ABSTRACT
Mitochondrial fusion and fission (mitochondrial dynamics) are homeostatic processes that safeguard normal cellular function. This relationship is especially strong in tissues with constitutively high energy demands, such as brain, heart and skeletal muscle. Less is known about the role of mitochondrial dynamics in developmental systems that involve changes in metabolic function. One such system is spermatogenesis. The first mitochondrial dynamics gene, Fuzzy onions (Fzo), was discovered in 1997 to mediate mitochondrial fusion during Drosophila spermatogenesis. In mammals, however, the role of mitochondrial fusion during spermatogenesis remained unknown for nearly two decades after discovery of Fzo. Mammalian spermatogenesis is one of the most complex and lengthy differentiation processes in biology, transforming spermatogonial stem cells into highly specialized sperm cells over a 5-week period. This elaborate differentiation process requires several developmentally regulated mitochondrial and metabolic transitions, making it an attractive model system for studying mitochondrial dynamics in vivo. We review the emerging role of mitochondrial biology, and especially its dynamics, during the development of the male germ line.
KEY WORDS: Membrane fission, Membrane fusion, Mitochondrial dynamics, Spermatogenesis
Summary: Spermatogenesis in mammals is associated with developmental changes in mitochondrial structure and metabolism. Mitochondrial dynamics has emerged as a key factor regulating mammalian spermatogenesis.
Introduction
Mitochondria are crucial to most eukaryotic cells, and their function is maintained by several quality control mechanisms. These include mitochondrial fusion and fission (mitochondrial dynamics) and mitophagy, the degradation of mitochondrial content by selective autophagy (Youle and Narendra, 2011). In addition to controlling organelle morphology, continuous fusion and fission events safeguard mitochondrial function by enabling the hundreds of mitochondria within a cell to mix, promoting homogeneity (Chan, 2012). Without fusion, mitochondrial heterogeneity increases, which compromises mitochondrial physiology and leads to cellular dysfunction. For example, in mice, whole-animal ablation of mitochondrial fusion causes embryonic lethality (Chen et al., 2003), and organ-specific ablation disrupts homeostasis in tissues such as brain (Chen et al., 2007), heart (Chen et al., 2011; Papanicolaou et al., 2012) and skeletal muscle (Chen et al., 2010; Mishra et al., 2015). Similarly, whole-animal ablation of mitochondrial fission results in midgestation lethality (Ishihara et al., 2009; Wakabayashi et al., 2009), whereas tissue-specific ablation perturbs homoeostasis in the brain (Wakabayashi et al., 2009), heart (Ikeda et al., 2015; Kageyama et al., 2014) and skeletal muscle (Favaro et al., 2019). Thus, dynamics serves as a crucial mitochondrial quality control mechanism to promote cell and tissue homeostasis.
Mitophagy is an additional layer of quality control that utilizes autophagy (Mizushima, 2007; Mizushima et al., 1998; Tsukada and Ohsumi, 1993) to remove excessive mitochondria or damaged mitochondria that are beyond repair (Pickles et al., 2018). During mitophagy, microtubule-associated protein 1A/1B light chain 3 (collectively LC3; also known as MAP1LC3A, MAP1LC3B and MAP1LC3C) is recruited to the autophagosomal membrane and binds to mitochondria that selectively express mitophagy receptors on their outer membrane. This molecular recognition designates damaged mitochondria as cargo for autophagosomes, and the resulting mitophagosomes subsequently fuse with lysosomes for degradation and recycling of the engulfed organelles. Much of the molecular workings of mitophagy have been parsed in cultured cells, and recently developed mouse models are enabling the analysis of mitophagy in vivo (Kuma et al., 2017; McWilliams et al., 2016; Sun et al., 2015).
Although mounting evidence suggests that mitochondrial dynamics and mitophagy maintain basal cellular homeostasis, much less is known about their role during differentiation and developmentally regulated mitochondrial and metabolic transitions. Mammalian spermatogenesis is a rich system in which to address this gap in knowledge because it requires several such transitions. In this article, we review key aspects of mitochondrial dynamics and mitophagy, and discuss the cellular and metabolic regulation of spermatogenesis. We then describe the various mouse models of mitochondrial dysfunction that exhibit male infertility and discuss the emerging role of mitochondrial dynamics during spermatogenesis. Our discussion centers on mouse spermatogenesis, but insights from other model organisms and clinical studies are discussed where appropriate.
Mitochondrial dynamics at a glance
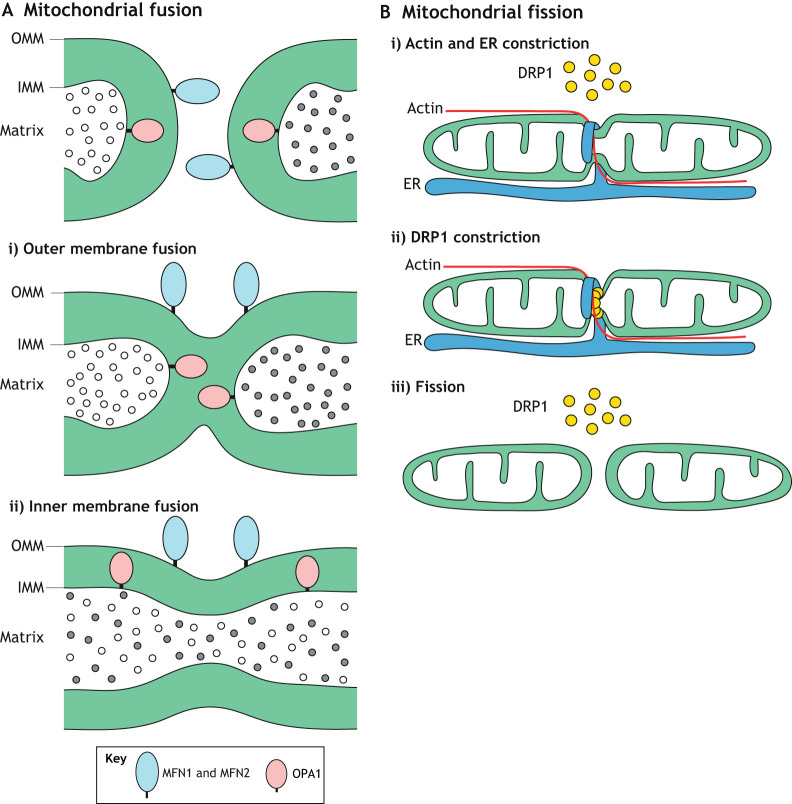
Despite their static and autonomous appearance in micrographs, mitochondria continuously fuse and divide, resulting in the circulation of their components throughout the entire organellar pool. Unlike most organelles that undergo fusion, mitochondria contain double membranes – a highly folded inner membrane (IMM) that harbors the oxidative phosphorylation (OXPHOS) complexes and a semi-permeable outer membrane (OMM) that encloses the organelle (Tzagoloff, 1982). Mitochondrial fusion therefore involves two separate fusion events, each requiring a unique machinery featuring large GTP-hydrolyzing enzymes of the dynamin superfamily. Outer membrane fusion, mediated by the mitofusins MFN1 and MFN2, is followed by inner membrane fusion, mediated by the dynamin-like 120 kDa protein (OPA1) (Fig. 1A). Mutations in mitofusins or Opa1 prevent fusion, which compromises OXPHOS and leads to cellular dysfunction and disease (Chan, 2020).
Fig. 1.
Mitochondrial fusion and fission. (A) Mitochondrial fusion occurs in two distinct steps, both mediated by large GTP-hydrolyzing enzymes of the dynamin superfamily. MFN1 and MFN2 mediate fusion of the mitochondrial outer membrane (OMM). Then, OPA1 mediates fusion of the inner membrane (IMM), which results in mixing of matrix components. Although OPA1 is present on opposing IMMs, it is not required to be present on both membranes. (B) Mitochondrial fission is a multistep process. In the initial phase, actin and the ER associate with the mitochondrial tubule. The ER wraps around and constricts the mitochondrion. Receptors on the mitochondrial outer membrane (not shown) recruit cytosolic DRP1 to this constriction site. Multiple DRP1 molecules oligomerize around the mitochondrion to form a ring-shaped structure that further constricts and severs the mitochondrial tubule.
Mitochondrial fusion is counter-balanced by fission. This balance maintains proper organellar size and morphology, which facilitates mitochondrial distribution and transport throughout the cell. Early steps in mitochondrial fission involve actin filaments and the endoplasmic reticulum (ER), which mark sites of fission by wrapping around and constricting mitochondria (Friedman et al., 2011; Korobova et al., 2013) (Fig. 1B). Subsequently, receptors on the OMM recruit cytosolic dynamin-related protein 1 (DRP1; encoded by Dnm1l), a mechanochemical enzyme that assembles into ring-like structures to further constrict and sever mitochondrial tubules. In mammals, four receptors recruit cytosolic DRP1 to the mitochondrial outer membrane: mitochondrial fission factor (MFF), mitochondrial dynamics proteins of 49 and 51 kDa (MID49 and MID51; encoded by Mief2 and Mief1, respectively), and mitochondrial fission 1 (FIS1) (Chan, 2012). Dynamin-2 (DNM2) has been proposed to act at a terminal step following DRP1 constriction (Lee et al., 2016), but this idea was recently challenged (Fonseca et al., 2019; Kamerkar et al., 2018; Nagashima et al., 2020). Lysosomes (Wong et al., 2018) and Golgi-derived vesicles (Nagashima et al., 2020) have also been implicated in mitochondrial fission, but how they interact with the fission machinery remains to be resolved.
