Abstracts
Background
Human epidermal growth factor receptor 2(HER2) status is a crucial predictive factor for prognostic assessment and targeted therapy selection, which may be influenced by intratumor heterogeneity and molecular divergence between the primary site and different metastases. Therefore, we performed a prospective study to confirm the concordance of HER2 amplification in circulating tumor DNA(ctDNA) with primary tumor tissue and verified its clinical implications.
Methods
A total of 105 breast cancer patients were enrolled, and dynamic monitoring of HER2 copy numbers in ctDNA was conducted in 31 participants during the treatment. Totally 186 plasma samples were prospectively obtained and blinded to test HER2 copy numbers in ctDNA based on low-coverage whole genome sequencing(WGS) by next-generation sequencing(NGS).
Results
Comparing HER2 copy numbers in ctDNA collected before the initiation of next line of anticancer treatment with primary tumor tissue, the concordant rate of HER2 amplification was 86.5%(χ2 = 52.901, p < 0.001), with a positive and negative predictive value of 94.9% and 80.7%, respectively. Histopathologically positive, high-level amplification of HER2 copy numbers in the baseline was significantly correlated with best objective response during the anticancer therapy(p = 0.010). Moreover, HER2 copy numbers fluctuated with HER2-targeted therapeutic response, and the patients with a constantly positive level after 6 weeks of treatment appeared to suffer from significantly reduced progression free survival(p < 0.001).
Conclusions
HER2 amplification in ctDNA, with a concordance rate of over 80% with primary tumors, may be a predictive index for prognostic evaluation and therapeutic response monitoring in a noninvasive, repeatable and practical method for breast cancer patients.
Keywords: Breast cancer, ctDNA, HER2 amplification, HER2 copy numbers
Highlights
-
•
The concordance between HER2 amplification in ctDNA and tumor tissues was 86.5%.
-
•
The positive and negative predictive value surpassed previous studies.
-
•
HER2 copy numbers in ctDNA may predict the prognosis of anti-HER2 therapy.
-
•
HER2 copy numbers in ctDNA may predict response in HER2 positive breast cancer.
-
•
The study of various-stage patients may be closer to real-world clinical practice.
1. Introduction
Human epidermal growth factor receptor 2 (HER2) plays a pivotal role in cancer cell survival and proliferation, and HER2 positivity accounts for approximately 15–20% of breast cancers, with aggressive tumor behavior and poor prognosis [[1], [2], [3]]. The introduction of HER2-targeted therapy, with trastuzumab as a representative, has resulted in dramatically improved response rates and prolonged survival in both primary and metastatic breast cancer [4], [5]. Since it is factored into all prognostic and treatment decisions [6], [7], the 8th edition of the American Joint Committee on Cancer staging system incorporates HER2 status into TNM-prognostic staging groups as Level I evidence [8].
In clinical practice, HER2 status is measured by immunohistochemistry (IHC) or fluorescence in situ hybridization (FISH) on a section of tumor tissue obtained by surgery or needle biopsy [9], which will inevitably be influenced by intratumor spatial heterogeneity (ITH) when tested in tissue gained from single tumor biopsy samples. Moreover, the prevalence of HER2 status may be discordant between the primary tumor and metastases in approximately 25% of cases, especially after chemotherapy [10], [11]. Although biopsy of newly diagnosed metastases from primary breast cancer for testing of ER, PR, and HER2 status to direct therapy is recommended by guidelines [6], [12], it may sometimes be impossible to determine the histopathological information of metastases. Liquid biopsy makes it possible to interrogate the genetic evolution of tumor, monitor treatment response, and assess the emergence of therapeutic resistance repeatedly with minimal invasiveness [13].Therefore, circulating tumor DNA (ctDNA) to evaluate HER2 amplification may be a repeatable and noninvasive approach for dynamic efficacy monitoring and treatment decision-making [14], [15].
Our previous study performed in gastric cancer suggested that the changes of HER2 copy numbers in ctDNA based on low-coverage whole genome sequencing (WGS) by next-generation sequencing (NGS) may monitor the treatment efficacy of trastuzumab superiorly to commonly used biomarkers, such as carcinoembryonic antigen (CEA) and CA199 [16], whereas there is scarcely any similar evidence in both early and metastatic breast cancer. Thus, we investigated a prospective study to confirm the concordance of HER2 amplification in plasma ctDNA with primary tumor tissue, as well as the clinical significance of the modification in HER2 copy numbers during the anti-HER2 therapy.
