Abstract
Introduction
The clinico-pathological and molecular factors that drive the prognosis of invasive lobular breast carcinoma (ILC) are not entirely explored. In this regard, the development and validation of a prognostic model for ILC and the investigation of the distribution of molecular abnormalities (focusing on CDK4/6 alterations) according to prognosis were the aims of this study.
Patients and methods
Two clinico-pathological multi-center data-sets of early-stage ILC patients (Training/Validation Set, TS/VS) were gathered. A 3-class model was developed according to the multivariate analysis for disease-free-survival (DFS) and externally validated. Mutational, copy number variation and transcriptomic analyses by targeted next generation sequencing (NGS) were performed (and validated with quantitative PCR) in an explorative cohort of patients with poor and good prognosis.
Results
Data from overall 773 patients (TS/VS: 491/282) were gathered. The developed model significantly discriminated low/intermediate/high risk in the TS (10-years DFS: 76.3%/67.6%/39.8%, respectively, p<0.0001) and in the VS (p<0.0001). In the explorative cohort for molecular analysis (34 patients), CDK4 gain was present exclusively in the poor prognosis group (35.0%, p = 0.03; OR 7.98, 95%CI 1.51–42.1, p = 0.014). Moreover, CDK4 and 6 overexpression showed a trend toward an association with poor prognosis (OR 2.7, 95%CI 0.4–18.1, p = 0.3; OR 3.29, 95%CI 0.56–19.25, p = 0.18).
Conclusions
A risk stratification model, able to accurately separate early-stage ILC patients’ prognosis into different risk classes according to clinico-pathological variables, allowed to investigate potential biomarkers of prognosis with targeted NGS. CDK4 gain is suggested for future validation as a prognostic biomarker and a potential therapeutic opportunity in ILC patients.
Keywords: Breast cancer, Lobular, Prognosis, Next-generation sequencing, Transcriptome analysis, CDK4
Highlights
-
•
The current multicenter analysis developed and validated a prognostic nomogram for early stage ILC.
-
•
A next-generation sequencing analysis was performed in prognostic ‘outlier’ patients.
-
•
CDK4 gain emerges as a potential negative prognostic factor in ILC patients. .
1. Introduction
Invasive lobular carcinoma (ILC), accounting for 5%–15% of all invasive breast tumours, represents the second most common histologic type of breast cancer (BC), after invasive carcinoma of no special type (NST) [1]. The lobular histotype is generally characterized by luminal phenotype and lower mitotic index and grading in comparison with NST [2]. Although these aspects are typically associated with a favorable prognosis, the overall long-term survival of ILC appears worse than that of invasive carcinoma of NST [3]. Even the recent genomic characterization of ILC underlined that this histotype represents a peculiar entity of BC [[4], [5], [6]].
However, despite the several dissimilarities between lobular and ductal histology, the prognostic and therapeutic aspects of ILC are currently borrowed from the ductal histotype. Thus, an issue for clinical practice is represented by the identification of predictors of prognosis for ILC to assist in the choice of a pertinent therapeutic decision [7]. Recently, the correlation between genomic alterations and the risk of recurrence suggested that chromosome 1q gain was predictor of a better outcome, whereas 11p gain was predictor of worse outcome [5]. However, the prognostic role of genomic alterations varies considerably and does not emerge equally from studies [8], providing limited insight into the prognosis of this histotype.
A possible strategy to identify potential prognostic factors consists in the early identification of patients characterized by ‘best’ and ‘worst’ prognosis. These patients, defined as ‘outliers’, may potentially be featured by peculiar molecular alterations accountable for their specific outcome. Moreover, this strategy may help to combine the clinical aspects with the recent technologies [9].
In this regard, the aim of this study was to develop and validate a prognostic nomogram for early stage ILC patients, combining clinico-pathological factors, to finally identify those prognostic ‘outliers’ candidate to undergo genomic analysis with targeted next generation sequencing (NGS).
2. Materials and methods
2.1. Patients’ population
Consecutive patients with diagnosis of early stage ILC underwent surgery at 3 Italian institutions (Verona and Padova providing data for the training set (TS) and Napoli for the validation set (VS) between 1990 and 2013 were considered eligible. Inclusion criteria were ‘pure’ ILC diagnosis (stage I-III), curative surgery and availability of clinico-pathological parameters.
2.2. End-points
The aims of this study were 1) to develop a clinico-pathological prognostic model in a multi-center population of ILC (TS), to finally recognize ‘outlier’ patients, in terms of disease-free survival (DFS) and overall survival (OS); 2) to validate the nomogram in an external patients’ cohort (VS); 3) to explore potential molecular drivers of prognosis with NGS in a subset of prognostic ‘outliers’.
2.3. Statistical analysis
Descriptive statistics were adopted to summarize appropriate study data. The Hazard Ratios (HR) and the 95% Confidence Interval (CI) were assessed for each variable applying the Cox univariate model [10]. A multivariate hazard model was calculated adopting the stepwise regression (forward selection, enter limit and remove limit, p = 0.10 and p = 0.15, respectively), to detect independent prognostic factors [11]. The outcomes were DFS and OS. To accomplish the multivariate model overfit, an internal cross-validation technique was performed [[12], [13], [14], [15]]. Further details are reported in Supplementary Material.
