Abstract
Reagents such as Amplex® Red have been developed for detecting hydrogen peroxide (H2O2) and are used to measure the release of H2O2 from biological samples such as mammalian leukocytes undergoing the oxidative burst. Caenorhabditis elegans is commonly used as a model host in the study of interactions with microbial pathogens and releases reactive oxygen species (ROS) as a component of its defense response. We adapted the Amplex® Red Hydrogen Peroxide/Peroxidase Assay Kit to measure H2O2 output from live Caenorhabditis elegans exposed to microbial pathogens. The assay differs from other forms of ROS detection in the worm, like dihydrofluorescein dyes and genetically encoded probes such as HyPer, in that it generally detects released, extracellular ROS rather than intracellular ROS, though the distinction between the two is blurred by the fact that certain species of ROS, including H2O2, can cross membranes. The protocol involves feeding C. elegans on a lawn of the pathogen of interest for a period of time. The animals are then rinsed off the plates in buffer and washed to remove any microbes on their cuticle. Finally, the animals in buffer are distributed into 96-well plates and Amplex® Red and horseradish peroxidase (HRP) are added. Any H2O2 released into the buffer by the worms will react with the Amplex® Red reagent in a 1:1 ratio in the presence of HRP to produce the red fluorescent excitation product resorufin that can be measured fluorometrically or spectrophotometrically, and the amount of H2O2 released can be calculated by comparison to a standard curve. The assay is most appropriate for studies focused on released ROS, and its advantages include ease of use, the ability to use small numbers of animals in a plate reader assay in which measurements can be taken either fluorometrically or spectrophotometrically.
Keywords: Amplex, Hydrogen peroxide, C. elegans, ROS, NADPH oxidase, Oxidative burst, Pathogen
Background
Interest in developing assays to study the amount and type of reactive oxygen species produced by C. elegans was primarily driven by the use of this model organism in studies focused on aging and infection. Oxidative damage caused by ROS has long been hypothesized to contribute to aging, and therefore measuring the intracellular oxidative environment and how it changes over time and in various aging mutants is relevant. Likewise, ROS are generated during the C. elegans immune response where they are capable of acting as signaling or effector molecules. Understanding the sources of ROS and being able to measure their magnitude is therefore of key importance (reviewed by [Labuschagne and Brenkman, 2013; McCallum and Garsin, 2016; Miranda-Vizuete and Veal, 2017 ]). Commonly used methods to measure ROS in C. elegans include dihydrofluorescein dyes, the genetically encoded HyPer probe, and the Amplex® Red assay. Additional methods with more specific applications include MioSox, a modified dihydroethidium (DHE) that is specific for measuring ROS in the mitochondria, and the Grx1-roGFP2 genetically-encoded probe that measures changes in redox potential (Labuschagne and Brenkman, 2013). All current methodologies that measure ROS have caveats that must be carefully considered when deciding what method to use and how to interpret the results.
The dihydrofluorescein dyes, including 2,7-dichlorodihydrofluorescein diacetate (H2DCFDA) commonly used in C. elegans, are colorless and membrane permeable in the reduced form, but become fluorescent and non-membrane permeable when oxidized. Therefore, these dyes can penetrate the cytoplasmic membrane and provide a decent measure of intracellular ROS (Labuschagne and Brenkman, 2013). However, to measure the fluorescence from C. elegans using a plate reader, a large number of animals (1,000) are required ( Schulz et al., 2007 ; Ewald et al., 2017 ). Alternatively, animals can be visualized using fluorescence microscopy (Sem and Rhen, 2012). Two disadvantages to H2DCFDA include the fact that it is not specific to H2O2, and it may exert free radical properties, contributing to the generation of ROS in addition to its measurement (Labuschagne and Brenkman, 2013; Dikalov and Harrison, 2014).
