5 genes were sequenced in case of mutation (25), and no mutations, including deletion or insertion, were detected.
FIG 1.
Identification of the isolated MERS-CoV ChinaGD01 strain. (A) Nested RT-PCR targeting the MERS-CoV RdRp and N genes was performed using primers recommended by the WHO. (B) Vero 81 cells were inoculated with virus-containing culture supernatant for 24 h, and sera from the MERS patient and a healthy donor (negative control) were used to detect viral proteins. (C) NGS of nucleic acid extracted from cell culture described above; clean data were mapped to the ChinaGD01 complete genome sequence with 100% coverage.
Phylogenetic and glycosylation analysis of ChinaGD01 and EMC/2012 sequences.
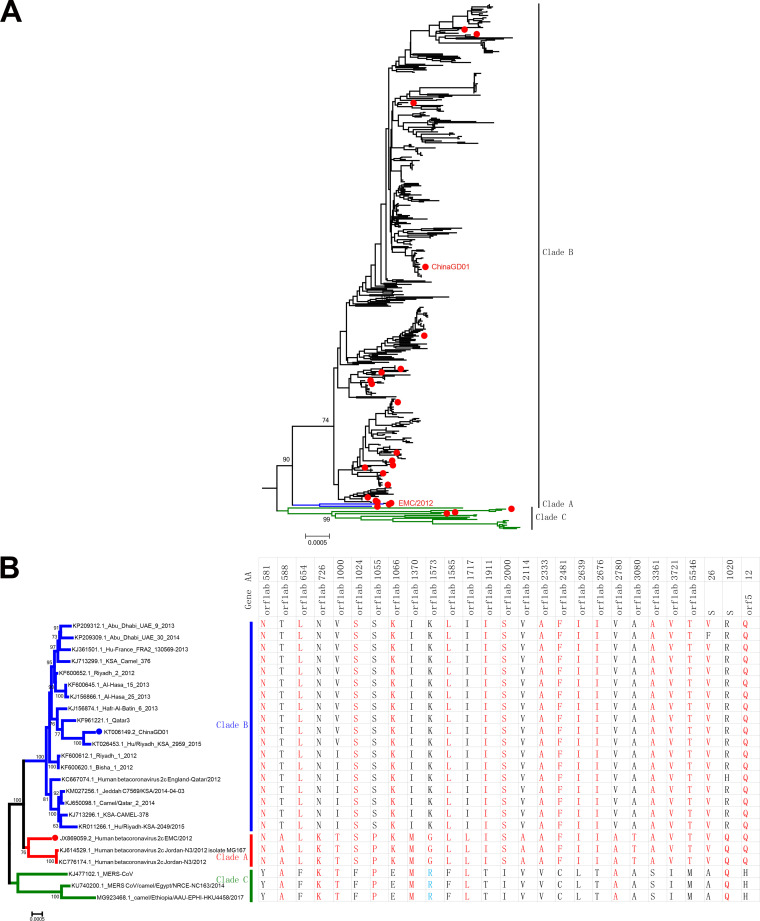
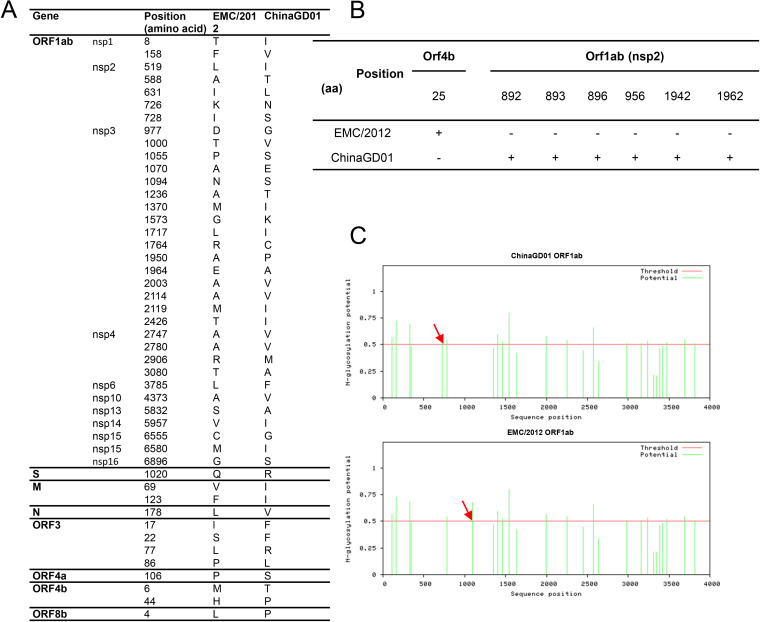
A phylogenetic tree was constructed using all available MERS-CoV complete genome sequences (Fig. 2A). There are three distinct clades of MERS-CoV, including clade A, clade B, and clade C. The early MERS cases were in clade A clusters, including the prototype EMC/2012, which was responsible for the early outbreaks of MERS. Recently, nearly all MERS-CoVs isolated from ongoing outbreaks belonged to clade B. Viruses in clade C were found only in Africa (26) and were phylogenetically distinct from contemporary viruses from the Arabian Peninsula (clades A and B). Comparison of all coding sequences and single amino acid polymorphism (SAP) analysis were performed using representative virus strains to identify clade-specific amino acid mutations. As summarized in Fig. 2B, most clade-specific mutations were identified in the orf1ab gene and, to a lesser extent, in the spike (S) and orf5 genes. Comparison of MERS-CoV ChinaGD01 and MERS-CoV EMC/2012 showed 46 unique amino acid substitutions; 34 of them were distributed in the orf1ab gene, 1 was in the spike gene (Q1020R), and the other mutations were scattered in M, N, and accessory protein genes (orf3, orf4a, orf4b, and orf8b) (Fig. 3A). As expected, hot spots for nonsynonymous substitutions of MERS-CoV were primarily located in the regions that encoded viral proteins with lower levels of functional constraint; also, larger genes were more susceptible to mutations than smaller genes (27).
FIG 2.
Phylogenetic analysis of ChinaGD01 and EMC/2012. (A) Phylogenetic relationship analysis and single amino acid comparison among different MERS-CoV clades based on all complete genomes available were performed. The complete genomes of 458 strains of MERS-CoV in GenBank were analyzed using the maximum-likelihood method implemented in PhyML. Numbers at the nodes represent bootstrap support. Twenty-five representative MERS-CoV genomes are highlighted with red dots (A); these were used for single amino acid difference comparison among clades (B). Corresponding amino acid mutations are presented on the right side, and the locations of amino acid substitutions are indicated.