2. Methods
2.1. Study design
A total of 105 female patients who were pathologically diagnosed with breast cancer under treatment in the National Cancer Center of China from January 2014 to December 2017 were recruited in the study and prospectively detected after an agreement from the Ethical committee. Among them, dynamic monitoring of HER2 copy numbers in ctDNA was conducted in 31 participants during the treatment, including both early breast cancer patients and metastatic breast cancer patients. All the blood samples were prospectively collected and blinded for detection in the absence of pathological information of the primary tumor. Afterwards, the clinicopathological information of the primary tissue and the demographic data of the patients were systematically collected and analyzed. The tissue HER2 status was evaluated using routine IHC and FISH methods according to the American Society of Clinical Oncology/College of American Pathologists clinical practice guidelines, in which HER2 positive is defined as HER2 protein overexpression measured by IHC status (IHC 3+) or by FISH measurement of a HER2 gene copy number of six or more or a HER2/CEP17 ratio of 2.0 or greater [17].
The study was approved by the institutional review board of National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College. Every participant signed written informed consent for serial blood collection and sample assay.
2.2. DNA extraction and whole genome sequencing
To detect the HER2 copy numbers in plasma for each sample, DNA was extracted from 500 μl of plasma and then subjected to WGS library protocol, as described previously [16]. The sequencing was performed on an Illumina HiSeq 2500 platform (Illumina, San Diego, CA), and approximately 5 M sequencing reads were obtained.
2.3. Copy number variation analysis
Gene copy number variation analysis was performed following the procedure previously described [18]. In brief, after the removal of the Illumina adaptors and low-quality bases, high-quality reads were mapped to the hg19 reference genome using BWA (Burrows-Wheeler Alignment Tool, version 0.7.12-r1039) with default parameters.
Uniquely mapped reads were extracted from the alignment reads. The whole reference genome was divided into nonoverlapping observation windows (bins) with the size of 1 Mb. Read numbers and GC content were calculated in each bin. To remove bias, the bin read count was normalized based on GC content, and a reference dataset was used to represent the relative copy number. The range of normal diploid HER2 copy number was 1.78–2.22 [16]. The relative reads number (RRN) > 2.22 for the bin covering the HER2 locus indicates HER2 amplification, while RRN≥4.0 indicates high-level amplification.
2.4. Statistical analyses
The relationship between plasma HER2 copy numbers in ctDNA and primary tumor tissue was assessed by χ 2 or Fisher’s exact test and was also used for the correlation between HER2 high-level amplification and the best objective response during anti-cancer treatment. Kaplan-Meier analysis was conducted to compare the survival outcomes of patients whose HER2 copy numbers remained positive after 6 weeks of treatment and those whose HER2 copy numbers descended to the level below amplification. All analyses were performed with SPSS software, version 22.0 (SPSS Inc., Chicago, IL, USA), and the differences with p < 0.05 were considered statistically significant.