HyPer, in contrast, is considered very specific to H2O2. It consists of a cpYFP (circularly permuted yellow fluorescent protein) inserted into the regulatory domain of OxyR-RD and placed under the control of a C. elegans promoter. Both tissue-specific promoters and non-specific promoters have been utilized in the worm depending on the application ( Back et al., 2012 ; Knoefler et al., 2012 ). The sensor works based on the formation of an intramolecular disulfide bond that occurs upon interaction with H2O2. The strength of the two fluorescent emission peaks released by HyPer changes upon formation of the disulfide bond; one decreases and the other increases in a proportionate manner ( Belousov et al., 2006 ). The advantage of such a ratiometric approach is that quantification can be done independently of the expression levels of the protein and therefore no normalization is required (Labuschagne and Brenkman, 2013). An additional advantage is the ability to monitor hydrogen peroxide levels over time and observe both increases and decreases. Disadvantages include the fact that HyPer is sensitive to pH changes and overexpression of these probes can significantly affect H2O2 redox signaling of the cell potentially confounding results. Furthermore, measuring the emission peaks requires sophisticated microscopy of individual animals or large numbers of animals (1,000/sample) to obtain accurate results from a plate reader (Labuschagne and Brenkman, 2013; Dikalov and Harrison, 2014; Ewald et al., 2017 ).
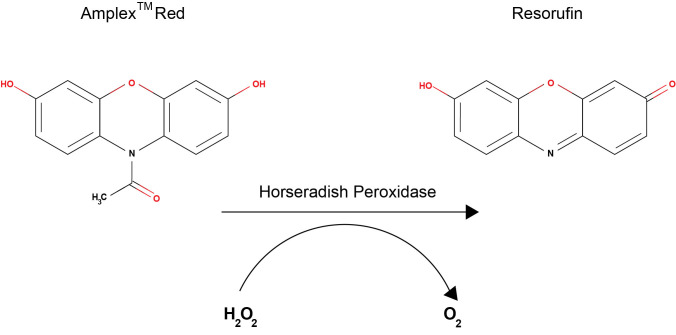
The Amplex® Red assay is based on the oxidation of 10-acetyl-3,7- dihydroxypenoxazine, which is catalyzed by horseradish peroxidase (HRP) in the presence of H2O2 to produce a red fluorescent oxidation product, resorufin, at a 1:1 ratio (Figure 1). Resorufin can be measured both fluorometrically and spectrophotometrically, though the fluorescent measurements are more sensitive and can detect lower levels of H2O2 ( Mohanty et al., 1997 ). The Amplex® Red Hydrogen Peroxide/Peroxidase Assay Kit was first adapted to C. elegans by our group to measure the output of ROS following exposure to microbial pathogens ( Chavez et al., 2007 ; Chavez et al., 2009 ; Tiller and Garsin, 2014; van der Hoeven, et al., 2015; Liu et al., 2019 ). The assay measures the amount of H2O2 excreted by the worm into the surrounding medium. The major source of ROS during this response appears to be an NADPH oxidase called BLI-3, which is found in the hypodermis (skin), intestine and pharynx ( Edens et al., 2001 ; Chavez et al., 2007 and 2019; van der Hoeven et al., 2015 ). BLI-3 is a member of the dual oxidase (DUOX) family of proteins, which all feature a peroxidase domain in addition to an NADPH oxidase domain that generates H2O2 rather than superoxide (Aguirre and Lambeth, 2010). Bioinformatic predictions and experimental evidence indicate that BLI-3 is located in the cytoplasmic membranes of the tissue in which it is produced and oriented in a manner such that H2O2 will be excreted into extracellular environments ( Edens et al., 2001 ; Chavez et al., 2009 ; Aguirre and Lambeth, 2010; Moribe et al., 2012 ; van der Hoeven et al., 2015 ). Based on the localization of BLI-3, H2O2 is released from the hypodermal surface and into the lumen of the GI tract whereupon it is excreted due to the defecation cycle that the animals continuously undergo. When measuring H2O2 from animals exposed to pathogen, the levels can be significantly, but not completely, suppressed by the addition of an NADPH oxidase inhibitor ( Chavez et al., 2007 and 2009). These results suggest that there are additional sources of ROS during infection that are contributing the pool of H2O2 that is being measured. A likely source is the mitochondria, which are known to become dysfunctional during pathogen assault ( Pellegrino et al., 2014 ), and ROS generated from this source in the form of H2O2 would be able to cross membranes and contribute to the extracellular pools. Advantages to using the Amplex® Red assay for measuring ROS in the worm include the fact that it is specific to H2O2, it can be done on samples containing as few as 30 worms in a plate reader, and it does not require sophisticated genetics or microscopy (Labuschagne and Brenkman, 2013; Tiller and Garsin, 2014). Disadvantages include the fact that there is no tissue specificity and one must be careful to maintain samples in the dark, as resorurfin is light sensitive (Labuschagne and Brenkman, 2013). Herein, we present a detailed protocol of our adaption of the Amplex® Red Hydrogen Peroxide/Peroxidase Assay Kit to measuring H2O2 output from the worm following pathogen exposure.
