Abstract
CD1d-restricted Vα14 invariant natural killer T (iNKT) cells play an important role in the regulation of diverse immune responses. MicroRNA mediated RNA interference is emerging as a crucial regulatory mechanism in the control of iNKT cells differentiation and function. Yet, roles of specific microRNAs in the development and function of iNKT cells remain to be further addressed. Here, we identified the gradually increased expression of miR-150 during the maturation of iNKT cells in thymus. Using miR-150 knockout (KO) mice, we found that miR-150 deletion resulted in an interruption of iNKT cell final maturation in both thymus and periphery. Upon activation, iNKT cells from miR-150KO mice showed significantly increased IFN-γ production compared to wild type (WT) iNKT cells. Bone marrow transferring experiments demonstrated the cell-intrinsic characteristics of iNKT cell maturation and functional defects in mice lacking miR-150. Furthermore, miR-150 target c-Myb was significantly up-regulated in miR-150KO iNKT cells, which potentially contribute to iNKT cell defects in miR-150KO mice. Our data define a specific role of miR-150 in the development and function of iNKT cells.
Introduction
CD1d-restricted Vα14 invariant natural killer T (iNKT) cells are a conserved lymphocyte lineage that co-express a T cell receptor (TCR) as well as several receptors identified on NK cells, such as NK1.1, interleukin-2 (IL-2)/15Rβ chain (CD122), and various Ly49 molecules (1–3). iNKT cells are developed from CD4+CD8+ double-positive (DP) thymus precursors and are characterized by the expression of the homologous invariant Vα14-Jα18/Vβ8 and Vα24-Jα18/Vβ11 TCR chains in mice and humans, respectively (4–6). Rare DP-precursor cells that express Vα14-Jα18 TCRα chain are first positively selected by CD1d, a non classical MHC-I like molecule. iNKT cells then undergo a development and maturation process involving the sequential down regulation of CD24 and up-regulation of CD44 and NK1.1 (7–9). The first stage of iNKT cells (stage1) are defined as CD24low, CD44low, and NK1.1–. Ontogeny and transfer studies indicated a developmental sequence from CD44low NK1.1– to CD44high NK1.1– (stage 2). At this latter stage, iNKT cells are still considered immature but can leave the thymus and colonize peripheral organs, where they reside mostly in the liver, spleen and bone marrow, and are less abundant in the lymph nodes. The final maturation stage, which occurs either in the thymus or in the peripheral organs, is defined by the expression of NK1.1 (stage 3) (8, 10). In addition to the acquisition of NK1.1 expression, the final maturation of iNKT cells is also accompanied by the up-regulation of several other cell-surface receptors, including CD122, CD69, Ly49C/I, and Ly49G2 (11, 12). Mature CD1d-dependent iNKT cells exhibit a memory phenotype (CD62L-CD69+ CD44hi) and can be either CD4+ single positive (SP) or CD4-CD8- double-negative (DN) cells (13). The CD1d-restricted glycosphingolipid antigen alpha-GalactosylCeramide (αGalCer) potently activates iNKT cells, triggering within hours the copious production of a wide range of Th1 and Th2 cytokines such as IFN-γ and IL-4 (14). The early and potent cytokine secretion by Vα14 iNKT cells following TCR stimulation provides an important link between the innate and adaptive immune systems. Consistent with this, Vα14 iNKT cells appear to play an important role in regulating early immune responses to infections, maintenance of self tolerance, and anti-tumor functions (15–19).
The development of iNKT cells is controlled by different transcription factors, cytokines, and signaling molecules involved in the SLAM/Fyn/SAP/PKC and NF-κB signaling pathways. Deleting just one of these molecules impairs iNKT cell generation and/or development, indicating that multiple regulation mechanisms concomitantly control the development of iNKT cells (10, 20–24). Small microRNAs (miRNAs) are short non-coding RNAs (20–23 nt) that negatively regulate gene expression by inducing the degradation or translational repression of the target protein-coding mRNAs (25). Recently, our laboratory and another group found that deletion of Dicer, the RNase III enzyme essential for the processing of mature functional miRNAs, results in a profound interruption of iNKT cell development, maturation and function (26–28). However, little insight has been made concerning the role of specific miRNAs in the development and function of iNKT cells. MiRNA miR-150 is selectively expressed in mature B and T cells, but not their progenitors. Deficiency of miR-150 blocks B1 cell expansion and enhances humoral immune response (29, 30). In this report, we found that the expression of miR-150 was up-regulated during iNKT cell maturation and activation. To test the potential role of miR-150 in iNKT cells, we compared iNKT cell development and function between miR-150 knockout (KO) and wild type (WT) mice. We found that miR-150 deletion impaired iNKT cell final maturation in both thymus and peripheral lymphoid organs, and that iNKT cells from miR-150KO mice showed increased cytokine production capacity. Furthermore, bone marrow transfer experiments indicated that the maturation and functional changes of iNKT cells in miR-150KO mice are intrinsic to iNKT cells. Together, our results define a specific role of miR-150 in the development and function of iNKT cells.
Materials and Methods
Mice
Conventional miR-150KO mice in C57BL/6 background and WT C57BL/6 mice were purchased from the Jackson Laboratory (Bar Harbor, ME) and bred in our animal facility. In this study, 6 to 10-wk-old age and sex-matched miR-150KO and WT mice were used. Handling of mice and experimental procedures were in accordance with requirements of the Institutional Animal Care and Use Committee.
Genotyping
MiR-150KO mice were genotyped using the following PCR primer pairs: 5 - CAAGGACAGGAACCCTTCAGCA-3 and 5-CCATGATGCCTGGAAGACATTTC-3. The miR-150 deletion allele produced a 262-bp PCR product; whereas the wild type allele resulted in an 866-bp product.
Flow cytometry
Single-cell suspensions were washed twice with staining buffer (PBS, 2% FCS) and incubated with Fc Block (clone 2.4G2). Cells were stained with PBS57–CD1d tetramers as described (31). The following conjugated monoclonal antibodies (mAbs) were used: NK1.1 (PK136), TCR-β (H57–597), CD44 (IM7), CD122 (5H4), CD69 (H1.2F3), B220 (RA3–6B2), LY49C(5E6), LY 49G2(4D11); CD1d (1B1), CD8 (53–6.7), CD4 (RM4–5), CD25 (PC61), FoxP3 (FJK-16s), IL-4 (11B11), and IFN-γ (XMG1.2). All mAbs were from BD Biosciences or eBioscience. Data were analyzed using CELLQuest Pro (BD Biosciences) or FlowJo software. Apoptosis assays were carried out by staining with Annexin V (BD Biosciences) according to the manufacturer’s instructions. For iNKT cell proliferation in vivo, as revealed by incorporation of Bromodeoxyuridine (BrdU), BrdU (BD Biosciences) was injected at 1 mg/mouse (i.p.). After 24 hours of BrdU injection, thymocytes were analyzed using FITC BrdU Flow Kit (BD Biosciences) as per manufacturer’s protocol.
In vivo α-GalCer-induced activation assay
Two micrograms of α-GalCer or vehicle in 100 ul of PBS were injected into the tail vein. For intracellular cytokine staining, spleen cells were collected at 2 hours after injection and cultured in T cell medium (RPMI 1640 with 10% FCS, Hepes, penicillin and streptomycin, pyruvate, nonessential amino acids, L-glutamine, and 2-ME). Golgistop was added to a final concentration of 3uM, and the cells were incubated for an additional hour. Cells were extracellularly stained with anti-TCRβ and CD1d-tetramer. After washing and fixing with 2% paraformaldehyde, cells were permeabilized with 0.1% saponin and stained with anti-IFN γ and anti-IL4 Abs and analyzed by flow cytometry.
In vitro PMA and inomycin activation assay
Spleen cells from WT and miR-150 KO mice were cultured in T cell medium (RPMI 1640 with 10% FCS, Hepes, penicillin and streptomycin, pyruvate, nonessential amino acids, Lglutamine, and2-ME) in the presence of PMA (50ng/ml) and Inomycin (1uM) for 1 hour, Golgistop was added to a final concentration of 3uM, and the cells were incubated for an additional 2 hours. IFN γ and anti-IL4 were detected by intracellular staining and flow cytometry.
INKT cell sorting
Total thymocytes and spleen cells from normal C57BL/6 mice were first stained with anti-mouse CD8-biotin (for thymocytes) or anti-mouse CD8-biotin and anti-mouse B220-biotin antibodies (for spleen cells), and CD8+ T cells and B220+ cells were then depleted with anti-biotin beads using autoMACS ™ (Miltenyi Biotec). Negatively selected cells were then stained with TCR-β antibody, α-GalCer/CD1d tetramers, anti-mouse NK1.1 and CD44 antibodies. Total or different stages of iNKT cells were then sorted by BD FACS AriaII™.
