Abstract
In patients with locally advanced breast cancer undergoing neoadjuvant chemotherapy (NAC), some patients achieve a complete pathologic response (pCR), some achieve a partial response, and some do not respond at all or even progress. Accurate prediction of treatment response has the potential to improve patient care by improving prognostication, enabling de-escalation of toxic treatment that has little benefit, facilitating upfront use of novel targeted therapies, and avoiding delays to surgery. Visual inspection of a patient’s tumor on multiparametric MRI is insufficient to predict that patient’s response to NAC. However, machine learning and deep learning approaches using a mix of qualitative and quantitative MRI features have recently been applied to predict treatment response early in the course of or even before the start of NAC. This is a novel field but the data published so far has shown promising results. We provide an overview of the machine learning and deep learning models developed to date, as well as discuss some of the challenges to clinical implementation.
Keywords: Artificial intelligence, Machine learning, Multiparametric MRI, Neoadjuvant chemotherapy
Abbreviations: 1H-MRS, proton magnetic resonance spectroscopy; 23 N MRS, sodium magnetic resonance spectroscopy; ADC, apparent diffusion coefficient; ANN, artificial neural network; AUC, area under the curve; CNN, convolutional neural network; DCE-MRI, dynamic contrast-enhanced magnetic resonance imaging; DWI, diffusion-weighted imaging; EF, enhancement fraction; FGT, fibroglandular tissue; LR, logistic regression; MB, Markov blanket; NAC, neoadjuvant chemotherapy; pCR, pathologic complete response; SVM, support vector machine; TN, triple negative
Highlights
-
•
Machine and deep learning have shown potential in predicting neoadjuvant treatment outcomes using multiparametric MRI data.
-
•
Machine and deep learning-based early identification of chemotherapy non-responders could improve patient management.
-
•
Deep learning techniques using CNN may prove to be more powerful and more robust than traditional machine learning classifiers.
1. Clinical background
In patients with locally advanced breast cancer, treatment has historically consisted of surgical resection followed by post-operative radiation and chemotherapy. Since clinical trials have demonstrated that neoadjuvant chemotherapy (NAC), or chemotherapy administered prior to surgery, is equivalent to chemotherapy administered after surgery, an increasing number of patients are receiving NAC prior to surgery. The primary goal of NAC is to decrease the size of the tumor, leading to downstaging or even pathologic complete response (pCR). This enables breast conservation surgery (BCS) in women who previously required a mastectomy as well as less extensive BCS; additionally, it also eliminates the need for axillary lymph node dissection in a subset of patients, saving them the long-term morbidity of associated lymphedema. In early-stage breast cancer, NAC has been proposed as a potential standard of care, and to date it is widely used to treat triple-negative and HER2+ subtypes of breast cancer, enabling increased rates of breast-conserving surgery and decreased axillary dissection [1]. A pCR to NAC is significantly associated with improved disease-free and overall survival in high-risk breast cancer subtypes [2], whereas a poor response to NAC is associated with an adverse prognosis [3]. However, pCR is only achieved in only 30–50% of breast cancer patients and therefore accurate and early predictors of treatment response are warranted. Early identification of treatment resistance would enable de-escalation of toxic treatment that has little benefit and could prompt initiation of alterative, more personalized neoadjuvant or post-neoadjuvant treatment strategies [4,5].
2. Imaging of treatment response
Although the assessment of tumor response to NAC may be measured with mammography, breast ultrasound, or molecular imaging [[6], [7], [8], [9], [10], [11]], magnetic resonance imaging (MRI) is the most sensitive imaging technique for the assessment and prediction of response [[12], [13], [14], [15], [16]]. In studies to date, tumor burden/tumor response has been assessed typically with multiparametric MRI prior to NAC, after NAC, and sometimes during NAC as well. Using imaging to identify a priori those who will not benefit from standard NAC can allow non-responders to be triaged to alternative treatments or immediate surgery, thus improving patient care. This would both expedite the delivery of effective treatment and eliminate the administration of potentially toxic and ineffective therapies.
Initial work on treatment response was focused on MRI measurements of tumor diameter, according to RECIST criteria [17], and tumor volume with dynamic contrast-enhanced MRI (DCE-MRI) [18]. However, changes in tumor size and volume usually occur later during treatment and so there is a need to better assess tumor response earlier during NAC. Multiparametric MRI of the breast, which combines morphological parameters from DCE-MRI with functional parameters from MRI techniques such as diffusion-weighted imaging (DWI) and 3D proton magnetic spectroscopic imaging (3D 1H-MRSI), enables the simultaneous assessment of qualitative and quantitative imaging biomarkers. Initial studies have shown that multiparametric MRI further improves the accuracy of treatment response assessment over DCE-MRI alone [19]. Changes in apparent diffusion coefficient (ADC) values reflect changes in tissue cellularity, which can be affected during treatment earlier than lesion size and therefore may be used for early prediction of treatment outcome [20]. Other studies have incorporated proton magnetic resonance spectroscopy (1H-MRS) or sodium MR spectroscopy (23N MRS), which can provide metabolic information on breast tumors [[21], [22], [23]]. While earlier studies have employed mainly univariate and multivariate regression models, recent work has adopted more sophisticated predictive modeling approaches using a variety of radiomics, machine learning, and deep learning techniques.
3. Advanced image analysis and artificial intelligence for response prediction
Radiomics is the conversion of medical images into high-dimensional mineable data [24,25]. In oncology, a tumor is segmented and hundreds or even thousands of quantitative imaging features, derived from tumor shape, texture, kinetics, etc, are extracted. These features encode both simple patterns within medical images but also many higher order patterns not apparent to the human eye. This collection of features is often referred to as a “radiomic signature.” Statistical or machine learning classifiers are then applied on the radiomics signatures to classify patients according to a predicted outcome (e.g., response to NAC). In supervised machine learning, the computer is presented with paired “radiomics signatures” and patient outcomes to learn patterns in the data such that for a given “radiomics signature” input, it is able to predict the patient outcome [25]. Many machine learning methods are available for this task including logistic regression, random forest/decision trees, and support vector machine (SVM). More recently, deep learning techniques using convolutional neural networks (CNNs) have been developed that are more powerful and more robust than traditional machine learning classifiers [26]. With deep learning, feature extraction and feature classification are performed in concert directly from the raw medical images. This eliminates the dependency on image pre-processing and allows for a less constrained learning process. However, it also vastly increases the search space of the model, and thus requires orders of magnitude more training data and more computing power for optimal performance.