Figure 1. Conversion of Amplex Red to Resorufin in the presence of H2O2 and horseradish peroxidase (HRP) .
Materials and Reagents
96-well microplate, black, clear bottom with lid (Corning costar, catalog number: 3603)
60 mm Petri dish (Fisher Scientific, catalog number: FB0875713A)
1.7 ml microcentrifuge tube
15 ml conical tube
Micropipette tips
PluriStrainer 10 μm (pluriSelect, catalog number: 43-50010-01)
Serological pipettes
Caenorhabditis elegans wild-type N2 strain (Caenorhabditis Genetics Center, University of Minnesota)
Escherichia coli OP50 strain (Caenorhabditis Genetics Center, University of Minnesota)
Enterococcus faecalis OG1RF strain (ATCC 47077)
Autoclaved distilled water
Cholesterol (Sigma-Aldrich, catalog number: C-8503)
Absolute ethanol (Fisher, catalog number: BP2818-4)
Sodium chloride (NaCl) (Fisher, catalog number: S271-500)
Peptone (Becton, Dickinson and Company, catalog number: 211677)
Agar (Becton, Dickinson and Company, catalog number: 214530)
Calcium chloride (CaCl2) (EMD, catalog number: 3000)
Magnesium sulfate (MgSO4) (EMD, catalog number: MX0070-1)
Potassium phosphate, dibasic (K2HPO4) (Sigma-Aldrich, catalog number: P3786)
Potassium phosphate, monobasic (KH2PO4) (Sigma-Aldrich, catalog number: 795488)
Sodium phosphate dibasic (Na2HPO4) (Sigma-Aldrich, catalog number: S3264)
Nystatin (EMD Millipore, catalog number: 475914-1GM)
Gentamicin sulfate (Sigma-Aldrich, catalog number: G3632-5G)
Horse radish peroxidase (part of kit: Invitrogen, catalog number: A22188)
Dimethyl sulfoxide
Amplex® Red hydrogen peroxide/peroxidase assay kit (Invitrogen, catalog number: A22188)
Hydrogen peroxide (VWR, catalog number: VW3540-2)
Brain heart infusion powder (Becton, Dickinson and Company, catalog number: 211059)
Luria-Bertani powder (Becton, Dickinson and Company, catalog number: 244620)
Brain heart infusion (BHI) agar plates with gentamicin (50 μg/ml), made from BHI powder with agar added. Autoclaved, cooled, gentamycin added before pouring
Brain heart infusion (BHI) broth, made from BHI powder and autoclaved.