FIG 3.
Genome comparison. (A) Three clades, A, B, and C, are circulating in the world, and most of the clade-specific mutations were found in the orf1ab, S, and orf5 genes. (B) O-linked glycosylation comparison analysis of all MERS-CoV coding genes using the NetOGlyc 4.0 server. (C) N-linked glycosylation comparison analysis of all MERS-CoV coding genes using the NetNglyc 1.0 server.
Glycosylation contributes to viral protein stability and functions, and also possible targets for neutralizing antibodies (28–30). Recently, the Veesler group showed that there was a large number and combinatorial diversity of N-linked glycans covering the surface of MERS-CoV and SARS-CoV S, which represented a challenge for antigen recognition by the immune system (31). Glycosylation site prediction analysis showed that indeed some mutations occurred within potential glycosylation sites in the orf1ab and orf4b genes, which may affect viral protein functions and MERS-CoV pathogenicity (Fig. 3B). Six glycosylation sites in orf1ab are predicted as O glycosylated for ChinaGD01, which were absent in EMC/2012 strain (Fig. 3B). One O-linked glycosylation site in orf4b was predicted in EMC/2012 but not in ChinaGD01. Two different potential N-glycosylation sites (amino acids [aa] 726 and 1094) were also present in the orf1ab genes of ChinaGD01 and EMC/2012 (Fig. 3C). Based on the analysis described above, more potential O-glycosylation sites existed within ChinaGD01. Their roles in virus infection and immune responses need to be further studied.
Kinetics of ChinaGD01 replication and sensitivity to type I interferon (IFN).
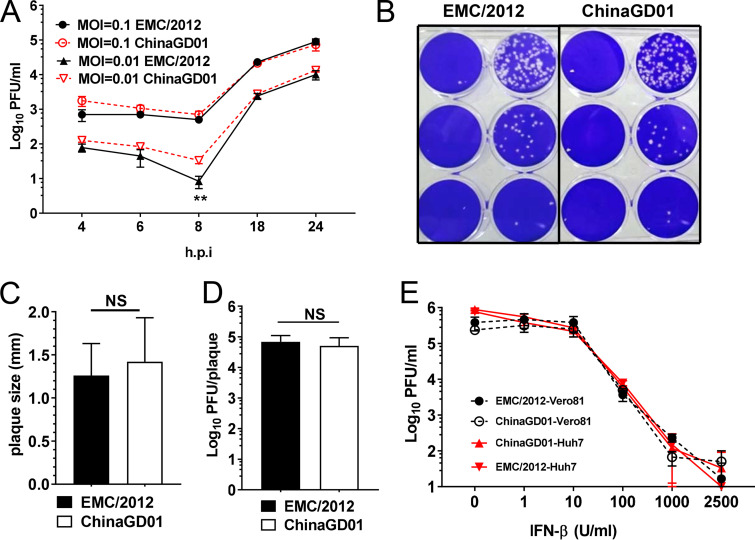
To investigate the growth kinetics and virulence of different MERS-CoV strains in vitro, Vero 81 cells were infected with ChinaGD01 or EMC/2012 which was rescued from a bacterial artificial chromosome (BAC) clone reverting all cell culture-adapted mutations at a multiplicity of infection (MOI) of 0.1 or 0.01. As shown in Fig. 4A, no difference in replication was observed at the high MOI, whereas ChinaGD01 replicated slightly to a higher level at an early time point (6 to 8 h postinfection [p.i.]) at an MOI of 0.01, although this difference disappeared at later time points. This suggests that ChinaGD01 could replicate faster at early stages of the replication cycle. Next, viral-plaque morphology was compared in Vero 81 cells. There was no difference in plaque shape or size (Fig. 4B and C). The diameters of plaques formed by EMC/2012 and ChinaGD01 both ranged from 1.5 to 2 mm. Plaques were also picked and were used to determine viral titer per plaque. There were about 5 × 104 PFU of infectious viral particles per plaque for both strains (Fig. 4D).
FIG 4.
Kinetics of ChinaGD01 replication and sensitivity to type I interferon in vitro. (A) Vero 81 cells were infected with different MERS-CoV strains at an MOI of 0.1 or 0.01, supernatant was harvested at the indicated time points, and viral titers were determined by plaque assay. Student’s t test was used to analyze differences in mean values between groups. All results are expressed as means ± standard errors of the means (SEM). A P value of <0.05 was considered to be statistically significant (*, P ≤ 0.05; **, P ≤ 0.01). (B) Plaque morphology in Vero 81 cells at day 3 p.i. (C) Plaque size comparison and plaque diameter. NS, not significant. (D) Comparison of infectious viral particle per plaque. Plaques were picked from MERS-CoV-infected Vero 81 cells. Viral titers were determined. (E) Vero 81 and Huh 7 cells in triplicate were treated with the indicated concentrations of human IFN-β 24 h prior to being infected with MERS-CoV ChinaGD01 or EMC/2012 at an MOI of 0.1. Cells were then incubated for another 24 h in the presence of the same concentration of IFN-β, supernatants were harvested, and virus titers were determined by plaque assay in Vero 81 cells. Data are representative of those from two independent experiments.
Type I interferon is a critical component of the antiviral innate immune response. It elicits an antiviral state in infected cells and contributes to early virus control (32). Due to accumulated mutations, different viral strains could possess differential sensitivities to interferon treatment. Sensitivity of EMC/2012 and ChinaGD01 to IFN-β was evaluated in Vero 81 cells and Huh 7 cells. Cells were pretreated with the desired dose of IFN-β for 24 h and were then infected with EMC/2012 or ChinaGD01 at an MOI of 0.1 and cultured for another 24 h. Viral titers in the supernatant were measured. As shown in Fig. 4E, EMC/2012 and ChinaGD01 were equally sensitive to IFN-β treatment, and no significant difference in the effect on viral replication was observed for the viruses. Overall, these results suggested that ChinaGD01 virus did not evolve new mechanisms to evade the innate immune response.
Increased pathogenicity in ChinaGD01-infected mice.