Mitochondrial quality control by mitophagy
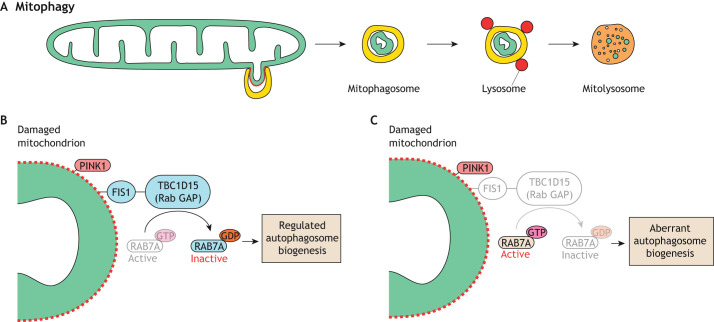
Mitophagy is the selective degradation of mitochondria by autophagy (Fig. 2A). The best characterized model of mitophagy is the phosphatase and tensin homolog (PTEN) induced kinase I (PINK1)–Parkin pathway. PINK1 is a serine/threonine kinase that is normally kept at low levels due to degradation by proteases upon import into the mitochondrial matrix. Insults that disrupt mitochondrial membrane potential prevent PINK1 import and degradation, leading to its stabilization on the mitochondrial outer membrane (Narendra et al., 2010). PINK1 then phosphorylates ubiquitin at the damaged mitochondrial surface, leading to recruitment of the E3 ubiquitin ligase Parkin, which adds additional ubiquitin molecules onto mitochondrial outer membrane proteins. Further phosphorylation of these ubiquitin chains by PINK1 recruits additional Parkin, generating a positive-feedback loop that activates the ubiquitin proteasome system (UPS) (Chan et al., 2011; Rakovic et al., 2019) and recruits autophagosomes for mitophagy (Sekine and Youle, 2018).
Fig. 2.
Mitophagy. (A) Overview of mitophagy. An autophagosome engulfs a damaged portion of a mitochondrion to form a mitophagosome that fuses with lysosomes. The mitochondrion is degraded in the resulting mitolysosome. (B) A model showing the role of FIS1 during PINK1-mediated mitophagy. FIS1 at the damaged mitochondrial surface interacts with TBC1D15, a mitochondrial Rab GAP that inactivates RAB7A, to regulate mitophagosome formation. (C) In the absence of FIS1, the uncontrolled action of RAB7A disrupts autophagosome membrane dynamics, resulting in aberrant mitophagy.
Mitophagy utilizes FIS1, which is the only receptor of DRP1 in Saccharomyces cerevisiae and is required for mitochondrial fission in this species (Jakobs et al., 2003; Mozdy et al., 2000; Tieu et al., 2002). In mammalian cells, however, FIS1 has a minor role in mitochondrial fission (Losón et al., 2013; Otera et al., 2010) and a more prominent role in mitophagy. During Parkin-mediated mitophagy, FIS1 interacts with the mitochondrial Rab GTPase-activating protein (GAP) TBC1D15 (Onoue et al., 2013), to inhibit RAB7A (Yamano et al., 2014, 2018). In its active state, RAB7A promotes growth of double membraned autophagosomes by mediating fusion of ATG9A-containing vesicles (Tan and Tang, 2019). It has been proposed that, without FIS1, RAB7A remains constitutively active at the mitochondrial surface, leading to dysregulated growth of autophagosomal membranes (Fig. 2B,C) (Yamano et al., 2014). This cellular defect is observed by light microscopy as LC3 tubulation and aggregation (Yamano et al., 2014). Similarly, large LC3 aggregates are found in fis-1-knockout nematodes treated with mitochondrial toxins (Shen et al., 2014). Consistent with its role in mitophagy, FIS1 is required for degradation of paternal mitochondria shortly after fertilization in mice (Rojansky et al., 2016). FIS1 has also been implicated in PINK1–Parkin-independent mitophagy (Yamashita et al., 2016). Thus, mammalian FIS1 plays a role in the clearance of mitochondria via mitophagy. However, a recent study reported that Fis1 knockout in skeletal muscle increases mitophagy (Zhang et al., 2019), indicating that its role in mitophagy might be tissue specific, warranting the study of FIS1 in multiple cell types.
Spermatogenesis overview
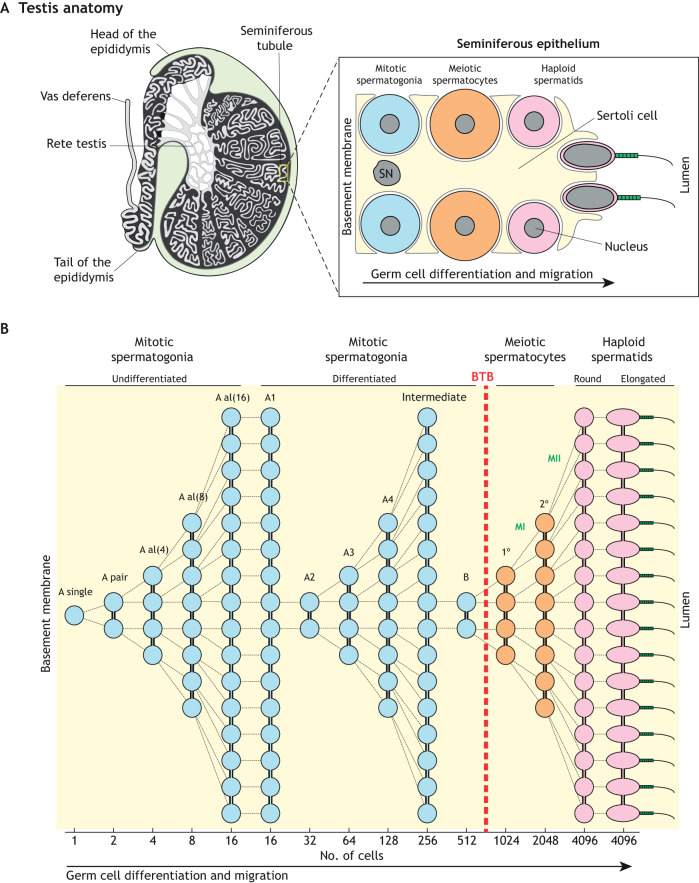
Male germ cell development is a promising system for studying mitochondrial dynamics and mitophagy because it involves drastic changes to mitochondrial shape, number and distribution (Hermo et al., 2010a). This complex and lengthy differentiation process occurs within the highly convoluted seminiferous tubules of the testes (Fig. 3A) and can be divided spatiotemporally into three major categories: (1) mitotic amplification of spermatogonia, (2) genome reduction in meiotic spermatocytes, and (3) morphological transformation of haploid spermatids into spermatozoa. Mitotically dividing spermatogonia reside near the basement membrane at the tubule periphery, and their differentiated descendants migrate towards the tubule lumen. This process depends critically on the intimately associated somatic Sertoli cells – nurse cells that provide structural support, metabolites and differentiation cues for the developing germ cells (Griswold, 1998). As germ cells divide, incomplete cytokinesis allows their daughter cells to remain connected via stable intercellular bridge structures (Greenbaum et al., 2011), enabling them to share gene products (Braun et al., 1989). Numerous such divisions result in the formation of long ‘chains’ of syncytial cells (Fig. 3B).
Fig. 3.
Spermatogenesis. (A) Left panel, anatomy of the mammalian testis highlighting the convoluted seminiferous tubules in which spermatogenesis takes place. Right panel, schematic illustration of the seminiferous epithelium highlighting the intimate association between somatic Sertoli cells and germ cells. For simplicity, only the major germ cell types are shown. (B) Cellular pedigree of a single undifferentiated spermatogonium, highlighting germ cell amplification. The theoretical number of syncytial cells at each stage is shown at the bottom. Note that meiotic spermatocytes and post-meiotic spermatids develop on the adluminal side of the blood-testis barrier (BTB). SN, Sertoli cell nucleus; A al, A aligned; 1°, primary spermatocyte; 2°, secondary spermatocyte; MI, meiosis I; MII, meiosis II.
In mice, spermatogenesis begins at birth with mitotic divisions of progenitor cells called Type A-single spermatogonia, which give rise to pairs of interconnected cells called Type A-paired spermatogonia (Fig. 3B). Further divisions produce chains of 4 to 16 cells called Type A-aligned spermatogonia. These divisions expand the pool of undifferentiated spermatogonia while maintaining a subset with spermatogonial stem cell (SSC) activity. However, the molecular identity of SSCs remains a matter of debate. The prevailing model for decades posited that only A-singles retain SSC activity. However, recent studies have challenged this model, and two additional models of SSC dynamics have emerged. The original and revised models are described in two excellent reviews (De Rooij, 2017; Lord and Oatley, 2017). Briefly, in one revised model, SSC activity is restricted to a small subset of A-singles that express stem markers, such as Id4 and Pax7 (Aloisio et al., 2014). In the other model, SSC activity is maintained by a much larger population of undifferentiated spermatogonia that express glial cell line-derived neurotrophic factor (GDNF) family receptor α1 (GFRα1, encoded by Gfra1) (Hara et al., 2014). Together with RET, GFRα1 binds GDNF (Naughton et al., 2006), which is secreted by Sertoli cells to regulate SSC activity (Hofmann, 2008). Chains of Gfra1-expressing spermatogonia are proposed to retain stemness by undergoing syncytial fragmentation to revert to smaller chains or A-singles. In contrast, SSCs in Drosophila (Spradling et al., 2001) and Caenorhabditis elegans (Kimble and White, 1981) divide asymmetrically, forming one daughter cell that differentiates and another that retains stemness (Oatley and Brinster, 2012).
Upon receiving cues from the surrounding Sertoli cells, chains of undifferentiated spermatogonia begin to differentiate and irreversibly commit to meiosis (Griswold, 2016). The sixth and final mitotic division of differentiating spermatogonia gives rise to spermatocytes, which traverse the blood–testis barrier (BTB), enter the adluminal compartment and initiate meiosis, a defining event in spermatogenesis (Cohen et al., 2006). In meiosis, a spermatocyte undergoes DNA duplication and two meiotic divisions to generate four haploid round spermatids.