Real-time RT-PCR
RNAs from total thymocytes, spleen T cells and iNKT cells from miR-150KO or WT control mice were purified, respectively, using the Ambion mirVana miRNA isolation kit (Ambion) according to the manufacturer’s instructions. The expression of miR-150 was examined using the Applied Biosystems TaqMan MicroRNA Assay kit (Applied Biosystems, Foster City, CA) according to the manufacturer’s instructions; snoRNU202 was used as endogenous control. c-Myb (Mm00501741) and GAPDH TaqMan real time PCR primers and probes were purchased from Applied Biosystems. PCR amplification was carried out on the Applied Biosystems 7900 Real-Time PCR system, relative quantization using the ĈT value in miR-150KO versus WT control mice was carried out and fold changes were calculated.
Mixed bone marrow transfer experiments
To generate bone marrow chimeras, 6–8 week-old B6.SJL recipient mice (CD45.1+) were lethally irradiated initially with 900 rads. Donor bone marrows was harvested from age- and sex-matched B6.SJL and miR-150KO mice (CD45.2+) by flushing with a syringe containing sterile basal tissue culture medium. After erythrocyte lysis, mature T cells were depleted by biotin-conjugated anti-mouse CD3 (BD Biosciences) mAbs and anti-biotin magnetic beads (Miltenyi Biotec) from each donor bone marrows, using an AutoMACS sorter (Miltenyi Biotec.). Over 90% of mature T cells depletion was confirmed by flow cytometry. CD45.1+ B6.SJL and CD45.2+ miR-150KO mice bone marrows were mixed at a 1:1 ratio and 10 million cells per mouse (in a volume of 100uL) were then injected into the irradiated recipients by tail vein. The chimeras were analyzed 8 weeks after reconstitution.
Statistical analysis
Statistical analysis was performed with Prism 5.0 (GraphPad Software). The two-tailed Student’s t test was used. Differences were considered statistically significant when P < 0.05.
Results
Stage-specific expression of miR-150 during iNKT cell development
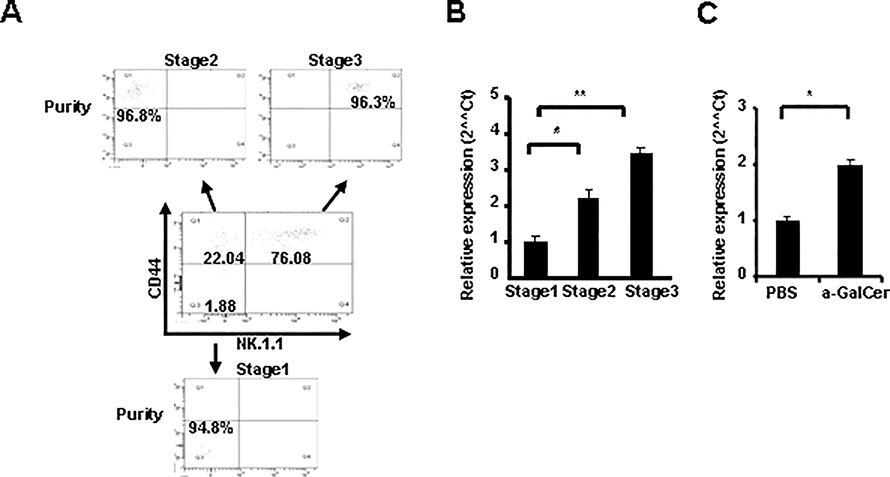
To detect miR-150 expression during the development of iNKT cell, thymocytes from normal C57BL/6 mice were stained with antibodies against TCRβ, PBS57-CD1d Tetramer, CD44 and NK1.1, different stage of iNKT cells were sorted based on CD44 and NK1.1 expression (Fig. 1A). MiR-150 expression was then detected by Taqman real-time PCR. As shown in Fig. 1B, miR-150 expression was detectable in all three stages of iNKT cells. However, miR-150 expression increased gradually during the maturation of iNKT cells and reached the highest level at the final maturation stage 3 (CD44+NK1.1+), consistent with recent findings by Bezman at al (32). INKT cells from spleen have a relatively higher miR-150 expression level compared to thymus iNKT cells, and miR-150 is also highly expressed in conventional thymic and splenic T cells (Supplemental Fig. 1A). This is consistent with previous report that miR-150 is highly expressed in mature and resting lymphocytes (29). Nevertheless, in contrast with the report showing down-regulated miR-150 in B cells after activation (29), in vivo stimulation of iNKT cells with a-GalCer for 3 days resulted in a significantly increased miR-150 expression in spleen iNKT cells (Fig. 1C). Thus, the stage-specific expression pattern of miR-150 during iNKT cell development and activation suggests its potential involvement in iNKT cell development and function.
FIGURE 1.
Expression of miR-150 is up-regulated during the development and activation of iNKT cells. A, Different developmental stages (CD44−NK1.1−, CD44+NK1.1− and CD44+NK1.1+) of thymic iNKT cells were sorted from C57BL/6 mice. B, Taqman real-time PCR analysis of miR-150 transcript in CD44−NK1.1−, CD44+NK1.1− and CD44+NK1.1+ iNKT cells from the thymuses of C57BL/6 mice. Results were the mean of triplicate and normalized to a control gene (snoRNU 202). Error bars are SD. * P <0.05; ** P <0.01. Results are representatives of three independent experiments. C, C57BL/6 mice were stimulated in vivo with α -GalCer for 3 days, spleen iNKT cells were sorted and RNA was extracted. MiR-150 expression was detected by Taqman real-time PCR. Results were the mean of triplicate and normalized to a control gene (snoRNU 202). Error bars are SD. Results are representatives of three independent experiments.
Defective iNKT cell development in miR-150KO mice
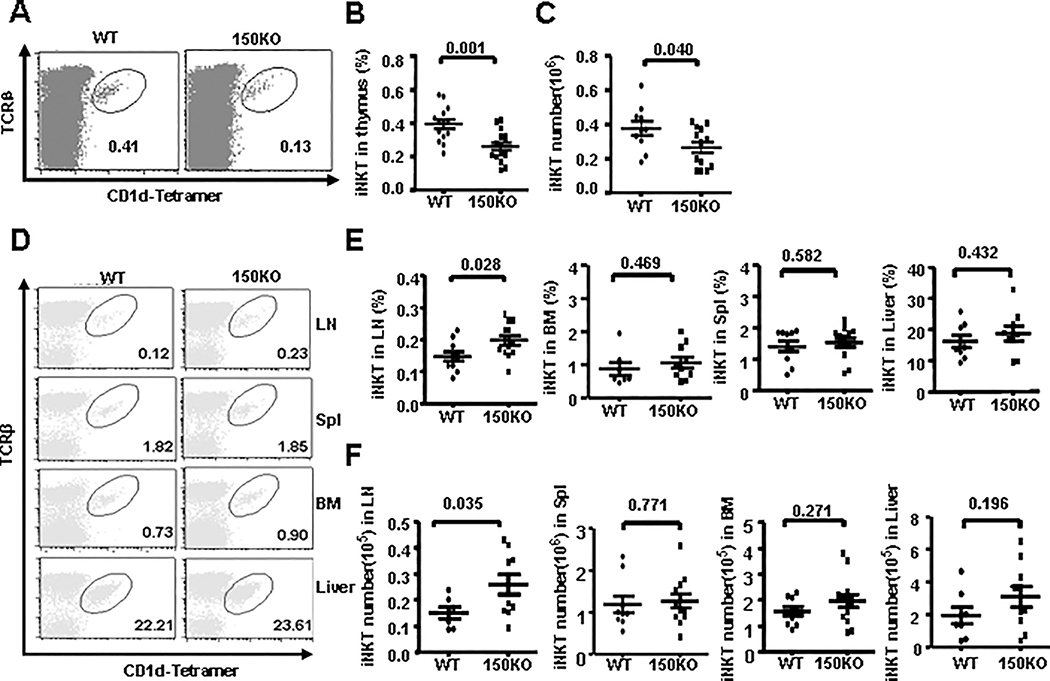
To assess the role of miR-150 in iNKT cell development, conventional miR-150KO mice were evaluated and compared with age and sex-matched WT B6 mice. Taqman real-time PCR analysis confirmed the substantial deletion of miR-150 in the thymus and spleen of miR-150KO mice compared with that of WT control mice (Supplemental Fig. 1B). To detect the possible involvement of miR-150 in conventional T cell development, we compared the ratio and number of conventional T cell subsets in the thymus including CD4+ and CD8+ single-positive (SP), CD4+CD8+ double-positive (DP) and CD4-CD8- double negative (DN) T cells between miR-150 KO and WT mice. No substantial difference was identified (Supplemental Fig. 2A–B). Early T cell precursors, CD4-CD8- DN T cells, progress through the CD44+CD25-(DN1) and the CD44+CD25+(DN2) stages to the CD44-CD25-(DN4) stage. To further dissect the early T cell development profile in miR-150KO mice, we gated DN thymocytes and analyzed their CD44 and CD25 expression profiles (Supplemental Fig. 2C). The miR-150KO mice showed a DN thymocyte developmental profile comparable to that of WT control mice, indicating that miR-150 deficiency did not affect conventional T cell development. In addition, we also found no significant changes of CD4+Foxp3+ regulatory T cell and γδ T cell ratios in the thymus of miR-150KO mice compared to that of WT control mice (Supplemental Fig. 2D–E). Interestingly, we found significant reduction of the percentage (P = 0.001) and cell number (P = 0.04) of thymus iNKT cells stained by anti-TCRβ and PBS57–CD1d tetramers in the miR-150KO mice compared to that in WT mice (Fig. 2A–C). The iNKT cells from spleen, lymph nodes, bone marrow, and liver were further analyzed. As shown in Fig. 2D–F, the frequencies and number of iNKT cells in the spleen, bone marrow, and liver from miR-150KO mice were comparable with that from WT control mice. However, significantly increased frequency (P = 0.028) and number (P = 0.035) of iNKT cells were observed in the lymph nodes from miR-150KO mice compared to that from WT control mice. Thus, deficiency of miR-150 causes an impairment of development in thymus iNKT cells, but not in conventional T cell, CD4+Foxp3+ regulatory T cells, and γδ T cells.