2.4. Prognostic score assessment
A step-by-step protocol was pursued consistent with the methodological approach for developing a nomogram for cancer prognosis proposed by Iasonos et al. [13]. The recommendations for tumor marker prognostic studies (REMARK) criteria were followed [16,17]. To develop the prognostic model [18], the log-hazard rates obtained from the Cox model were adopted to derive weighting factors of a prognostic index, aimed to recognize differential risks of death. Coefficients estimates were ‘normalised’ dividing by the smallest one and rounding the resulting ratios to the nearest integer value [19]. A continuous score, assigning to patients an ‘individualized’ risk, was generated. Patients’ outcomes, according to the prognostic score, were exhibited by distributing patients into three risk classes. The cut-offs were selected at nearly equal distance along the range of values [20]. Finally, an external validation of the DFS model was explored in the VS. The receiver operating characteristic analysis allowed to estimate the accuracy of the prognostic model, by the area under the curve (AUC) determination with standard error (SE).
2.5. Samples and molecular analysis
Formalin-fixed and paraffin-embedded (FFPE) ILC samples from patients at poor prognosis (defined with the prognostic model) were selected from the University of Verona according to the availability of a tissue sample from the surgical specimen of the primary tumor. In order to compare the molecular pattern of these samples at poor prognosis with those at good, we selected a subgroup of cases at good prognosis according to the developed prognostic model. The collected material has been subjected to targeted NGS analysis for somatic mutation (SM), copy number variation (CNV) and transcriptomic analysis. In addition, quantitative-PCR, immunohistochemistry (IHC) and fluorescent in situ hybridization (FISH) for the validation of gene alterations of interest were performed. Finally, stromal tumor-infiltrating lymphocytes (sTIL) were also evaluated. Fisher’s exact test and Peto Odds Ratio (OR) for estimating the risk of association of a given biomarker with each prognostic class were used as appropriate. Finally, in order to identify potential biological profiles associated with the prognosis, the multiple correspondence analysis (MCA) was accomplished [21].
2.6. Mutational, CNV and transcriptomic analyses
Matched tumor/normal DNA and RNA from all FFPE samples was subjected to NGS. To analyse DNA, an Ion Ampliseq custom panel was used to investigated SM and CNV status of all exons of 26 selected genes upon the results of published whole genome sequencing and exome breast cancer data, as described in the supplementary material [4,22]. Mutational load and chromosome integrity number (CIN) were also evaluated as illustrated in previous studies [23]. Available RNA was converted in cDNA and subjected to analysis using Ion Ampliseq Transcriptome Human Gene Expression Kit [24].
2.7. Immunohistochemistry, FISH and sTIL assessment
Immunohistochemistry for PD1, PD-L1, phmTOR, CDK4 and CDK6 was performed on surgical specimens using 4 μm FFPE tissue. Fluorescent in situ hybridization for PD-L1, CDK4, ESR1 and mTOR was performed as well. Stromal TIL were assessed on hematoxylin and eosin-stained sections according to the International TILs Working Group 2014 recommendations [25].
3. Results
3.1. Patients’ characteristics
Clinico-pathological information from 773 patients (491 for TS and 282 for VS) with ILC who underwent surgery was gathered [26]. Patients’ characteristics of the TS are reported in Table 1. At a median follow-up of 77 months (range 1–396), median DFS was 175 months (95%CI 153–197), with a 5-/10-year rate of 82.4%/70.5%, respectively. Median OS was 213 months (95%CI 190–236), with a 5-/10-year rate of 91.8%/82.2%, respectively.
Table 1.
Clinico-pathological and therapeutic characteristics in patients with invasive lobular carcinoma (Training Set, N = 491).
| Characteristics | Subcategories | ILC [TS] Patients N (%) |
|---|---|---|
| Menopausal status | Premenopausal | 142 (28.9) |
| Postmenopausal | 349 (71.1) | |
| Grading | 1 | 124 (25.3) |
| 2 | 155 (31.6) | |
| 3 | 75 (15.3) | |
| Unknown | 137 (27.9) | |
| Oestrogen Receptor status | Positive | 460 (93.7) |
| Negative | 17 (3.5) | |
| Unknown | 14 (2.8) | |
| Progesterone Receptor status | Positive | 412 (83.9) |
| Negative | 52 (10.6) | |
| Unknown | 27 (5.5) | |
| Ki67 | <5% | 136 (27.7) |
| ≥5% | 313 (63.7) | |
| Unknown | 42 (8.6) | |
| HER2 status | Positive | 27 (5.5) |
| Negative | 353 (71.9) | |
| Unknown | 111 (22.6) | |
| T category according to TNM [7° Edition] | 1 | 276 (56.2) |
| 2 | 148 (30.1) | |
| 3 | 40 (8.1) | |
| 4 | 23 (4.7) | |
| Unknown | 4 (0.9) | |
| Lymph-nodal status | Positive | 180 (36.7) |
| Negative | 297 (60.5) | |
| Unknown | 14 (2.8) | |
| Vascular Invasion | Present | 87 (17.7) |
| Absent | 290 (59.1) | |
| Unknown | 114 (23.2) | |
| Multifocality | Present | 93 (18.9) |
| Absent | 375 (76.4) | |
| Unknown | 23 (4.7) | |
| Type of surgery | Tumorectomy | 130 (26.5) |
| Quadrantectomy | 165 (33.6) | |
| Mastectomy | 196 (39.9) | |
| Adjuvant hormonal therapy | Yes | 432 (88.0) |
| No | 534 (11.0) | |
| Unknown | 5 (1.0) | |
| Adjuvant chemotherapy | Yes | 199 (40.5) |
| No | 292 (59.5) | |
| Adjuvant Trastuzumab | Yes | 11 (2.2) |
| No | 480 (97.8) | |
| Adjuvant radiotherapy | Yes | 301 (61.3) |
| No | 177 (36.0) | |
| Unknown | 13 (2.6) |
Legend-Table 1. ILC, invasive lobular carcinoma; TS, training set; N, number.