Initially small and inconspicuous, round spermatids undergo spermiogenesis, a dramatic morphological transformation to become polarized sperm cells with a head, midpiece and tail (Hermo et al., 2010b). This morphological transformation requires culling of excess cellular components into residual bodies for phagocytic degradation by Sertoli cells. Furthermore, the nuclear genetic material is repackaged by protamines – small, arginine-rich, DNA-binding proteins – into a slender and compacted nucleus (Bao and Bedford, 2016), which is capped by the acrosome, a unique lysosome-related organelle that releases digestive enzymes to enable fertilization.
OXPHOS fuels spermatogenesis
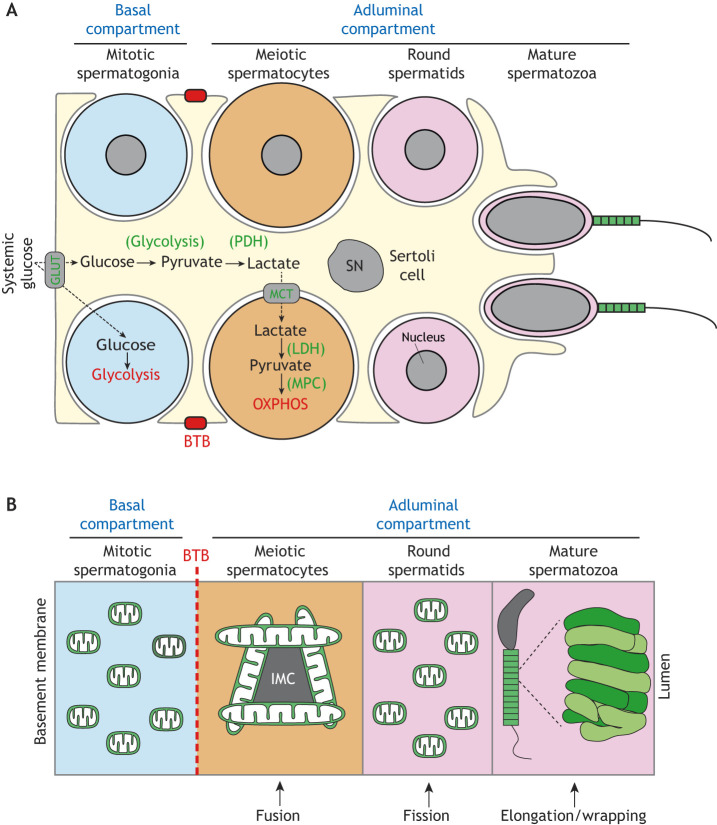
These highly coordinated germ cell differentiation events depend on the surrounding Sertoli cells. In addition to providing metabolites and differentiation cues for the developing germ cells, Sertoli cells form the BTB that divides the seminiferous epithelium into the basal and adluminal compartments, secluding the latter from the interstitial space and vasculature (Stanton, 2016) (Fig. 4A). As a result, during their progression from spermatogonial stem cells into sperm, germ cells encounter unique microenvironments with varying availability to glucose and other metabolites. Spermatogonia and SSCs reside in the basal compartment, where they have access to the vasculature and interstitial fluid, whereas the more-advanced spermatocytes and spermatids in the adluminal compartment have limited access.
Fig. 4.
Mitochondrial respiration and dynamics during spermatogenesis. (A) Mitochondrial respiration during spermatogenesis. Spermatogonia in the basal compartment have direct access to systemic glucose, which they use for glycolysis. Spermatocytes and spermatids in the adluminal compartment, however, are separated from the vasculature and interstitial space by the BTB, and thus rely on Sertoli cells for a carbon source. Sertoli cells take up systemic glucose via glucose transporters (GLUTs) and glycolytically convert it into pyruvate, which is converted into lactate via pyruvate dehydrogenase (PDH). Lactate is then shuttled by monocarboxylate transporters (MCT) into spermatocytes, which convert it back into pyruvate via lactate dehydrogenase (LDH). Finally, pyruvate is imported into mitochondria by the mitochondrial pyruvate carrier (MPC) for fueling oxidative phosphorylation (OXPHOS). The nuclei of the lower spermatogonium and spermatocyte are omitted for clarity. (B) Mitochondrial dynamics during spermatogenesis. Mitochondria are generally small and spherical in spermatogonia, which reside in the basal compartment. Upon traversing the BTB (dashed red line) and entering the adluminal compartment, mitochondria elongate and cluster around the nuage, also referred to as intermitochondrial cement (IMC). In post-meiotic spermatids, mitochondria fragment. Finally, near the end of spermiogenesis, mitochondria elongate and tightly pack around the sperm midpiece. SN, Sertoli cell nucleus.
Male germ cells have distinct metabolic requirements depending on their differentiation state (Fig. 4A). Spermatogonia are generally believed to rely on glucose for energy production by glycolysis (Rato et al., 2012). In contrast, spermatocytes and spermatids require lactate and pyruvate for survival (Bajpai et al., 1998; Grootegoed et al., 1984; Nakamura et al., 1982, 1984). This distinction likely reflects the fact that spermatogonia in the basal compartment have direct access to systemic glucose, whereas spermatocytes and spermatids in the adluminal compartment are separated from the vasculature by the BTB. Thus, spermatocytes and spermatids rely on the surrounding Sertoli cells for secretion of lactate as a carbon source (Boussouar and Benahmed, 2004). This idea is further supported by expression of lactate dehydrogenase (Ldhc) selectively in advanced germ cells in the adluminal compartment (Goldberg et al., 2010). Because lactate is converted into pyruvate by LDHC, and pyruvate can be converted to acetyl-coenzyme A to fuel OXPHOS, these data indicate that meiotic and post meiotic cells in the adluminal compartment likely rely more heavily on mitochondrial OXPHOS activity.
The requirement for lactate and pyruvate by meiotic spermatocytes is consistent with the high energy demands associated with meiosis and, in particular, meiotic prophase I (MPI). This lengthy process constitutes ∼90% of meiosis and a fourth of the entire spermatogenic process, and is generally subdivided into four stages – leptotene, zygotene, pachytene and diplotene (Handel and Schimenti, 2010). The leptotene stage marks the formation of the synaptonemal complex – a proteinaceous structure that enables interaction between the homologous chromosomes – and initiation of genome-wide programmed double-strand breaks (DSBs). In zygotene, the synaptonemal complex grows along the homologous chromosomes, enabling them to synapse. During pachytene, synapsis is completed, and ATP-dependent biochemical reactions drive homologous recombination and resolution of DSBs. Finally, in diplotene, the recombined chromosomes detach for segregation into daughter cells.
Classic electron microscopy (EM) analyses of mitochondrial ultrastructure in rodent testes provide support for the increased OXPHOS activity during MPI. Mitochondria in spermatogonia are generally small and spherical, and contain ‘orthodox’ cristae (De Martino et al., 1979; Meinhardt et al., 1999; Seitz et al., 1995), an ultrastructural conformation associated with low OXPHOS activity (Hackenbrock, 1966; Mannella, 2006a,b). Conversely, mitochondria in pachytene spermatocyte are elongated and contain ‘condensed’ cristae, an ultrastructural conformation associated with high OXPHOS utilization. In post-meiotic spermatids, mitochondria fragment and their cristae return to an intermediate state between orthodox and condensed, suggesting a shift back to glycolysis (Meinhardt et al., 1999).
Several mouse models of mitochondrial dysfunction have corroborated the critical contribution of OXPHOS to spermatogenesis and, in particular, to meiosis. For example, mice with error-prone mtDNA replication, due to a mutation in the proofreading subunit of the mtDNA polymerase γ (PolgD257A), exhibit male infertility (Kujoth et al., 2005; Trifunovic et al., 2004). Owing to accumulation of mtDNA point mutations and deletions, these ‘mtDNA mutator’ mice have early degeneration of multiple organ systems – a phenotype interpreted as accelerated aging. The testes of mtDNA mutator mice have severe degeneration of seminiferous tubules and depletion of germ cells by 10 months of age (Kujoth et al., 2005). Increasing or decreasing expression of mitochondrial transcription factor A (Tfam), which regulates mtDNA levels, mitigates or exacerbates the infertility phenotype in mtDNA mutator mice, respectively (Jiang et al., 2017). Together, these data suggest that spermatogenesis is highly sensitive to perturbations in OXPHOS.
Mouse models also indicate that MPI is highly susceptible to mitochondrial dysfunction. Mice with a large-scale (4696 bp) pathogenic mtDNA deletion have reduced OXPHOS activity and exhibit meiotic arrest during the zygotene to pachytene transition (Nakada et al., 2006). Similarly, mice with genetic ablation of the testis-specific adenine nucleotide translocator (Ant4; encoded by Slc25a31) are less efficient at utilizing mitochondrial ATP and exhibit spermatogenic arrest during leptotene of MPI (Brower et al., 2007, 2009). These studies indicate that mitochondrial function is particularly important during MPI.
Clinical studies also suggest a link between mtDNA integrity and male fertility. Male patients with infertility are sometimes found to have mtDNA mutations (Baklouti-Gargouri et al., 2014; Carra et al., 2004; Kao et al., 1995; Lestienne et al., 1997), and some patients with mitochondrial disease caused by mtDNA mutations are infertile (Demain et al., 2017; Folgerø et al., 1993; Spiropoulos et al., 2002). Furthermore, mutations in the mitochondrial polymerase γ (POLG) contribute to male infertility (Demain et al., 2017; Luoma et al., 2004; Rovio et al., 2001).
The emerging role of mitochondrial dynamics during spermatogenesis
As detailed above, mitochondrial function is essential for spermatogenesis. Because mitochondrial dynamics safeguards mitochondrial function, perturbations in dynamics could be expected to disrupt spermatogenesis. Indeed, Hales and Fuller discovered the first mitochondrial fusion gene during a Drosophila mutagenesis screen for male sterility (Hales and Fuller, 1997). During Drosophila spermatid development, mitochondria aggregate and fuse to form the Nebenkern, which resembles an onion slice by EM due to the concentric wrapping of two giant mitochondria around each other (Demarco et al., 2014; Fuller, 1993). Hales and Fuller found that mutations in Fuzzy onions (Fzo), a Drosophila homolog of mitofusin, cause fragmentation of the Nebenkern (giving it the appearance of ‘fuzzy onions’) and male sterility (Hales and Fuller, 1997). Since then, emerging evidence indicates that other mitochondrial dynamics factors also promote spermatogenesis in the fly. Drosophila mitofusin (Marf), the other homolog of mammalian mitofusin, as well as Opa1 and Drp1, maintain male germline stem cells (Demarco and Jones, 2019; Demarco et al., 2019). Thus, in flies, spermatogenesis requires both mitochondrial fusion and fission.