Nematode growth medium agar plates (see Recipes)
M9W buffer (see Recipes)
50 mg/ml gentamicin sulfate (see Recipes)
10 mg/ml nystatin (see Recipes)
5 mg/ml cholesterol (see Recipes)
Potassium Phosphate Buffer (1 M, pH 6.0) (see Recipes)
Equipment
Micropipettes
Fluorescence and absorbance microplate reader (Biotek, Cytation 5 multimode reader)
Incubators for stable temperatures; 20 °C, 25 °C, 37 °C
Tabletop centrifuge
Stereo microscope (Zeiss, Stemi 2000)
Vortex mixer
Autoclave
Software
Gen5 (Biotek, https://www.biotek.com/products/software-robotics-software/gen5-microplate-reader-and-imager-software/ )
Microsoft Excel (Microsoft, https://products.office.com/en-us/excel )
GraphPad Prism (GraphPad, https://www.graphpad.com/scientific-software/prism/ )
Procedure
-
Worm synchronization and growth
Transfer 10-15 N2 wild-type adult worms from a crowded non-starved NGM plate to a fresh 100 mm NGM plate.
Let the worms lay eggs overnight at 20 °C.
Next day, collect all the worms in the plate in 5 ml of M9W buffer.
Filter the worm suspension through a 10 μm pluriStrainer into a 50 ml conical tube. Only L1-stage worms can pass through the filter.
Count the number of L1-stage worms that pass through the filter.
Put approximately 500 of L1-staged worms on a fresh 100 mm NGM agar plate with an OP50 bacterial lawn.
Allow the worms to develop to the late L3 to early L4 stage at 20 °C, about 36 h, before starting the next step at which you will need worms at the young-adult stage.
-
Preparation of E. faecalis plates and control E. coli plates for worm exposure
The night before the worms reach the proper stage (young-adult), start an overnight culture of E. faecalis OG1RF in 2 ml of BHI broth and E. coli OP50 in 2 ml of LB broth at 37 °C with shaking.
Add 400 μl of the E. faecalis culture to 100 mm BHI agar plates containing 50 μg/ml of gentamycin and spread to create a lawn. Gentamycin prevents the growth of any E. coli transferred over with the worms, as E. faecalis is inherently resistant. Grow at 37 °C for 4-6 h. Additionally, add 400 μl of the E. coli culture to 100 mm NGM agar plates and grow at 37 °C overnight. Make one plate each for every condition you wish to test.
Remove the E. faecalis plates from the incubator and allow them to cool to room temperature.
Wash the worms (young-adult stage) off from the NGM agar plate by adding 5 ml of M9W buffer. Transfer the suspended worms into 15 ml conical tubes. Wash the remaining worms off the plates using extra M9W buffer, if necessary.
Either let the worms settle to the bottom of the tube or spin them down for one minute at 200 × g. Aspirate the liquid, being careful not to disturb the collected worms. Wash the worms twice with M9W buffer. Transfer worms to E. faecalis (OG1RF) or control E. coli (OP50) plates.
Let the plates incubate overnight at 25 °C (about 16 h). This is the standard temperature used when exposing C. elegans to pathogenic bacteria.
-
Preparation of reagents for Amplex Red Assay (enough for 20 reactions)
Dilute the 5x reaction buffer that comes with the Amplex® Red assay kit to make a total of 25 ml of working buffer.
Thaw a tube of the horse radish peroxidase (HRP) and aliquot 100 μl into 1.7 ml microcentrifuge tubes. One tube should then be aliquoted further into 20 μl volumes, also in microcentrifuge tubes. Keep one 20 μl volume out for immediate use and freeze the rest at -20 °C. Avoid repeated freezing and thawing.
Using the dimethyl sulfoxide (DMSO) that comes with the kit, add 60 μl to the Amplex® Red reagent provided in the kit (154 μg) and dissolve. Aliquot the dissolved reagent into 11 μl volumes in 1.7 ml microcentrifuge tubes. Leave one tube out for immediate use and freeze the rest at -20 °C.
Right before you are ready to begin the assay, add the reagents together with the following volumes: 970 μl 1x reaction buffer, 20 μl HRP, 10 μl Amplex® Red. Mix well and keep in the dark until use.
-
Amplex® Red assay for C. elegans
Wash the worms off the E. faecalis and control plates and use 1x reaction buffer to transfer.
Collect the worms in 15 ml tubes suspended in reaction buffer.