FIGURE 2.
Defective iNKT cell development in miR-150KO mice. A, Representative dot plots of thymus iNKT cells stained by anti- TCRβ Ab and CD1d-Tetramer from miR-150KO and WT mice. B and C, The frequency (B) and number(C) of TCRβ+CD1d-Tetramer+ iNKT cells in the thymus of miR-150KO and WT mice. Each point represents one individual mouse and the mean values are indicated by the middle horizontal lines from 3–5 independent experiments (3–5 mice/experiment). D, The representative staining of iNKT cells (gated on B220 negative cells) in lymph nodes, spleen, bone marrow, and liver from miR-150KO and WT mice. E and F, The frequency (E) and cell number (F) of iNKT cells in the lymph nodes, spleen, bone marrow and liver of miR-150KO and WT mice, Each point represents one individual mouse and the mean values are indicated by middle horizontal lines from 3–5 independent experiments (3–5 mice/experiment). Statistical analysis was performed with Prism 5.0 (GraphPad Software). Differences were considered statistically significant when P < 0.05.
Defective iNKT cell maturation in miR-150KO mice
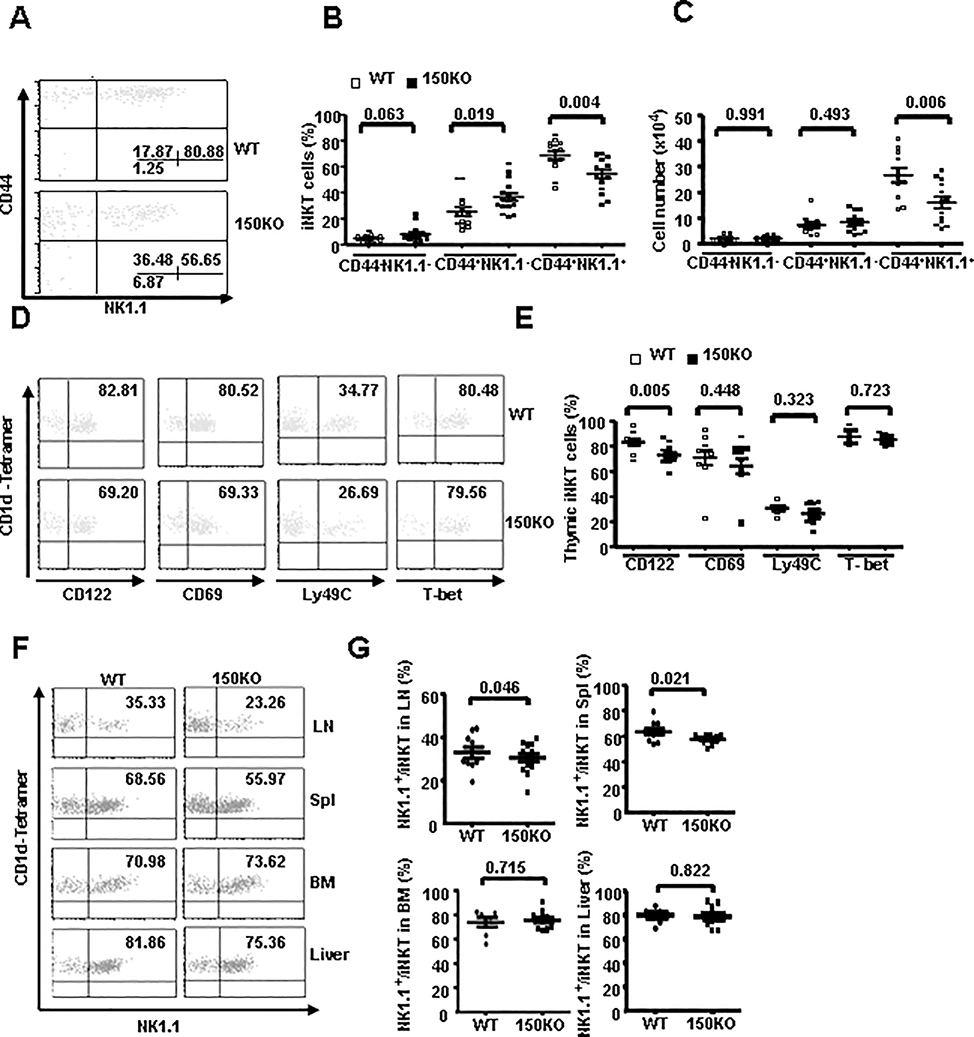
Accumulated studies have indicated that iNKT cells undergo a series of maturation stages in the thymus immediately after positive selection, based on their CD44 and NK1.1 expressions. Given that iNKT cells exhibit a stage-specific miR-150 expression pattern, the CD44/NK1.1 profile of CD1d-tetramer+ thymocytes was analyzed to test the potential role of miR-150 in iNKT cell maturation. We found that the frequency of mature NK1.1+CD44+ iNKT cells deceased significantly (P = 0.004), while the semi-mature NK1.1-CD44+ stage 2 iNKT cells increased significantly (P = 0.019), but the early immature NK1.1-CD44- iNKT cells remain unchanged in miR-150KO mice compare to WT mice (Fig. 3A–B). Furthermore, the number of mature NK1.1+CD44+ iNKT cells decreased significantly (P = 0.006) in miR-150KO mice compare to that of WT mice, while the number of NK1.1-CD44- and NK1.1-CD44+ iNKT cells was comparable between miR-150KO and WT control mice (Fig. 3C). The development and maturation of iNKT cells is also accompanied by the up-regulation of several other cell-surface receptors, such as CD122, CD69 and Ly-49 in addition to NK1.1, and the final steps of iNKT cell development also need the transcription factor T-bet and cytokine signaling initiated by IL-15 (8, 33, 34). To further dissect the developmental impairment and the potential molecular mechanisms involved in iNKT cell development in miR-150KO mice, the expression levels of these molecules in iNKT cells were evaluated and compared between miR-150KO and WT control mice. As shown in Fig. 3D–E, the frequency of CD122 positive iNKT cells was significantly decreased in miR-150KO mice compared to WT control mice (P = 0.005), consistent with the defective iNKT cell maturation based on NK1.1 expression. Interestingly, the expressions of CD69, Ly49C, and T-bet in thymus iNKT cells were comparable between miR-150KO and WT control mice. Thus, miR-150KO mice have a partial defect in iNKT cell maturation at the late stage, suggesting that miR-150 regulates thymus iNKT cell development and maturation.
FIGURE 3.
Defective iNKT cell maturation in miR-150KO mice. A, Dot plots depict CD44 and NK1.1 expression in thymus iNKT cells of miR-150KO and WT control mice. B-C, events showed the percentage (B) and number (C) of iNKT cell sub-populations based on their CD44 and NK1.1 expression patterns gated on TCRβ and CD1d-Tetramer double-positive iNKT cells in the thymus. D, Dot plots depict CD122, CD69, ly49C and T-bet expression in the thymus iNKT cells of miR-150KO and WT control mice. E, Percentage of CD122, CD69, Ly49c and T-bet positive cells gated on TCRβ and CD1d-Tetramer double-positive iNKT cells in the thymus iNKT cells of miR-150KO and WT control mice. Each point represents one individual mouse and the mean values are indicated by the middle horizontal lines from 3–5 independent experiments (3–5 mice/experiment). F, Representative dot plots of NK1.1 staining (gated on B200 negative iNKT cells) in lymph nodes, spleen, bone marrow and liver of miR-150KO and WT control mice. G, Percentage of NK1.1 positive cells gated on lymph nodes, spleen, bone marrow and liver of miR-150KO and WT control mice Each point represents one individual mouse and the mean values are indicated by horizontal lines from 3–5 independent experiments (3–5 mice/experiment).