3.2. Survival analysis
The univariate analysis was reported in Table S1. At the multivariate analysis, T-category (1–2) and negative nodal status were independent predictors for longer DFS; age at diagnosis <60 years, negative nodal status and Ki67 < 5% were independent factors for longer OS. A Ki67 threshold of 5% was adopted as optimal prognostic cut-off considering the previously reported and validated results in a large series of ILC [2]. At the internal cross-validation analysis, all variables were confirmed as independent factors (Table 2).
Table 2.
Multivariate analysis in patients with invasive lobular carcinoma (Training Set, N = 491).
| Predictors | DFS HR (95% CI) [p-value] |
Replication Rate [Internal Validation] | OS HR (95% CI) [p-value] |
Replication Rate [Internal Validation] |
|---|---|---|---|---|
| T-category according to TNM (7° Edition) [3–4 vs. 1–2] | 1.78 (0.97–3.25) [0.062] | 60% | – | – |
| Nodal Status [Positive vs. Negative] | 2.46 (1.50–4.05) [<0.0001] | 95% | 3.32 (1.71–6.45) [<0.0001] | 100% |
| Age [>60 years vs. ≤60 years] | – | – | 2.19 (1.14–4.21) [0.019] | 90% |
| Ki67 [≥5% vs. <5%] | – | – | 2.47 (1.02–5.94) [0.044] | 80% |
Legend-Table 2. DFS, disease-free survival; HR, hazard ratio; CI, confidence interval; OS, overall survival, T, tumor.
3.3. Prognostic score assessment
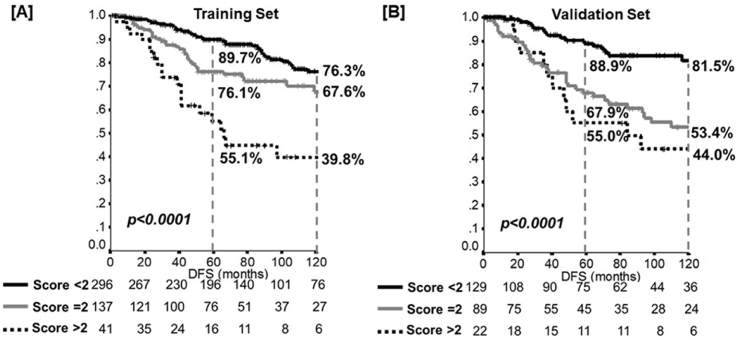
All the covariates showing independent prognostic role in the Cox model (T-category, nodal status, age at diagnosis and Ki67) were included in the prognostic index. According to the HR achieved at the multivariate analysis, a prognostic scoring index was allocated to each patient in order to categorize the individual risk of recurrence and death (Table S2). Based on the prognosis, patients were separated into three risk groups for DFS and OS: 1) low risk of recurrence and death: score<2 (ie, the best outcome estimate); 2) intermediate risk of recurrence and death: score = 2; 3) high risk of recurrence and death: score >2 (ie, worst outcome estimate). According to the prognostic model, a significant prognostic difference between patients at low, intermediate, and high risk was obtained for DFS (10-year: 76.3%/67.6%/39.8%, p<0.0001; AUC 0.60 (SE, 0.03)); Fig. 1, Panel A) and OS (10-year: 92.7%/82.7%/67.1%, p<0.0001; AUC 0.66 (SE, 0.03)).
Fig. 1.
Disease-free survival (DFS) according to the risk-class model in the Training set [Panel A] and in the Validation set [Panel B]. p-value: log-rank analysis.
3.4. External validation analysis
The VS consisted of 282 patients (Table S3); median follow-up 86 months (range 1–348). The model, developed in the TS, has been proven to be likewise able to discriminate the DFS in the VS (Fig. 1, Panel B). Indeed, a significant prognostic difference between patients at low, intermediate, and high risk was obtained (10-year: 81.5%/53.4%/44.0%, p<0.0001; AUC 0.70 (SE, 0.03)). Considering the overall population (TS plus VS; Table S4 and Additional File 1), the performance of the model adjusting for the set was confirmed (p<0.001), without difference in terms of DFS between the TS and VS (HR 1.02, 95%CI 0.77–1.37, p = 0.88).
3.5. Patients’ sample
Patient’s samples for molecular analysis were selected based on the inclusion criteria: 1) selection according to the prognostic model (patients with the ‘best’ and ‘worst’ outcome, excluding patients with intermediate outcome), and 2) tissue availability at the coordinating center (University of Verona). These tissue samples were available for 20 and 14 patients scored at poor and good prognosis class according to the DFS model, respectively (Table S5, Fig. S1, Fig. S2).