In rodents, dramatic morphological transformation of mitochondria during spermatogenesis (Fig. 4B) suggests an evolutionarily conserved role for mitochondrial dynamics. Mitochondria in spermatogonia and early MPI are generally small and spherical. They elongate during pachytene and then fragment again in post-meiotic spermatids (De Martino et al., 1979). In maturing spermatids, small mitochondrial spheres line the midpiece in highly coordinated arrays and elongate while wrapping around the midpiece (Ho and Wey, 2007). Thus, spermatogenesis carefully regulates mitochondrial morphology to support the physiological requirements of the developing germ cells. However, despite these long-known mitochondrial transitions, the role of the major mitochondrial dynamics factors during mammalian spermatogenesis remained unknown until recently.
The first mitochondrial dynamics factor examined during mouse spermatogenesis was MFN1. Zhang and colleagues removed Mfn1 from the male germline using the Vasa-Cre driver, which expresses at around embryonic day 15 (E15) (Zhang et al., 2016). Mfn1 mutants were infertile, had reduced testis size and failed to produce sperm. These defects coincided with a reduction in spermatocytes, suggesting a defect during meiosis. Undifferentiated spermatogonia were not reduced during the first round of spermatogenesis, indicating that Mfn1 is dispensable for the formation of these progenitor cells (Zhang et al., 2016).
Our recent study expanded on the role of mitochondrial fusion during mouse spermatogenesis (Varuzhanyan et al., 2019). We observed that Mfn1 Mfn2 double mutants failed to produce any sperm, indicating an absolute requirement for mitochondrial fusion during spermatogenesis. Histological analysis revealed a reduction in post-meiotic spermatids in fusion-deficient mice, indicating a defect during meiosis. Consistent with the known energetic demands of meiosis, the zygotene to pachytene transition in wild-type mice was associated with upregulation of OXPHOS. Mfn1 Mfn2 double mutants exhibited meiotic arrest during this developmental transition, forming fewer pachytene cells that had reduced OXPHOS activity. These data indicate meiosis as the most susceptible stage to loss of mitochondrial fusion. Furthermore, long-term loss of mitochondrial fusion additionally depleted differentiated spermatogonia (Varuzhanyan et al., 2019), which is consistent with upregulation of OXPHOS during spermatogonial differentiation (Lord and Nixon, 2020).
After meiosis, mitochondria undergo robust fragmentation, representing a developmentally regulated mitochondrial fission event. A role for mitochondrial fission in spermatogenesis was shown using mice with a homozygous gene-trap allele of Mff (Mffgt), which have reduced sperm count and subfertility (Chen et al., 2015). In addition, we recently found that round spermatids in Mffgt mice have elongated mitochondria with constrictions indicative of failed fission events (Varuzhanyan et al., 2020). Near the end of spermiogenesis in wild-type mice, small mitochondrial spheres line the sperm axoneme and wrap around the midpiece to form the mitochondrial sheath. This observation suggests that mitochondrial fission in spermatids may facilitate formation of the mitochondrial sheath. Consistent with this idea, Mffgt spermatids have disjointed mitochondrial sheaths with large regions lacking mitochondria, suggesting poor recruitment, or wrapping, of mitochondria around the spermatid midpiece (Varuzhanyan et al., 2020, in press).
The role of mitochondrial dynamics during human spermatogenesis remains largely unknown. A clinical study found that low MFN2 expression in sperm is associated with asthenozoospermia (reduced sperm motility) and reduced sperm mitochondrial membrane potential (Fang et al., 2018). There is no evidence that MFF or other mitochondrial fission factors are important for male fertility in humans. Fetal and adult testis-expressed 1 (FATE1) is a testis-specific protein with some sequence similarity to MFF (Olesen et al., 2001), but its role in human fertility remains inconclusive (Olesen et al., 2003).
Autophagy and mitophagy during spermatogenesis
Recent studies indicate that autophagy is vital for cellular remodeling in post-meiotic spermatids (Shang et al., 2016; Wang et al., 2014). A role for autophagy in spermiogenesis is not surprising given the culling of excess cellular components during this process (Fig. 5). Removal of the core autophagy gene Atg7 (Komatsu et al., 2005) from primordial germ cells diminished autophagic flux in spermatids and blocked acrosome biogenesis (Wang et al., 2014). More recently, Atg7 was found to be important for spermatid polarization and cytoplasmic removal during spermiogenesis (Shang et al., 2016). In elongating spermatids, autophagy was required for the degradation of PDLIM1, a regulator of cytoskeletal dynamics. Removal of Atg7 did not affect development of earlier germ cell types indicating that autophagy is less active in these cells (Shang et al., 2016). This notion was corroborated by a more recent report that found the highest abundance of autophagosomes in spermatids (Yang et al., 2017). Taken together, these data indicate that autophagy contributes to acrosome maintenance, spermatid polarization and degradation of cytoplasmic components during spermiogenesis.
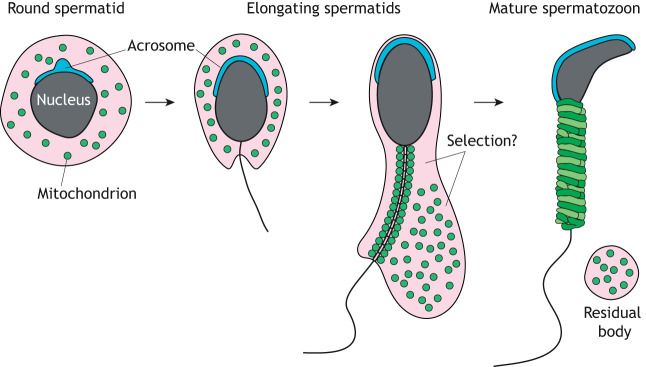
Fig. 5.
Mitochondrial reorganization during spermiogenesis. Schematic of spermiogenesis highlighting the formation of the acrosome and the reorganization of mitochondria. During spermatid elongation, a subset of mitochondria line the sperm midpiece, while the rest are transferred into residual bodies for phagocytic degradation in Sertoli cells. It is unknown how the cell determines the fate of these two mitochondrial populations. The acrosome (blue) is another organelle that undergoes drastic reorganization during spermiogenesis.
Spermiogenesis also removes excess mitochondria, raising the issue of whether mitophagy is involved. In Pink1 mutant flies, spermatids have aberrant mitochondria and defects in individualization (Clark et al., 2006). Furthermore, the UPS, which drives Parkin-mediated mitophagy (Chan and Chan, 2011; Chan et al., 2011; Rakovic et al., 2019), is highly active during mammalian spermiogenesis (Bose et al., 2014; Hermo et al., 2010c). These observations raise the possibility that Parkin-mediated mitophagy is involved in degrading mitochondria during spermiogenesis. Just before spermiation (sperm release), excess mitochondria and other cellular components agglomerate into residual bodies for phagocytic degradation by Sertoli cells (Dietert, 1966) (Fig. 5). However, it is unknown whether spermiogenesis employs a selective or nonselective method for removing mitochondria. Future studies should examine whether mitophagy contributes to the removal of mitochondria from germ cells and/or residual bodies, which notably contain lysosomes (Dietert, 1966).
A key feature of spermatid development is formation of the acrosome (Fig. 5). The acrosome has classically been characterized as a Golgi-derived, lysosome-related organelle (Khawar et al., 2019). Vesicles budding from the trans-Golgi network fuse to each other to form a large proacrosomal granule that attaches to the spermatid nucleus. With continual fusion of vesicles, the acrosomal granule grows and flattens around the spermatid nucleus, eventually covering most of its surface. Besides the Golgi, other sources of membranes can contribute to acrosome biogenesis (Berruti and Paiardi, 2011; Khawar et al., 2019), indicating an essential role for vesicular trafficking during this process. Recently, it was shown that mitochondrial cardiolipin localizes to the acrosome (Ren et al., 2019), suggesting that mitochondria might also provide membranes to the acrosome. Additional evidence also implicates mitochondria in acrosome biogenesis. As described above for cultured cells, FIS1 facilitates mitophagy by interacting with TBC1D15, a RabGAP for RAB7A. TBC1D15 (Zhang et al., 2005) and other TBC domain-containing proteins are highly expressed in the testis and some of them are implicated in acrosome biogenesis. For example, the testis-specific male germ cells Rab GTPase-activating protein (MgcRabGap; encoded by Tbc1d21) colocalizes with RAB3A in the acrosome (Lin et al., 2011). Furthermore, spermatocytes express TBC1D9 (Nakamura et al., 2015) and spermatozoa express a whole host of Rab proteins (Bae et al., 2019). Thus, future studies should explore whether mitochondrial FIS1 regulates vesicular trafficking and mitophagy in post-meiotic spermatids.
Conclusions and perspectives
Mitochondrial dynamics is now well appreciated to maintain basal cellular homeostasis. However, its role in regulating developmentally regulated metabolic transitions remains less well understood. Mammalian spermatogenesis is a promising system for exploring this issue because it requires several such transitions. Furthermore, throughout germ cell development, mitochondrial morphology changes dramatically, culminating with the highly coordinated wrapping of mitochondria around the sperm midpiece. While posttranslational modifications are known to regulate the major mitochondrial fusion and fission factors (Hoppins, 2014), the regulatory networks that orchestrate mitochondrial dynamics during spermatogenesis remain unknown. Because mitochondrial dynamics is intricately linked to cellular metabolism (Mishra and Chan, 2016), these mitochondrial transitions likely reflect the changing metabolic requirements of germ cells as they traverse the compartmentalized seminiferous epithelium.