Immature iNKT cells also leave the thymus and are matured in the peripheral immune organs. Using NK1.1 as a peripheral iNKT cell maturation marker, peripheral iNKT cells maturation status were further evaluated in miR-150KO mice. As shown in Fig. 3F–G, iNKT cell maturation was slightly but significantly interrupted in spleen and lymph nodes in miR-150KO mice (P = 0.021 and 0.046, respectively). Together, our data suggest that miR-150 also regulates peripheral iNKT cell maturation.
MiR-150 contributes to thymus iNKT cell survival during iNKT cell development
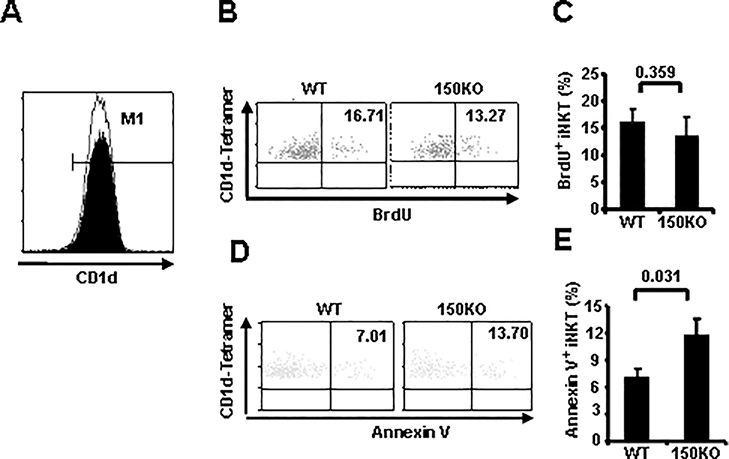
CD1d expressed in CD4+CD8+ double-positive thymocytes controls iNKT selection and development, and mice lacking CD1d have severe defects in iNKT development (35). To ascertain that the impaired iNKT cell development was not due to defective CD1d expression, thymocytes from miR-150KO and WT control mice were compared for CD1d expressions. As shown in Fig. 4A, CD1d expressions were found to be almost equivalent between miR-150KO and WT mice, ruling out the possibility of CD1d deficiency as a cause of the thymic iNKT developmental defects in miR-150KO mice.
FIGURE 4.
MiR-150 contributes to thymus iNKT cell survival during iNKT cell development.
A, Histogram plots depict CD1d expression in CD4+CD8+ (DP) thymocytes of WT control (filled histogram) and miR-150KO mice (open histogram). Data are representative of three independent experiments (3–5 mice/experiment). B, WT control and miR-150KO mice were injected with BrdU, 24 hours later, thymocytes were isolated and stained for TCRβ, CD1d-Tetramer, and BrdU Abs. BrdU positive cells in gated TCRβ+-Tetramer+ double positive (iNKT) cells are shown. C, Percentage of BrdU positive cells in the thymus iNKT cells of WT control and miR-150 KO mice are shown. Results are representatives of three independent experiments (3–5 mice/experiment). D, Thymocytes were stained for TCRβ, CD1d-Tetramer, and Annexin V. Gated TCRβ+ -Tetramer+ double positive (iNKT) cells are shown. E, Percentage of Annexin V positive cells in gated thymus iNKT cells of WT control and miR-150KO mice. Data are representative of two independent experiments (3–5 mice/experiment).
To further dissect the mechanisms involved in thymus iNKT cell homeostasis, proliferation and apoptosis of thymus iNKT cells were evaluated in miR-150KO mice. To examine the former possibility, we injected miR-150KO and WT mice with BrdU and determined the rate of iNKT cell proliferation in the thymus. As shown in Fig. 4B–C, thymus iNKT cells from miR-150KO mice incorporated BrdU at a comparable rate compared to WT control mice. Thus, the disparity between the comparable proliferation observed and the overall decreased numbers of iNKT cells in the thymus suggested that increased cell death might be occurring in iNKT cells from miR-150KO mice. Therefore, fresh thymocytes from WT and miR-150KO mice were stained with the apoptotic marker Annexin V to assess levels of cell apoptosis. As shown in Fig. 4D–E, miR-150KO mice showed a higher frequency of Annexin V positive iNKT cells than that of WT control mice (P = 0.031). Collectively, these data suggest that miR-150 regulates thymus iNKT cell survival, and may not be required for iNKT cell selection and proliferation.
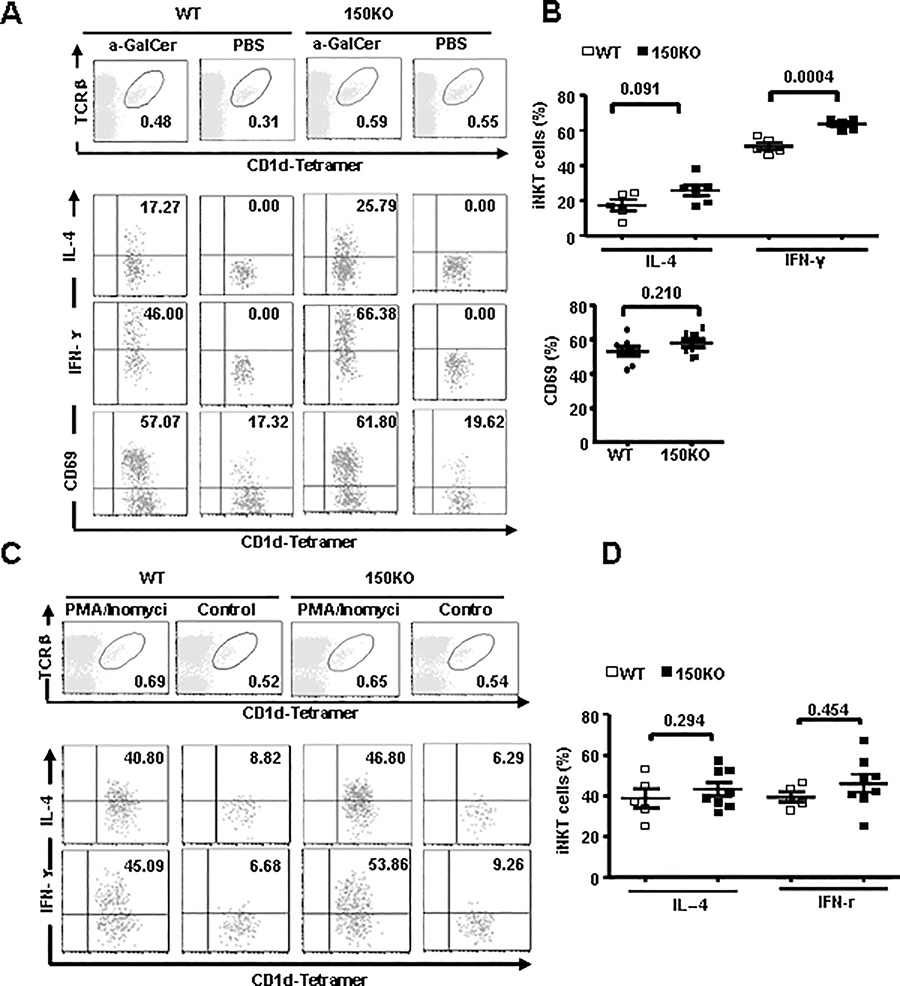
Increased α-GalCer-mediated iNKT cell IFN-γ production in miR-150KO mice
The prompt production of large amounts of cytokines in response to TCR signaling is a unique feature of iNKT cells, which is closely related to their potent immune regulatory functions (36). To investigate the role of miR-150 in iNKT cell function, we injected mice with α-GalCer and evaluated the production of cytokines by iNKT cells with intracellular cytokine staining. As showed in Fig. 5A–B, two hours after in vivo α-GalCer treatment, more spleen iNKT cells from miR-150KO mice produced IL-4 and IFN-γ compared to that from WT control mice, especially for IFN-γ producing NKT cells (P = 0.0004), suggesting the increased iNKT cell responsiveness to a-GalCer stimulation in the miR-150KO mice. However, CD69 expression in iNKT cells from miR-150 KO and WT mice were comparable. Interestingly, when spleen iNKT cells were stimulated in vitro with phorbol myristate acetate (PMA) and ionomycin, which by pass proximal TCR-mediated signaling events, comparable numbers of both IFN-γ and IL-4 producing iNKT cells were detected in miR-150KO and WT control mice (Fig. 5C–D). These results suggest that miR-150 may regulate iNKT cell cytokine production, possibly through targeting the upstream TCR-mediated signaling molecular pathways.
FIGURE 5.