3.6. Molecular features according to prognosis
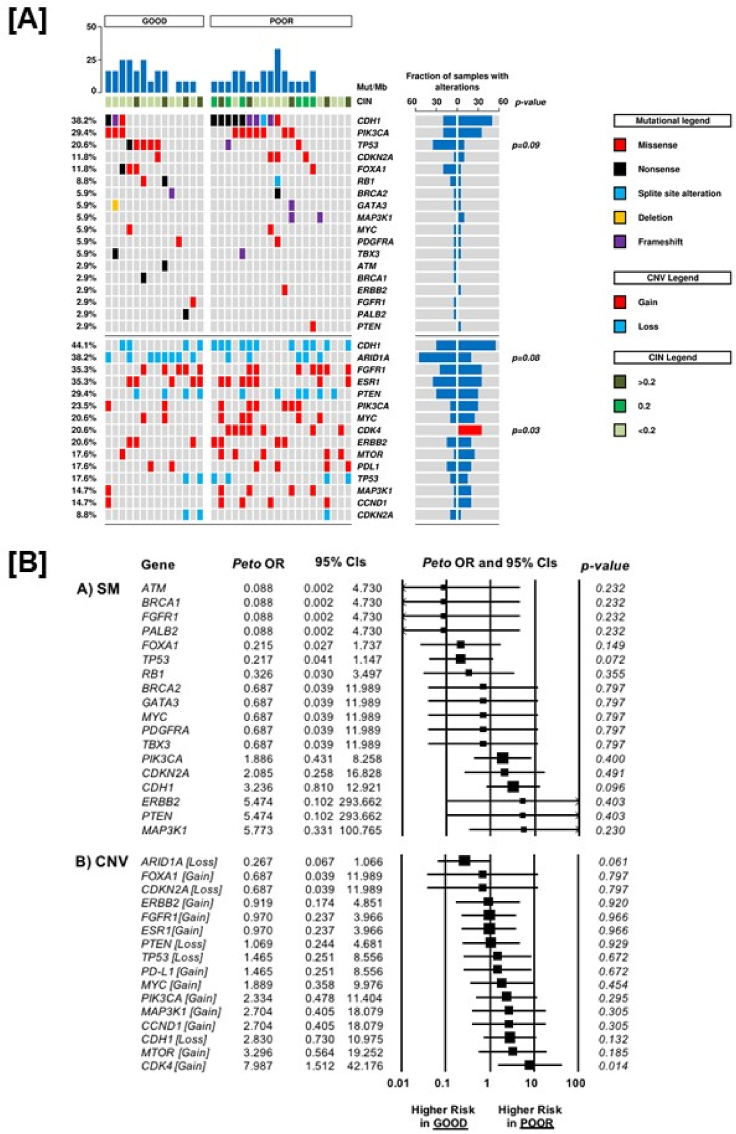
Mutations were observed in 29 cases of all series for 26 genes analysed. In detail: one mutation was observed in 10/34 cases (29.4%), more than one in 19/34 (55.9%) cases while no alteration in 5/34 cases (14.7%). A mean of 12.4 mutations/Mb was achieved. The most commonly mutated gene in the whole cohort was CDH1 (38.2%), followed by PIK3CA (29.4%) and TP53 (20.6%) (Fig. 2 (Panel A), Table 3, Table S6). Loss of heterozygosis of CDH1 (44.1%) and ARID1A (38.2%) were the most frequent CNV events, followed by gain in FGFR1 and ESR1 (each 12/34; 35.3%).
Fig. 2.
Comparison of mutational load, chromosome integrity number, somatic mutations and copy number variation between patients with poor prognosis and patients with good prognosis [Panel A]. Odds Ratio analysis [Panel B] of somatic mutations (Panel A) and copy-number variation (Panel B) according to prognosis: an OR<1 indicates a higher chance to be associated with good prognosis; an OR>1 indicates a higher chance to be associated with poor prognosis. CIN, chromosome integrity number; CNV, copy number variation; OR, odds Ratio; CI, confidence interval; SM, somatic mutation; CNV, copy number variation.
Table 3.
Prevalence of somatic mutations and copy number variations analysis of the 26 genes in the 34 invasive lobular carcinoma patients according to prognostic groups.
| Gene | Alteration | Poor Group Patients N (%) |
Good Group Patients N (%) |
Total N (%) |
p-value* |
|---|---|---|---|---|---|
| AKT1 | – | – | – | – | - |
| ARID1A | Loss | 5 (25.0) | 8 (57.1) | 13 (38.2) | 0.08 |
| ATM | SM | 0 | 1 (7.1) | 1 (2.9) | - |
| BRCA1 | SM | 0 | 1 (7.1) | 1 (2.9) | - |
| BRCA2 | SM | 1 (5.0) | 1 (7.1) | 2 (5.9) | - |
| CCND1 | Gain | 4 (20.0) | 1 (7.1) | 5 (14.7) | - |
| CDH1 | SM | 10 (50.0) | 3 (21.4) | 13 (38.2) | - |
| Loss | 11 (55.0) | 4 (28.6) | 15 (44.1) | - | |
| CDK4 | Gain | 7 (35.0) | 0 | 7 (20.6) | 0.03 |
| CDKN2A | SM | 3 (15.0) | 1 (7.1) | 4 (11.8) | - |
| Loss | 1 (5.0) | 2 (14.3) | 3 (8.8) | - | |
| ERBB2 | SM | 1 (5.0) | 0 | 1 (2.9) | - |
| Gain | 4 (20.0) | 3 (21.4) | 7 (20.6) | - | |
| ESR1 | Gain | 7 (35.0) | 5 (35.7) | 12 (35.3) | - |
| FGFR1 | SM | 0 | 1 (7.1) | 1 (2.9) | - |
| Gain | 7 (35.0) | 5 (35.7) | 12 (35.3) | - | |
| FOXA1 | SM | 1 (5.0) | 3 (21.4) | 4 (11.8) | - |
| Gain | 1 (5.0) | 1 (7.1) | 2 (5.9) | - | |
| GATA3 | SM | 1 (5.0) | 1 (7.1) | 2 (5.9) | - |
| MAP3K1 | SM | 2 (10.0) | 0 | 2 (5.9) | - |
| Gain | 4 (20.0) | 1 (7.1) | 5 (14.7) | - | |
| MTOR | Gain | 5 (25.0) | 1 (7.1) | 6 (17.6) | - |
| MYC | SM | 1 (5.0) | 1 (7.1) | 2 (5.9) | - |
| Gain | 5 (25.0) | 2 (14.3) | 7 (20.6) | - | |
| PALB2 | SM | 0 | 1 (7.1) | 1 (2.9) | - |
| PDGFRA | SM | 1 (5.0) | 1 (7.1) | 2 (5.9) | - |
| PDL1 | Gain | 4 (20.0) | 2 (14.3) | 6 (17.6) | - |
| PGR | – | – | – | – | - |
| PIK3CA | SM | 7 (35.0) | 3 (21.4) | 10 (29.4) | - |
| Gain | 6 (30.0) | 2 (14.3) | 8 (23.5) | - | |
