Abstract
Diagnosis of early invasive breast cancer relies on radiology and clinical evaluation, supplemented by biopsy confirmation. At least three issues burden this approach: a) suboptimal sensitivity and suboptimal positive predictive power of radiology screening and diagnostic approaches, respectively; b) invasiveness of biopsy with discomfort for women undergoing diagnostic tests; c) long turnaround time for recall tests. In the screening setting, radiology sensitivity is suboptimal, and when a suspicious lesion is detected and a biopsy is recommended, the positive predictive value of radiology is modest. Recent technological advances in medical imaging, especially in the field of artificial intelligence applied to image analysis, hold promise in addressing clinical challenges in cancer detection, assessment of treatment response, and monitoring disease progression. Radiomics include feature extraction from clinical images; these features are related to tumor size, shape, intensity, and texture, collectively providing comprehensive tumor characterization, the so-called radiomics signature of the tumor. Radiomics is based on the hypothesis that extracted quantitative data derives from mechanisms occurring at genetic and molecular levels. In this article we focus on the role and potential of radiomics in breast cancer diagnosis and prognostication.
Keywords: Breast cancer, Prediction, Digital breast tomosynthesis, Radiomics, Magnetic resonance imaging, Artificial intelligence
Highlights
-
•
In the screening setting, radiology sensitivity is suboptimal.
-
•
Artificial intelligence hold promise in cancer diagnosis and prognostication.
-
•
Radiomics include feature extraction from clinical images.
Abbreviations
- BC
breast cancer
- MRI
magnetic resonance imaging
- AI
artificial intelligence
- RQS
radiomics quality score
- ROI
region of interest
- DBT
digital breast tomosynthesis
- AUC
area under the curve
- TRIPOD
Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis
- DWI
diffusion weighted imaging
1. Introduction
In 2018, breast cancer (BC) was the leading cancer among women in every European country and the leading overall cause of death from cancer in women in Europe [1]. It has been estimated that variations observed in breast cancer incidence across European countries can at least in part be due to differences in organized and opportunistic screening activities and differences in the prevalence and distribution of the major risk factors [1]. It is likely that the combined effects of earlier detection and improvements in treatment options underpinned declines in BC mortality rates in most European countries, with greater decreases in Northern and Western European countries relative to Central and Eastern Europe [2]. Therefore, the early diagnosis of BC remains a crucial cancer control strategy. Detection of BC at an early-stage contributes to better outcomes in treated patients because small non-metastatic (early) disease can be effectively treated, with potential to obtain a survival gain at 5 years from the diagnosis [3]. BC screening strategies are implemented to improve patient outcome [4,5]. In addition, early detection and treatment of BC also improve women’s quality of life due to the adoption of less invasive surgical procedures [6]. Many countries adopts population-wide BC screening, initially performed with film-screen and then with digital mammography, aiming to lower mortality from BC by earlier detection of the disease. However, DM has moderate sensitivity, for which estimates vary from 67.3% to 93.3% [7]. Digital Breast Tomosynthesis enabling pseudo-3D imaging of the breast result in better discrimination of tissue structures and improves visualization of cancer. In women with mammographically dense breasts, digital breast tomosynthesis increased sensitivity but not specificity in diagnosis [7]. Contemporary diagnosis of early invasive BC relies on radiology and clinical evaluation, supplemented by biopsy confirmation. Unfortunately, at least three issues burden this approach: a) suboptimal sensitivity and suboptimal positive predictive power of radiology screening and diagnostic approaches, respectively; b) invasiveness of biopsy with discomfort for women undergoing diagnostic tests; c) long turnaround time for recall tests [7]. In particular, diagnosis of early BC relies on the combined use of mammography or tomosynthesis, ultrasound with different bioptical approaches to reach a gold standard for definitive confirmation of malignancy. Such an approach has some limitations: in the screening setting, radiology sensitivity is suboptimal, and when a suspicious lesion is detected and a biopsy is recommended, the positive predictive value of radiology is limited, implying that biopsies often yield negative results [7,8]. Increasing accuracy in the diagnostic setting of early BC is an unmet need of oncology. In addition, the biology of cancer is complex: cancer is a self-sustaining and adaptive process interacting dynamically with its microenvironment, which continues to challenge researchers, and clinicians despite significant progress in understanding its biological keystones [9]. Recent technological advances in medical imaging, especially in the field of artificial intelligence applied to image analysis, hold promise in addressing clinical challenges in cancer detection, treatment assessment, and monitoring of disease progression [10,11]. One of the ultimate goals of medical imaging is to detect a neoplastic lesion as early as possible, then to classify the lesion and predict its clinical course and its biological aggressiveness to optimize the type and intensity (appropriateness) of treatment.