Let the worms settle to the bottom of the tube or spin them down for one minute at 200 × g. Aspirate the liquid, being careful not to disturb the collected worms.
Wash the worms 3-4 times in 1 ml of reaction buffer to get rid of the bacteria.
For each sample, transfer the 1 ml of worms and reaction buffer to a 60 mm empty Petri plate. Cut the end of a pipette tip to make the opening larger and, looking under the stereomicroscope, suck up about 15 worms in 25 μl of reaction buffer and transfer to a well of a 96-well plate. Do this twice to obtain 30 +/- 2 worms per 50 μl of reaction buffer/well. Count animals and adjust if necessary. Repeat twice more to generate technical triplicates of the sample.
Prepare dilutions in 1x reaction buffer of H2O2 for the standard curve as described in the Amplex® Red kit literature. We use 0 μM, 1 μM, 2 μM, 3 μM, 4 μM and 5 μM as final concentrations. Dispense 50 μl of 2x concentrated H2O2 standards to 96-well plate in duplicate. Desired final concentrations of H2O2 standards will be achieved after adding the same volume of Amplex® red reagent mix. It is recommended that the H2O2-standard wells should be as far as possible from the sample wells in order to prevent interference of volatile H2O2 from high-concentration standard wells into the sample wells.
Mix the Amplex® Red reagent, HRP and buffer (Step C4) and add 50 μl to each well of the 96-well plate with the worm samples, controls and standards. Record the time assay begins.
-
Reading/recording assay
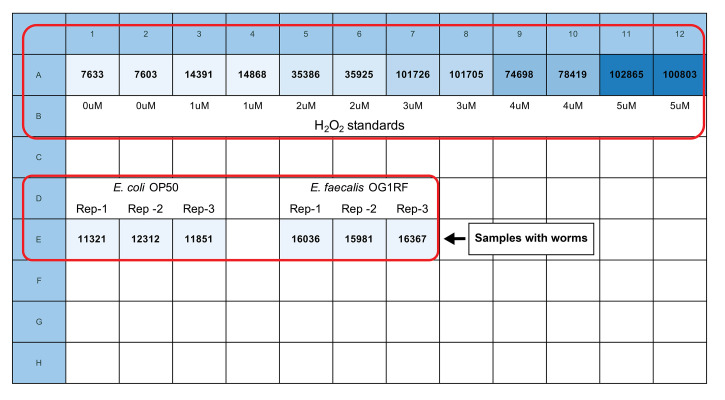
Using a microplate reader, read the fluorescence using 540 +/-20 nm excitation and 590 +/-20 nm emission, bottom optics position and “extended” gain. We use a Biotek Cytation 5 microplate reader with Gen5 software to obtain the data. Measurements are expressed as Relative Fluorescence Units (RFU). See Figure 2 for plate organization and fluorescence readings of a sample experiment designed to test the difference of H2O2 production by worms in the presence of non-pathogenic (E. coli OP50 strain) and pathogenic (E. faecalis OG1RF strain) bacteria as food. Take time points at 30, 60, 120 and 180 min. Generally, the 60-min reading demonstrates the best dynamic range.
Figure 2. Sample plate organization and fluorescence readings.
Row “A” has duplicates of increasing concentrations (0-5 μM) of H2O2 for creating the standard curve. Row “E” has technical triplicates of the samples with the worms exposed to E. coli OP50 and E. faecalis OG1RF bacteria. Note that the samples are placed in the wells relatively far from the H2O2 standards. The reading was taken 60 min after mixing the samples with the Amplex Red reagent.
Data Analysis
-
Calculation of the amount of H2O2 produced
Calculate the refined RFU (rRFU) of the samples by subtracting the measurement of the 0 μM H2O2 standard (the average of the technical duplicates) from the readings. The results represent the RFU generated by the worms in the samples during the measured time period.
Calculate the rRFU of the standards by subtracting the measurement of 0 μM H2O2 standard (background fluorescence of the reaction buffer) from the measurements of all of the standards.