3. Results
3.1. Patient characteristics
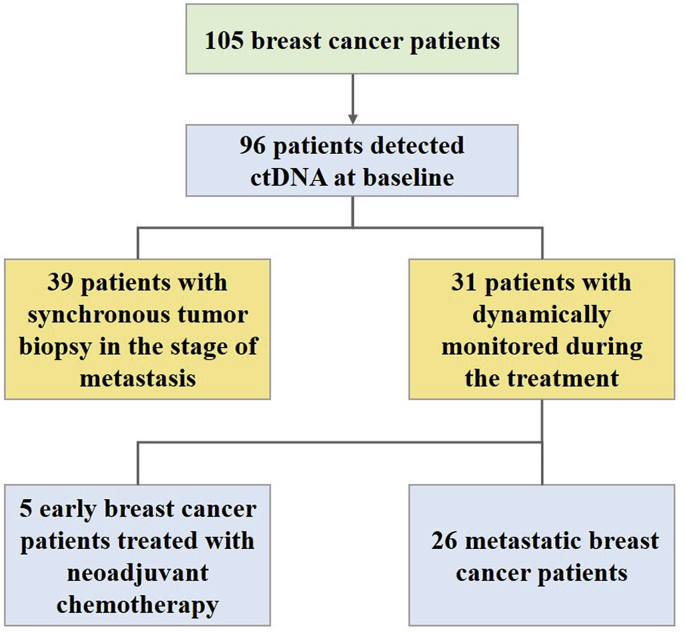
The analyses contained a total of 105 female breast cancer patients with a median age of 49 years, ranging from 24 to 73 years. The clinicopathological characteristics of the enrolled patients were presented in Table 1. Among them, 65 participants (61.9%) were hormone receptor (HR)-positive, and 57 (54.3%) were HER2-positive in the primary tumor. Most of the patients (94.3%) were stage IV, and approximately two-thirds of them had visceral metastases. Nearly three-tenths of the patients were available for two or more times serial assessments of plasma ctDNA, and 96 samples in the cohort were collected before the initiation of a new line of treatment (Fig. 1).
Table 1.
The clinicopathological characteristics of the enrolled patients.
| Patients (n) | Percentage (%) | |
|---|---|---|
| Age | ||
| <40 | 19 | 18.1% |
| ≥40 to <60 | 80 | 76.2% |
| ≥60 | 6 | 5.7% |
| Menstrual status | ||
| Premenopausal | 57 | 54.3% |
| Menopause | 48 | 45.7% |
| HR status | ||
| Positive | 65 | 61.9% |
| Negative | 40 | 38.1% |
| HER2 status | ||
| Positive | 57 | 54.3% |
| Negative | 48 | 45.7% |
| TNM staging | ||
| II-III | 6 | 5.7% |
| IV | 99 | 94.3% |
| Position of metastatic site | ||
| Nonvisceral | 38 | 36.2% |
| Visceral | 67 | 63.8% |
| Detection times | ||
| Once | 74 | 70.5% |
| Twice or more | 31 | 29.5% |
| Treatment mode | ||
| Neoadjuvant chemotherapy | 5 | 4.8% |
| Adjuvant chemotherapy | 1 | 1.0% |
| Palliative chemotherapy | 99 | 94.2% |
HR: hormone receptor; HER2: human epidermal growth factor receptor 2.
Fig. 1.
Study flowchart.
3.2. The concordance of HER2 status between plasma ctDNA and primary tumor tissue
In total, 186 plasma samples from 105 patients were included in the analyses, which were prospectively obtained and blinded to test the HER2 copy numbers in ctDNA. Afterwards, tissue HER2 status by IHC or FISH in the primary tumor were unblinded for analysis and compared with the results of serial plasma HER2 copy numbers by low-coverage WGS from the pretreatment to progressive timepoints. Comparing the HER2 copy numbers in plasma ctDNA acquired from the baseline with the primary tissue pathological results, the concordant rate was 86.5% (χ2 = 52.901, p < 0.001, Table 2). For patients with HER2 positivity in the primary tumor, 77.1% of the samples presented an amplification in plasma HER2 copy numbers of ctDNA, while for those that were HER2 negative in the primary tumor, a high concordant rate of 95.8% was observed in HER2 status between tissue and ctDNA. The positive and negative predictive values of HER2 amplification in ctDNA were 94.9% and 80.7%, respectively. In the cohort with plasma ctDNA before the initiation of a new line of treatment, 39 of the metastatic breast cancer patients underwent synchronous tumor biopsy in the stage of metastasis to evaluate the HER2 status, and the concordant rate of HER2 amplification between ctDNA and synchronous tumor tissue reached 89.7%, with the positive and negative predictive values of 94.4%(17/18) and 85.7%(18/21), respectively.
Table 2.
The concordance of HER2 status between plasma ctDNA and primary tissue.
| Tissue HER2 status |
||||
|---|---|---|---|---|
| positive | negative | |||
| plasma HER2 amplification in ctDNA | positive | 37 (77.1%) | 2(4.2%) | χ2 = 52.901 (p < 0.001) |
| negative | 11(22.9%) | 46(95.8%) | ||
HER2: human epidermal growth factor receptor 2; ctDNA: circulating tumor DNA.