| PTEN | SM | 1 (5.0) | 0 | 1 (2.9) | - |
| Loss | 6 (30.0) | 4 (28.6) | 10 (29.4) | - | |
| RB1 | SM | 1 (5.0) | 2 (14.3) | 3 (8.8) | - |
| TBX3 | SM | 1 (5.0) | 1 (7.1) | 2 (5.9) | - |
| TP53 | SM | 1 (5.0) | 5 (35.7) | 7 (20.6) | 0.09 |
| Loss | 4 (20.0) | 2 (14.3) | 6 (17.6) | - |
Legend-Table 3. N, Number; SM, somatic mutation; p-value* according to Fisher’s exact test (only p<0.10 are reported).
The prevalence of gene SM and CNV according to prognostic groups was reported in Table 3 and Supplementary Material. Interestingly, gain of CDK4 (7/34; 21.2%) was exclusively present in this poor prognosis group, whereas no good prognosis case showed this alteration (p = 0.03). Moreover, CDK4 gain resulted in a statistically significant higher chance to be associated with poor prognosis (OR 7.98, 95%CI 1.51–42.1, p = 0.014) (Fig. 2, Panel B). Regarding the association of CDK4 gain and clinico-pathological characteristics, this alteration was more frequent in node positive (p = 0.03) and high proliferative tumours, as defined by Ki67, despite a non-significant statistically difference (p = 0.14).
When grouping the molecular alterations involved in the regulation of G1/S phase cell cycle progression herein evaluated (CDK4 and CCND1 gain or CDKN2A mutation), this signature was significantly more associated with poor prognosis in the whole patients’ sample (OR 6.24, 95%CI 1.59–24.5, p = 0.009), and in the RB1 wild type population (32 patients, OR 5.33, 95%CI 1.33–21.28, p = 0.018).
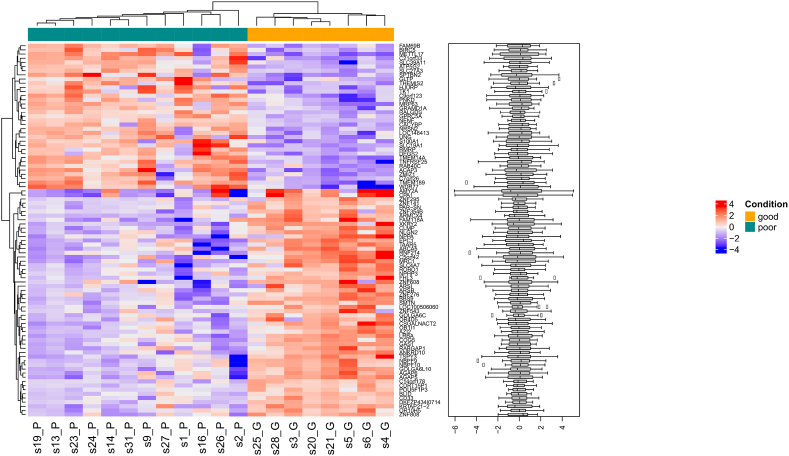
RNA was available for 20 samples (12 poor and 8 good). Ninety genes (as reported in Fig. 3) were differentially expressed between the two prognostic groups (Table S7, Supplementary Results, Table S8).
Fig. 3.
Heatmap displaying normalised expression values of the 90 differentially expressed genes between the two prognostic groups at a p-value cutoff of 0.05. Hierarchical clustering correctly separates the ‘good’ and the ‘poor’ samples.
According to the FISH analysis, CDK4 gain (Fig. S3, Panel A) was detected in the 10% of patients at poor prognosis (those presenting also CDK4 gain at NGS), while no CDK4 gain was detected in patients at good, without a significant difference between the two groups. According to the IHC analysis, nuclear CDK4 overexpression (score 3+) (Fig. S3, Panel B) was detected in the 20% and 7.1% of patients at poor and good prognosis, respectively (p = 0.62). No association between CDK4 gain at NGS and nuclear CDK4 overexpression at IHC was observed. CDK4 gain at FISH and CDK4 overexpression were more frequently associated with poor prognosis, despite a non-statistically significant difference, with a similar trend for CDK6 overexpression as well (Fig. S3, Panel C; Fig. S4). Patients with poor prognosis resulted to have a significantly higher chance to be cumulatively associated with abnormalities in CDK4/CDK6 overall expression or CNV (Fig. S5, Supplementary Material).