4. Clinical implementation of machine learning with MRI for response prediction
Several studies have evaluated the potential of machine learning with multiparametric MRI to predict response to NAC at an early stage, when adaptive treatment can be established.
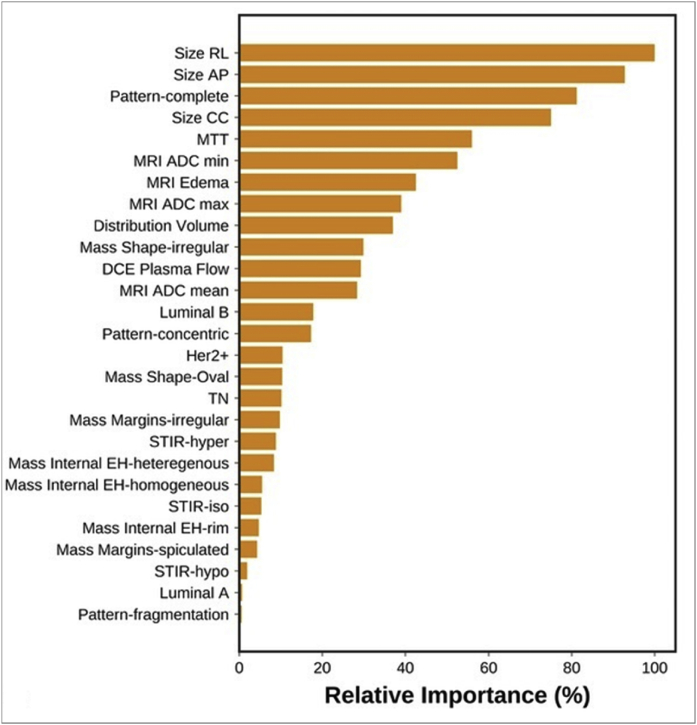
In a study by Tahmassebi et al. [27], 38 patients were scanned before and after two cycles of NAC with a 3T multiparametric MRI scan. Qualitative features were extracted from T2-weighted images (e.g., signal intensity and presence of edema) and from DCE images (e.g., tumor size, pattern of shrinkage, mass or non-mass enhancement, shape, margins, internal enhancement characteristics, distribution, and symmetry). Quantitative features were extracted from DCE images (e.g., mean plasma flow, volume distribution, and mean transit time) and DWI images (e.g., minimum, maximum, and mean ADC values). Twenty-three quantitative and qualitative features were fed to machine learning classifiers to predict residual cancer burden (classified as complete pathologic response with no evidence of residual disease, minimal residual disease, moderate residual disease burden, and extensive residual cancer burden). Eight machine learning classifiers were used to predict residual cancer burden, recurrence-free survival, and disease-specific survival, namely linear support vector machine, linear discriminant analysis, logistic regression, random forests, stochastic gradient descent, decision tree, adaptive boosting, and extreme gradient boosting (XGBoost). Each specific learning algorithm was designed to provide the best model to fit the input data and predict the class labels correctly. Features were ranked based on their importance in the model using recursive feature elimination. Four-fold cross-validation was used to prevent overfitting. Area under the curve (AUC) was the classification metric. Fig. 1 summarizes the feature importance based on recursive feature elimination, for prediction of response to NAC. The most relevant features for prediction of residual cancer burden included change in lesion size, complete pattern of shrinkage, mean transit time, peritumoral edema, and minimum ADC value. Out of the eight machine learning classifiers, XGBoost outperformed other classifiers for prediction of response to NAC (AUC = 0.86).
Fig. 1.
Feature importance of mpMRI model in prediction of RCB class. RCB, Residual cancer burden. Reprinted with permission from: Tahmassebi A, Wengert GJ, Helbich TH, Bago-Horvath Z, Alaei S, Bartsch R, Dubsky P, Baltzer P, Clauser P, Kapetas P, Morris EA, Meyer-Baese A, Pinker K. Impact of Machine Learning With Multiparametric Magnetic Resonance Imaging of the Breast for Early Prediction of Response to Neoadjuvant Chemotherapy and Survival Outcomes in Breast Cancer Patients. Invest Radiol. 2019; 54(2):110–117.
In another study, O’Flynn et al. [28] investigated the role of multiparametric MRI to predict response to NAC in 32 women with locally advance breast cancer who were scanned before and after two cycles of NAC. For this study, treatment response was evaluated on final surgical histology, pCR was classified as “no invasive and no in situ residual disease in the breast or nodes” and near pCR was classified as presence of “non-measurable isolated microscopic foci of residual invasive or in situ disease”. Non-responders had measurable residual invasive and in situ disease. Enhancement fraction (EF), tumor volume, initial area under the gadolinium curve, and quantitative pharmacokinetic parameters (Ktrans, kep, ve) were recorded. ADC and R2* values were recorded pixel-by-pixel. The percentage change in overall mean values for all parameters before and after two cycles of chemotherapy according to pCR status was evaluated using a paired t-test. Linear discriminant analysis determined the most important parameter in predicting pCR. A reduction in the EF (−41% ± 38%) and tumor volume (−80% ± 25%) after two cycles of NAC were significantly greater in those achieving pCR (p = 0.025, p = 0.011 respectively). A reduction in the EF of 7% after two cycles of NAC identified those more likely to achieve pCR with a sensitivity of 63% and specificity of 77% (AUC 0.76). Tumor volume required a much greater percentage decrease (71%) to yield an equivalent specificity of 77%. Other parameters were not contributory to predict response to NAC. Contrary to the aforementioned study, ADC measurements from this multi-parametric model showed no impact in differentiating responders from non-responders. ADC values in fact demonstrated a small fall in those achieving pCR and a rise in the non-responders.