Among the patients that were histopathologically HER2-positive in the primary tumor, high-level amplification in HER2 copy numbers of ctDNA collected before the initiation of the next line of treatment was significantly correlated with the best objective response (BOR) in the subsequent anti-cancer therapy (p = 0.010, Table 3). The 19 patients with plasma HER2 high-level amplification all achieved partial response, while only 15/22 attained partial response in those without high-level amplification. However, no parallel results can be summarized from the patients that were histopathologically HER2-negative in the primary tumor.
Table 3.
The correlation between high-level amplification in HER2 copy numbers of ctDNA and best objective response.
| Best Objective Response |
||||
|---|---|---|---|---|
| PR | SD | |||
| plasma HER2 copy number | high-level amplification | 19 | 0 | p=0.010 |
| without high-level amplification | 15 | 7 | ||
HER2: human epidermal growth factor receptor 2; ctDNA: circulating tumor DNA; PR: partial response; SD: stable disease.
3.3. Dynamics of the plasma HER2 copy numbers during therapeutic courses
Among them, 31 patients, including 26 metastatic breast cancer patients and 5 patients at the early stage treated with neoadjuvant chemotherapy, were dynamically monitored during the treatment to investigate the predictive value for therapeutic response.
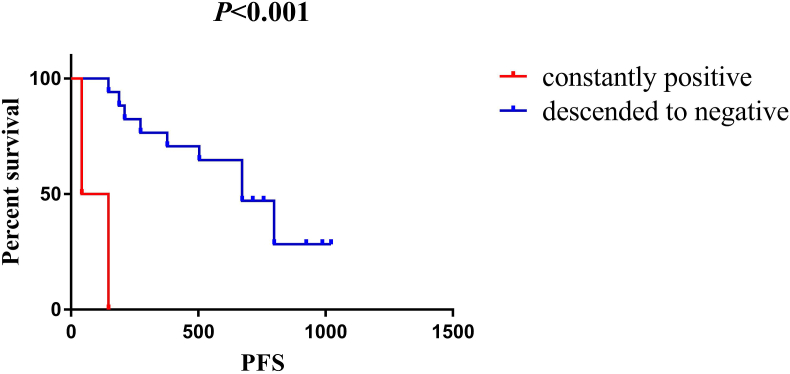
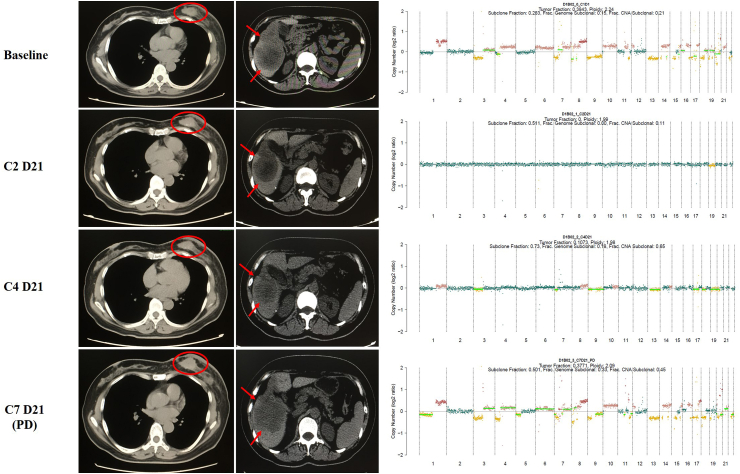
In the cohort of 26 metastatic patients whose HER2 status was positive in the primary tissue, their plasma HER2 copy numbers were tracked in a real-time manner when receiving combination therapy with pyrotinib (an oral tyrosine kinase inhibitor of HER2 [19]) and capecitabine, and a total of 102 plasma samples were analyzed. 19 of the 26 candidates were identified to have HER2 amplification in ctDNA before the medication and the level of HER2 copy numbers in each patient declined dramatically after 6 weeks of HER2-targeted treatment. Although the HER2 copy numbers failed to drop below the level of amplification in only two candidates, statistically significantly decreased progression-free survival (PFS) was observed in these two patients compared with others in which HER2 copy numbers descended to the level below amplification (average PFS 3.2 months vs 21.9 months, p < 0.001, Fig. 2). Furthermore, via synchronously comparing the dynamics of HER2 copy numbers with response evaluation by means of imaging at each follow-up timepoint, we found that the level of HER2 copy numbers in plasma ctDNA fluctuated corresponding to treatment response, as depicted in Fig. 3. At the timepoint of image-confirmed progressive disease, HER2 copy numbers increased obviously compared with the beneficial timepoints, and 7/9 of them reached the amplification criteria again. In some individuals, HER2 copy numbers ascended for several weeks before the establishment of image-confirmed progressive disease, which made it possible to monitor disease status prior to imaging.