Increased α-GalCer-dependent iNKT cell function in the miR-150 KO iNKT cells. A, Intracellular staining analysis of IL-4, IFN-γ and CD69 expression from spleen iNKT cells of WT control and miR-150KOmice after in vivo α-GalCer stimulation for 2 hours. B, Percentage of IL-4 and IFN-γ positive iNKT cells, and CD69 positive iNKT cells in the WT and miR-150KO mice after in vivo α-GalCer stimulation for 2 hours. Results are representative of 3 independent experiments (3–5 mice/experiment). C, Whole splenocytes from WT control and miR-150KO mice were treated with PMA and Ionomycin in vitro for 3 hours. IL-4 and INF-γ expression in spleen iNKT cells were analyzed by intracellular staining. D, Percentage of IL-4 and IFN-γ positive iNKT cells in the WT control and miR-150KOmice after PMA and Ionomycin treatment in vitro for 3 hours are shown. Results are representative of 3–5 independent experiments (3–5 mice/experiment).
MiR-150 deficiency affects iNKT cell maturation and function via a cell-autonomous mechanism
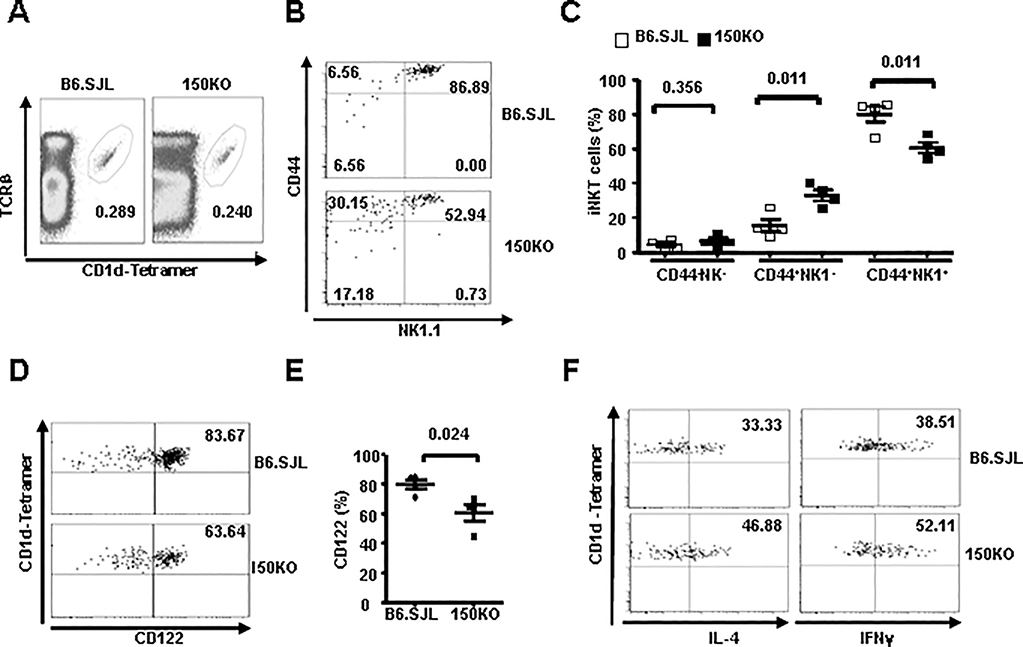
Accumulated studies suggest that iNKT cell development and function are regulated by both the bone marrow and microenvironments. To determine whether the altered iNKT cell development and cytokine secretion in miR-150KO mice is cell intrinsic or related to the defects in their environment, we used a mixed bone marrow transfer model, in which both WT and miR-150KO iNKT precursors develop in the same WT microenvironment. B6.SJL (CD45.1+) mice were lethally irradiated and reconstituted with a 1:1 mixture of bone marrow cells from the age- and sex-matched B6.SJL and miR-150KO mice (CD45.2+). iNKT cells development, maturation and cytokine production capacity were analyzed after 8 weeks post the reconstitution. As shown in Fig. 6A, the bone marrow from miR-150KO mice reconstituted iNKT cells comparably to that from bone marrow of B6.SJL mice. However, significantly decreased mature CD44+NK1.1+ and increased immature CD44+NK1.1- iNKT cell frequency (P = 0.011) appeared in miR-150KO bone marrow-derived iNKT cells compared to that form B6.SJL mice (Fig. 6B–C). In addition, miR-150KO bone marrow-derived iNKT cells showed significantly reduced CD122 expression compared to that from B6.SJL mice (P = 0.024) (Fig. 6D–E). Furthermore, as shown in Fig. 6F, more IFN-γ and IL-4 producing iNKT cells were observed in miR-150KO bone marrow derived iNKT cells compare to that from B6.SJL bone marrow after in vivo α-GalCer stimulation. Thus, the iNKT cell phenotypes in the bone marrow chimeras, including interrupted iNKT cell final stage maturation, reduced CD122 expression, and elevated α-GalCer-induced cytokine production, are consistent with the iNKT phenotypes identified in miR-150 KO mice. These observations therefore indicate the cell-intrinsic nature of the maturation and functional changes of iNKT cells in mice lacking miR-150.
FIGURE 6.
Defective iNKT cell development but increased iNKT cell function is cell intrinsic in miR-150KO mice. CD45.2+ miR-150KO and CD45.1+ B6.SJL bone marrows were mixed at a 1:1 ratio and transferred into lethally irradiated B6.SJL hosts. Eight weeks later, reconstituted animals were analyzed by flow cytometry. A, Frequency of thymus iNKT cells originated from the CD45.1+ B6.SJL bone marrows and CD45.2+ miR-150KO bone marrows. B, Flow cytometry analysis of iNKT developmental subsets gated from the B6.SJL and miR-150KO derived iNKT cells. C, Percentage of each iNKT developmental subset in the total iNKT cells of B6.SJL and miR-150KO mice are shown. D, Flow cytometry analysis of CD122 expression in the B6.SJL and miR-150KO derived iNKT cells. E, Percentage of CD122 positive cell in the B6.SJL and miR-150KO derived iNKT cells. F, Intracellular staining analysis of IL-4 and IFNγ expression from B6.SJL and miR-150KO derived spleen iNKT cells after in vivo α-GalCer stimulation for 2 hours. Data are representative of two independent experiments with the same trend (3–4 mice/experiment).
Up-regulation of c-Myb expression in the thymus iNKT cells of miR-150KO mice
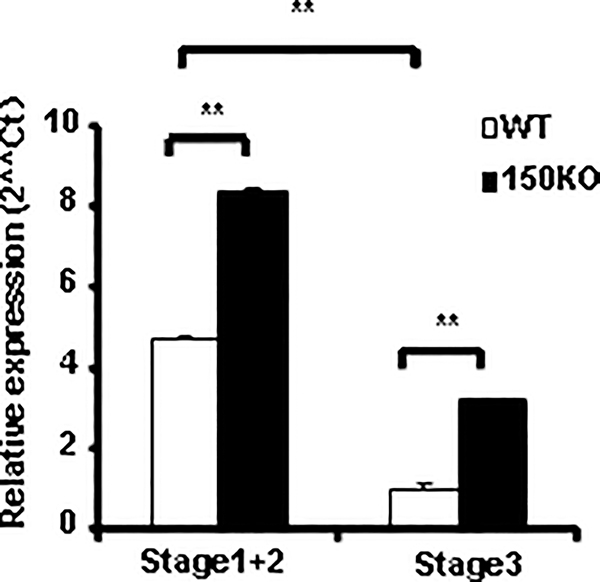
A top predicted target of miR-150 is c-Myb, a transcription factor controlling lymphocyte differentiation and proliferation. Combining loss- and gain-of-function gene targeting approaches for miR-150 with conditional and partial ablation of c-Myb, Xiao et al has recently confirmed that miR-150 indeed controls c-Myb expression in vivo, through which B cell development and differentiation are regulated (30). The c-Myb is highly expressed in lymphocyte progenitors and down regulated upon maturation, and ectopic expression of c-Myb blocked the differentiation of hematopoietic cells. Thus, the appropriate levels of c-Myb are strictly defined at distinct differentiation steps of each hematopoietic cell lineage (30, 37, 38). To investigate the potential involvement of c-Myb in miR-150 mediated iNKT cell regulation, we first evaluated c-Myb expression in thymus immature (stage 1&2) and mature (stage 3) iNKT cells by a TaqMan real-time RT-PCR. As shown in Fig. 7, consistent with the observation in other lymphocytes, c-Myb is also highly expressed in immature iNKT cells and down regulated in mature iNKT cells. Thus, during iNKT cell development in thymus, the dynamic expression pattern of c-Myb is in mirror contrast with that of miR-150 (Fig. 1A and Fig. 7), which may be required for iNKT cell maturation. Furthermore, c-Myb expression was significantly up-regulated in both immature (stage 1&2) and mature (stage 3) iNKT cells from miR-150KO mice compared to that in WT mice (Fig. 7). Thus, these data indicate that c-Myb may serves as one of the potential targets in miR-150-mediated late stage iNKT cell development defect in the thymus.
FIGURE 7.