Regarding the sTIL assessment, the median value was 1% (range 0–30%), with only 6% of cases presenting a TILs percentage ≥10%. No significant association between TILs level (considered as a categorical variable, <1% and ≥1%) and prognosis was found (OR 1.75, 95%CI 0.45–6.8, p = 0.41). In addition, no association between TILs level and CDK4 gains was identified.
Finally, the MCA suggests that patients with good prognosis (upper left quadrant) and poor prognosis (lower right quadrant) seem to be correlated with distinct molecular alterations as defined by the distribution of some biomarkers, that appear far from the origin, and diagonally opposite (Fig. S6).
4. Discussion
The current multicenter study suggests that the combination of clinico-pathological parameters seems able to stratify the prognosis of patients affected by early-stage ILC. Moreover, the external validation supported the discrimination performance of the prognostic model, highlighting the potential ability of this model to accurately separate ILC patients into three risk classes according to their individual risk of recurrence.
The risk classification system for DFS comprises the two traditional clinico-pathological parameters adopted in daily clinical practice: T-category and nodal status. These factors represent the milestone variables driving the prognosis and the choice of adjuvant treatment in the context of early-stage BC, together with the hormone receptor and HER2 status [7]. Regarding OS, age and Ki67 represent the variables included in the prognostic model, besides the nodal status. Previous retrospective series reported other potential prognostic factors for ILC, such as the tumor grade; however, they did not consider the role of Ki67, a proliferation marker usually involved in the treatment decision of early BC, and they also included metastatic tumours [27]. Conversely (and similarly to our data) tumor histological grade did not result to be an independent prognostic factor for early-stage ILC in a different retrospective series [28]. With regard to the Ki67 role in the lobular histotype, we previously suggested that a very low cut-off of Ki67 (4%–5%) may be able to significantly discriminate the prognosis of patients with ‘pure’ ILC [2,29].
Our analysis does not represent a simple investigation of prognostic factors for ILC (as previously reported by different retrospective studies); the novelty of this study consists in the development and validation of a prognostic tool (consisting of the combination of reliable clinico-pathological factors with different prognostic weight according to the model’s score assessment) easy to adopt in the clinical practice. These results may help the clinicians to estimate the DFS and OS of ILC, a histotype where several aspects (including the prognosis) represent a matter of research for the personalized medicine. Indeed, although the common prognostic factors are indiscriminately applied for both lobular and ductal histology, determining that ILC patients are substantially treated in the same way as those affected by invasive carcinoma of NST, the overall prognosis of the two histotypes appears different [3,27]. Thus, this peculiar prognostic aspect supports the hypothesis that specific molecular features might drive the ILC prognosis. In this regard, the selection of ‘exceptional’ responders may increase the likelihood of finding a molecular characteristic that could account for the outcome [9].
Based on this approach, we performed the molecular analysis in an explorative subset of ‘worst’ prognostic performers, comparing the results with those of a subset of ‘best’ performers. The most interesting finding of our analysis is represented by the potential negative prognostic role of CDK4 gain, detected by NGS analysis. Moreover, the CDK4 amplification, detected by FISH analysis, and the nuclear CDK4 overexpression displayed a trend toward an association with poor prognosis.
The involvement of CDK4 amplification/gain in tumorigenesis has been found in various tumours including malignant gliomas and sarcomas [30,31]. In the context of BC, a previous analysis, conducted in 95 patients undergone surgery (80 carcinoma of NST and 15 ILC), suggested that CDK4 amplification (detected in 15.8% of patients) was associated with high tumor cell proliferation [32]. In another study, included 102 cases of carcinoma of NST, the immunohistochemical nuclear expression of CDK4, detected in about 70% of tumor samples, did not show any significant correlations with the main clinical prognostic factors nor with survival [33].
However, no previous studies reported the prognostic role of CDK4 in the specific context of ILC. According to the TCGA data, no CDK4 alterations was detected in 127 ILC. On the contrary, the genomic characterization of ILC conducted by Desmedt et al. identified CDK4 gain in 1.7% of the 170 lobular tumours with CNA data. The discrepancy in CDK4 gain prevalence between our series and the others may be explained by differences in term of prognostic characteristics, considering that we detected CDK4 gain only in patients selected for their extremely poor prognosis. Thus, this potential prognostic role deserves to be further explored in the featured context of ILC with worse outcome.
The combined subset analysis of the molecular alterations involved in the G1/S phase regulation strengthens a potential role of this cell cycle pathway in the prognosis of ILC. However, the low number of analysed samples and the criteria in the selection of patients, limit the possible implication of such data, given the risk of a false positive result.
As expected in the lobular histotype, the most frequent alteration is represented by the CDH1 mutation and loss of heterozygosity, without a different distribution in the two prognostic groups. Similarly, mutations in PIK3CA, TP53, CDKN2A and FOXA1 does not present a different distribution according to prognosis.
The incidence of FOXA1 mutation and ESR1 gain are similar to that reported by Desmedt et al. (11.8% vs. 9% and 35.3% vs. 25.3%, respectively) [5]. Considering that FOXA1 plays a key role in the endocrine signaling, these gene alterations need to be further explored in the context of ILC, where the endocrine therapy represents the main therapeutic strategy [34].