Fig. 2.
Kaplan-Meier analysis-based estimation of probabilities of PFS in metastatic breast cancer patients in accordance with the modification of HER2 copy numbers during HER2-targeted therapy (constantly positive group: the patients whose HER2 copy numbers remained positive after 6 weeks of HER2-targeted treatment; descended to negative group: the patients whose HER2 copy numbers descended to the level below amplification after 6 weeks of HER2-targeted treatment).
Fig. 3.
The level of HER2 copy numbers in plasma ctDNA fluctuated along with treatment response.
For early breast cancer patients, we analyzed the plasma ctDNA collected before the neoadjuvant chemotherapy and after two cycles of chemotherapy in 5 patients, and the concordance rate between the histopathological HER2 status in tumor biopsy and synchronous plasma ctDNA was 80%. Similarly, the level of HER2 copy numbers dropped rapidly down to the level below amplification after two cycles of chemotherapy.
4. Discussion
HER2 protein overexpression and gene amplification remain the primary predictors of responsiveness to HER2-targeted therapies in breast cancer and gastric cancer [20], [21]. However, malignant tumors are highly heterogeneous in multiple aspects, including intratumor heterogeneity and the molecular divergence between primary site and different metastases, which may not be completely presented in morphology-based pathological classifications of the primary tumor site at diagnosis [22], [23], [24], [25]. Due to the critical role of HER2 status in the selection of HER2-directed targeted therapy, we designed a repeatable, noninvasive and practical method for the real-time tracing of HER2 amplification in ctDNA based on low-coverage genome-wide sequencing and verified its clinical implications in therapeutic effect monitoring and prognosis prediction.
In our present study, HER2 copy number variations measured in plasma ctDNA had a concordant rate of 86.5% compared with primary tumor tissues in the cohort that included both metastatic breast cancer patients and those of early stage according to our single-blind evaluation, which surpassed both the positive predictive value and negative predictive value of previous results by ddPCR or NGS [15,[26], [27], [28]]. Apart from that aspect, our detecting technique by NGS makes it possible to depict the genomic profiling of chromosomes, including the HER2 gene and other related genes, while ddPCR merely detects partial fragment of HER2. Our prior study in advanced gastric cancer achieved even higher concordance, of which the rate compared with synchronous tumor tissues by dual in situ hybridization (DISH) reached 91.07% [16]. However, this investigation in breast cancer contained breast cancer candidates of various stages, and some of them received three-line or more chemotherapy and targeted therapy, which may be closer to real-world clinical practice in breast cancer. In addition, owing to the fact that the loss of HER2-positive status in metastatic sites can occur in more than 20% of patients with primary HER2-positive breast cancer [10], the discordance between the primary sites and metastases may contribute to relatively lower accordance between the primary tumor and metastatic plasma ctDNA. Accordingly, HER2 copy numbers measured in plasma ctDNA may reflect the status of the recurrent tumor and serve as a real-time predictive biomarker for potential responsiveness to HER2-targeted therapies, especially when it is not feasible to conduct repeated biopsies in advanced breast cancer. For the patients whose HER-2 status was negative in preliminary tumor tissue, further study should focus on whether the acquisition of HER2 gene amplification in ctDNA would benefit from HER2-targeted therapies, which may provide a rational therapeutic option and improve the prognosis of this heavily pretreated population.