OXPHOS drives mammalian spermatogenesis and, in particular, meiosis. Emerging evidence indicates that mitochondrial fusion is also critical for spermatogenesis and meiosis by enabling spermatogonial differentiation and a metabolic shift during meiosis (Varuzhanyan et al., 2019; Zhang et al., 2016). OPA1, which plays a central role in mitochondrial fusion and regulates OXPHOS, would be expected to play a similar role during male germ cell development, but this should be tested experimentally. In elongating spermatids, mitochondria elongate as they wrap around the developing midpiece, raising the possibility that these mitochondria undergo fusion. Analysis of static EM images seems to suggest that mitochondria elongate without undergoing fusion (Ho and Wey, 2007). However, future studies should address this genetically by removing mitofusins or Opa1 from post-meiotic spermatids.
Our recent data show that Mff is required in post-meiotic spermatids for developmentally regulated mitochondrial fission and formation of the mitochondrial sheath (Varuzhanyan et al., 2020). Thus, spermiogenesis may be a promising system for studying the regulatory networks that drive mitochondrial fission in vivo. Future studies should test whether other mitochondrial fission factors, such as MID49, MID50 and DRP1, are also required for mitochondrial sheath formation. Removal of Mfn1 does not rescue the spermatogenesis defect of Mffgt mice (Chen et al., 2015), suggesting a specific requirement for mitochondrial fission in spermatids. Taken together, these data suggest that round spermatids acutely downregulate fusion and upregulate fission to ensure robust mitochondrial fragmentation, which may help reorganize mitochondria during spermatid polarization and recruit them to the developing midpiece for formation of the mitochondrial sheath.
Near the end of spermiogenesis, spermatid mitochondria face two distinct fates. A group of ∼50 mitochondria line the midpiece to form the mitochondrial sheath, while the remaining mitochondria, along with other cytosolic components, are transferred into residual bodies destined for phagocytic degradation in Sertoli cells (Fig. 5). It remains unknown whether this mitochondrial segregation happens at random or involves an active selection process. Future studies should explore whether mitophagy is involved during this process. Because mitochondrial fission has been linked to mitophagy (Burman et al., 2017; Tanaka et al., 2010), it should be examined whether mitochondrial fission facilitates culling of excess mitochondria during spermiogenesis. Mitophagy can also occur independently of mitochondrial fission (Yamashita et al., 2016). In this model, small regions of mitochondria bud directly into autophagosomes in a Drp1-independent manner.
The product of spermatogenesis is a highly motile sperm cell capable of a long journey to fertilization. Flagellar motility depends on cellular ATP, which is converted into mechanical work by dynein motors (Serohijos et al., 2006). The source of this ATP has been studied extensively in various species (Storey, 2004). Both glycolysis and OXPHOS are active in spermatozoa, but the primary source of ATP for sperm motility remains unclear (du Plessis et al., 2015). Given the prominence of mitochondria at the sperm midpiece and their relatively high efficiency of generating ATP, it is likely that OXPHOS plays the major role in fueling flagellar locomotion. Indeed, in the genus Mus, the species M. spretus and M. spicilegus, which exhibit higher sperm motility and ATP production, are associated with higher OXPHOS utilization (Tourmente et al., 2015). Thus, it would be interesting to determine whether mitochondrial morphology and ultrastructure correlate with the high OXPHOS utilization in these species.
Mitochondrial dynamics drives spermatogenesis in multiple species. In Drosophila, many of the major mitochondrial dynamics genes – Dmfn, Opa1 and Drp1 – are essential for male germline stem cells, and Fzo is required at the post-meiotic spermatid stage. In mice, fusion is dispensable for maintenance of spermatogonial stem cells but required for spermatogonial differentiation and meiosis. Clinical studies further suggest a role for mitochondrial dynamics in human spermatogenesis. These observations indicate that mitochondrial dynamics may serve as a unique control point for regulating spermatogenesis, which may aid in the development of novel contraceptives and treatments for male infertility.
Acknowledgements
We thank all members of the Chan laboratory for helpful discussions and for comments on the manuscript.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
Work in the laboratory of D.C.C. is funded by the National Institutes of Health (R35GM127147A). Deposited in PMC for release after 12 months.
References
- Aloisio G. M., Nakada Y., Saatcioglu H. D., Peña C. G., Baker M. D., Tarnawa E. D., Mukherjee J., Manjunath H., Bugde A., Sengupta A. L. et al. (2014). PAX7 expression defines germline stem cells in the adult testis. J. Clin. Invest. 124, 3929-3944. 10.1172/JCI75943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae J.-W., Kim S.-H., Kim D.-H., Ha J. J., Yi J. K., Hwang S., Ryu B.-Y., Pang M.-G. and Kwon W.-S. (2019). Ras-related proteins (Rab) are key proteins related to male fertility following a unique activation mechanism. Reprod. Biol. 19, 356-362. 10.1016/j.repbio.2019.10.001 [DOI] [PubMed] [Google Scholar]
- Bajpai M., Gupta G. and Setty B. S. (1998). Changes in carbohydrate metabolism of testicular germ cells during meiosis in the rat. Eur. J. Endocrinol. 138, 322-327. 10.1530/eje.0.1380322 [DOI] [PubMed] [Google Scholar]
- Baklouti-Gargouri S., Ghorbel M., Ben Mahmoud A., Mkaouar-Rebai E., Cherif M., Chakroun N., Sellami A., Fakhfakh F. and Ammar-Keskes L. (2014). Identification of a novel m.9588G>a missense mutation in the mitochondrial COIII gene in asthenozoospermic Tunisian infertile men. J. Assist. Reprod. Genet. 31, 595-600. 10.1007/s10815-014-0187-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao J. and Bedford M. T. (2016). Epigenetic regulation of the histone-to-protamine transition during spermiogenesis. Reproduction 151, R55-R70. 10.1530/REP-15-0562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berruti G. and Paiardi C. (2011). Acrosome biogenesis. Spermatogenesis 1, 95-98. 10.4161/spmg.1.2.16820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose R., Manku G., Culty M. and Wing S. S. (2014). Ubiquitin–Proteasome System in Spermatogenesis. In Posttranslational Protein Modifications in the Reproductive System (ed. Sutovsky P.), pp. 181-213. New York, NY: Springer. [DOI] [PubMed] [Google Scholar]
- Boussouar F. and Benahmed M. (2004). Lactate and energy metabolism in male germ cells. Trends Endocrinol. Metab. 15, 345-350. 10.1016/j.tem.2004.07.003 [DOI] [PubMed] [Google Scholar]
- Braun R. E., Behringer R. R., Peschon J. J., Brinster R. L. and Palmiter R. D. (1989). Genetically haploid spermatids are phenotypically diploid. Nature 337, 373-376. 10.1038/337373a0 [DOI] [PubMed] [Google Scholar]
- Brower J. V., Rodic N., Seki T., Jorgensen M., Fliess N., Yachnis A. T., McCarrey J. R., Oh S. P. and Terada N. (2007). Evolutionarily conserved mammalian adenine nucleotide translocase 4 is essential for spermatogenesis. J. Biol. Chem. 282, 29658-29666. 10.1074/jbc.M704386200 [DOI] [PubMed] [Google Scholar]
- Brower J. V., Lim C. H., Jorgensen M., Oh S. P. and Terada N. (2009). Adenine nucleotide translocase 4 deficiency leads to early meiotic arrest of murine male germ cells. Reproduction 138, 463-470. 10.1530/REP-09-0201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burman J. L., Pickles S., Wang C., Sekine S., Vargas J. N. S., Zhang Z., Youle A. M., Nezich C. L., Wu X., Hammer J. A. et al. (2017). Mitochondrial fission facilitates the selective mitophagy of protein aggregates. J. Cell Biol. 216, 3231-3247. 10.1083/jcb.201612106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carra E., Sangiorgi D., Gattuccio F. and Rinaldi A. M. (2004). Male infertility and mitochondrial DNA. Biochem. Biophys. Res. Commun. 322, 333-339. 10.1016/j.bbrc.2004.07.112 [DOI] [PubMed] [Google Scholar]
- Chan D. C. (2012). Fusion and fission: interlinked processes critical for mitochondrial health. Annu. Rev. Genet. 46, 265-287. 10.1146/annurev-genet-110410-132529 [DOI] [PubMed] [Google Scholar]
- Chan D. C. (2020). Mitochondrial dynamics and its involvement in disease. Annu. Rev. Pathol. Mech. Dis. 15, 235-259. 10.1146/annurev-pathmechdis-012419-032711 [DOI] [PubMed] [Google Scholar]
- Chan N. C. and Chan D. C. (2011). Parkin uses the UPS to ship off dysfunctional mitochondria. Autophagy 7, 771-772. 10.4161/auto.7.7.15453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan N. C., Salazar A. M., Pham A. H., Sweredoski M. J., Kolawa N. J., Graham R. L. J., Hess S. and Chan D. C. (2011). Broad activation of the ubiquitin–proteasome system by Parkin is critical for mitophagy. Hum. Mol. Genet. 20, 1726-1737. 10.1093/hmg/ddr048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Detmer S. A., Ewald A. J., Griffin E. E., Fraser S. E. and Chan D. C. (2003). Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 160, 189-200. 10.1083/jcb.200211046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., McCaffery J. M. and Chan D. C. (2007). Mitochondrial fusion protects against neurodegeneration in the cerebellum. Cell 130, 548-562. 10.1016/j.cell.2007.06.026 [DOI] [PubMed] [Google Scholar]