Mani et al. [29] investigated the early prediction of response in 20 patients after just one cycle of NAC, analyzing not only functional information retrieved from DCE and DWI but also ultrasonographic, clinical, and histopathological information. They used a representative set of machine learning and feature selection algorithms including three linear classifiers (Gaussian Naïve Bayes, logistic regression (LR), and Bayesian LR), two decision tree-based classifiers (CART36 and RF), one kernel-based classifier (SVM) and one rule learner (Ripper). A small number of features was selected, and irrelevant features were excluded to reduce risk overfitting. Datasets with 13 imaging variables, 12 clinical variables, and 25 combined imaging plus clinical variables in addition to the outcome variable were assessed (Table 1). Thirteen imaging features from quantitative DCE-MRI and 11 clinical variables were relevant. Imaging and clinical parameters separately had similar overall performance; imaging and clinical variables together boosted the performance of Bayesian LR considerably, resulting in an accuracy of 0.9 and an AUC of 0.96.
Table 1.
List of clinical and imaging variables used. Reprinted from: Mani S, Chen Y, Li X, Arlinghaus L, Chakravarthy AB, Abramson V, Bhave SR, Levy MA, Xu H, Yankeelov TE. Machine learning for predicting the response of breast cancer to neoadjuvant chemotherapy. J Am Med Inform Assoc. 2013; 20(4):688-95.
| Clinical Variable | Description | Imaging Variable | Key Term | Description |
|---|---|---|---|---|
| Age | Age at the time of diagnosis | Delta ADC | Delta | t1, t2 difference |
| ER+ | Estrogen receptor | Delta Ktrans FXL | Ktrans | Pharmocokinetic transfer constant |
| PR+ | Progesterone receptor | Delta Ktrans FXLvp | FXL | Fast exchange limit |
| HER2+ | Human epidermal growth factor receptor | Delta Ktrans FXR | FXR | Fast exchange regime |
| Clinical Grade | Pretreatment clinical grade | Delta ve FXL | vp | Blood plasma volume fraction |
| Proliferative rate | Delta ve FXvp | ve | Extravascular extracellular volume fraction | |
| Pre-treatment nodal status | Pathologically confirmed by fine needle aspiration or sentinel node evaluation | Delta ve FXR | ti | Intra cellular water lifetime of wated molecule |
| Clinical-T | Pretreatment clinical size based on clinical findings judged most accurate for that case (physical exam, ultrasound, mammogram, conventional MRI) | Delta vp FXL | ||
| Clinical-N | Pretreatment nodal stage based on pathologically confirmed by fine needle aspiration of node or sentinel evaluation | Delta ti FXR | ||
| Pre-treatment clinical stage | Staging of the breast cancer prior to initiation of systemic chemotherapy | Ktrans, t1 FXL | ||
| Pre-treatment physical exam | Longest diameter by physical exam (CM) | Ktrans, t1 FXLvp | ||
| Pre-treatment longest diameter (ultra sound) | Longest dimension (CM) Clinical judgment is used to determine the modality most accurate for that case (physical exam, ultrasound, mammogram, conventional MRI) | Ktrans, t1 FXR | ||
| Delta tumor volume |
In a follow-up study [30], the authors developed a predictive model with an increased number of imaging features (118 instead of 13), which were derived from semiquantitative and quantitative DCE-MRI and DWI-MRI parameters. The imaging parameters were combined with 11 clinical variables. With a sample size of 28 patients, they achieved similar results to the prior study (AUC = 0.86) (Table 2). The authors used Bayesian LR with feature selection within a machine learning framework to capture non-linear relationships between variables and outcome and integrated clinical and imaging data obtained before and after one cycle of NAC to predict response in breast cancer patients undergoing NAC. To increase predictive performance and decrease overfitting, feature selection algorithms were used to select only a small number of features that were highly predictive of response to NAC. The feature selections algorithms included HITON-Markov blanket (MB), Gram-Schmidt (GS) orthogonalization with a maximum number of 10 features output, and BLCD-MB. The MB-based feature selection algorithms selected only two clinical and two imaging features (ER+, PR+, mean ADC after one cycle of treatment, and mean of the change of the top 15% of kep), generating an accuracy of 0.82 (95% CI 0.68–0.96). When clinical and imaging features were combined, they generated an accuracy of 0.86 (95% CI 0.71–0.96), a sensitivity of 0.88 (95% CI 0.71–1) and a specificity of 0.82 (95% CI 0.56–1), which were higher compared to the accuracy, sensitivity, and specificity yielded by the current RECIST approach which amounted respectively to 0.71, 0.82, and 0.65. The Gram-Schmidt-based algorithm performed more poorly and selected all the 11 clinical variables (range 15–28 folds), 58 imaging variables (range 1–24 folds) and 60 (range 1–27 folds) when clinical and imaging variables were combined.
Table 2.
List of pretreatment clinical variables with a short description. NAC, neoadjuvant chemotherapy. Reprinted from: Mani S, Chen Y, Arlinghaus LR, Li X, Chakravarthy AB, Bhave SR, Welch EB, Levy MA, Yankeelov TE. Early prediction of the response of breast tumors to neoadjuvant chemotherapy using quantitative MRI and machine learning. AMIA Annu Symp Proc. 2011; 2011:868–77.