Serial ctDNA tracking makes it possible to reasonably forecast the therapeutic response and to timely detect treatment resistance in a noninvasive, repeatable and practical approach. With regard to plasma ctDNA in the baseline, our analysis on the correlation between high-level amplification in HER2 copy numbers and clinical best objective response indicated that HER2 high-level amplification in ctDNA may be a reliable predictive index for the evaluation of therapeutic responses during anticancer therapy. Moreover, sequential HER2 copy numbers fluctuated along with therapeutic response, and patients with the status of constantly positive levels after 6 weeks of treatment seemed to suffer from significantly reduced PFS, which has similar clinical significance as the clearance of EGFR mutations in ctDNA for lung adenocarcinoma after 8 weeks treatment with gefitinib or erlotinib [29,30]. Although the study by Bechmann T [26] failed to demonstrate a relationship between HER2 gene copy numbers and treatment effects during neoadjuvant chemotherapy in primary breast cancer, our previous study in gastric cancer revealed that the fluctuations of HER2 copy number in ctDNA could efficiently monitor trastuzumab efficacy and was even superior to commonly used biomarkers CEA and CA199 [16]. Other former studies on HER2 amplification using ddPCR also suggested its role in monitoring the effects of treatments and forecasting the prognosis in gastric cancer [14], [31].
In this research, we investigated the potential clinical significance of HER2 amplification in plasma ctDNA based on WGS in both early and metastatic breast cancer. For HER2-positive breast cancer patients, HER2 copy numbers in ctDNA before another line of treatment can be regarded as a predictive factor in clinical best objective response, while the modification of HER2 copy numbers may be closely associated with prognosis for HER2-targeted therapy and sensitively reflect current therapeutic resistance. With regard to HER2-negative metastatic breast cancer patients in the primary tumor, the acquisition of HER2 gene amplification in ctDNA may furnish an opportunity for HER2-targeted therapies, which may offer more therapeutic options for heavily pretreated patients and satisfy the requirements of individualized treatment. However, as this was a proof-of-concept study with a relatively small sample size, some limitations cannot be neglected, and the conclusions require further validation in a larger population. Due to multigene mechanisms involved in the resistance to HER2 blockade agents [32], a multiparameter model may be considered in detecting resistance to HER2-targeted therapy, and our previous studies focused on the criteria to predict the efficacy and determine the resistance to HER2-targeted therapies via ctDNA based on the resistance-related tumor genetic alterations in TP53/PIK3CA/MTOR/PTEN [19], [28]. Further large-scale studies may take the conclusions in these proof-of-concept studies into account and establish a multiparameter model with which to maximize the ability of predicting resistance to HER2-targeted therapies in breast cancer patients.
5. Conclusions
In conclusion, our findings suggested that the level of HER2 amplification in plasma ctDNA based on WGS, confirmed with a relatively high concordance with primary tumor, may forecast the potential treatment response before the initiation of another line of anticancer therapy in breast cancer patients. Furthermore, longitudinal tracing of HER2 copy numbers could be a predictive index for the prognosis of anti-HER2 therapy and be sensitive to current therapeutic resistance in a noninvasive, repeatable and practical method.
Ethical approval
The study was approved by the institutional review board of National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College.
Funding statement
This study was funded by the National Natural Science Foundation of China (81472453, 81874122), and the CAMS Initiative for Innovative Medicine (2017-I2M-3–004).
Declaration of competing interest
The authors declare that they have no conflicts of interest.
Acknowledgements
We thank all the participants involved in the study for their contributions.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.breast.2019.12.010.
Contributor Information
Sijia Lu, Email: lusijia@yikongenomics.com.