- Chen H., Vermulst M., Wang Y. E., Chomyn A., Prolla T. A., McCaffery J. M. and Chan D. C. (2010). Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell 141, 280-289. 10.1016/j.cell.2010.02.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Liu Y. and Dorn G. W. (2011). Mitochondrial fusion is essential for organelle function and cardiac homeostasis. Circ. Res. 109, 1327-1331. 10.1161/CIRCRESAHA.111.258723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Ren S., Clish C., Jain M., Mootha V., McCaffery J. M. and Chan D. C. (2015). Titration of mitochondrial fusion rescues Mff-deficient cardiomyopathy. J. Cell Biol. 211, 795-805. 10.1083/jcb.201507035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark I. E., Dodson M. W., Jiang C., Cao J. H., Huh J. R., Seol J. H., Yoo S. J., Hay B. A. and Guo M. (2006). Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature 441, 1162 10.1038/nature04779 [DOI] [PubMed] [Google Scholar]
- Cohen P. E., Pollack S. E. and Pollard J. W. (2006). Genetic analysis of chromosome pairing, recombination, and cell cycle control during first meiotic prophase in mammals. Endocr. Rev. 27, 398-426. 10.1210/er.2005-0017 [DOI] [PubMed] [Google Scholar]
- De Martino C., Floridi A., Marcante M. L., Malorni W., Barcellona P. S., Bellocci M. and Silvestrini B. (1979). Morphological, histochemical and biochemical studies on germ cell mitochondria of normal rats. Cell Tissue Res. 196, 1-22. 10.1007/BF00236345 [DOI] [PubMed] [Google Scholar]
- De Rooij D. G. (2017). The nature and dynamics of spermatogonial stem cells. Development 144, 3022-3030. 10.1242/dev.146571 [DOI] [PubMed] [Google Scholar]
- Demain L. A. M., Conway G. S. and Newman W. G. (2017). Genetics of mitochondrial dysfunction and infertility. Clin. Genet. 91, 199-207. 10.1111/cge.12896 [DOI] [PubMed] [Google Scholar]
- Demarco R. S. and Jones D. L. (2019). Mitochondrial fission regulates germ cell differentiation by suppressing ROS-mediated activation of Epidermal Growth Factor Signaling in the Drosophila larval testis. Sci. Rep. 9, 19695 10.1038/s41598-019-55728-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demarco R. S., Eikenes Å. H., Haglund K. and Jones D. L. (2014). Investigating spermatogenesis in Drosophila melanogaster. Methods 68, 218-227. 10.1016/j.ymeth.2014.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demarco R. S., Uyemura B. S., D'Alterio C. and Jones D. L. (2019). Mitochondrial fusion regulates lipid homeostasis and stem cell maintenance in the Drosophila testis. Nat. Cell Biol. 21, 710 10.1038/s41556-019-0332-3 [DOI] [PubMed] [Google Scholar]
- Dietert S. E. (1966). Fine structure of the formation and fate of the residual bodies of mouse spermatozoa with evidence for the participation of lysosomes. J. Morphol. 120, 317-346. 10.1002/jmor.1051200402 [DOI] [Google Scholar]
- du Plessis S. S., Agarwal A., Mohanty G. and Van der Linde M. (2015). Oxidative phosphorylation versus glycolysis: what fuel do spermatozoa use? Asian J. Androl. 17, 230 10.4103/1008-682X.135123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang F., Ni K., Shang J., Zhang X., Xiong C. and Meng T. (2018). Expression of mitofusin 2 in human sperm and its relationship to sperm motility and cryoprotective potentials. Exp. Biol. Med. 243, 963-969. 10.1177/1535370218790919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaro G., Romanello V., Varanita T., Desbats M. A., Morbidoni V., Tezze C., Albiero M., Canato M., Gherardi G., De Stefani D. et al. (2019). DRP1-mediated mitochondrial shape controls calcium homeostasis and muscle mass. Nat. Commun. 10, 1-17. 10.1038/s41467-019-10226-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folgerø T., Bertheussen K., Lindal S., Torbergsen T. and Øian P. (1993). Mitochondrial disease and reduced sperm motility. Hum. Reprod. 8, 1863-1868. 10.1093/oxfordjournals.humrep.a137950 [DOI] [PubMed] [Google Scholar]
- Fonseca T. B., Sánchez-Guerrero Á., Milosevic I. and Raimundo N. (2019). Mitochondrial fission requires DRP1 but not dynamins. Nature 570, E34-E42. 10.1038/s41586-019-1296-y [DOI] [PubMed] [Google Scholar]
- Friedman J. R., Lackner L. L., West M., DiBenedetto J. R., Nunnari J. and Voeltz G. K. (2011). ER tubules mark sites of mitochondrial division. Science 334, 358-362. 10.1126/science.1207385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller M. T. (1993). Spermatogenesis. In The Development of Drosophila melanogaster, (Eds M. Bate; A. Martinez Arias) pp. 71-147. New York: Cold Spring Harbor. [Google Scholar]
- Goldberg E., Eddy E. M., Duan C. and Odet F. (2010). LDHC: the ultimate testis-specific gene. J. Androl. 31, 86-94. 10.2164/jandrol.109.008367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum M. P., Iwamori T., Buchold G. M. and Matzuk M. M. (2011). Germ Cell Intercellular Bridges. Cold Spring Harb. Perspect. Biol. 3, a005850 10.1101/cshperspect.a005850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griswold M. D. (1998). The central role of Sertoli cells in spermatogenesis. Semin. Cell Dev. Biol. 9, 411-416. 10.1006/scdb.1998.0203 [DOI] [PubMed] [Google Scholar]
- Griswold M. D. (2016). Spermatogenesis: the commitment to meiosis. Physiol. Rev. 96, 1-17. 10.1152/physrev.00013.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grootegoed J. A., Jansen R. and Van der Molen H. J. (1984). The role of glucose, pyruvate and lactate in ATP production by rat spermatocytes and spermatids. Biochim. Biophys. Acta 767, 248-256. 10.1016/0005-2728(84)90194-4 [DOI] [PubMed] [Google Scholar]
- Hackenbrock C. R. (1966). Ultrastructural bases for metabolically linked mechanical activity in mitochondria I. Reversible Ultrastructural Changes with Change in Metabolic Steady State in Isolated Liver Mitochondria. J. Cell Biol. 30, 269-297. 10.1083/jcb.30.2.269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hales K. G. and Fuller M. T. (1997). Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell 90, 121-129. 10.1016/S0092-8674(00)80319-0 [DOI] [PubMed] [Google Scholar]
- Handel M. A. and Schimenti J. C. (2010). Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nat. Rev. Genet. 11, 124-136. 10.1038/nrg2723 [DOI] [PubMed] [Google Scholar]
- Hara K., Nakagawa T., Enomoto H., Suzuki M., Yamamoto M., Simons B. D. and Yoshida S. (2014). Mouse spermatogenic stem cells continually interconvert between equipotent singly isolated and syncytial states. Cell Stem Cell 14, 658-672. 10.1016/j.stem.2014.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermo L., Pelletier R.-M., Cyr D. G. and Smith C. E. (2010a). Surfing the wave, cycle, life history, and genes/proteins expressed by testicular germ cells. Part 1: background to spermatogenesis, spermatogonia, and spermatocytes. Microsc. Res. Tech. 73, 241-278. 10.1002/jemt.20783 [DOI] [PubMed] [Google Scholar]
- Hermo L., Pelletier R.-M., Cyr D. G. and Smith C. E. (2010b). Surfing the wave, cycle, life history, and genes/proteins expressed by testicular germ cells. Part 2: changes in spermatid organelles associated with development of spermatozoa. Microsc. Res. Tech. 73, 279-319. 10.1002/jemt.20787 [DOI] [PubMed] [Google Scholar]
- Hermo L., Pelletier R.-M., Cyr D. G. and Smith C. E. (2010c). Surfing the wave, cycle, life history, and genes/proteins expressed by testicular germ cells. Part 4: intercellular bridges, mitochondria, nuclear envelope, apoptosis, ubiquitination, membrane/voltage-gated channels, methylation/acetylation, and transcription factors. Microsc. Res. Tech. 73, 364-408. 10.1002/jemt.20785 [DOI] [PubMed] [Google Scholar]
- Ho H.-C. and Wey S. (2007). Three dimensional rendering of the mitochondrial sheath morphogenesis during mouse spermiogenesis. Microsc. Res. Tech. 70, 719-723. 10.1002/jemt.20457 [DOI] [PubMed] [Google Scholar]
- Hofmann M.-C. (2008). Gdnf signaling pathways within the mammalian spermatogonial stem cell niche. Mol. Cell. Endocrinol. 288, 95-103. 10.1016/j.mce.2008.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppins S. (2014). The regulation of mitochondrial dynamics. Curr. Opin. Cell Biol. 29, 46-52. 10.1016/j.ceb.2014.03.005 [DOI] [PubMed] [Google Scholar]
- Ikeda Y., Shirakabe A., Maejima Y., Zhai P., Sciarretta S., Toli J., Nomura M., Mihara K., Egashira K., Ohishi M. et al. (2015). Endogenous Drp1 mediates mitochondrial autophagy and protects the heart against energy stress. Circ. Res. 116, 264-278. 10.1161/CIRCRESAHA.116.303356 [DOI] [PubMed] [Google Scholar]
- Ishihara N., Nomura M., Jofuku A., Kato H., Suzuki S. O., Masuda K., Otera H., Nakanishi Y., Nonaka I., Goto Y.-I. et al. (2009). Mitochondrial fission factor Drp1 is essential for embryonic development and synapse formation in mice. Nat. Cell Biol. 11, 958-966. 10.1038/ncb1907 [DOI] [PubMed] [Google Scholar]
- Jakobs S., Martini N., Schauss A. C., Egner A., Westermann B. and Hell S. W. (2003). Spatial and temporal dynamics of budding yeast mitochondria lacking the division component Fis1p. J. Cell. Sci. 116, 2005-2014. 10.1242/jcs.00423 [DOI] [PubMed] [Google Scholar]