| Clinical variable | Description |
|---|---|
| Age | Age at the time of diagnosis |
| ER+ | Estrogen receptor |
| PR+ | Progesterone receptor |
| HER2+ | Human epidermal growth factor receptor |
| Clinical Grade | Pretreatment clinical grade |
| Proliferative rate | No of cells in mitosis per 10 high power fields |
| Nodal status | Pathologically confirmed by fine needle aspiration or sentinel node evaluation |
| Clinical-T | Pretreatment clinical size based on clinical imaging (ie, physical examination, ultrasound, mammogram, conventional MRI) judged to be most accurate for each case. In patients in whom these measurements were discordant, the most reliable measurement (as deemed by the treating physician) was utilized to determine tumor size before chemotherapy |
| Clinical-N | Pretreatment nodal stage based on pathologically confirmed by fine needle aspiration of node or sentinel evaluation |
| Clinical stage | Staging of the breast cancer before initiation of NAC. Clinical staging includes physical examination as well as standard imaging including ultrasound, mammogram and clinical MRI |
| Physical examination | Longest diameter by physical examination (cm) |
Some studies have attempted to predict response to NAC with pretreatment imaging alone. For example, Cain et al. [31] used pretreatment MRI performed in 288 patients to predict response to NAC using a multivariate machine learning-based model (LR and an SVM). This study analyzed computer-extracted features solely from pretreatment MRI and did not evaluate differences between pre- and post- (1 or 2 cycles of NAC) treatment MRI. The larger dataset size allowed the creation of an independent validation cohort for each of the following subpopulations: 1) all neoadjuvant therapy (NAT) patients, 2) NAC patients, and 3) triple-negative or HER2+ (TN/HER2+) patients treated with NAT. The entire cohort was equally divided into a training set, which was used to generate the machine learning models, and a test set. A stepwise multilinear regression-based feature selection procedure was used to select features from the training set for predicting pCR. The initial set of features comprised 529 features that were used to train a multivariate logistic regression classifier and a support vector machine classifier. The trained models were used to predict pCR in the test set. Feature selection and training classifiers in the training set was done for all patients and was then repeated for two subpopulations (i.e., NAC patients and TN/HER2+ patients treated with NAT). Twelve features were selected from the training set for the three cohorts: six were extracted from tumor alone, five were extracted from FGT alone, and one was extracted from both tumor and FGT. Only two were significant for TN/HER2+ patients who received NAT. One was “change in variance of uptake”, a tumor-based feature which quantifies the change in variance of tumor uptake by finding the minimum ratio of the variances of tumor voxels in two consecutive time points. This feature had the highest AUC (0.71, 95% CI 0.58–0.83) among the 12 features selected in all subpopulations evaluated: lower values were predictive of pCR. An additional feature, ‘SER Partial tissue vol cu mm T1,’ extracted using fibroglandular tissue (FGT), was also selected and found to be significant in TN/HER2+ patient subpopulation. This feature is a volumetric measure of FGT enhancement (extracted from T1 non-fat-saturated sequences) using the signal enhancement ratio of FGT voxels. For this feature, higher values predicted a lower chance of achieving pCR. This study demonstrates that while multivariate models (e.g., SVM, LR) were prognostic of pCR in the TN/HER2+ patient subgroup (p < 0.002), the prognostic value of the model in predicting pCR across the entire cohort was significant, but to a lesser extent (p = 0.01).
In another example of pretreatment MRI-based predictive modeling, Aghaei et al. [32] studied quantitative kinetic imaging features to predict response to NAC from the pretreatment MRI scan of 68 cancer patients. Tumors were segmented using computer aided detection and 39 kinetic image features were extracted from both the segmented tumor and background parenchyma. Features are summarized in Table 3. Only a small set of non-redundant and highly performing imaging features were selected. Two approaches were used to analyze the data. First, individual features were analyzed with a simple feature fusion method (average, weighted combination, and selection of the maximum or minimum feature value) that combined classification results from multiple features; the correlation coefficients of individual image features were also computed and compared to identify non-redundant image features. Second, a statistical machine learning classifier-based method selected optimal features and predicted tumor response to NAC using an artificial neural network (ANN) as a base classifier integrated with a wrapper subset evaluator. The base classifier was trained with a leave-one-case-out validation method where each case was selected as an independent testing case and the remaining cases in the dataset were used to form a training dataset. The ANN was subsequently applied to the testing case and used to generate a classification score. Using the feature fusion method, ten features yielded AUC >0.6 in classifying between the complete response and the partial and nonresponse case groups: 1) average intensity and 2) maximum pixel intensity from the entire tumor region, 3) volume, 4) average intensity and 5) standard deviation from active tumor region, excluding necrotic region, 6) volume and 7) skewness of low-enhanced pixel intensity from the necrotic area, 8) average intensity, 9) standard deviation from the background parenchyma, and 10) average intensity from the absolute bilateral feature difference of BPE between the left and right breasts. From the comparison results, five final low-redundancy image features [2,3] were selected with correlation less than 0.5. These five features were used to classify responders and non-responders. This simple feature fusion method achieved an AUC = 0.85 ± 0.05, which was significantly higher than the AUC using each individual feature (which ranged from 0.604 ± 0.072 to a maximum of 0.713 ± 0.065). The ANN-based classifier selected 11 features. The five most relevant features were: 1) average contrast enhancement and 2) standard deviation of contrast enhancement inside an entire tumor region, 3) standard deviation of contrast enhancement in the enhanced area, 4) average pixel value of necrotic regions, and 5) ratio of necrotic volume over tumor volume. The ANN-based classifier proved more accurate, with an AUC = 0.96 ± 0.03, which was significantly higher than that of the simple fusion method (p < 0.01). These results highlight the idea that quantitative imaging feature analysis has higher discriminatory power and is better able to predict outcome compared to visually assessable features (e.g., tumor size, average contrast enhancement). For example, the heterogeneity of tumor contrast enhancement represented by the standard deviation of the contrast enhancement on the active tumor region had the highest discriminatory power (AUC 0.778 ± 0.066), and this marker cannot be accurately and reliably evaluated using a visual or subjective evaluation method.
Table 3.