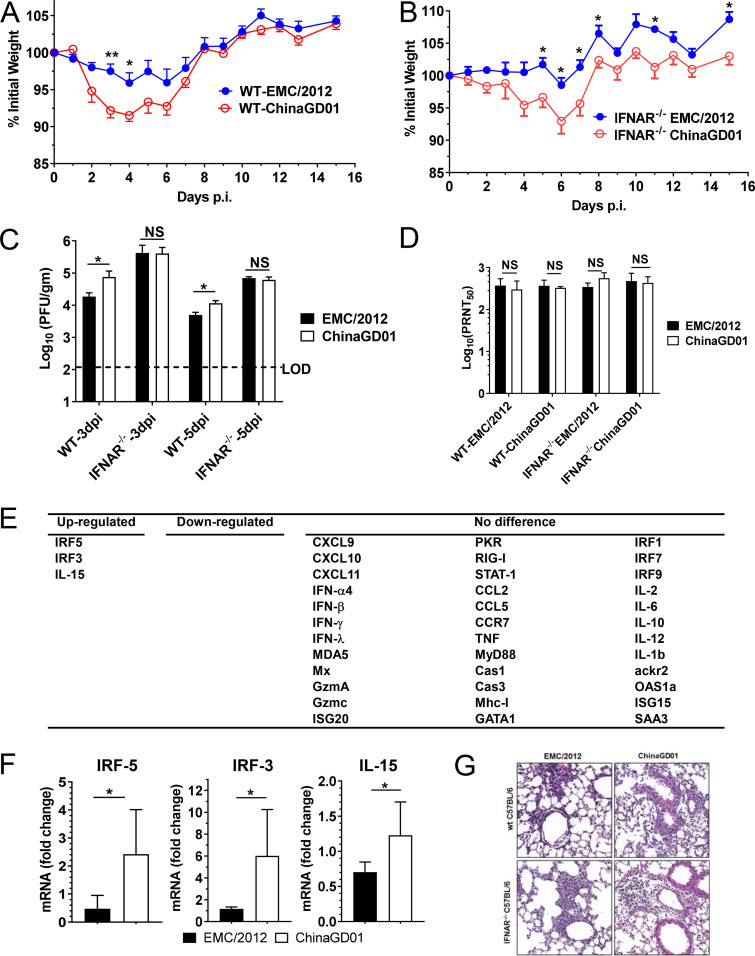
To determine if clade B virus ChinaGD01 had different pathogenicity in vivo, a well-defined MERS mouse model was used (33). Mice were transduced with a recombinant, nonreplicating adenovirus expressing the hDPP4 receptor for MERS-CoV. Five days after transduction, mice were intranasally infected with 1 × 105 PFU of EMC/2012 or ChinaGD01. Weight changes were monitored daily. Virus titers in the lungs were measured at day 5 p.i. As shown in Fig. 5A and B, both wild-type (WT) and IFNAR−/− mice showed significantly more weight loss after ChinaGD01 infection. Both strains replicated efficiently in the lungs of infected mice (Fig. 5C). However, WT mice that had an intact immune system, but not IFNAR−/− mice, showed delayed viral clearance after ChinaGD01 infection, suggesting that ChinaGD01 replicated to higher levels and was more virulent in vivo (Fig. 5C).
FIG 5.
Increased pathogenicity in ChinaGD01-infected mice. (A and B) Weight loss in Ad5-hDPP4-transduced WT C57BL/6 mice (A) and IFNAR−/− C57BL/6 mice (B) infected intranasally with 105 PFU of MERS-CoV ChinaGD01 or EMC/2012. (C) Lungs were harvested at days 3 and 5 p.i. Virus titers were determined by plaque assay. Student’s t test was used to analyze differences in mean values between groups. All results are expressed as means ± standard errors of the means (SEM). A P value of <0.05 was considered to be statistically significant (*, P ≤ 0.05; **, P ≤ 0.01). (D) Two-way antigenic cross-PRNT assay between ChinaGD01 and EMC/2012 was performed using sera harvested from mice challenged with different MERS-CoVs 15 days p.i. (E and F) A total of 39 cytokines, chemokines, transcription factors, and two housekeeping genes (hypoxanthine phosphoribosyltransferase [HPRT] and beta-actin) were selected for qRT-PCR. Representative genes are shown. (G) Histopathological damage. Lungs from B6 mice were removed at the indicated time points 3 p.i., fixed in zinc formalin, and embedded in paraffin. Sections were stained with hematoxylin and eosin (20×).
MERS-CoV has circulated in human populations for more than 6 years. A critical question is whether antibodies raised from different MERS-CoV strain infections are cross protective. To address this question, plaque reduction neutralization tests (PRNT) were performed using sera harvested from mice infected with EMC/2012 or ChinaGD01 15 days p.i. As shown in Fig. 5D, mice infected with both strains produced high titers of neutralizing antibodies. Antibodies raised from the different viral strain infections could comparably cross neutralize each other (Fig. 5D), suggesting that there was no antigenic difference between the viruses.
To compare cytokine and chemokine production in EMC/2012- and ChinaGD01-infected lungs, a panel of 39 cytokine, chemokine, and transcription factor genes associated (Fig. 5E) with the proinflammatory response, anti-inflammatory response, apoptosis, and the interferon pathway were evaluated at day 3 p.i. and 5 p.i. by quantitative RT-PCR (qRT-PCR). As shown in Fig. 5E and F, genes, such as the IRF3, IRF5, and interleukin 15 (IL-15) genes, which are related to innate and inflammation immune responses were upregulated in ChinaGD01-infected mice at day 5 p.i. compared to EMC/2012-infected mice, but not at day 3 p.i. (data not shown). Lungs were removed at day 3 p.i., fixed in zinc formalin, and embedded in paraffin. Sections were stained with hematoxylin and eosin. Perivascular and peribronchial lymphoid cell infiltration and interstitial pneumonia in both ChinaGD01- and EMC/2012-infected mice were observed (Fig. 5G). All the results indicated that ChinaGD01 infection induces more robust inflammation in the lungs, which could contribute to the more severe pathological changes described above (Fig. 5A and B).
ChinaGD01 infection did not induce robust virus-specific T cell responses in mice.