In current radiological practice, mammographic, ultrasonographic, or MRI evaluation of tumor is largely qualitative including subjective evaluations such as tumor aspect (e. g., spiculated, rounded, with necrosis, microcalcification), density, type of enhancement, anatomic relationship to the surrounding tissues in order to inform further treatment. However, in recent years, it has become evident that the future of medicine lies not only in early diagnosis of disease, but also in individually tailored treatments. This concept that has been designated as ‘personalized medicine’ (PM), aims to deliver the right treatment to the right patient at the right time [12]. In this scenario, quantitative evaluation of clinical medical images is the natural consequence of the path towards personalized medicine. Recent significant advancements within the field of medical image analysis relies on the application of Artificial Intelligence (AI) methods for the processing of large quantities of iconographic data from different imaging modalities. In this framework three different broad approaches can be identified, whose tentative definitions, although continuously updated, can be currently outlined as follows:
-
•
Radiomics is the process of extracting quantitative properties, named features, from an image (or from a specific Region of Interest (ROI) identified in an image). This feature extraction activity is typically realized by means of pattern recognition algorithms and provides, as a result, a set of numbers, each one representing a quantitative description of a specific either geometrical or physical property of the image portion under consideration. In oncological applications, examples of features are tumor size, shape, intensity, and texture, collectively providing a comprehensive tumor characterization, called the radiomics signature of the tumor [13]. From an epistemological perspective, radiomics is based on the hypothesis that the extracted features reflect mechanisms occurring at genetic and molecular levels [[13], [14], [15], [16], [17]]. Coherently, the suffix “-omics” is a term that originated in molecular biology to characterize DNA (genomics), RNA (transcriptomics), proteins (proteomics), and metabolites (metabolomics).
-
•
Machine learning indicates those computational algorithms that utilize as input the image features extracted by radiomics in order to provide as output predictions concerning disease outcomes on follow-up. Unsupervised machine learning classifies the radiomics features without using any information provided by or determined by an available historical set of imaging data of the same kind of the one under investigation. Supervised machine learning methods are first trained by means of an available data archive, i.e. all parameters in the algorithm are tuned until the method provides an optimal trade-off between its ability to fit the training set and its generalization power when a new data example arrives. In the world of supervised machine learning, sparsity-enhancing regularization networks are able to make the prediction while, at the same time, identifying the extracted features that mostly impact such prediction.
-
•
Deep learning is an extreme modification of machine learning, in which the image is directly given as input to a multi-layer neural network whose task is to sequentially modify the image and reduce its size until a set of numbers is automatically produced. These numbers represent the set of features to give as input to a supervised machine learning method that performs the prediction task.
According to a rather simplified perspective, the advantage of using unsupervised learning methods is that they do not need the training phase to work (and the availability of an appropriate historical database with which to realize such phase) but are typically used just for classification purposes. Supervised methods require the training phase to work but have more general applicability conditions and, for example, can be used for regression and to realize multi-task prediction. Supervised methods may utilize features as input data, which are typically image properties with a specific meaning in the application framework; but, in a deep learning approach, the methods may learn these features during the prediction process. This latter approach avoids the feature extraction step, which may be critical, but introduces an uncertainty element in the interpretation of the automatically extracted features.
2. Technical issues
At the present time, radiomics is a complex process that involves several steps and, generally, the application of radiomics to standard clinical images is not fully automated. For example, a Radiomics Quality Score (RQS) [17] can be assigned as an indicator of study factors that imply reliable results related to radiomics application in medical imaging. RQS is determined according to a set of recommendations established for the reporting of studies developing, validating, or updating a prediction model using radiomics, regardless of whether the model serves diagnostic or prognostic purposes, However, a review of the literature (to February 2018) identifying 17 retrospective studies, all published after 2015, that provided BC-related radiomics data on 3928 patients, found that the overall RQS was relatively low [18]. Specifically, it was reported that for RQS estimated on a 36-points scale, mean RQS score was only 11.88 ± 5.8 [18] across studies, reflecting a 32.73% strength of the overall quality of studies, which is limited. The low values of RQS reported in previous studies, suggest that a careful look at the overall quality of the study is mandatory to overcome or minimize some known limitations of radiomics feature extraction. To reach a clinical utility for BC-related radiomics studies, research with potential to influence treatment, patient outcome, and social impact has to be strongly encouraged. Indeed, high-quality studies have the potential to place radiology (and radiomics related researchers) at the pinnacle of quality in evidence-based practice [19]. To help researchers and clinicians to improve the overall study quality in this field, a modified RQS description is reported in Table 1 for BC-related radiomics studies, adapted and simplified from the one elaborated by Lambin et al. [17].