- Jiang M., Kauppila T. E. S., Motori E., Li X., Atanassov I., Folz-Donahue K., Bonekamp N. A., Albarran-Gutierrez S., Stewart J. B. and Larsson N.-G. (2017). Increased total mtDNA copy number cures male infertility despite unaltered mtDNA mutation load. Cell Metab. 26, 429-436.e4. 10.1016/j.cmet.2017.07.003 [DOI] [PubMed] [Google Scholar]
- Kageyama Y., Hoshijima M., Seo K., Bedja D., Sysa-Shah P., Andrabi S. A., Chen W., Höke A., Dawson V. L., Dawson T. M. et al. (2014). Parkin-independent mitophagy requires Drp1 and maintains the integrity of mammalian heart and brain. EMBO J. 33, 2798-2813. 10.15252/embj.201488658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamerkar S. C., Kraus F., Sharpe A. J., Pucadyil T. J. and Ryan M. T. (2018). Dynamin-related protein 1 has membrane constricting and severing abilities sufficient for mitochondrial and peroxisomal fission. Nat. Commun. 9, 5239 10.1038/s41467-018-07543-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao S.-H., Chao H.-T. and Wei Y.-H. (1995). Mitochondrial deoxyribonucleic acid 4977-bp deletion is associated with diminished fertility and motility of human sperm. Biol. Reprod. 52, 729-736. 10.1095/biolreprod52.4.729 [DOI] [PubMed] [Google Scholar]
- Khawar M. B., Gao H. and Li W. (2019). Mechanism of acrosome biogenesis in mammals. Front. Cell Dev. Biol. 7, 195 10.3389/fcell.2019.00195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble J. E. and White J. G. (1981). On the control of germ cell development in Caenorhabditis elegans. Dev. Biol. 81, 208-219. 10.1016/0012-1606(81)90284-0 [DOI] [PubMed] [Google Scholar]
- Komatsu M., Waguri S., Ueno T., Iwata J., Murata S., Tanida I., Ezaki J., Mizushima N., Ohsumi Y., Uchiyama Y. et al. (2005). Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 169, 425-434. 10.1083/jcb.200412022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korobova F., Ramabhadran V. and Higgs H. N. (2013). An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science 339, 464-467. 10.1126/science.1228360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujoth G. C., Hiona A., Pugh T. D., Someya S., Panzer K., Wohlgemuth S. E., Hofer T., Seo A. Y., Sullivan R., Jobling W. A. et al. (2005). Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science 309, 481-484. 10.1126/science.1112125 [DOI] [PubMed] [Google Scholar]
- Kuma A., Komatsu M. and Mizushima N. (2017). Autophagy-monitoring and autophagy-deficient mice. Autophagy 13, 1619-1628. 10.1080/15548627.2017.1343770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. E., Westrate L. M., Wu H., Page C. and Voeltz G. K. (2016). Multiple dynamin family members collaborate to drive mitochondrial division. Nature 540, 139-143. 10.1038/nature20555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lestienne P., Reynier P., Chretien M. F., Penisson-Besnier I., Malthiery Y. and Rohmer V. (1997). Oligoasthenospermia associated with multiple mitochondrial DNA rearrangements. Mol. Hum. Reprod. 3, 811-814. 10.1093/molehr/3.9.811 [DOI] [PubMed] [Google Scholar]
- Lin Y.-H., Lin Y.-M., Kuo Y.-C., Wang Y.-Y. and Kuo P.-L. (2011). Identification and characterization of a novel Rab GTPase-activating protein in spermatids. Int. J. Androl. 34, e358-e367. 10.1111/j.1365-2605.2010.01126.x [DOI] [PubMed] [Google Scholar]
- Lord T. and Nixon B. (2020). Metabolic Changes Accompanying Spermatogonial Stem Cell Differentiation. Dev. Cell 52, 399-411. 10.1016/j.devcel.2020.01.014 [DOI] [PubMed] [Google Scholar]
- Lord T. and Oatley J. M. (2017). A revised Asingle model to explain stem cell dynamics in the mouse male germline. Reproduction 154, R55-R64. 10.1530/REP-17-0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losón O. C., Song Z., Chen H. and Chan D. C. (2013). Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Mol. Biol. Cell 24, 659-667. 10.1091/mbc.e12-10-0721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luoma P., Melberg A., Rinne J. O., Kaukonen J. A., Nupponen N. N., Chalmers R. M., Oldfors A., Rautakorpi I., Peltonen L., Majamaa K. et al. (2004). Parkinsonism, premature menopause, and mitochondrial DNA polymerase γ mutations: clinical and molecular genetic study. The Lancet 364, 875-882. 10.1016/S0140-6736(04)16983-3 [DOI] [PubMed] [Google Scholar]
- Mannella C. A. (2006a). The relevance of mitochondrial membrane topology to mitochondrial function. Biochim. Biophys. Acta 1762, 140-147. 10.1016/j.bbadis.2005.07.001 [DOI] [PubMed] [Google Scholar]
- Mannella C. A. (2006b). Structure and dynamics of the mitochondrial inner membrane cristae. Biochim. Biophys. Acta Mol. Cell Res. 1763, 542-548. 10.1016/j.bbamcr.2006.04.006 [DOI] [PubMed] [Google Scholar]
- McWilliams T. G., Prescott A. R., Allen G. F. G., Tamjar J., Munson M. J., Thomson C., Muqit M. M. K. and Ganley I. G. (2016). mito-QC illuminates mitophagy and mitochondrial architecture in vivo. J. Cell Biol. 214, 333-345. 10.1083/jcb.201603039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhardt A., Wilhelm B. and Seitz J. (1999). Mini symposium. New aspects of spermatogenesis. Expression of mitochondrial marker proteins during spermatogenesis. Hum. Reprod. Update 5, 108-119. 10.1093/humupd/5.2.108 [DOI] [PubMed] [Google Scholar]
- Mishra P. and Chan D. C. (2016). Metabolic regulation of mitochondrial dynamics. J. Cell Biol. 212, 379-387. 10.1083/jcb.201511036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra P., Varuzhanyan G., Pham A. H. and Chan D. C. (2015). Mitochondrial dynamics is a distinguishing feature of skeletal muscle fiber types and regulates organellar compartmentalization. Cell Metab. 22, 1033-1044. 10.1016/j.cmet.2015.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N. (2007). Autophagy: process and function. Genes Dev. 21, 2861-2873. 10.1101/gad.1599207 [DOI] [PubMed] [Google Scholar]
- Mizushima N., Sugita H., Yoshimori T. and Ohsumi Y. (1998). A new protein conjugation system in human the counterpart of the yeast Apg12p conjugation system essential for autophagy. J. Biol. Chem. 273, 33889-33892. 10.1074/jbc.273.51.33889 [DOI] [PubMed] [Google Scholar]
- Mozdy A. D., McCaffery J. M. and Shaw J. M. (2000). Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J. Cell Biol. 151, 367-380. 10.1083/jcb.151.2.367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima S., Tábara L.-C., Tilokani L., Paupe V., Anand H., Pogson J. H., Zunino R., McBride H. M. and Prudent J. (2020). Golgi-derived PI(4)P-containing vesicles drive late steps of mitochondrial division. Science 367, 1366-1371. 10.1126/science.aax6089 [DOI] [PubMed] [Google Scholar]
- Nakada K., Sato A., Yoshida K., Morita T., Tanaka H., Inoue S.-I., Yonekawa H. and Hayashi J.-I. (2006). Mitochondria-related male infertility. Proc. Natl. Acad. Sci. USA 103, 15148-15153. 10.1073/pnas.0604641103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M., Fujiwara A., Yasumasu I., Okinaga S. and Arai K. (1982). Regulation of glucose metabolism by adenine nucleotides in round spermatids from rat testes. J. Biol. Chem. 257, 13945-13950. [PubMed] [Google Scholar]
- Nakamura M., Okinaga S. and Arai K. (1984). Metabolism of pachytene primary spermatocytes from rat testes: pyruvate maintenance of adenosine triphosphate level. Biol. Reprod. 30, 1187-1197. 10.1095/biolreprod30.5.1187 [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Asano A., Hosaka Y., Takeuchi T., Iwanaga T. and Yamano Y. (2015). Expression and intracellular localization of TBC1D9, a Rab GTPase-accelerating protein, in mouse testes. Exp. Anim. 64, 415-424. 10.1538/expanim.15-0016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D. P., Jin S. M., Tanaka A., Suen D.-F., Gautier C. A., Shen J., Cookson M. R. and Youle R. J. (2010). PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 8, e1000298 10.1371/journal.pbio.1000298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naughton C. K., Jain S., Strickland A. M., Gupta A. and Milbrandt J. (2006). Glial cell-line derived neurotrophic factor-mediated RET signaling regulates spermatogonial stem cell fate. Biol. Reprod. 74, 314-321. 10.1095/biolreprod.105.047365 [DOI] [PubMed] [Google Scholar]
- Oatley J. M. and Brinster R. L. (2012). The germline stem cell niche unit in mammalian testes. Physiol. Rev. 92, 577-595. 10.1152/physrev.00025.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olesen C., Larsen N. J., Byskov A. G., Harboe T. L. and Tommerup N. (2001). Human FATE is a novel X-linked gene expressed in fetal and adult testis. Mol. Cell. Endocrinol. 184, 25-32. 10.1016/S0303-7207(01)00666-9 [DOI] [PubMed] [Google Scholar]
- Olesen C., Silber J., Eiberg H., Ernst E., Petersen K., Lindenberg S. and Tommerup N. (2003). Mutational analysis of the human FATE gene in 144 infertile men. Hum. Genet. 113, 195-201. 10.1007/s00439-003-0974-9 [DOI] [PubMed] [Google Scholar]
- Onoue K., Jofuku A., Ban-Ishihara R., Ishihara T., Maeda M., Koshiba T., Itoh T., Fukuda M., Otera H., Oka T. et al. (2013). Fis1 acts as a mitochondrial recruitment factor for TBC1D15 that is involved in regulation of mitochondrial morphology. J. Cell. Sci. 126, 176-185. 10.1242/jcs.111211 [DOI] [PubMed] [Google Scholar]