Antiviral T cell responses are responsible for virus clearance (34, 35). We also found that T cells are required for MERS-CoV clearance from infected lungs (36). Development of robust and functional MERS-CoV-specific T cells is important to clear MERS-CoV and reduce pathological damage induced by viral replication. Bronchoalveolar lavage fluid (BALF) from ChinaGD01- or EMC/2012-infected mice was harvested at day 7 p.i. Virus-specific T cells in BALF were stimulated with a CD4 epitope peptide (N98 [unpublished data]) or a CD8 T cell epitope peptide (S1165) for MERS-CoV (described previously) in the presence of brefeldin A (BFA) (33). The CD4 and CD8 epitopes used in this study were conserved in MERS-CoV, with 100% identity between EMC/2012 and ChinaGD01 strains. IFN-γ and tumor necrosis factor (TNF) productions were measured by intracellular cytokine staining (ICS). As shown in Fig. 6A and B, anti-MERS-CoV CD4+ and CD8+ T cell responses were generated in mice infected with both strains. These T cells were multifunctional, as they were able to produce more than one cytokine, such as IFN-γ and TNF, upon stimulation. However, after ChinaGD01 infection, there were significantly lower virus-specific T cell responses in the lung than in EMC/2012-infected mice (Fig. 6C and D), suggesting that ChinaGD01 and EMC/2012 differentially regulated antiviral T cell responses. Considering the increased weight loss, increased virus replication, and decreased T cell response in ChinaGD01-infected mice, the clade B ChinaGD01 stain exhibited enhanced pathogenicity of MERS-CoV and was more virulent than early epidemic clade A virus.
FIG 6.
ChinaGD01 infection did not induce robust virus-specific T cell responses in mice. BALFs from ChinaGD01 or EMC/2012 infected mice were harvested at day 7 p.i. Virus-specific T cells were stimulated with T cell epitope peptides. IFN-γ and TNF productions were measured by intracellular cytokine staining. Representative flow plots for virus-specific CD4+ and CD8+ T cells are shown (A and B). Frequencies of IFN-γ+ CD4+ or CD8+ T cells are shown (C and D). Data are representative of those from two independent experiments.
DISCUSSION
In this study, we isolated the first imported MERS-CoV ChinaGD01 strain in Guangdong, China. Genetic, virological, and immunological comparisons between MERS-CoV early epidemic clade A strain EMC/2012 and ChinaGD01 showed changes in genomic evolution and related enhancement in pathogenicity and virulence in vivo.
Genomic comparison analysis between clade A and clade B viruses showed that clade-specific mutations are mainly located in regions (orf1ab) with a lower level of functional constraint. Nonsynonymous mutations rarely occurred in the spike protein, which has receptor binding and contains dominant antigenic sites. Unexpectedly, quite a few mutations in orf1ab and orf4b are predicted to be potential O-glycosylated or N-glycosylated sites in the ChinaGD01 strain. It is well known that glycosylation is involved in maintaining protein conformation, contributes to a protein’s biological activity, and is a major target for neutralizing antibodies (31). Glycosylation and its potential impact on pathogenic mechanisms were lacking. Further studies are required to address the roles of these additional glycosylation sites in clade B virus replication and pathogenicity.
MERS-CoV was found to evade and inhibit the host innate immune response (37–39). Several groups determined that MERS-CoV accessory proteins were antagonists to innate immune response and played critical roles in MERS pathogenesis (40). orf4a and orf4b were demonstrated to block the roles of interferon-related genes (ISG), such as RIG-I, MDA5, and PKR (37, 39, 41, 42). Little is known regarding whether viruses from different clades of MERS-CoV exhibit differential sensitivity to IFN treatment (26). In the current study, ChinaGD01 and EMC/2012 were found to be equally sensitive to high-dose and low-dose IFN-β treatment in vitro, indicating that type I interferon treatment of patients infected with different strains of MERS-CoV was still an important way to control virus infection (43). Although there was no difference in IFN sensitivity observed, clade B virus infection did induce higher expression of inflammation-related molecules in the lungs of infected animals, such as IRF3, IRF5, and IL-15. IRF3 and IRF5 are members of the interferon regulatory factor family, which mediate type I IFN production in response to viral infections (44). IL-15 is a cytokine with a broad range of biological functions in many diverse cell types, including T cells, NK cells, dendritic cells (DC), macrophages, and even granulocytes (45). In addition to promoting T cell proliferation, it also plays a major role in the development of inflammation in viral infection settings, as reviewed previously (46). Higher expression levels of these molecules in ChinaGD01-infected mice could be associated with dysregulated innate immune responses and increased pathological damage, as determined by increased weight loss (Fig. 5A and B).
In addition, MERS-CoV ChinaGD01 infection led to decreased virus-specific T cell responses in mice, which could be responsible for delayed virus clearance and increased weight loss. Our previous study showed that T cells are required for SARS-CoV clearance (47) in mice, and MERS-CoV-specific T cell responses in patients were also correlated with disease severity and outcomes. Patients with higher CD8+ T cell responses tended to have mild diseases (48). Mutations in the ChinaGD01 genome could potentially dysregulate innate and inflammatory responses, which, in turn, inhibit T cell responses as we published previously (36, 49–52), which requires further studies.
In summary, MERS-CoV strains from early epidemic clade A and contemporary epidemic clade B have significant genetic and phenotypic differences. Clade B virus ChinaGD01 also caused more severe disease in mice with delayed virus clearance, increased inflammatory cytokines, and decreased antiviral T cell responses compared to the early clade A virus EMC/2012. Although the differences described in this report in virulence and pathogenicity between MERS-CoV ChinaGD01 and EMC/2012 were modest, frequent epidemiological and phylogenetic studies are required to monitor the virulence and disease severity of contemporary MERS-CoVs. Such studies may inform risk assessment of ongoing MERS-CoV evolution.
MATERIALS AND METHODS
Specimens, mice, and cells.
Nasopharyngeal swabs, sputum, and serum were collected from an imported MERS case from South Korea (53) and stored in –80°C in June 2015. Healthy donor serum was collected from Guangzhou Blood Center as a negative control. IFNAR−/− mice were purchased from the Jackson Laboratory (ME), and WT C57BL/6 mice were purchased from Vital River Laboratories Co., Ltd. (Beijing, China). All of the mice were housed in a specific-pathogen-free animal care facility at Guangzhou Medical University. All studies described here were approved by the Research Ethics Committee and Institutional Animal Care and Use Committee of Guangzhou Medical University. Huh 7 cells were purchased from Creative Biolabs, and Vero 81 (CCL-81) cells were obtained from the ATCC. All work with MERS-CoV was conducted in the joint biosafety level 3 laboratory of Guangdong Inspection and Quarantine Technology Center, Guangzhou Custom.
Isolation and identification of the MERS-CoV ChinaGD01 strain.