Fei Ma, Email: drmafei@126.com.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- 1.Yarden Y., Sliwkowski M.X. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2(2):127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 2.Harbeck N., Gnant M. Breast cancer. Lancet. 2017;389(10074):1134–1150. doi: 10.1016/S0140-6736(16)31891-8. [DOI] [PubMed] [Google Scholar]
- 3.Loibl S., Gianni L. HER2-positive breast cancer. Lancet. 2017;389(10087):2415–2429. doi: 10.1016/S0140-6736(16)32417-5. [DOI] [PubMed] [Google Scholar]
- 4.Slamon D.J., Leyland-Jones B., Shak S., Fuchs H., Paton V., Bajamonde A. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344(11):783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 5.Slamon D., Eiermann W., Robert N., Pienkowski T., Martin M., Press M. Adjuvant trastuzumab in HER2-positive breast cancer. N Engl J Med. 2011;365(14):1273–1283. doi: 10.1056/NEJMoa0910383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Poznak C., Somerfield M.R., Bast R.C., Cristofanilli M., Goetz M.P., Gonzalez-Angulo A.M. Use of biomarkers to guide decisions on systemic therapy for women with metastatic breast cancer: American society of clinical Oncology clinical practice guideline. J Clin Oncol. 2015;33(24):2695–2704. doi: 10.1200/JCO.2015.61.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gradishar W.J., Anderson B.O., Balassanian R., Blair S.L., Burstein H.J., Cyr A. Breast cancer, version 4.2017, NCCN clinical practice guidelines in Oncology. J Natl Compr Cancer Netw. 2018;16(3):310–320. doi: 10.6004/jnccn.2018.0012. [DOI] [PubMed] [Google Scholar]
- 8.Amin M.B. Springer; 2018. AJCC cancer staging manual. eighth ed. The American College of Surgeons (ACS) [Google Scholar]
- 9.Zhang S., Li L., Wang T., Bian L., Hu H., Xu C. Real-time HER2 status detected on circulating tumor cells predicts different outcomes of anti-HER2 therapy in histologically HER2-positive metastatic breast cancer patients. BMC Canc. 2016;16:526. doi: 10.1186/s12885-016-2578-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Niikura N., Liu J., Hayashi N., Mittendorf E.A., Gong Y., Palla S.L. Loss of human epidermal growth factor receptor 2 (HER2) expression in metastatic sites of HER2-overexpressing primary breast tumors. J Clin Oncol. 2012;30(6):593–599. doi: 10.1200/JCO.2010.33.8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niikura N., Tomotaki A., Miyata H., Iwamoto T., Kawai M., Anan K. Changes in tumor expression of HER2 and hormone receptors status after neoadjuvant chemotherapy in 21,755 patients from the Japanese breast cancer registry. Ann Oncol. 2016;27(3):480–487. doi: 10.1093/annonc/mdv611. [DOI] [PubMed] [Google Scholar]
- 12.NCCN guidelines insights: breast cancer, version 1. 2018. http://www.nccn.org
- 13.Siravegna G., Marsoni S., Siena S., Bardelli A. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol. 2017;14(9):531–548. doi: 10.1038/nrclinonc.2017.14. [DOI] [PubMed] [Google Scholar]
- 14.Shoda K., Ichikawa D., Fujita Y., Masuda K., Hiramoto H., Hamada J. Monitoring the HER2 copy number status in circulating tumor DNA by droplet digital PCR in patients with gastric cancer. Gastric Cancer. 2017;20(1):126–135. doi: 10.1007/s10120-016-0599-z. [DOI] [PubMed] [Google Scholar]
- 15.Page K., Hava N., Ward B., Brown J., Guttery D.S., Ruangpratheep C. Detection of HER2 amplification in circulating free DNA in patients with breast cancer. Br J Canc. 2011;104(8):1342–1348. doi: 10.1038/bjc.2011.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang H., Li B., Liu Z., Gong J., Shao L., Ren J. HER2 copy number of circulating tumour DNA functions as a biomarker to predict and monitor trastuzumab efficacy in advanced gastric cancer. Eur J Cancer. 2018;88:92–100. doi: 10.1016/j.ejca.2017.10.032. [DOI] [PubMed] [Google Scholar]
- 17.Wolff A.C., Hammond M.E., Hicks D.G., Dowsett M., McShane L.M., Allison K.H. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol. 2013;31(31):3997–4013. doi: 10.1200/JCO.2013.50.9984. [DOI] [PubMed] [Google Scholar]