Summary of computed kinetic image features in five groups.a These features are computed from three different regions—background parenchymal region of the whole (left and right) breast regions, left breast and right breast.b Absolute bilateral feature difference of BPE between the left and right breasts. Reprinted with permission from: Aghaei F, Tan M, Hollingsworth AB, Qian W, Liu H, Zheng B. Computer-aided breast MR image feature analysis for prediction of tumor response to chemotherapy. Med Phys. 2015; 42(11):6520-8.
| Feature group | Feature number | Description |
|---|---|---|
| Tumor area | 1–7 | Volume, average intensity, maximum pixel intensity, standard deviation, and skewness of tumor pixel intensity, maximum value of tumor radius, and shape factor |
| Enhanced area | 8–11 | Volume, average intensity, standard deviation, and skewness of contrast-enhanced pixel intensity |
| Necrotic area | 12–16 | Volume, average intensity, standard deviation, and skewness of low-enhanced pixel intensity, ratio of necrotic volume over tumor volume |
| Background parenchymal areaa | 17–34 | Average intensity, standard deviation, skewness, maximum pixel intensity, average value of top 1%, and average value of top 5% of pixel values |
| Absolute bilateral difference of BP areab | 35–39 | Average intensity, standard deviation, skewness, average value of top 1%, and average value of top 5% of pixel values |
5. Deep learning with MRI for response prediction
Recently, deep learning methods have been proposed for prediction response to NAC using pretreatment MRI alone. Ha et al. [33] trained a CNN to take tumor regions of interest from the pretreatment MRI and predict whether the patient would achieve a complete pathologic response considered as no residual invasive disease in the breast and lymph nodes (ypT0/Tis ypN0), partial pathologic response, or no response/progression. The study was performed using 141 patients with locally advanced breast cancer. The CNN consisted of ten convolutional layers, four max pooling layers, and a fully connected layer. Data augmentation, 50% dropout, and L2 regularization were used to prevent overfitting. The CNN achieved an overall mean accuracy of 88% in three-class prediction of NAC (i.e., discriminating one class from the other two). The complete response group had a specificity of 95.1% ± 3.1%, a sensitivity of 73.9% ± 4.5%, and an accuracy of 87.7% ± 0.6%. The partial response group had a specificity of 91.6% ± 1.3%, a sensitivity of 82.4% ± 2.7%, and an accuracy of 87.7% ± 0.6%. The non-responder group had a specificity of 93.4% ± 2.9%, a sensitivity of 76.8% ± 5.7%, and an accuracy of 87.8% ± 0.6%. The dataset size in this study – 141 patients – is not large enough to fully harness the potential of deep learning for treatment response prognostication. The 88% accuracy achieved in this study is therefore especially encouraging, since it can be expected that future work with larger datasets will achieve even higher predictive accuracy. Key findings from the above mentioned studies are summarized on Table 4.
Table 4.
Summary of findings across key articles. The machine learning classifiers in bold characters represent those that yielded the most significant results and the AUC values are related to the results from those classifiers in bold characters.
| Study | Analyzed images | Machine learning classifiers | Most relevant selected features | AUC |
|---|---|---|---|---|
| Tahmassebi et al. | DCE, DWI T2 | Linear support vector machine Linear discriminant analysis logistic regression Random forests Stochastic gradient descent Decision tree Adaptive boosting Extreme gradient boosting (XGBoost) |
Change in lesion size Complete pattern of shrinkage Mean transit time Peritumoral edema Minimum ADC value |
0.86 |
| O’Flynn et al. | DCE, DWI, T2 | Linear discriminant analysis | Enhancement fraction (EF) Tumor volume |
0.76 |
| Mani et al. | DCE, DWI | Linear classifiers (Gaussian Naïve Bayes, Logistic Regression, and Bayesian Logistic Regression) decision tree-based classifiers (CART and Random Forests) Kernel based classifier (Support Vector Machine) Rule learner (Ripper) |
See Table 1. | 0.96 |
| Mani et al. | DCE, DWI | GS-10 HITON-MB BLCD-MB |
Mean ADC post one cycle of treatment Mean of the change of the top 15% of kep as estimated by the TK model |
0.86 |
| Cain et al. | T1 non-fat sat, DCE |
Multivariate logistic regression classifier (fitglm) Support vector machine classifier (fitcsvm and fitSVMposterior) |
Change in variance of uptake | 0.71 |
| Aghaei et al. | DCE | Simple feature fusion method Artificial neural network (ANN) with a wrapper subset evaluator |
Average contrast enhancement Standard deviation of contrast enhancement inside an entire tumor region Standard deviation of contrast enhancement in the enhanced area Average pixel value of necrotic regions Ratio of necrotic volume over tumor volume |
0.96 |
| Ha et al. | First T1 postcontrast dynamic images | Convolutional neural networks (CNN) | Not specified | 0.88 |
6. Challenges
Despite these encouraging results, the field of machine learning using multiparametric breast MRI for early prediction of NAC treatment response is still in its infancy. To date, studies have been retrospective, single-institutional, and have included relatively small numbers of patients, which limits the statistical power of the studies and may compromise the generalizability of the results. Additionally, multiparametric MRI has been performed using a wide range of MRI hardware; as well as varied scan protocols, sequence parameters, and post-processing steps. Rigorous standardization of MRI hardware and software is needed. Ideally, quantitative MRI techniques should also be used to further improve repeatability and reproducibility. Deep learning is a particularly promising technique for early prediction of treatment response, but to avoid overfitting, it is necessary to train models on extremely large datasets that are large and diverse enough to span the biological heterogeneity of the diseases and outcomes they seek to classify. Breast cancer is a highly heterogeneous disease and so models with real potential for clinical translation must be orders of magnitude larger than all studies to date. The curation of highly standardized, large, multi-institutional MRI datasets is a herculean task, but it is a prerequisite to building robust machine learning models that will work across patients and across institutions and that have real potential for clinical use. Finally, it is also necessary to establish more standardized and transparent ways to validate the machine learning models being developed. Rigorous testing by third parties in prospective studies is essential to guarantee a model’s diagnostic accuracy and is needed prior to implementation in the clinical setting.