Specimens from the MERS patient were inoculated into Huh 7 cells in the presence of ampicillin (1,000 U/ml), streptomycin (1,000 U/ml), gentamicin (10 μg/ml), and amphotericin B (0.25 μg/ml) and cultured at 37°C, and supernatants were harvested and passaged to new Vero 81 cells every 3 days until cytopathic effects (CPE) were obvious. The presence of MERS-CoV in the culture was identified and confirmed by RT-PCR and indirect immunofluorescence (IFA), followed by next-generation whole-genome sequencing. For RT-PCR, viral RNA was extracted from infected cells using a QIAamp viral RNA minikit (Qiagen, Hilden, Germany) and the MERS-CoV genome was amplified by using specific primer sets as recommended by the WHO. For IFA, Vero 81 cells were grown on coverslips overnight and then infected by the supernatant from viral-isolation culture; 24 h later, viral proteins were detected using the MERS patient’s serum, followed by Alexa Fluor 488-labeled goat anti-human IgG (Jackson, West Grove, PA). Serum from a healthy donor served as a negative control. 4′,6-Diamidino-2-phenylindole (DAPI) was used for nuclear staining. Finally, to obtain the full genome sequence, next-generation sequencing (NGS) was conducted using a BGISEQ-500 sequencing instrument and the NGS data were aligned to the MERS-CoV ChinaGD01 sequence (accession number KT006149.2) in GenBank. ChinaGD01 was plaque purified twice in Vero 81 cells before use.
Rescue of MERS-CoV EMC/2012 from a BAC infectious clone.
A BAC clone carrying the MERS-CoV EMC/2012 infectious cDNA was kindly provided by Luis Enjuanes. EMC/2012 virus was rescued by transfection of Vero 81 cells with pBAC-EMC/2012 plasmid as previously described, and virus was plaque purified in Vero 81 cells twice before use (54).
Phylogenetic analysis and single amino acid polymorphism analysis.
To determine the evolutionary relationship between MERS-CoV clade A and clade B, all available MERS-CoV (Table S1) complete genomes and phylogenetic analyses of MERS-CoV whole genomes were collected and separate genes were also analyzed using the PhyML program (55) with the GTR model. Single amino acid polymorphism (SAP) were identified and analyzed between clade A and clade B to find clade-specific SAPs which could potentially associate with pathogenicity changes.
Glycosylation analyses of viral proteins.
The NetNglyc 1.0 server (56) was used to predict N-glycosylation sites using artificial neural networks. The NetOglyc server (57) was used to produce neural-network predictions of mucin type GalNAc O-glycosylation sites in mammalian proteins.
Sensitivity to type I interferon.
Vero 81 cells in triplicate were treated with the desired concentrations of human IFN-β (Sino Biological, Inc., Beijing, China) 24 h prior to being infected with MERS-CoV at an MOI of 0.1. Cells were then incubated for another 24 h in the presence of the same concentration of IFN-β. Supernatants were harvested, and virus titers were determined by plaque assay.
Ad5-hDPP4 transduction and infection of mice.
Mice were sensitized to MERS-CoV infection after prior transduction with adenovirus 5 expressing human DPP4 (Ad5-hDPP4) as previously described (33). Briefly, mice were transduced with Ad5-hDPP4 5 days before intranasal challenge with 1 × 105 PFU of MERS-CoV. Lungs were removed into phosphate-buffered saline (PBS) at days 5 p.i. and homogenized. Virus titers of clarified supernatants were assayed in Vero 81 cells and expressed as PFU per gram of tissue.
Plaque reduction neutralization test.
Two weeks following MERS-CoV infection of the hDPP4-transduced mice, serum samples were harvested, 10-fold serially diluted in Dulbecco’s modified Eagle medium (DMEM), and mixed 1:1 with 100 PFU of MERS-CoV. After 1 h of incubation at 37°C, the mixture was added to Vero 81 cells for an additional 1 h to permit absorption. Cells were then overlaid with 1.2% agarose (containing 2% fetal bovine serum [FBS] and DMEM). After further incubation for 3 days, agarose plugs were removed for collection of virus. The remaining plaques were visualized by 0.1% crystal violet staining.
RT-qPCR analysis.
RNA was extracted from infected lungs using TRIzol (Invitrogen, Carlsbad, CA) and used as a template for cDNA synthesis. A set of 39 cytokines, chemokines, and transcription factors was selected for qRT-PCR analysis. qRT-PCR was performed using a previously described set of primers (58).
Preparation of cells from BALF and ICS.
Mice were infected with MERS-CoV and sacrificed at the desired time points. Bronchoalveolar lavage fluid (BALF) was acquired by inflating lungs with 1 ml of complete RPMI 1640 medium via cannulation of the trachea followed by lavage four times. Cells in the BALF were collected by centrifugation. For intracellular cytokine staining (ICS), cells from BALF were cultured in 96-well dishes at 37°C for 5 to 6 h in the presence of 1 to 5 μM peptide (GL Biochem Inc., Shanghai, China), brefeldin A (BFA; BD Biosciences, San Jose, CA), and antigen-presenting cells (CHB3 cells). Cells were then labeled for cell surface markers, fixed and permeabilized with Cytofix/Cytoperm solution (BD Biosciences), and labeled with anti-intracellular cytokine antibodies. All antibodies were purchased from BD Biosciences, eBioScience, or BioLegend (San Diego, CA). In addition, the CD4 epitope (nucleocapsid protein epitope N98-112; residues 98 to 112) and CD8 epitope (spike protein epitope S1165-1173; residues 1165 to 1173) used in this study were conserved in MERS-CoV, with 100% identity between EMC/2012 and ChinaGD01.
Statistical analysis.
Student’s t test was used to analyze differences in mean values between groups. All results are expressed as means ± standard errors of the means (SEM). P values of <0.05 were considered statistically significant.
Supplementary Material
ACKNOWLEDGMENTS
We thank Luis Enjuanes for providing pBAC-MERS-EMC/2012 plasmid. We thank Stanley Perlman for a critical review of the manuscript.
This research was supported in part by grants from the Guangdong Provincial Department of Science and Technology (2018B020207013), the National Key Research and Development Program of China (2018YFC1200100 and 2016YFC1202701), the National Science and Technology Major Project (2018ZX10301403-001-003 and 2018ZX10101002), the National Natural Science Foundation of China (81772191, 81702047, 91842106, and 8181101118), the Thousand Talents Plan Award of China 2015 and the State Key Laboratory of Respiratory Disease (SKLRD-QN-201715 and SKLRD-QN-201912), the Guangzhou Medical University High-level University Innovation Team Training Program (Guangzhou Medical University release [2017] no. 159), the National Key Technology R&D Program (2018YFC1311900), and the Guangdong Science and Technology Foundation (2019B030316028).