- 18.Zong C., Lu S., Chapman A.R., Xie X.S. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science. 2012;338(6114):1622–1626. doi: 10.1126/science.1229164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma F., Li Q., Chen S., Zhu W., Fan Y., Wang J. Phase I study and biomarker analysis of pyrotinib, a novel irreversible pan-ErbB receptor tyrosine kinase inhibitor, in patients with human epidermal growth factor receptor 2-positive metastatic breast cancer. J Clin Oncol. 2017;35(27):3105–3112. doi: 10.1200/JCO.2016.69.6179. [DOI] [PubMed] [Google Scholar]
- 20.Wolff A.C., Hammond M.E.H., Allison K.H., Harvey B.E., McShane L.M., Dowsett M. HER2 testing in breast cancer: American society of clinical oncology/college of American Pathologists clinical practice guideline focused update summary. J Oncol Pract. 2018:JOP1800206. doi: 10.1200/JOP.18.00206. [DOI] [PubMed] [Google Scholar]
- 21.Bartley A.N., Washington M.K., Colasacco C., Ventura C.B., Ismaila N., Benson A.B., 3rd HER2 testing and clinical decision making in gastroesophageal adenocarcinoma: guideline from the College of American Pathologists, American society for clinical pathology, and the American society of clinical Oncology. J Clin Oncol. 2017;35(4):446–464. doi: 10.1200/JCO.2016.69.4836. [DOI] [PubMed] [Google Scholar]
- 22.Gerlinger M., Rowan A.J., Horswell S., Math M., Larkin J., Endesfelder D. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang J., Fujimoto J., Zhang J., Wedge D.C., Song X., Zhang J. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science. 2014;346(6206):256–259. doi: 10.1126/science.1256930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Turajlic S., Swanton C. Metastasis as an evolutionary process. Science. 2016;352(6282):169–175. doi: 10.1126/science.aaf2784. [DOI] [PubMed] [Google Scholar]
- 25.Yang M., Forbes M.E., Bitting R.L., O’Neill S.S., Chou P.C., Topaloglu U. Incorporating blood-based liquid biopsy information into cancer staging: time for a TNMB system? Ann Oncol. 2018;29(2):311–323. doi: 10.1093/annonc/mdx766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bechmann T., Andersen R.F., Pallisgaard N., Madsen J.S., Maae E., Jakobsen E.H. Plasma HER2 amplification in cell-free DNA during neoadjuvant chemotherapy in breast cancer. J Cancer Res Clin Oncol. 2013;139(6):995–1003. doi: 10.1007/s00432-013-1413-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gevensleben H., Garcia-Murillas I., Graeser M.K., Schiavon G., Osin P., Parton M. Noninvasive detection of HER2 amplification with plasma DNA digital PCR. Clin Cancer Res. 2013;19(12):3276–3284. doi: 10.1158/1078-0432.CCR-12-3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma F., Zhu W., Guan Y., Yang L., Xia X., Chen S. ctDNA dynamics: a novel indicator to track resistance in metastatic breast cancer treated with anti-HER2 therapy. Oncotarget. 2016;7(40):66020–66031. doi: 10.18632/oncotarget.11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Z., Cheng Y., An T., Gao H., Wang K., Zhou Q. Detection of EGFR mutations in plasma circulating tumour DNA as a selection criterion for first-line gefitinib treatment in patients with advanced lung adenocarcinoma (BENEFIT): a phase 2, single-arm, multicentre clinical trial. The Lancet Respiratory Medicine. 2018 doi: 10.1016/S2213-2600(18)30264-9. [Online First)] [DOI] [PubMed] [Google Scholar]
- 30.Mok T., Wu Y.L., Lee J.S., Yu C.J., Sriuranpong V., Sandoval-Tan J. Detection and dynamic changes of EGFR mutations from circulating tumor DNA as a predictor of survival outcomes in NSCLC patients treated with first-line intercalated erlotinib and chemotherapy. Clin Cancer Res. 2015;21(14):3196–3203. doi: 10.1158/1078-0432.CCR-14-2594. [DOI] [PubMed] [Google Scholar]
- 31.Kinugasa H., Nouso K., Tanaka T., Miyahara K., Morimoto Y., Dohi C. Droplet digital PCR measurement of HER2 in patients with gastric cancer. Br J Canc. 2015;112(10):1652–1655. doi: 10.1038/bjc.2015.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garrett J.T., Arteaga C.L. Resistance to HER2-directed antibodies and tyrosine kinase inhibitors: mechanisms and clinical implications. Cancer Biol Ther. 2011;11(9):793–800. doi: 10.4161/cbt.11.9.15045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.