The unsupervised clustering analysis of the transcriptome evidenced differences between the two prognostic groups in terms of gene expression level. Unlike a previous integrative study [4], our analysis showed that a series of genes, overexpressed in the poor prognosis group, may play a relevant role in the oncogenesis. For example, the METTL17 interacts with both the AF1 and AF2 domains of ERα/β. In a recent study, the observation that knockdown of METTL17 reduces BC cell growth suggests that METTL17 regulates the cancer cell growth possibly through modulation of ERα function as well as the expression of ERα/β target genes [35].
Concerning the presence of sTIL, our analysis showed that most of ILCs are characterized by low sTIL levels, without differences between the two prognostic cohorts. These results are consistent with a recent analysis reporting that the percentage of sTIL in ILC was lower compared to that in invasive carcinoma of NST and that the sTIL level did not represent an independent prognostically variable [36].
The most notable limit of this study, beyond the retrospective design and the absence of a central pathology review, is represented by the small sample size for the molecular analysis. Therefore, the estimation in the frequency of genomic alterations is imprecise. A larger cohort is needed to validate the results in independent datasets. Despite this, the stratification according to prognosis, based on a large cohort of ILC patients (more than 750), offers the possibility to better identify potential molecular differences associated with outcome.
5. Conclusions
A risk stratification model, able to potential separate early-stage ILC patients’ prognosis into different risk classes according to clinico-pathological variables, allowed to investigate potential biomarkers of prognosis with targeted NGS in an explorative cohort. The reported molecular analysis needs to be interpreted as hypothesis generating. CDK4 gain is suggested for future validation as a prognostic biomarkers and potential therapeutic opportunity.
Ethical approval
The study was approved by the local Ethics Committees (Coordinating Center [University of Verona]: Prot. CESC n° 24,163, May 20th, 2014).
Acknowledgments
E.B. is supported by a grant of the Italian Association for Cancer Research (AIRC) Investigator Grant n. 20583. E.B. is currently supported by Institutional Funds (Università Cattolica del Sacro Cuore - UCSC Project D1.2019). P.D. is supported by Fondazione Nadia Valsecchi Onlus.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.breast.2020.01.034.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.McCart Reed A.E., Kutasovic J.R., Lakhani S.R., Simpson P.T. Invasive lobular carcinoma of the breast: morphology, biomarkers and ’omics. Breast Cancer Res. 2015;17:12. doi: 10.1186/s13058-015-0519-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carbognin L., Sperduti I., Fabi A. Prognostic impact of proliferation for resected early stage ’pure’ invasive lobular breast cancer: cut-off analysis of Ki67 according to histology and clinical validation. Breast. 2017;35:21–26. doi: 10.1016/j.breast.2017.06.005. [DOI] [PubMed] [Google Scholar]
- 3.Pestalozzi B.C., Zahrieh D., Mallon E. Distinct clinical and prognostic features of infiltrating lobular carcinoma of the breast: combined results of 15 International Breast Cancer Study Group clinical trials. J Clin Oncol. 2008;26:3006–3014. doi: 10.1200/JCO.2007.14.9336. [DOI] [PubMed] [Google Scholar]
- 4.Ciriello G., Gatza M.L., Beck A.H. Comprehensive molecular portraits of invasive lobular breast cancer. Cell. 2015;163:506–519. doi: 10.1016/j.cell.2015.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desmedt C., Zoppoli G., Gundem G. Genomic characterization of primary invasive lobular breast cancer. J Clin Oncol. 2016;34:1872–1881. doi: 10.1200/JCO.2015.64.0334. [DOI] [PubMed] [Google Scholar]
- 6.Michaut M., Chin S.F., Majewski I. Integration of genomic, transcriptomic and proteomic data identifies two biologically distinct subtypes of invasive lobular breast cancer. Sci Rep. 2016;6:18517. doi: 10.1038/srep18517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Curigliano G., Burstein H.J., E P W De-escalating and escalating treatments for early-stage breast cancer: the st. Gallen international expert consensus conference on the primary therapy of early breast cancer 2017. Ann Oncol. 2017;28:1700–1712. doi: 10.1093/annonc/mdx308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desmedt C., Zoppoli G., Sotiriou C., Salgado R. Transcriptomic and genomic features of invasive lobular breast cancer. Semin Canc Biol. 2017;44:98–105. doi: 10.1016/j.semcancer.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 9.Kummar S., Williams P.M., Lih C.J. Application of molecular profiling in clinical trials for advanced metastatic cancers. J Natl Cancer Inst. 2015;107 doi: 10.1093/jnci/djv003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hess K.R. Graphical methods for assessing violations of the proportional hazards assumption in Cox regression. Stat Med. 1995;14:1707–1723. doi: 10.1002/sim.4780141510. [DOI] [PubMed] [Google Scholar]
- 11.Armstrong K. Methods in comparative effectiveness research. J Clin Oncol. 2012;30:4208–4214. doi: 10.1200/JCO.2012.42.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sauerbrei W., Schumacher M. A bootstrap resampling procedure for model building: application to the Cox regression model. Stat Med. 1992;11:2093–2109. doi: 10.1002/sim.4780111607. [DOI] [PubMed] [Google Scholar]