J.Z., J.H., and J.Z. designed and coordinated the study. N.Z., M.P., R.C., D.-Q.S., Z.W., L.C., Y.L., Z.Y., W.T., and C.K.P.M. codesigned and contributed reagents, materials, and analysis tools. Y.W., J.S., X.L., A.Z., L.Z., W.G., M.G., X.N., J.D., Z.Z., Y.S., and S.H. performed the experiments. J.Z., Y.W., J.S., and A.Z. wrote the manuscript. All authors contributed to the interpretation and conclusions presented.
We declare no competing financial interests.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Zaki AM, Van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. 2012. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 2.Azhar EI, El-Kafrawy SA, Farraj SA, Hassan AM, Al-Saeed MS, Hashem AM, Madani TA. 2014. Evidence for camel-to-human transmission of MERS coronavirus. N Engl J Med 370:2499–2505. doi: 10.1056/NEJMoa1401505. [DOI] [PubMed] [Google Scholar]
- 3.Reusken CBEM, Haagmans BL, Müller MA, Gutierrez C, Godeke G-J, Meyer B, Muth D, Raj VS, Smits-De Vries L, Corman VM, Drexler J-F, Smits SL, El Tahir YE, De Sousa R, van Beek J, Nowotny N, van Maanen K, Hidalgo-Hermoso E, Bosch B-J, Rottier P, Osterhaus A, Gortázar-Schmidt C, Drosten C, Koopmans MPG. 2013. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet Infect Dis 13:859–866. doi: 10.1016/S1473-3099(13)70164-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Memish ZA, Mishra N, Olival KJ, Fagbo SF, Kapoor V, Epstein JH, Alhakeem R, Durosinloun A, Al Asmari M, Islam A. 2013. Middle East respiratory syndrome coronavirus in bats, Saudi Arabia. Emerg Infect Dis 19:1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang Y, Du L, Liu C, Wang L, Ma C, Tang J, Baric RS, Jiang S, Li F. 2014. Receptor usage and cell entry of bat coronavirus HKU4 provide insight into bat-to-human transmission of MERS coronavirus. Proc Natl Acad Sci U S A 111:12516–12521. doi: 10.1073/pnas.1405889111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Assiri A, Al-Tawfiq JA, Al-Rabeeah AA, Al-Rabiah FA, Al-Hajjar S, Al-Barrak A, Flemban H, Al-Nassir WN, Balkhy HH, Al-Hakeem RF, Makhdoom HQ, Zumla AI, Memish ZA. 2013. Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: a descriptive study. Lancet Infect Dis 13:752–761. doi: 10.1016/S1473-3099(13)70204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu G, Hu Y, Wang Q, Qi J, Gao F, Li Y, Zhang Y, Zhang W, Yuan Y, Bao J, Zhang B, Shi Y, Yan J, Gao GF. 2013. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature 500:227–231. doi: 10.1038/nature12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raj VS, Mou H, Smits SL, Dekkers DHW, Müller MA, Dijkman R, Muth D, Demmers JAA, Zaki A, Fouchier RAM, Thiel V, Drosten C, Rottier PJM, Osterhaus ADME, Bosch BJ, Haagmans BL. 2013. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Widagdo W, Raj VS, Schipper D, Kolijn K, van Leenders GJLH, Bosch BJ, Bensaid A, Segalés J, Baumgärtner W, Osterhaus ADME, Koopmans MP, van den Brand JMA, Haagmans BL. 2016. Differential expression of the Middle East respiratory syndrome coronavirus receptor in the upper respiratory tracts of humans and dromedary camels. J Virol 90:4838–4842. doi: 10.1128/JVI.02994-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meyerholz DK, Lambertz AM, McCray PB. 2016. Dipeptidyl peptidase 4 distribution in the human respiratory tract: implications for the Middle East respiratory syndrome. Am J Pathol 186:78–86. doi: 10.1016/j.ajpath.2015.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Widagdo W, Sooksawasdi Na Ayudhya S, Hundie GB, Haagmans BL. 2019. Host determinants of MERS-CoV transmission and pathogenesis. Viruses 11:280. doi: 10.3390/v11030280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharif-Yakan A, Kanj SS. 2014. Emergence of MERS-CoV in the Middle East: origins, transmission, treatment, and perspectives. PLoS Pathog 10:e1004457. doi: 10.1371/journal.ppat.1004457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ng DL, Al Hosani F, Keating MK, Gerber SI, Jones TL, Metcalfe MG, Tong S, Tao Y, Alami NN, Haynes LM, Mutei MA, Abdel-Wareth L, Uyeki TM, Swerdlow DL, Barakat M, Zaki SR. 2016. Clinicopathologic, immunohistochemical, and ultrastructural findings of a fatal case of Middle East respiratory syndrome coronavirus infection in the United Arab Emirates, April 2014. Am J Pathol 186:652–658. doi: 10.1016/j.ajpath.2015.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alsaad KO, Hajeer AH, Al Balwi M, Al Moaiqel M, Al Oudah N, Al Ajlan A, AlJohani S, Alsolamy S, Gmati GE, Balkhy H, Al-Jahdali HH, Baharoon SA, Arabi YM. 2018. Histopathology of Middle East respiratory syndrome coronovirus (MERS‐CoV) infection—clinicopathological and ultrastructural study. Histopathology 72:516–524. doi: 10.1111/his.13379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ki M. 2015. 2015 MERS outbreak in Korea: hospital-to-hospital transmission. Epidemiol Health 37:e2015033. doi: 10.4178/epih/e2015033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guan WD, Mok CKP, Chen ZL, Feng LQ, Li ZT, Huang JC, Ke CW, Deng X, Ling Y, Wu SG, Niu XF, Perera RA, Da Xu Y, Zhao J, Zhang LQ, Li YM, Chen RC, Peiris M, Chen L, Zhong NS. 2015. Characteristics of traveler with Middle East respiratory syndrome, China, 2015. Emerg Infect Dis 21:2278–2280. doi: 10.3201/eid2112.151232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bak SL, Jun KI, Jung J, Kim J-H, Kang CK, Park WB, Kim N-J, Oh M-D. 2018. An atypical case of Middle East respiratory syndrome in a returning traveler to Korea from Kuwait, 2018. J Korean Med Sci 33:e348. doi: 10.3346/jkms.2018.33.e348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geoghegan JL, Holmes EC. 2018. The phylogenomics of evolving virus virulence. Nat Rev Genet 19:756–769. doi: 10.1038/s41576-018-0055-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim Y, Cheon S, Min C-K, Sohn KM, Kang YJ, Cha Y-J, Kang J-I, Han SK, Ha N-Y, Kim G, Aigerim A, Shin HM, Choi M-S, Kim S, Cho H-S, Kim Y-S, Cho N-H. 2016. Spread of mutant Middle East respiratory syndrome coronavirus with reduced affinity to human CD26 during the South Korean outbreak. mBio 7:e00019-16. doi: 10.1128/mBio.00019-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim D-W, Kim Y-J, Park SH, Yun M-R, Yang J-S, Kang HJ, Han YW, Lee HS, Kim HM, Kim H, Kim A-R, Heo DR, Kim SJ, Jeon JH, Park D, Kim JA, Cheong H-M, Nam J-G, Kim K, Kim SS. 2016. Variations in spike glycoprotein gene of MERS-CoV, South Korea, 2015. Emerg Infect Dis 22:100–104. doi: 10.3201/eid2201.151055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Volk WA, Gebhardt B, Hammaskjold M, Kaomer R. 1995. Medical microbiology. Lippincott-Raven, Philadelphia, PA. [Google Scholar]