Up-regulation of c-Myb expression in the thymus iNKT cells of miR-150KO mice. Different stage of iNKT cells were sorted from the thymus of miR-150KO and WT control mice according to their CD44 and NK1.1 expression. To get enough cells for RNA analysis, stage 1 and stage 2 iNKT cells were combined as immature iNKT cells and stage 3 iNKT cells as mature iNKT cells. RNA was extracted from immature and mature iNKT cells, and c-Myb expression was analyzed by Taqman real-time PCR. Results were the representative of two independent experiments.
Discussion
INKT cells are a separate lineage of T lymphocyte that go through a distinct developmental pathway controlled by an unique gene expression pattern (10). In this study, we demonstrate an important role for miR-150 in directing the maturation and function of iNKT cells. Absence of miR-150 results in an impairment of iNKT cell maturation in both thymus and peripheral lymphoid organs and thymus iNKT cell homeostasis, but up-regulates IFN-γ cytokine production capacity. The maturation and functional changes of iNKT cells in miR-150KO mice are iNKT cell intrinsic. As miR-150 deletion resulted in up-regulated c-Myb expression in both immature and mature thymus iNKT cells, c-Myb may be a major target through which miR-150 regulate NKT cell development, maturation and function. During the submission, Lanier’s group (32) reported that miR-150 regulates the development of NK and NKT cells. In line with their findings, we also found the reduced frequency of NK cells in thymus and spleen from miR-150KO mice (data not shown), indicating that appropriate miR-150 expression is necessary for both NK and iNKT cell development. Furthermore, using a mixed bone-marrow transferring chimeras model, they also found that the frequency of thymic miR-150KO iNKT cells from WT:miR150KO chimeras was mildly decreased (1.5 ± 0.2-fold compared to WT iNKT cells), and that loss of miR-150 in iNKT cells revealed a mild reduction in the population of thymic mature NK1.1+ iNKT cells. However, the reductions they detected were not statistically significant. One possible reason is that in their transferring experiments, 4–5 mice per group may not have enough power to detect a difference for this mild reduction, compared to the almost complete blocking of early iNKT cell development in the mice with miR-150 overexpression. Compared to iNKT cells in bone marrow- or thymus-specific total miRNA deletion mouse models (26–28), miR-150 deletion alone is unlikely accountable for the profound interruption of iNKT cell development, maturation and function identified in those mice. This may suggest the potential involvement of multiple miRNAs or groups of miRNAs in iNKT cell developmental and functional regulation, which is also supported by our recent miRNA expression profiles of iNKT cells (unpublished data).
INKT cells differentiate from double positive conventional T cells and develop in thymus, with the final maturation step (NK1.1 expression) occurring in both the thymus and the periphery (1). Numerous transcription factors, cytokines and signaling molecules have been identified as unique requirements for iNKT cell maturation once commitment to the NKT lineage has been confirmed. In particular, transcription factor T-bet is essential for final iNKT cell maturation (10, 34). However, a difference in T-bet expression was not identified in the iNKT cells of miR-150KO mice compared to that of WT control mice, indicating that defective iNKT cell maturation in miR-150KO mice is not T-bet-dependent. NF-kB, IL-15, IL-15Rβ (CD122) and IL-2Rβ also play an essential role in the maturation and overall population size of iNKT cells (39–41). CD122 is up-regulated between stages 2 and 3 of thymus iNKT development. Whether the immature iNKT cell phenotype in miR-150KO mice is induced by a failure to up-regulate CD122 or a developmental blockage needs to be further identified. CD122 is involved in inducing expression of anti-apoptotic proteins Bcl-2 and Bcl-xl and is important for NKT cell homeostasis (42). We have found that thymus iNKT cells from miR-150KO mice showed increased apoptosis compared to that from WT controls, which could result in a decreased iNKT cell ratio and number in miR-150KO mice.
Spleen and lymph nodes iNKT cells from miR-150KO mice showed a decrease in NK1.1 expression akin to the defective maturation of thymus iNKT. Consistent with previous studies showing that immature CD44+NK1.1- iNKT cells preferably accumulate in the lymph nodes (43, 44), lymph nodes of miR-150KO mice have increased iNKT cell frequency compared to that in WT control mice. As thymus and peripheral microenvironments differentially mediated development and maturation of iNKT cells, these results suggest that miR-150 is involved in both thymus and peripheral iNKT cells maturation. Given that iNKT cell maturation defects appear to be cell intrinsic, as demonstrated by our bone marrow chimera experiment (Fig. 6), the differential iNKT cell maturation phenotype in individual peripheral lymphoid organs may come from different migration capacities of iNKT cells toward individual peripheral lymphoid organs.
INKT cells rapidly secrete cytokines upon TCR stimulation, thus modulating acquired immune responses. In this study, we show that in addition to a role in iNKT cell maturation, miR-150 indeed negatively regulate iNKT cell production of IFN-γ and for a lesser extent, IL-4 upon TCR stimulation. It is well known that immature CD44+NK1.1- iNKT cells produce more IL-4, while mature CD44+NK1.1+ iNKT cells produce more IFN-γ (8). Considering that the lower ratio of mature CD44+NK1.1+ iNKT cells present in the miR-150KO mice, the increased IFN-γ production of iNKT cells in miR-150KO mice can not be explained by the defective iNKT cell maturation. Other mechanisms must be involved in miR-150 regulating iNKT cell IFN-γ production. Several transcription factors known to regulate cytokine gene transcription, such as T-bet, GATA-3, and NF-κB have been implicated in iNKT cells (34, 39, 44, 45). In addition, epigenetic regulation of IL-4 and IFN-γ expression is reported to occur during iNKT cell development (46). It was known that Notch signaling regulates IL-4 expression, while GM-CSF signaling controls IL-4 and IFN-γ secretion without disturbing the transcription and translation of these two cytokines (47, 48). Previous studies showed that c-Myb mRNA is induced upon B and T cell activation, while miR-150 expression was reduced upon B cell activation, suggesting the involvement of miR-150 and its target, c-Myb, in T and B cell activation and function (29, 49, 50). In contrast with these results identified in T and B cells, however, iNKT cells stimulated with α-GalCer for 3 days showed increased miR-150 expression (Fig. 1B), suggesting the potentially discrete mechanisms that exist in miR-150-mediated activation and functional regulation of iNKT cells versus T and B cells. The increased iNKT cytokine production upon α-GalCer stimulation instead of PMA/ionomycin stimulation in miR-150KO mice suggest that miR-150 controls IFN-γ production through targeting proximal components of TCR signaling pathway. The detailed mechanisms of miR-150 mediated iNKT cell activation and cytokine production remain to be defined.
The transctiption factor c-Myb is expressed in hematopoietic stem cells and progenitors of all hematopoietic lineages and is required for differentiation along individual hematopoietic cell lineages. In the thymus, DN and DP thymocyte subsets have the highest expression of c-Myb and its expression decreases after positive selection (51, 52). Conditional deletion of c-Myb at various stages of T cell development has suggested that c-Myb influences the DN to DP transition, the survival of DP thymocytes, and the differentiation of CD4+ thymocytes (51). AIberola-Ila’s group recently identified the central role of c-Myb in priming DP thymocytes to enter the iNKT lineage by simultaneously regulating CD1d expression, the half-life of DP cells and expression of SLAMF1, SLAMF6 and SAP (21). MiR-150 is specifically expressed in mature lymphcytes, but not their progenitors, which is opposite to the pattern of c-Myb expression. Combining loss- and gain-of-function gene targeting approaches for miR-150, studies from Rajewsky’s group showed miR-150 controls B cell differentiation through targeting c-Myb (30). In consistent with up-regulated c-Myb expression in early iNKT cells from miR-150KO mice (Fig. 7), we do not expect the abnormal iNKT cell selection and early develop in miR-150KO mice. Most interestingly, Lanier’s group recently reported that over expression of miR-150 dramatically reduced iNKT cell number and blocked iNKT cell development in the early stage, which is related to down-regulated expression of c-Myb (32), further indicating the importance of down-regulation of miR-150 and up-regulation of c-Myb for the early development of iNKT cells.
On the other hand, constitutive expression of c-Myb has been reported to block erythrocyte maturation (53, 54). A more recent study using a dose- and timing-controlled gene expression system showed over expression of c-Myb, however, prevented the terminal differentiation of erythrocytes and megakaryocytes (37), indicating that appropriate levels of c-Myb protein are strictly defined at distinct development and differentiation steps of each hematopoietic cell lineage. In agreement with this notion, our study showed that miR-150 deletion, resulting in up-regulated c-Myb expression level, mediated defective iNKT final maturation and changed function. This suggests the target role of c-Myb for miR-150 in iNKT cells and the necessity of miR-150-mediated c-Myb down-regulation in the terminal maturation of iNKT cells. Therefore, the expression of the correct level at the appropriate time during differentiation seems to be important for c-Myb to regulate development and maturation of iNKT cells, in which miR-150 may be a critical player. However, we can not exclude that other miR-150 potential target genes may also contribute to the iNKT cell phenotypes observed in miR-150 KO mice. For examples, miR-150 was demonstrated to target the transcription factor early growth response 2 (Egr2) and the purinergic P2X7 receptor, which has been suggested in controlling iNKT cell proliferation, apoptosis, activation (55–58). Therefore, more credential evidence is needed to further confirm the target role of c-Myb in miR-150-mediated iNKT cell regulation.