- 13.Iasonos A., Schrag D., Raj G.V., Panageas K.S. How to build and interpret a nomogram for cancer prognosis. J Clin Oncol. 2008;26:1364–1370. doi: 10.1200/JCO.2007.12.9791. [DOI] [PubMed] [Google Scholar]
- 14.Bria E., Di Modugno F., Sperduti I. Prognostic impact of alternative splicing-derived hMENA isoforms in resected, node-negative, non-small-cell lung cancer. Oncotarget. 2014;5:11054–11063. doi: 10.18632/oncotarget.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harrell F.E., Jr., Lee K.L., Califf R.M. Regression modelling strategies for improved prognostic prediction. Stat Med. 1984;3:143–152. doi: 10.1002/sim.4780030207. [DOI] [PubMed] [Google Scholar]
- 16.Alonzo T.A. Standards for reporting prognostic tumor marker studies. J Clin Oncol. 2005;23:9053–9054. doi: 10.1200/JCO.2005.04.3778. [DOI] [PubMed] [Google Scholar]
- 17.Simon R.M., Paik S., Hayes D.F. Use of archived specimens in evaluation of prognostic and predictive biomarkers. J Natl Cancer Inst. 2009;101:1446–1452. doi: 10.1093/jnci/djp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bria E., De Manzoni G., Beghelli S. A clinical-biological risk stratification model for resected gastric cancer: prognostic impact of Her2, Fhit, and APC expression status. Ann Oncol. 2013;24:693–701. doi: 10.1093/annonc/mds506. [DOI] [PubMed] [Google Scholar]
- 19.Di Maio M., Lama N., Morabito A. Clinical assessment of patients with advanced non-small-cell lung cancer eligible for second-line chemotherapy: a prognostic score from individual data of nine randomised trials. Eur J Canc. 2010;46:735–743. doi: 10.1016/j.ejca.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 20.Carbognin L., Sperduti I., Ciccarese M. Prognostic model for advanced breast carcinoma with luminal subtype and impact of hormonal maintenance: implications for post-progression and conditional survival. Breast. 2016;29:24–30. doi: 10.1016/j.breast.2016.06.021. [DOI] [PubMed] [Google Scholar]
- 21.Novelli F., Milella M., Melucci E. A divergent role for estrogen receptor-beta in node-positive and node-negative breast cancer classified according to molecular subtypes: an observational prospective study. Breast Cancer Res. 2008;10:R74. doi: 10.1186/bcr2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nik-Zainal S., Davies H., Staaf J. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campesato L.F., Barroso-Sousa R., Jimenez L. Comprehensive cancer-gene panels can be used to estimate mutational load and predict clinical benefit to PD-1 blockade in clinical practice. Oncotarget. 2015;6:34221–34227. doi: 10.18632/oncotarget.5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salgado R., Denkert C., Demaria S. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Ann Oncol. 2015;26:259–271. doi: 10.1093/annonc/mdu450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carbognin L., Simbolo M., Vicentini C. Next-generation targeted sequencing (NGTS) investigating CDK4 as a prognostic driver in pure invasive lobular breast carcinoma (ILC): preliminary results in early-stage patients (pts) stratified according to a validated clinico-pathological model. J Clin Oncol. 2018;36 doi: 10.1016/j.breast.2020.01.034. 542-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Z., Yang J., Li S. Invasive lobular carcinoma of the breast: a special histological type compared with invasive ductal carcinoma. PloS One. 2017;12 doi: 10.1371/journal.pone.0182397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adachi Y., Ishiguro J., Kotani H. Comparison of clinical outcomes between luminal invasive ductal carcinoma and luminal invasive lobular carcinoma. BMC Canc. 2016;16:248. doi: 10.1186/s12885-016-2275-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carbognin L., Sperduti I., Brunelli M. Subpopulation Treatment Effect Pattern Plot (STEPP) analysis of Ki67 assay according to histology: prognostic relevance for resected early stage ’pure’ and ’mixed’ lobular breast cancer. J Exp Clin Canc Res. 2016;35:50. doi: 10.1186/s13046-016-0325-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qi Q., Kang S.S., Zhang S. Co-amplification of phosphoinositide 3-kinase enhancer A and cyclin-dependent kinase 4 triggers glioblastoma progression. Oncogene. 2017;36:4562–4572. doi: 10.1038/onc.2017.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee S.E., Kim Y.J., Kwon M.J. High level of CDK4 amplification is a poor prognostic factor in well-differentiated and dedifferentiated liposarcoma. Histol Histopathol. 2014;29:127–138. doi: 10.14670/HH-29.127. [DOI] [PubMed] [Google Scholar]
- 32.An H.X., Beckmann M.W., Reifenberger G. Gene amplification and overexpression of CDK4 in sporadic breast carcinomas is associated with high tumor cell proliferation. Am J Pathol. 1999;154:113–118. doi: 10.1016/S0002-9440(10)65257-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peurala E., Koivunen P., Haapasaari K.M. The prognostic significance and value of cyclin D1, CDK4 and p16 in human breast cancer. Breast Cancer Res. 2013;15:R5. doi: 10.1186/bcr3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ross-Innes C.S., Stark R., Teschendorff A.E. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012;481:389–393. doi: 10.1038/nature10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Du P., Yuan B., Cao J. Methyltransferase-like 17 physically and functionally interacts with estrogen receptors. IUBMB Life. 2015;67:861–868. doi: 10.1002/iub.1444. [DOI] [PubMed] [Google Scholar]
- 36.Desmedt C., Salgado R., Fornili M. Immune infiltration in invasive lobular breast cancer. J Natl Cancer Inst. 2018;110(7):768–776. doi: 10.1093/jnci/djx268. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.