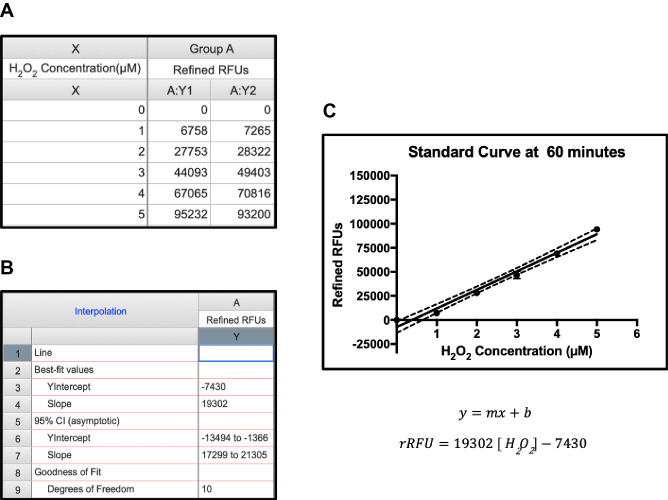
Plot the rRFU data of the standards using GraphPad Prism or similar software to create a linear standard curve. See Figure 3 for a sample standard curve created using GraphPad Prism.
Calculate the slope (m) and the y-intercept (b) of the standard curve equation (rRFU = m[H2O2] + b). GraphPad Prism software automatically calculates the values.
-
Use the formula below to calculate the amount of H2O2 produced per minute for each sample. H2O2 production:
Note: Below is the detailed derivation of the formula above from H2O2 concentration.
H2O2 concentration in each well:
[
Total H2O2 amount produced in each well with 100 μl volume:
H2O2 production rate
H2O2 production rate
-
Statistical analysis of the data
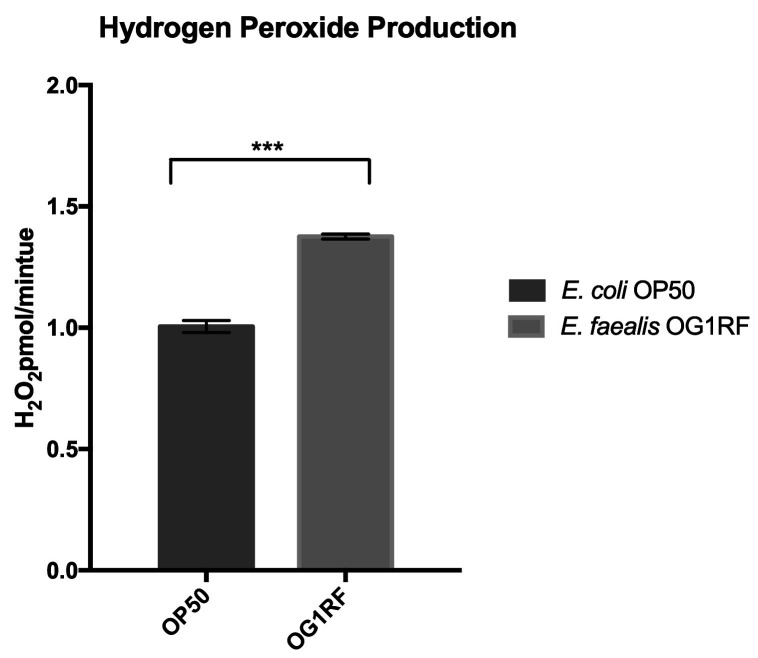
Each sample has three technical replicates. GraphPad Prism or similar software is used to analyze the data. Create a bar graph by averaging the H2O2 production for each sample. Error bars represent the standard error of mean (SEM) for the three replicates. See Figure 4 for a sample bar graph of H2O2 production in the presence of non-pathogenic and pathogenic bacteria; E. coli (OP50 strain) and E. faecalis (OG1RF), respectively.
The unpaired two-tailed t-test is used to test the statistical significance of the data.