- Otera H., Wang C., Cleland M. M., Setoguchi K., Yokota S., Youle R. J. and Mihara K. (2010). Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J. Cell Biol. 191, 1141-1158. 10.1083/jcb.201007152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papanicolaou K. N., Kikuchi R., Ngoh G. A., Coughlan K. A., Dominguez I., Stanley W. C. and Walsh K. (2012). Mitofusins 1 and 2 are essential for postnatal metabolic remodeling in heart. Circ. Res. 111, 1012-1026. 10.1161/CIRCRESAHA.112.274142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickles S., Vigié P. and Youle R. J. (2018). Mitophagy and quality control mechanisms in mitochondrial maintenance. Curr. Biol. 28, R170-R185. 10.1016/j.cub.2018.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakovic A., Ziegler J., Mårtensson C. U., Prasuhn J., Shurkewitsch K., König P., Paulson H. L. and Klein C. (2019). PINK1-dependent mitophagy is driven by the UPS and can occur independently of LC3 conversion. Cell Death Differ. 26, 1428-1441. 10.1038/s41418-018-0219-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rato L., Alves M. G., Socorro S., Duarte A. I., Cavaco J. E. and Oliveira P. F. (2012). Metabolic regulation is important for spermatogenesis. Nat. Rev. Urol. 9, 330-338. 10.1038/nrurol.2012.77 [DOI] [PubMed] [Google Scholar]
- Ren M., Xu Y., Erdjument-Bromage H., Donelian A., Phoon C. K. L., Terada N., Strathdee D., Neubert T. A. and Schlame M. (2019). Extramitochondrial cardiolipin suggests a novel function of mitochondria in spermatogenesis. J. Cell Biol. 218, 1491-1502. 10.1083/jcb.201808131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojansky R., Cha M.-Y. and Chan D. C. (2016). Elimination of paternal mitochondria in mouse embryos occurs through autophagic degradation dependent on PARKIN and MUL1. eLife 5, e17896 10.7554/eLife.17896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovio A. T., Marchington D. R., Donat S., Schuppe H.-C., Abel J., Fritsche E., Elliott D. J., Laippala P., Ahola A. L., McNay D. et al. (2001). Mutations at the mitochondrial DNA polymerase (POLG) locus associated with male infertility. Nat. Genet. 29, 261-262. 10.1038/ng759 [DOI] [PubMed] [Google Scholar]
- Seitz J., Möbius J., Bergmann M. and Meinhardt A. (1995). Mitochondrial differentiation during meiosis of male germ cells. Int. J. Androl. 18 Suppl. 2, 7-11. [PubMed] [Google Scholar]
- Sekine S. and Youle R. J. (2018). PINK1 import regulation; a fine system to convey mitochondrial stress to the cytosol. BMC Biol. 16, 2 10.1186/s12915-017-0470-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serohijos A. W. R., Chen Y., Ding F., Elston T. C. and Dokholyan N. V. (2006). A structural model reveals energy transduction in dynein. Proc. Natl. Acad. Sci. USA 103, 18540-18545. 10.1073/pnas.0602867103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y., Wang H., Jia P., Zhao H., Liu C., Liu W., Song Z., Xu Z., Yang L. and Wang Y. (2016). Autophagy regulates spermatid differentiation via degradation of PDLIM1. Autophagy 12, 1575-1592. 10.1080/15548627.2016.1192750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q., Yamano K., Head B. P., Kawajiri S., Cheung J. T. M., Wang C., Cho J.-H., Hattori N., Youle R. J. and van der Bliek A. M. (2014). Mutations in Fis1 disrupt orderly disposal of defective mitochondria. Mol. Biol. Cell 25, 145-159. 10.1091/mbc.e13-09-0525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiropoulos J., Turnbull D. M. and Chinnery P. F. (2002). Can mitochondrial DNA mutations cause sperm dysfunction? Mol. Hum. Reprod. 8, 719-721. 10.1093/molehr/8.8.719 [DOI] [PubMed] [Google Scholar]
- Spradling A., Drummond-Barbosa D. and Kai T. (2001). Stem cells find their niche. Nature 414, 98-104. 10.1038/35102160 [DOI] [PubMed] [Google Scholar]
- Stanton P. G. (2016). Regulation of the blood-testis barrier. Semin. Cell Dev. Biol. 59, 166-173. 10.1016/j.semcdb.2016.06.018 [DOI] [PubMed] [Google Scholar]
- Storey B. T. (2004). Mammalian sperm metabolism: oxygen and sugar, friend and foe. Int. J. Dev. Biol. 52, 427-437. 10.1387/ijdb.072522bs [DOI] [PubMed] [Google Scholar]
- Sun N., Yun J., Liu J., Malide D., Liu C., Rovira I. I., Holmström K. M., Fergusson M. M., Yoo Y. H., Combs C. A. et al. (2015). Measuring In Vivo Mitophagy. Mol. Cell 60, 685-696. 10.1016/j.molcel.2015.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan E. H. N. and Tang B. L. (2019). Rab7a and Mitophagosome Formation. Cells 8, 224 10.3390/cells8030224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A., Cleland M. M., Xu S., Narendra D. P., Suen D.-F., Karbowski M. and Youle R. J. (2010). Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J. Cell Biol. 191, 1367-1380. 10.1083/jcb.201007013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieu Q., Okreglak V., Naylor K. and Nunnari J. (2002). The WD repeat protein, Mdv1p, functions as a molecular adaptor by interacting with Dnm1p and Fis1p during mitochondrial fission. J. Cell Biol. 158, 445-452. 10.1083/jcb.200205031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourmente M., Villar-Moya P., Rial E. and Roldan E. R. S. (2015). Differences in ATP Generation Via Glycolysis and Oxidative Phosphorylation and Relationships with Sperm Motility in Mouse Species. J. Biol. Chem. 290, 20613-20626. 10.1074/jbc.M115.664813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifunovic A., Wredenberg A., Falkenberg M., Spelbrink J. N., Rovio A. T., Bruder C. E., Bohlooly Y.-M., Gidlöf S., Oldfors A., Wibom R. et al. (2004). Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature 429, 417-423. 10.1038/nature02517 [DOI] [PubMed] [Google Scholar]
- Tsukada M. and Ohsumi Y. (1993). Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 333, 169-174. 10.1016/0014-5793(93)80398-E [DOI] [PubMed] [Google Scholar]
- Tzagoloff A. (1982). Mitochondrial structure and compartmentalization. In Mitochondria (ed. Tzagoloff A.), pp. 15-38. Boston, MA: Springer US. [Google Scholar]
- Varuzhanyan G., Rojansky R., Sweredoski M. J., Graham R. L. J., Hess S., Ladinsky M. S. and Chan D. C. (2019). Mitochondrial fusion is required for spermatogonial differentiation and meiosis. eLife 8, e51601 10.7554/eLife.51601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varuzhanyan G., Chen H., Rojansky R., Ladinsky M. S., McCaffery M. and Chan D. (2020). Mitochondrial fission is required for organization of the mitochondrial sheath in spermatids. Biochim. Biophy. Acta General Subjects. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi J., Zhang Z., Wakabayashi N., Tamura Y., Fukaya M., Kensler T. W., Iijima M. and Sesaki H. (2009). The dynamin-related GTPase Drp1 is required for embryonic and brain development in mice. J. Cell Biol. 186, 805-816. 10.1083/jcb.200903065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Wan H., Li X., Liu W., Chen Q., Wang Y., Yang L., Tang H., Zhang X., Duan E. et al. (2014). Atg7 is required for acrosome biogenesis during spermatogenesis in mice. Cell Res. 24, 852 10.1038/cr.2014.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong Y. C., Ysselstein D. and Krainc D. (2018). Mitochondria-lysosome contacts regulate mitochondrial fission via RAB7 GTP hydrolysis. Nature 554, 382-386. 10.1038/nature25486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano K., Fogel A. I., Wang C., van der Bliek A. M. and Youle R. J. (2014). Mitochondrial Rab GAPs govern autophagosome biogenesis during mitophagy. eLife 3, e01612 10.7554/eLife.01612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano K., Wang C., Sarraf S. A., Münch C., Kikuchi R., Noda N. N., Hizukuri Y., Kanemaki M. T., Harper W., Tanaka K. et al. (2018). Endosomal Rab cycles regulate Parkin-mediated mitophagy. eLife 7, e31326 10.7554/eLife.31326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita S.-I., Jin X., Furukawa K., Hamasaki M., Nezu A., Otera H., Saigusa T., Yoshimori T., Sakai Y., Mihara K. et al. (2016). Mitochondrial division occurs concurrently with autophagosome formation but independently of Drp1 during mitophagy. J. Cell Biol. 215, 649-665. 10.1083/jcb.201605093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P., Ahmed N., Wang L., Chen H., Waqas Y., Liu T., Haseeb A., Bangulzai N., Huang Y. and Chen Q. (2017). In vivo autophagy and biogenesis of autophagosomes within male haploid cells during spermiogenesis. Oncotarget 8, 56791-56801. 10.18632/oncotarget.18221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle R. J. and Narendra D. P. (2011). Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 12, 9-14. 10.1038/nrm3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.-M., Walsh B., Mitchell C. A. and Rowe T. (2005). TBC domain family, member 15 is a novel mammalian Rab GTPase-activating protein with substrate preference for Rab7. Biochem. Biophys. Res. Commun. 335, 154-161. 10.1016/j.bbrc.2005.07.070 [DOI] [PubMed] [Google Scholar]
- Zhang J., Wang Q., Wang M., Jiang M., Wang Y., Sun Y., Wang J., Xie T., Tang C., Tang N. et al. (2016). GASZ and mitofusin-mediated mitochondrial functions are crucial for spermatogenesis. EMBO Rep. 17, 220-234. 10.15252/embr.201540846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Sliter D. A., Bleck C. K. E. and Ding S. (2019). Fis1 deficiencies differentially affect mitochondrial quality in skeletal muscle. Mitochondrion 49, 217-226. 10.1016/j.mito.2019.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]