7. Summary
Several large randomized trials have demonstrated that achieving pCR after neoadjuvant treatment for locally advanced breast cancer not only decreases patient morbidity by facilitating less invasive surgery but also aids in predicting patient mortality, as pCR is a marker for improved disease-free and overall survival [34,35]. However, only 30–50% [35] of patients undergoing neoadjuvant treatment achieve pCR, and it would be clinically advantageous to identify those patients for optimal triage of care. To date, traditional machine learning approaches have been applied to predict treatment response using a mix of qualitative and quantitative multiparametric MRI features early in the course of, or even before the start of, neoadjuvant treatment. Incorporating clinical data into these models further improves accuracy [36,37]. More recently, deep learning using CNNs have been used to predict pCR and have achieved results similar to the more traditional machine learning methods. However, the datasets used were not large enough to evaluate the full potential of the CNN approach and it is expected that future work with larger numbers of patients will demonstrate the superiority of a deep learning over traditional machine learning.
In conclusion, machine learning and deep learning using breast MRI enable the early prediction of pCR to neoadjuvant treatment with high accuracy. The integration of machine and deep learning has the potential to provide valuable predictive information on treatment outcomes and risk of recurrence and thus improve clinical management by minimizing toxicities from ineffective therapies, avoiding delays to surgery in non-responders, and facilitating upfront use of novel targeted therapies.
Funding
The project described was supported by RSNA Research & Education Foundation, through grant number RF1905. The content is solely the responsibility of the authors and does not necessarily represent the official views of the RSNA R&E Foundation. The project was also supported by the European School of Radiology and the National Institutes of Health/National Cancer Institute (NIH/NCI) Cancer Center Support Grant (Grant ID: P30 CA008748), USA. The content is solely the responsibility of the authors and does not necessarily represent the official views of the RSNA R&E Foundation. The funding sources were not involved in the study design; in the collection, analysis and interpretation of data; in the writing of the report; and in the decision to submit the article for publication.
Declaration of interest
Katja Pinker received payment for activities not related to the present article including lectures including service on speakers bureaus and for travel/accommodations/meeting expenses unrelated to activities listed from the European Society of Breast Imaging (MRI educational course, annual scientific meeting) and the IDKD 2019 (educational course). Elizabeth A Morris has received a grant from GRAIL Inc. The rest of the authors declare no potential competing interests.
Acknowledgements
We thank Joanne Chin and Johanna Goldberg for their assistance in this article.
References
- 1.(a) Curigliano G., Burstein H., Winer E.P., Gnant M., Dubsky P., Loibl S., Colleoni M., Regan M.M., Piccart-Gebhart M., Senn H.J., Thürlimann B. 2017. St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer. [DOI] [PubMed] [Google Scholar]; (b) André F., Baselga J., Bergh J., Bonnefoi H., Brucker S.Y., Cardoso F., Carey L., Ciruelos E., Cuzick J., Denkert C., Di Leo A., Ejlertsen B., Francis P., Galimberti V., Garber J., Gulluoglu B., Goodwin P., Harbeck N., Hayes D.F., Huang C.S., Huober J., Hussein K., Jassem J., Jiang Z., Karlsson P., Morrow M., Orecchia R., Osborne K.C., Pagani O., Partridge A.H., Pritchard K., Ro J., Rutgers E.J.T., Sedlmayer F., Semiglazov V., Shao Z., Smith I., Toi M., Tutt A., Viale G., Watanabe T., Whelan T.J., Xu B. De-escalating and escalating treatments for early-stage breast cancer: the St. Gallen international expert consensus conference on the primary therapy of early breast cancer 2017. Ann Oncol. 2019 Jul 1;30(7):1181. doi: 10.1093/annonc/mdy537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cortazar P., Zhang L., Untch M., ehta K., Costantino J.P., Wolmark N., Bonnefoi H., Cameron D., Gianni L., Valagussa P., Swain S.M., Prowell T., Loibl S., Wickerham D.L., Bogaerts J., Baselga J., Perou C., Blumenthal G., Blohmer J., Mamounas E.P., Bergh J., Semiglazov V., Justice R., Eidtmann H., Paik S., Piccart M., Sridhara R., Fasching P.A., Slaets L., Tang S., Gerber B., Geyer C.E., Jr., Pazdur R., Ditsch N., Rastogi P., Eiermann W., von Minckwitz G. Pathological complete response and long-term clinical benefit in breast cancer: the CTNeoBC pooled analysis. Lancet. 2014 Jul 12;384(9938):164–172. doi: 10.1016/S0140-6736(13)62422-8. [DOI] [PubMed] [Google Scholar]
- 3.Cortazar P., Geyer C.E., Jr. Pathological complete response in neoadjuvant treatment of breast cancer. Ann Surg Oncol. 2015 May;22(5):1441–1446. doi: 10.1245/s10434-015-4404-8. [DOI] [PubMed] [Google Scholar]
- 4.Pusztai L., Foldi J., Dhawan A., DiGiovanna M.P., Mamounas E.P. Changing frameworks in treatment sequencing of triple-negative and HER2-positive, early-stage breast cancers. Lancet Oncol. 2019 Jul;20;(7):e390–e396. doi: 10.1016/S1470-2045(19)30158-5. [DOI] [PubMed] [Google Scholar]