Table 1.
RQS for BC-related studies (adapted from Lambin et al: [17]). Ten items for a maximum value of 12 points, representing 100% of quality.
| Criteria | Points | |
|---|---|---|
| 1 | Image protocol quality: well-documented image protocols (for example, contrast, slice thickness, sequences etc., timing) and/or usage of public image protocols | +1 (if protocols are well-documented) +1 (if public protocol is used) |
| 2 | Segmentation procedure well documented (segmentation by different physicians/algorithms/software) | +1 |
| 3 | Feature reduction or adjustment for multiple testing. N.B.: Overfitting is inevitable if the number of features exceeds the number of samples | +1 |
| 4 | Multivariable analysis with non radiomics features. It permits correlating/inferencing between radiomics and non radiomics features | +1 |
| 5 | Assessment of reproducibility/replicability | +1 only internal +2 also external |
| 6 | Discrimination statistics - report discrimination statistics (for example, C-statistic, ROC curve, AUC) and their statistical significance (for example, p-values, confidence intervals). | +1 |
| 7 | Prospective study registered in a trial database | +1 |
| 8 | Comparison to ‘gold standard’ | +1 |
| 9 | External Validation | +1 |
| 10 | Open science and data (scans, region of interest segmentations, code, radiomics features calculated on a set of representative ROIs) | +1 |
3. Steps of radiomics workflow
Before analyzing radiomics-derived data, a typical workflow in breast imaging includes first the acquisition of high-quality images and the selection of a region of interest (ROI), identified and segmented either manually or automatically. The ROI selection process is not standardized and introduces variability, since it can include either the whole tumor or some parts of it [[13], [14], [15], [16], [17]]. In Fig. 1 a schematic representation of a typical radiomics workflow is presented. After the segmentation is completed, the selected regions are converted into three dimensions, to become volumes. Then, dedicated open source or in-house developed software extract quantitative features from the obtained volumes to produce a report, which is inserted into a database and could be integrated with other data (clinical information, genomic profiles, serum markers, and/or histology data) [20]. The database can be shared across different centers or institutions [13].
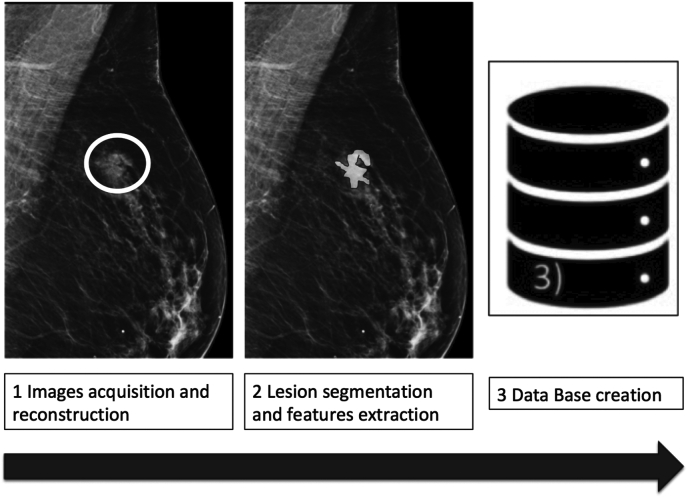
Fig. 1.
Radiomics flow-chart. Radiomics analysis can be divided into distinct processes. The first step included image acquisition and reconstruction with download of radiological images, generally in DICOM format. After image adjustment, the second step includes segmentation and features extraction. Finally, data are organized and collected in the database before analysis. Example of ROI positioning of a nodular spiculated lesion on mammography is shown.
4. Machine learning and deep learning in the radiomics workflow
Radiomics and, specifically, radiomics for BC, relies on the use of machine learning methods to provide predictive models based on the analysis of the features extracted from radiological data [[15], [16], [17], [18], [19], [20]].