To verify results with independent biological replicates, the experiment should be replicated a total of at least three times on different days with independent batches of worms. In our experience, the exact amount of ROS produced by a given batch of worms on a given day differs depending on slight differences in developmental timing and other variables. Therefore, it is not feasible to average experimental values obtained on different days. However, the PATTERNS are consistent. For example, in this experiment the animals exposed to E. faecalis ALWAYS produce significantly more ROS than animals exposed to non-pathogenic E. coli. For publication purposes, a representative experiment can be shown as primary data with biological replicates added to a supplement, if necessary.
Figure 3. Sample standard curve and equation.
A. Refined relative fluorescence units (rRFU) were plotted against concentration. B and C. Best-fit linear curve (B) and equation (C) were generated by Graphpad Prism.
Figure 4. Sample bar graph showing the rate of H2O2 production.
By each sample as picomoles per minute. Error bars represent the standard error of mean (SEM) from technical triplicates of each sample (P = 0.0002).
Recipes
-
50 mg/ml gentamicin sulfate
50 mg gentamicin sulfate is dissolved in 1 ml of distilled water and stored at -20 °C
-
10 mg/ml nystatin
10 mg nystatin is suspended in 1 ml 100% ethanol and stored at 4 °C
-
5 mg/ml cholesterol
5 mg cholesterol is dissolved in 1 ml of 100% ethanol and stored at room temperature
-
Nematode growth medium (NGM) agar plates (per liter)
-
Autoclave and cool to 50 °C
3 g NaCl
2.5 g peptone
18 g agar
975 ml distilled water
-
Add following while stirring with the same order:
25 ml of potassium phosphate buffer (1 M, pH 6.0)
1 ml of MgSO4 (1 M)
1 ml of CaCl2 (1 M)
1 ml of nystatin (10 mg/ml)
1 ml of cholesterol (5 mg/ml)
Pour 10 ml or 25 ml of NGM into 60 mm or 100 mm Petri dish, respectively. Let plates solidify overnight
-
-
M9W buffer
3 g of KH2PO4
6 g of Na2HPO4
5 g of NaCl
1 L distilled water
Autoclave
Add 1 ml of sterile MgSO4 (1 M)
-
Potassium Phosphate Buffer (1 M, pH 6.0) per liter
30.2 g of K2HPO4
118.2 g of KH2PO4
Complete volume to 1 L with distilled water
Autoclave and dispense in 50 ml conical tubes for future use
Acknowledgments
This work was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award number R01AI076406 to D. A. G. The content is solely the responsibility of the authors and does represent the official views of the National Institutes of Health. Also acknowledged, are the previous publications from on which this protocol was based ( Chavez et al., 2007 and 2009; Liu et al., 2019 ; Tiller and Garsin, 2014; van der Hoeven et al., 2015 ).