- 22.Chan JF, Lau SK, To KK, Cheng VC, Woo PC, Yuen K-Y. 2015. Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin Microbiol Rev 28:465–522. doi: 10.1128/CMR.00102-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Corman VM, Müller MA, Costabel U, Timm J, Binger T, Meyer B, Kreher P, Lattwein E, Eschbach-Bludau M, Nitsche A, Bleicker T, Landt O, Schweiger B, Drexler JF, Osterhaus AD, Haagmans BL, Dittmer U, Bonin F, Wolff T, Drosten C. 2012. Assays for laboratory confirmation of novel human coronavirus (hCoV-EMC) infections. Eurosurveillance 17:20334. doi: 10.2807/ese.17.49.20334-en. [DOI] [PubMed] [Google Scholar]
- 24.Lu R, Wang Y, Wang W, Nie K, Zhao Y, Su J, Deng Y, Zhou W, Li Y, Wang H. 2015. Complete genome sequence of Middle East respiratory syndrome coronavirus (MERS-CoV) from the first imported MERS-CoV case in China. Genome Announc 3:e00818-15. doi: 10.1128/genomeA.00818-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Durai P, Batool M, Shah M, Choi S. 2015. Middle East respiratory syndrome coronavirus: transmission, virology and therapeutic targeting to aid in outbreak control. Exp Mol Med 47:e181. doi: 10.1038/emm.2015.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chu DKW, Hui KPY, Perera RAPM, Miguel E, Niemeyer D, Zhao J, Channappanavar R, Dudas G, Oladipo JO, Traoré A, Fassi-Fihri O, Ali A, Demissié GF, Muth D, Chan MCW, Nicholls JM, Meyerholz DK, Kuranga SA, Mamo G, Zhou Z, So RTY, Hemida MG, Webby RJ, Roger F, Rambaut A, Poon LLM, Perlman S, Drosten C, Chevalier V, Peiris M. 2018. MERS coronaviruses from camels in Africa exhibit region-dependent genetic diversity. Proc Natl Acad Sci U S A 115:3144–3149. doi: 10.1073/pnas.1718769115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heddle JA, Athanasiou K. 1975. Mutation rate, genome size and their relation to the rec concept. Nature 258:359–361. doi: 10.1038/258359a0. [DOI] [PubMed] [Google Scholar]
- 28.Raman R, Tharakaraman K, Sasisekharan V, Sasisekharan R. 2016. Glycan–protein interactions in viral pathogenesis. Curr Opin Struct Biol 40:153–162. doi: 10.1016/j.sbi.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ning Y-J, Deng F, Hu Z, Wang H. 2017. The roles of ebolavirus glycoproteins in viral pathogenesis. Virol Sin 32:3–15. doi: 10.1007/s12250-016-3850-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vigerust DJ, Shepherd VL. 2007. Virus glycosylation: role in virulence and immune interactions. Trends Microbiol 15:211–218. doi: 10.1016/j.tim.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walls AC, Xiong X, Park Y-J, Tortorici MA, Snijder J, Quispe J, Cameroni E, Gopal R, Dai M, Lanzavecchia A, Zambon M, Rey FA, Corti D, Veesler D. 2019. Unexpected receptor functional mimicry elucidates activation of coronavirus fusion. Cell 176:1026–1039.e15. doi: 10.1016/j.cell.2018.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Samuel CE. 2001. Antiviral actions of interferons. Clin Microbiol Rev 14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao J, Li K, Wohlford-Lenane C, Agnihothram SS, Fett C, Zhao J, Gale MJ, Baric RS, Enjuanes L, Gallagher T, McCray PB, Perlman S. 2014. Rapid generation of a mouse model for Middle East respiratory syndrome. Proc Natl Acad Sci U S A 111:4970–4975. doi: 10.1073/pnas.1323279111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sant AJ, McMichael A. 2012. Revealing the role of CD4+ T cells in viral immunity. J Exp Med 209:1391–1395. doi: 10.1084/jem.20121517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmidt ME, Varga SM. 2018. The CD8 T cell response to respiratory virus infections. Front Immunol 9:678. doi: 10.3389/fimmu.2018.00678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao J, Zhao J, Mangalam AK, Channappanavar R, Fett C, Meyerholz DK, Agnihothram S, Baric RS, David CS, Perlman S. 2016. Airway memory CD4+ T cells mediate protective immunity against emerging respiratory coronaviruses. Immunity 44:1379–1391. doi: 10.1016/j.immuni.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rabouw HH, Langereis MA, Knaap RCM, Dalebout TJ, Canton J, Sola I, Enjuanes L, Bredenbeek PJ, Kikkert M, de Groot RJ, van Kuppeveld FJM. 2016. Middle East respiratory coronavirus accessory protein 4a inhibits PKR-mediated antiviral stress responses. PLoS Pathog 12:e1005982. doi: 10.1371/journal.ppat.1005982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siu K-L, Yeung ML, Kok K-H, Yuen K-S, Kew C, Lui P-Y, Chan C-P, Tse H, Woo PCY, Yuen K-Y, Jin D-Y. 2014. Middle East respiratory syndrome coronavirus 4a protein is a double-stranded RNA-binding protein that suppresses PACT-induced activation of RIG-I and MDA5 in the innate antiviral response. J Virol 88:4866–4876. doi: 10.1128/JVI.03649-13. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Yang Y, Zhang L, Geng H, Deng Y, Huang B, Guo Y, Zhao Z, Tan W. 2013. The structural and accessory proteins M, ORF 4a, ORF 4b, and ORF 5 of Middle East respiratory syndrome coronavirus (MERS-CoV) are potent interferon antagonists. Protein Cell 4:951–961. doi: 10.1007/s13238-013-3096-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Menachery VD, Mitchell HD, Cockrell AS, Gralinski LE, Yount BL, Graham RL, McAnarney ET, Douglas MG, Scobey T, Beall A, Dinnon K, Kocher JF, Hale AE, Stratton KG, Waters KM, Baric RS. 2017. MERS-CoV accessory ORFs play key role for infection and pathogenesis. mBio 8:e00665-17. doi: 10.1128/mBio.00665-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakagawa K, Narayanan K, Wada M, Makino S. 2018. Inhibition of stress granule formation by Middle East respiratory syndrome coronavirus 4a accessory protein facilitates viral translation, leading to efficient virus replication. J Virol 92:e00902-18. doi: 10.1128/JVI.00902-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Canton J, Fehr AR, Fernandez-Delgado R, Gutierrez-Alvarez FJ, Sanchez-Aparicio MT, García-Sastre A, Perlman S, Enjuanes L, Sola I. 2018. MERS-CoV 4b protein interferes with the NF-κB-dependent innate immune response during infection. PLoS Pathog 14:e1006838. doi: 10.1371/journal.ppat.1006838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Falzarano D, de Wit E, Rasmussen AL, Feldmann F, Okumura A, Scott DP, Brining D, Bushmaker T, Martellaro C, Baseler L, Benecke AG, Katze MG, Munster VJ, Feldmann H. 2013. Treatment with interferon-α2b and ribavirin improves outcome in MERS-CoV–infected rhesus macaques. Nat Med 19:1313–1317. doi: 10.1038/nm.3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nehyba J, Hrdličková R, Bose HR. 2009. Dynamic evolution of immune system regulators: the history of the interferon regulatory factor family. Mol Biol Evol 26:2539–2550. doi: 10.1093/molbev/msp167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ferlazzo G, Pack M, Thomas D, Paludan C, Schmid D, Strowig T, Bougras G, Muller WA, Moretta L, Münz C. 2004. Distinct roles of IL-12 and IL-15 in human natural killer cell activation by dendritic cells from secondary lymphoid organs. Proc Natl Acad Sci U S A 101:16606–16611. doi: 10.1073/pnas.0407522101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perera PY, Lichy JH, Waldmann TA, Perera LP. 2012. The role of interleukin-15 in inflammation and immune responses to infection: implications for its therapeutic use. Microbes Infect 14:247–261. doi: 10.1016/j.micinf.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao J, Zhao J, Perlman S. 2010. T cell responses are required for protection from clinical disease and for virus clearance in severe acute respiratory syndrome coronavirus-infected mice. J Virol 84:9318–9325. doi: 10.1128/JVI.01049-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao J, Alshukairi AN, Baharoon SA, Ahmed WA, Bokhari AA, Nehdi AM, Layqah LA, Alghamdi MG, Al Gethamy MM, Dada AM, Khalid I, Boujelal M, Al Johani SM, Vogel L, Subbarao K, Mangalam A, Wu C, Ten Eyck P, Perlman S, Zhao J. 2017. Recovery from the Middle East respiratory syndrome is associated with antibody and T-cell responses. Sci Immunol 2:eaan5393. doi: 10.1126/sciimmunol.aan5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao J, Zhao J, Van Rooijen N, Perlman S. 2009. Evasion by stealth: inefficient immune activation underlies poor T cell response and severe disease in SARS-CoV-infected mice. PLoS Pathog 5:e1000636. doi: 10.1371/journal.ppat.1000636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu WJ, Lan J, Liu K, Deng Y, Yao Y, Wu S, Chen H, Bao L, Zhang H, Zhao M, Wang Q, Han L, Chai Y, Qi J, Zhao J, Meng S, Qin C, Gao GF, Tan W. 2017. Protective T cell responses featured by concordant recognition of Middle East respiratory syndrome coronavirus-derived CD8+ T cell epitopes and host MHC. J Immunol 198:873–882. doi: 10.4049/jimmunol.1601542. [DOI] [PubMed] [Google Scholar]
- 51.Channappanavar R, Fett C, Zhao J, Meyerholz DK, Perlman S. 2014. Virus-specific memory CD8 T cells provide substantial protection from lethal severe acute respiratory syndrome coronavirus infection. J Virol 88:11034–11044. doi: 10.1128/JVI.01505-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Channappanavar R, Zhao J, Perlman S. 2014. T cell-mediated immune response to respiratory coronaviruses. Immunol Res 59:118–128. doi: 10.1007/s12026-014-8534-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Y, Liu D, Shi W, Lu R, Wang W, Zhao Y, Deng Y, Zhou W, Ren H, Wu J, Wang Y, Wu G, Gao GF, Tan W. 2015. Origin and possible genetic recombination of the Middle East respiratory syndrome coronavirus from the first imported case in China: phylogenetics and coalescence analysis. mBio 6:e01280-15. doi: 10.1128/mBio.01280-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Almazán F, DeDiego ML, Sola I, Zuñiga S, Nieto-Torres JL, Marquez-Jurado S, Andrés G, Enjuanes L. 2013. Engineering a replication-competent, propagation-defective Middle East respiratory syndrome coronavirus as a vaccine candidate. mBio 4:e00650-13. doi: 10.1128/mBio.00650-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 56.Gupta R, Jung E, Brunak S. 2004. Prediction of N-glycosylation sites in human proteins. DTU Health Tech, Technical University of Denmark, Lyngby, Denmark: https://www.cbs.dtu.dk/services/NetNGlyc/. [Google Scholar]
- 57.Steentoft C, Vakhrushev SY, Joshi HJ, Kong Y, Vester-Christensen MB, Schjoldager KT-BG, Lavrsen K, Dabelsteen S, Pedersen NB, Marcos-Silva L, Gupta R, Bennett EP, Mandel U, Brunak S, Wandall HH, Levery SB, Clausen H. 2013. Precision mapping of the human O‐GalNAc glycoproteome through SimpleCell technology. EMBO J 32:1478–1488. doi: 10.1038/emboj.2013.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trandem K, Jin Q, Weiss KA, James BR, Zhao J, Perlman S. 2011. Virally expressed interleukin-10 ameliorates acute encephalomyelitis and chronic demyelination in coronavirus-infected mice. J Virol 85:6822–6831. doi: 10.1128/JVI.00510-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.