- 5.Gnant M., Harbeck N., Thomssen C. St. Gallen/vienna 2017: a brief summary of the consensus discussion about escalation and de-escalation of primary breast cancer treatment. Breast Care. 2017 May;12;(2):102–107. doi: 10.1159/000475698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drew P.J., Kerin M.J., Mahapatra T., Malone C., Monson J.R., Turnbull L.W., Fox J.N. Evaluation of response to neoadjuvant chemoradiotherapy for locally advanced breast cancer with dynamic contrast-enhanced MRI of the breast. Eur J Surg Oncol. 2001 Nov;27(;(7):617–620. doi: 10.1053/ejso.2001.1194. [DOI] [PubMed] [Google Scholar]
- 7.Mumtaz H., Davidson T., Hall-Craggs M.A., Payley M., Walmsley K., Cowley G., Taylor I. Comparison of magnetic resonance imaging and conventional triple assessment in locally recurrent breast cancer. Br J Surg. 1997 Aug;84;(8):1147–1151. [PubMed] [Google Scholar]
- 8.Partridge S.C., Gibbs J.E., Lu Y., Esserman L.J., Sudilovsky D., Hylton N.M. Accuracy of MR imaging for revealing residual breast cancer in patients who have undergone neoadjuvant chemotherapy. AJR Am J Roentgenol. 2002 Nov;179;(5):1193–1199. doi: 10.2214/ajr.179.5.1791193. [DOI] [PubMed] [Google Scholar]
- 9.Yeh E., Slanetz P., Kopans D.B., Rafferty E., Georgian-Smith D., Moy L., Halpern E., Moore R., Kuter I., Taghian A. Prospective comparison of mammography, sonography, and MRI in patients undergoing neoadjuvant chemotherapy for palpable breast cancer. AJR Am J Roentgenol. 2005 Mar;184;(3):868–877. doi: 10.2214/ajr.184.3.01840868. [DOI] [PubMed] [Google Scholar]
- 10.Park J., Chae E.Y., Cha J.H., Shin H.J., Choi W.J., Choi Y.W., Kim H.H. Comparison of mammography, digital breast tomosynthesis, automated breast ultrasound, magnetic resonance imaging in evaluation of residual tumor after neoadjuvant chemotherapy. Eur J Radiol. 2018 Nov;108:261–268. doi: 10.1016/j.ejrad.2018.09.032. [DOI] [PubMed] [Google Scholar]
- 11.Raccagni I., Belloli S., Valtorta S., Stefano A., Presotto L., Pascali C., Bogni A., Tortoreto M., Zaffaroni N., Daidone M.G., Russo G., Bombardieri E., Moresco R.M. [18F] FDG and [18F] FLT PET for the evaluation of response to neo-adjuvant chemotherapy in a model of triple negative breast cancer. PLoS One. 2018 May 23;13(5) doi: 10.1371/journal.pone.0197754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abramson R.G., Li X., Hoyt T.L., Su P.F., Arlinghaus L.R., Wilson K.J., Abramson V.G., Chakravarthy A.B., Yankeelov T.E. Early assessment of breast cancer response to neoadjuvant chemotherapy by semi-quantitative analysis of high-temporal resolution DCE-MRI: preliminary results. Magn Reson Imaging. 2013 Nov;31(9):1457–1464. doi: 10.1016/j.mri.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arlinghaus L.R., Li X., Levy M., Smith D., Welch E.B., Gore J.C., Yankeelov T.E. Current and future trends in magnetic resonance imaging assessments of the response of breast tumors to neoadjuvant chemotherapy. J Oncol. 2010;2010:919620. doi: 10.1155/2010/919620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li X., Abramson R.G., Arlinghaus L.R., Kang H., Chakravarthy A.B., Abramson V.G., Farley J., Mayer I.A., Kelley M.C., Meszoely I.M., Means-Powell J., Grau A.M., Sanders M., Yankeelov T.E. Multiparametric magnetic resonance imaging for predicting pathological response after the first cycle of neoadjuvant chemotherapy in breast cancer. Investig Radiol. 2015 Apr;50:195–204. doi: 10.1097/RLI.0000000000000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu L.A., Chang R.F., Huang C.S., Lu Y.S., Chen H.H., Chen J.Y., Chang Y.C. Evaluation of the treatment response to neoadjuvant chemotherapy in locally advanced breast cancer using combined magnetic resonance vascular maps and apparent diffusion coefficient. J Magn Reson Imaging. 2015 Nov;42:1407–1420. doi: 10.1002/jmri.24915. [DOI] [PubMed] [Google Scholar]
- 16.Minarikova L., Bogner W., Pinker K., Valkovič L., Zaric O., Bago-Horvath Z., Bartsch R., Helbich T.H., Trattnig S., Gruber S. Investigating the prediction value of multiparametric magnetic resonance imaging at 3 T in response to neoadjuvant chemotherapy in breast cancer. Eur Radiol. 2017 May;27:1901–1911. doi: 10.1007/s00330-016-4565-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eisenhauer E.A. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 18.Hylton N.M., Blume J.D., Bernreuter W.K., Pisano E.D., Rosen M.A., Morris E.A., Weatherall P.T., Lehman C.D., Newstead G.M., Polin S., Marques H.S., Esserman L.J., Schnall M.D. ACRIN 6657 Trial Team and I-SPY 1 TRIAL Investigators. Locally advanced breast cancer: MR imaging for prediction of response to neoadjuvant chemotherapy--results from ACRIN 6657/I-SPY TRIAL. Radiology. 2012 Jun;263(3):663–672. doi: 10.1148/radiol.12110748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marinovich M.L.1, Sardanelli F., Ciatto S., Mamounas E., Brennan M., Macaskill P., Irwig L., von Minckwitz G., Houssami N. Early prediction of pathologic response to neoadjuvant therapy in breast cancer: systematic review of the accuracy of MRI. Breast. 2012 Oct;21(5):669–677. doi: 10.1016/j.breast.2012.07.006. Epub 2012 Aug 3. [DOI] [PubMed] [Google Scholar]
- 20.Fujimoto H., Kazama T., Nagashima T., Sakakibara M., Suzuki T.H., Okubo Y., Shiina N., Fujisaki K., Ota S., Miyazaki M. Diffusion-weighted imaging reflects pathological therapeutic response and relapse in breast cancer. Breast Canc. 2014 Nov;21;(6):724–731. doi: 10.1007/s12282-013-0449-3. [DOI] [PubMed] [Google Scholar]