In conclusions, we demonstrated a role for miR-150 in regulating the maturation and function of iNKT cells. Absence of miR-150 results in a moderate interruption of iNKT cell maturation in both thymus and peripheral lymphoid organs and thymus iNKT cell homeostasis, and up-regulates their IFN-γ production capacity. MiR-150-mediated iNKT maturation and function is cell intrinsic. Given the opposite expression levels of miR-150 and c-Myb in iNKT cells during their development and the critical role of miR-150 and c-Myb in iNKT cell development and maturation, it’s likely that the dynamic expression of miR-150 is required for normal NKT cell development through controlling the proper levels of c-Myb expression.
Supplementary Material
Acknowledgements
We thank the National Institutes of Health tetramer facility for CD1d- Tetramer, Matthew Weiland and Min Liu for maintaining mouse colonies and mouse genotyping, and all members of our laboratory for their advice and encouragement.
Source of support
This study was supported by Juvenile Diabetes Research Foundation International Grants 1-2005-039, 5-2006-918, and 5-2006-403, American Diabetes Association Grant 7-05-JF-30, and Henry Ford Immunology Program start-up (T71016 and T71017)
Abbreviations
- α-GalCer
alpha-galactosylceramide
- iNKT
invariant natural killer T cells
- MiR-150
MicroRNA-150
- KO
knock-out
- WT
wild-type
Footnotes
Disclosures
The authors have no financial conflicts of interest
References
- 1.Bendelac A, Savage PB, and Teyton L 2007. The biology of NKT cells. Annu Rev Immunol 25:297–336. [DOI] [PubMed] [Google Scholar]
- 2.Bendelac A, Rivera MN, Park SH, and Roark JH 1997. Mouse CD1-specific NK1 T cells: development, specificity, and function. Annu Rev Immunol 15:535–562. [DOI] [PubMed] [Google Scholar]
- 3.Ho EL, Carayannopoulos LN, Poursine-Laurent J, Kinder J, Plougastel B, Smith HR, and Yokoyama WM 2002. Costimulation of multiple NK cell activation receptors by NKG2D. J Immunol 169:3667–3675. [DOI] [PubMed] [Google Scholar]
- 4.Egawa T, Eberl G, Taniuchi I, Benlagha K, Geissmann F, Hennighausen L, Bendelac A, and Littman DR 2005. Genetic evidence supporting selection of the Valpha14i NKT cell lineage from double-positive thymocyte precursors. Immunity 22:705–716. [DOI] [PubMed] [Google Scholar]
- 5.Dellabona P, Padovan E, Casorati G, Brockhaus M, and Lanzavecchia A 1994. An invariant V alpha 24-J alpha Q/V beta 11 T cell receptor is expressed in all individuals by clonally expanded CD4–8- T cells. J Exp Med 180:1171–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tilloy F, Treiner E, Park SH, Garcia C, Lemonnier F, de la Salle H, Bendelac A, Bonneville M, and Lantz O 1999. An invariant T cell receptor alpha chain defines a novel TAP-independent major histocompatibility complex class Ib-restricted alpha/beta T cell subpopulation in mammals. J Exp Med 189:1907–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gapin L, Matsuda JL, Surh CD, and Kronenberg M 2001. NKT cells derive from double-positive thymocytes that are positively selected by CD1d. Nat Immunol 2:971–978. [DOI] [PubMed] [Google Scholar]
- 8.Benlagha K, Kyin T, Beavis A, Teyton L, and Bendelac A 2002. A thymic precursor to the NK T cell lineage. Science 296:553–555. [DOI] [PubMed] [Google Scholar]
- 9.Pellicci DG, Hammond KJ, Uldrich AP, Baxter AG, Smyth MJ, and Godfrey DI 2002. A natural killer T (NKT) cell developmental pathway iInvolving a thymus-dependent NK1.1(−)CD4(+) CD1d-dependent precursor stage. J Exp Med 195:835–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsuda JL, Zhang Q, Ndonye R, Richardson SK, Howell AR, and Gapin L 2006. T-bet concomitantly controls migration, survival, and effector functions during the development of Valpha14i NKT cells. Blood 107:2797–2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gadue P, and Stein PL 2002. NK T cell precursors exhibit differential cytokine regulation and require Itk for efficient maturation. J Immunol 169:2397–2406. [DOI] [PubMed] [Google Scholar]
- 12.Matsuda JL, Gapin L, Sidobre S, Kieper WC, Tan JT, Ceredig R, Surh CD, and Kronenberg M 2002. Homeostasis of V alpha 14i NKT cells. Nat Immunol 3:966–974. [DOI] [PubMed] [Google Scholar]
- 13.Lantz O, and Bendelac A 1994. An invariant T cell receptor alpha chain is used by a unique subset of major histocompatibility complex class I-specific CD4+ and CD4–8- T cells in mice and humans. J Exp Med 180:1097–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kronenberg M 2005. Toward an understanding of NKT cell biology: progress and paradoxes. Annu Rev Immunol 23:877–900. [DOI] [PubMed] [Google Scholar]
- 15.Tupin E, Kinjo Y, and Kronenberg M 2007. The unique role of natural killer T cells in the response to microorganisms. Nat Rev Microbiol 5:405–417. [DOI] [PubMed] [Google Scholar]
- 16.Lehuen A, Diana J, Zaccone P, and Cooke A 2010. Immune cell crosstalk in type 1 diabetes. Nat Rev Immunol 10:501–513. [DOI] [PubMed] [Google Scholar]
- 17.Aktan I, Chant A, Borg ZD, Damby DE, Leenstra PC, Lilley GW, Petty J, Suratt BT, Teuscher C, Wakeland EK, Poynter ME, and Boyson JE 2010. Slam haplotypes modulate the response to lipopolysaccharide in vivo through control of NKT cell number and function. J Immunol 185:144–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tahir SM, Cheng O, Shaulov A, Koezuka Y, Bubley GJ, Wilson SB, Balk SP, and Exley MA 2001. Loss of IFN-gamma production by invariant NK T cells in advanced cancer. J Immunol 167:4046–4050. [DOI] [PubMed] [Google Scholar]
- 19.Dhodapkar MV, Geller MD, Chang DH, Shimizu K, Fujii S, Dhodapkar KM, and Krasovsky J 2003. A reversible defect in natural killer T cell function characterizes the progression of premalignant to malignant multiple myeloma. J Exp Med 197:1667–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kovalovsky D, Alonzo ES, Uche OU, Eidson M, Nichols KE, and Sant’Angelo DB 2010. PLZF induces the spontaneous acquisition of memory/effector functions in T cells independently of NKT cell-related signals. J Immunol 184:6746–6755. [DOI] [PubMed] [Google Scholar]
- 21.Hu T, Simmons A, Yuan J, Bender TP, and Alberola-Ila J 2010. The transcription factor c-Myb primes CD4+CD8+ immature thymocytes for selection into the iNKT lineage. Nat Immunol 11:435–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwartzberg PL, Mueller KL, Qi H, and Cannons JL 2009. SLAM receptors and SAP influence lymphocyte interactions, development and function. Nat Rev Immunol 9:39–46. [DOI] [PubMed] [Google Scholar]
- 23.Cen O, Ueda A, Guzman L, Jain J, Bassiri H, Nichols KE, and Stein PL 2009. The adaptor molecule signaling lymphocytic activation molecule-associated protein (SAP) regulates IFN-gamma and IL-4 production in V alpha 14 transgenic NKT cells via effects on GATA-3 and T-bet expression. J Immunol 182:1370–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elewaut D, Shaikh RB, Hammond KJ, De Winter H, Leishman AJ, Sidobre S, Turovskaya O, Prigozy TI, Ma L, Banks TA, Lo D, Ware CF, Cheroutre H, and Kronenberg M 2003. NIK-dependent RelB activation defines a unique signaling pathway for the development of V alpha 14i NKT cells. J Exp Med 197:1623–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Winter J, Jung S, Keller S, Gregory RI, and Diederichs S 2009. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol 11:228–234. [DOI] [PubMed] [Google Scholar]
- 26.Fedeli M, Napolitano A, Wong MP, Marcais A, de Lalla C, Colucci F, Merkenschlager M, Dellabona P, and Casorati G 2009. Dicer-dependent microRNA pathway controls invariant NKT cell development. J Immunol 183:2506–2512. [DOI] [PubMed] [Google Scholar]