Competing Interests
The authors declare no competing interests.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Aguirre J. and Lambeth J. D.(2010). Nox enzymes from fungus to fly to fish and what they tell us about Nox function in mammals. Free Radic Biol Med 49(9): 1342-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Back P., De Vos W. H., Depuydt G. G., Matthijssens F., Vanfleteren J. R. and Braeckman B. P.(2012). Exploring real-time in vivo redox biology of developing and aging Caenorhabditis elegans . Free Radic Biol Med 52(5): 850-859. [DOI] [PubMed] [Google Scholar]
- 3. Belousov V. V., Fradkov A. F., Lukyanov K. A., Staroverov D. B., Shakhbazov K. S., Terskikh A. V. and Lukyanov S.(2006). Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nat Methods 3(4): 281-286. [DOI] [PubMed] [Google Scholar]
- 4. Chavez V., Mohri-Shiomi A. and Garsin D. A.(2009). Ce-Duox1/BLI-3 generates reactive oxygen species as a protective innate immune mechanism in Caenorhabditis elegans . Infect Immun 77(11): 4983-4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chavez V., Mohri-Shiomi A., Maadani A., Vega L. A. and Garsin D. A.(2007). Oxidative stress enzymes are required for DAF-16-mediated immunity due to generation of reactive oxygen species by Caenorhabditis elegans . Genetics 176(3): 1567-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dikalov S. I. and Harrison D. G.(2014). Methods for detection of mitochondrial and cellular reactive oxygen species. Antioxid Redox Signal 20(2): 372-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Edens W. A., Sharling L., Cheng G., Shapira R., Kinkade J. M., Lee T., Edens H. A., Tang X., Sullards C., Flaherty D. B., Benian G. M. and Lambeth J. D.(2001). Tyrosine cross-linking of extracellular matrix is catalyzed by Duox, a multidomain oxidase/peroxidase with homology to the phagocyte oxidase subunit gp91phox. J Cell Biol 154(4): 879-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ewald C. Y., Hourihan J. M., Bland M. S., Obieglo C., Katic I., Moronetti Mazzeo L. E., Alcedo J., Blackwell T. K. and Hynes N. E.(2017). NADPH oxidase-mediated redox signaling promotes oxidative stress resistance and longevity through memo-1 in C. elegans . Elife 6: e19493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Knoefler D., Thamsen M., Koniczek M., Niemuth N. J., Diederich A. K. and Jakob U.(2012). Quantitative in vivo redox sensors uncover oxidative stress as an early event in life . Mol Cell 47(5): 767-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Labuschagne C. F. and Brenkman A. B.(2013). Current methods in quantifying ROS and oxidative damage in Caenorhabditis elegans and other model organism of aging. Ageing Res Rev 12(4): 918-930. [DOI] [PubMed] [Google Scholar]
- 11. Liu Y., Kaval K. G., van Hoof A. and Garsin D. A.(2019). Heme peroxidase HPX-2 protects Caenorhabditis elegans from pathogens . PLoS Genet 15(1): e1007944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McCallum K. C. and Garsin D. A.(2016). The role of reactive oxygen species in modulating the Caenorhabditis elegans immune response . PLoS Pathog 12(11): e1005923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Miranda-Vizuete A. and Veal E. A.(2017). Caenorhabditis elegans as a model for understanding ROS function in physiology and disease . Redox Biol 11: 708-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mohanty J. G., Jaffe J. S., Schulman E. S. and Raible D. G.(1997). A highly sensitive fluorescent micro-assay of H2O2 release from activated human leukocytes using a dihydroxyphenoxazine derivative . J Immunol Methods 202(2): 133-141. [DOI] [PubMed] [Google Scholar]
- 15. Moribe H., Konakawa R., Koga D., Ushiki T., Nakamura K. and Mekada E.(2012). Tetraspanin is required for generation of reactive oxygen species by the dual oxidase system in Caenorhabditis elegans . PLoS Genet 8(9): e1002957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pellegrino M. W., Nargund A. M., Kirienko N. V., Gillis R., Fiorese C. J. and Haynes C. M.(2014). Mitochondrial UPR-regulated innate immunity provides resistance to pathogen infection. Nature 516(7531): 414-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schulz T. J., Zarse K., Voigt A., Urban N., Birringer M. and Ristow M.(2007). Glucose restriction extends Caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress . Cell Metab 6(4): 280-293. [DOI] [PubMed] [Google Scholar]
- 18. Sem X. and Rhen M.(2012). Pathogenicity of Salmonella enterica in Caenorhabditis elegans relies on disseminated oxidative stress in the infected host . PLoS One 7(9): e45417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tiller G. R. and Garsin D. A.(2014). The SKPO-1 peroxidase functions in the hypodermis to protect Caenorhabditis elegans from bacterial infection . Genetics 197(2): 515-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. van der Hoeven R., Cruz M. R., Chavez V. and Garsin D. A.(2015). Localization of the dual oxidase BLI-3 and characterization of its NADPH oxidase domain during infection of Caenorhabditis elegans . PLoS One 10(4): e0124091. [DOI] [PMC free article] [PubMed] [Google Scholar]