- 21.Jagannathan N.R., Kumar M., Seenu V., Coshic O., Dwivedi S.N., Julka P.K., Srivastava A., Rath G.K. Evaluation of total choline from in-vivo volume localized proton MR spectroscopy and its response to neoadjuvant chemotherapy in locally advanced breast cancer. Br J Canc. 2001 Apr 20;84(8):1016–1022. doi: 10.1054/bjoc.2000.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manton D.J., Chaturvedi A., Hubbard A., Lind M.J., Lowry M., Maraveyas A., Pickles M.D., Tozer D.J., Turnbull L.W. Neoadjuvant chemotherapy in breast cancer: early response prediction with quantitative MR imaging and spectroscopy. Br J Canc. 2006 Feb 13;94(3):427–435. doi: 10.1038/sj.bjc.6602948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tozaki M., Oyama Y., Fukuma E. Preliminary study of early response to neoadjuvant chemotherapy after the first cycle in breast cancer: comparison of 1H magnetic resonance spectroscopy with diffusion magnetic resonance imaging. Jpn J Radiol. 2010 Feb;28(2):101–109. doi: 10.1007/s11604-009-0391-7. [DOI] [PubMed] [Google Scholar]
- 24.Gillies R.J., Kinahan P.E., Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016 Feb;278(2):563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Avanzo M., Stancanello J., El Naqa I. Beyond imaging: the promise of radiomics. Phys Med. 2017 Jun;38:122–139. doi: 10.1016/j.ejmp.2017.05.071. [DOI] [PubMed] [Google Scholar]
- 26.LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015 May 28;521(7553):436–444.25. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 27.Tahmassebi A., Wengert G.J., Helbich T.H. Impact of machine learning with multiparametric magnetic resonance imaging of the breast for early prediction of response to neoadjuvant chemotherapy and survival outcomes in breast cancer patients. Investig Radiol. 2019 Feb;54(2):110–117. doi: 10.1097/RLI.0000000000000518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Flynn E.A., Collins D., D’Arcy J., Schmidt M., de Souza N.M. Multi-parametric MRI in the early prediction of response to neo-adjuvant chemotherapy in breast cancer: value of non-modelled parameters. Eur J Radiol. 2016 Apr;85(4):837–842. doi: 10.1016/j.ejrad.2016.02.006. [DOI] [PubMed] [Google Scholar]
- 29.Mani S., Chen Y., Arlinghaus L.R., Li X., Chakravarthy A.B., Bhave S.R., Welch E.B., Levy M.A., Yankeelov T.E. Early prediction of the response of breast tumors to neoadjuvant chemotherapy using quantitative MRI and machine learning. AMIA Annu Symp Proc. 2011;2011:868–877. [PMC free article] [PubMed] [Google Scholar]
- 30.Mani S., ChenY, Li X., Arlinghaus L., Chakravarthy A.B., Abramson V., Bhave S.R., Levy M.A., Xu H., Yankeelov T.E. Machine learning for predicting the response of breast cancer to neoadjuvant chemotherapy. J Am Med Inform Assoc. 2013 July-Aug;20(4):688–695. doi: 10.1136/amiajnl-2012-001332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cain E.H., Saha A., Harowicz M.R., Marks J.R., Marcom P.K., Mazurowski M.A. Multivariate machine learning models for prediction of pathologic response to neoadjuvant therapy in breast cancer using MRI features: a study using an independent validation set. Breast Canc Res Treat. 2019 Jan;173(2):455–463. doi: 10.1007/s10549-018-4990-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aghaei F., Tan M., Hollingsworth A.B., Qian W., Liu H., Zheng B. Computer-aided breast MR image feature analysis for prediction of tumor response to chemotherapy. Med Phys. 2015 Nov;42(11):6520–6528. doi: 10.1118/1.4933198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ha R., Chin C., Karcich J., Liu M.Z., Chang P., Mutasa S., Pascual Van Sant E., Wynn R.T., Connolly E., Jambawalikar S. Prior to initiation of chemotherapy. Can we predict breast tumor response? Deep learning convolutional neural networks approach using a breast MRI tumor dataset. J Digit Imaging. 2019 Oct;32(5):693–701. doi: 10.1007/s10278-018-0144-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gianni L., Pienkowski T., Im Y.H., Tseng L.M., Liu M.C., Lluch A., Starosławska E., de la Haba-Rodriguez J., Im S.A., Pedrini J.L., Poirier B., Morandi P., Semiglazov V., Srimuninnimit V., Bianchi G.V., Magazzù D., McNally V., Douthwaite H., Ross G., Valagussa P. 5-year analysis of neoadjuvant pertuzumab and trastuzumab inpatients with locally advanced, inflammatory, or early-stage HER2-positive breast cancer (NeoSphere): a multicentre, open-label, phase 2 randomised trial. Lancet Oncol. 2016 Jun;17(6):791–800. doi: 10.1016/S1470-2045(16)00163-7. [DOI] [PubMed] [Google Scholar]
- 35.Cortazar P., Zhang L., Untch M., Mehta K., Costantino J.P., Wolmark N., Bonnefoi H., Cameron D., Gianni L., Valagussa P., Swain S.M., Prowell T., Loibl S., Wickerham D.L., Bogaerts J., Baselga J., Perou C., Blumenthal G., Blohmer J., Mamounas E.P., Bergh J., Semiglazov V., Justice R., Eidtmann H., Paik S., Piccart M., Sridhara R., Fasching P.A., Slaets L., Tang S., Gerber B., Geyer C.E., Jr., Pazdur R., Ditsch N., Rastogi P., Eiermann W., von Minckwitz G. Pathological complete response and long-term clinical benefit in breast cancer: the CTNeoBC pooled analysis. Lancet. 2014 Jul 12;384(9938):164–172. doi: 10.1016/S0140-6736(13)62422-8. [DOI] [PubMed] [Google Scholar]
- 36.Weis J.A., Miga M.I., Yankeelov T.E. Three-dimensional image-based mechanical modeling for predicting the response of breast cancer to neoadjuvant therapy. Comput Methods Appl Mech Eng. 2017 Feb 1;314:494–512. doi: 10.1016/j.cma.2016.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yankeelov T.E. Integrating imaging data into predictive biomathematical and biophysical models of cancer. ISRN Biomath. 2012;2012:287394. doi: 10.5402/2012/287394. pii. [DOI] [PMC free article] [PubMed] [Google Scholar]