- 27.Zhou L, Seo KH, He HZ, Pacholczyk R, Meng DM, Li CG, Xu J, She JX, Dong Z, and Mi QS 2009. Tie2cre-induced inactivation of the miRNA-processing enzyme Dicer disrupts invariant NKT cell development. Proc Natl Acad Sci U S A 106:10266–10271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seo KH, Zhou L, Meng D, Xu J, Dong Z, and Mi QS 2010. Loss of microRNAs in thymus perturbs invariant NKT cell development and function. Cell Mol Immunol 7:447–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou B, Wang S, Mayr C, Bartel DP, and Lodish HF 2007. miR-150, a microRNA expressed in mature B and T cells, blocks early B cell development when expressed prematurely. Proc Natl Acad Sci U S A 104:7080–7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, Wang J, Rajewsky N, Bender TP, and Rajewsky K 2007. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell 131:146–159. [DOI] [PubMed] [Google Scholar]
- 31.Sharif S, Arreaza GA, Zucker P, Mi QS, Sondhi J, Naidenko OV, Kronenberg M, Koezuka Y, Delovitch TL, Gombert JM, Leite-De-Moraes M, Gouarin C, Zhu R, Hameg A, Nakayama T, Taniguchi M, Lepault F, Lehuen A, Bach JF, and Herbelin A 2001. Activation of natural killer T cells by alpha-galactosylceramide treatment prevents the onset and recurrence of autoimmune Type 1 diabetes. Nat Med 7:1057–1062. [DOI] [PubMed] [Google Scholar]
- 32.Bezman NA, Chakraborty T, Bender T, and Lanier LL 2011. miR-150 regulates the development of NK and iNKT cells. J Exp Med Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kennedy MK, Glaccum M, Brown SN, Butz EA, Viney JL, Embers M, Matsuki N, Charrier K, Sedger L, Willis CR, Brasel K, Morrissey PJ, Stocking K, Schuh JC, Joyce S, and Peschon JJ 2000. Reversible defects in natural killer and memory CD8 T cell lineages in interleukin 15-deficient mice. J Exp Med 191:771–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Townsend MJ, Weinmann AS, Matsuda JL, Salomon R, Farnham PJ, Biron CA, Gapin L, and Glimcher LH 2004. T-bet regulates the terminal maturation and homeostasis of NK and Valpha14i NKT cells. Immunity 20:477–494. [DOI] [PubMed] [Google Scholar]
- 35.Chen YH, Chiu NM, Mandal M, Wang N, and Wang CR 1997. Impaired NK1+ T cell development and early IL-4 production in CD1-deficient mice. Immunity 6:459–467. [DOI] [PubMed] [Google Scholar]
- 36.Matsuda JL, Naidenko OV, Gapin L, Nakayama T, Taniguchi M, Wang CR, Koezuka Y, and Kronenberg M 2000. Tracking the response of natural killer T cells to a glycolipid antigen using CD1d tetramers. J Exp Med 192:741–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sakamoto H, Dai G, Tsujino K, Hashimoto K, Huang X, Fujimoto T, Mucenski M, Frampton J, and Ogawa M 2006. Proper levels of c-Myb are discretely defined at distinct steps of hematopoietic cell development. Blood 108:896–903. [DOI] [PubMed] [Google Scholar]
- 38.Allen RD 3rd, Bender TP, and Siu G 1999. c-Myb is essential for early T cell development. Genes Dev 13:1073–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stanic AK, Bezbradica JS, Park JJ, Matsuki N, Mora AL, Van Kaer L, Boothby MR, and Joyce S 2004. NF-kappa B controls cell fate specification, survival, and molecular differentiation of immunoregulatory natural T lymphocytes. J Immunol 172:2265–2273. [DOI] [PubMed] [Google Scholar]
- 40.Vallabhapurapu S, Powolny-Budnicka I, Riemann M, Schmid RM, Paxian S, Pfeffer K, Korner H, and Weih F 2008. Rel/NF-kappaB family member RelA regulates NK1.1- to NK1.1+ transition as well as IL-15-induced expansion of NKT cells. Eur J Immunol 38:3508–3519. [DOI] [PubMed] [Google Scholar]
- 41.Ohteki T, Ho S, Suzuki H, Mak TW, and Ohashi PS 1997. Role for IL-15/IL-15 receptor beta-chain in natural killer 1.1+ T cell receptor-alpha beta+ cell development. J Immunol 159:5931–5935. [PubMed] [Google Scholar]
- 42.Cipres A, Gala S, Martinez AC, Merida I, and Williamson P 1999. An IL-2 receptor beta subdomain that controls Bcl-X(L) expression and cell survival. Eur J Immunol 29:1158–1167. [DOI] [PubMed] [Google Scholar]
- 43.Savage AK, Constantinides MG, Han J, Picard D, Martin E, Li B, Lantz O, and Bendelac A 2008. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity 29:391–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim PJ, Pai SY, Brigl M, Besra GS, Gumperz J, and Ho IC 2006. GATA-3 regulates the development and function of invariant NKT cells. J Immunol 177:6650–6659. [DOI] [PubMed] [Google Scholar]
- 45.Au-Yeung BB, and Fowell DJ 2007. A key role for Itk in both IFN gamma and IL-4 production by NKT cells. J Immunol 179:111–119. [DOI] [PubMed] [Google Scholar]
- 46.Stetson DB, Mohrs M, Reinhardt RL, Baron JL, Wang ZE, Gapin L, Kronenberg M, and Locksley RM 2003. Constitutive cytokine mRNAs mark natural killer (NK) and NK T cells poised for rapid effector function. J Exp Med 198:1069–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tanaka S, Tsukada J, Suzuki W, Hayashi K, Tanigaki K, Tsuji M, Inoue H, Honjo T, and Kubo M 2006. The interleukin-4 enhancer CNS-2 is regulated by Notch signals and controls initial expression in NKT cells and memory-type CD4 T cells. Immunity 24:689–701. [DOI] [PubMed] [Google Scholar]
- 48.Bezbradica JS, Gordy LE, Stanic AK, Dragovic S, Hill T, Hawiger J, Unutmaz D, Van Kaer L, and Joyce S 2006. Granulocyte-macrophage colony-stimulating factor regulates effector differentiation of invariant natural killer T cells during thymic ontogeny. Immunity 25:487–497. [DOI] [PubMed] [Google Scholar]
- 49.Golay J, Cusmano G, and Introna M 1992. Independent regulation of c-myc, B-myb, and c-myb gene expression by inducers and inhibitors of proliferation in human B lymphocytes. J Immunol 149:300–308. [PubMed] [Google Scholar]
- 50.Pauza CD 1987. Regulation of human T-lymphocyte gene expression by interleukin 2: immediate-response genes include the proto-oncogene c-myb. Mol Cell Biol 7:342–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bender TP, Kremer CS, Kraus M, Buch T, and Rajewsky K 2004. Critical functions for c-Myb at three checkpoints during thymocyte development. Nat Immunol 5:721–729. [DOI] [PubMed] [Google Scholar]
- 52.Maurice D, Hooper J, Lang G, and Weston K 2007. c-Myb regulates lineage choice in developing thymocytes via its target gene Gata3. EMBO J 26:3629–3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Clarke MF, Kukowska-Latallo JF, Westin E, Smith M, and Prochownik EV 1988. Constitutive expression of a c-myb cDNA blocks Friend murine erythroleukemia cell differentiation. Mol Cell Biol 8:884–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bartunek P, Kralova J, Blendinger G, Dvorak M, and Zenke M 2003. GATA-1 and c-myb crosstalk during red blood cell differentiation through GATA-1 binding sites in the c-myb promoter. Oncogene 22:1927–1935. [DOI] [PubMed] [Google Scholar]
- 55.Wu Q, Jin H, Yang Z, Luo G, Lu Y, Li K, Ren G, Su T, Pan Y, Feng B, Xue Z, Wang X, and Fan D MiR-150 promotes gastric cancer proliferation by negatively regulating the pro-apoptotic gene EGR2. Biochem Biophys Res Commun 392:340–345. [DOI] [PubMed] [Google Scholar]
- 56.Lazarevic V, Zullo AJ, Schweitzer MN, Staton TL, Gallo EM, Crabtree GR, and Glimcher LH 2009. The gene encoding early growth response 2, a target of the transcription factor NFAT, is required for the development and maturation of natural killer T cells. Nat Immunol 10:306–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou L, Qi X, Potashkin JA, Abdul-Karim FW, and Gorodeski GI 2008. MicroRNAs miR-186 and miR-150 down-regulate expression of the pro-apoptotic purinergic P2X7 receptor by activation of instability sites at the 3’-untranslated region of the gene that decrease steady-state levels of the transcript. J Biol Chem 283:28274–28286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kawamura H, Aswad F, Minagawa M, Govindarajan S, and Dennert G 2006. P2X7 receptors regulate NKT cells in autoimmune hepatitis. J Immunol 176:2152–2160. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.