At a first, pre-processing level, unsupervised machine learning algorithms allow the automatic stratification of BC patients realized by clustering the feature sets according to specific similarity measure [26]. In fact, this data partition is obtained through the minimization of a cost function involving distances between data and cluster prototypes, and optimal partitions are obtained through iterative optimization procedures that start from a random initialization and move from one cluster to another until no further improvement in the cost function optimization is noticed [27]. In classical approaches, each feature sample may belong to a unique cluster [28], while in fuzzy and possibilistic clustering formulations [18], [29] a different degree of membership is assigned to each sample with respect to each cluster and a probabilistic interpretation of such degree of membership is rather straightforward.
Supervised frameworks require a more sophisticated data preparation process and the use of prediction algorithms that need the realization of a training phase to work properly. Typical supervised schemes for radiomics (as well as for most applications) rely on the following ingredients:
-
•
A historical archive of feature sets extracted from the imaging data by means of pattern recognition algorithms.
-
•
A set of labels, each one associated to a feature set and encoding information concerning the follow-up of the disease (this information may either be binary or include different quantitative parameters characterizing the disease evolution, like its latency or aggressiveness).
-
•
A machine learning algorithm, which is trained by means of the historical database and the corresponding set of labels.
When a new set of un-labelled features becomes at disposal, the trained machine learning method is now able to generalize and provide a probabilistic prediction of the unknown labels (i.e., absence or presence, or level of, disease or outcome).
Supervised machine learning methods for radiomics include, for example, standard feature-based multi-layer perceptrons [29], [30], [31] and regularization networks that require the optimal minimization of functionals made of two terms: one describing the ability of the algorithm to fit the historical data and the other one describing its generalization power [21], [22], [23]. Ensemble methods represent a more modern approach to radiomics and, in particular, Random Forest [25,33] works as a large collection of decorrelated decision trees (a decision tree classifier organizes a series of test questions and conditions in a tree structure, recursively splitting training samples into subsets based on the value of a single attribute). One of the technical problems concerning both regularization networks and Random Forest is the optimal selection of the many input parameters characterizing these algorithms. This optimization step is typically addressed by means of methods borrowed from regularization theory [34], although more recently hybrid approaches that utilize unsupervised clustering for accomplishing optimization tasks showed a notable effectiveness [35]. Both regularization networks and decision trees compute a quantitative output parameter for each feature that can be identified as the relative importance of that feature with respect to the predictability of the target variable (or variables). These quantitative predictors can be therefore ranked by means of specific techniques that, for example, may work in a recursive manner, i.e. they train the classifier, compute the ranking for all features, remove the feature with the smallest ranking and go back to the training phase [17], [36], [37], [38], [39]. One of the critical aspects of machine learning for radiomics is related to the robustness of the forecasting procedure with respect to the feature extraction process. In fact, such a process obviously depends on the kind of pattern recognition method applied to the acquired images: different methods typically provide different image properties, which impacts both the effectiveness of the training process and the reliability of the prediction outcome. Deep learning overcomes this drawback by using Convolutional Neural Networks (CNNs) that directly use as input the reconstructed images and process them through several image processing layers [[32], [33], [34]]. The image features segmented by this iterative process are fed to a standard supervised machine learning method that performs the probabilistic prediction. The notable automation degree of these approaches and their flexibility for application to multi-modal imaging clearly make them the best candidate for next generation radiomics, although the prediction effectiveness of CNNs (for this specific application and, in particular for BC radiomics), and the correct interpretation of the segmented features are still open issues to investigate.
5. Radiomics in breast cancer diagnosis
A radiomics methodology, first applied to head and neck and to lung cancer imaging, has been more recently applied to breast imaging [8,9]. One of the possible applications of radiomics is motivated by the necessity to increase the accuracy of standard radiological techniques such as mammography, tomosynthesis and MRI [40,41]. Several research studies have investigated the usefulness and reliability of radiomics in distinguishing benign breast lesions or normal breast parenchyma from cancers. In general these studies demonstrate that, by adding radiomics to the standard radiological workflow, it would be possible to improve diagnostic accuracy of breast imaging [13]. In an MRI-based radiomics study, entropy of malignant lesions was found to be a useful parameter: entropy values were higher in malignant lesions compared to benign lesions, reflecting the tumoral heterogeneity and its vascular status [42]. Another study based on dynamic contrast-enhanced-MRI tried to identify a set of quantitative features extracted from MR images to differentiate luminal breast cancers from benign breast lesions [43]. The retrospective analysis of dynamic contrast-enhanced-MRI images of 508 lesions and 38 extracted features gave an area under the curve (AUC) for maximum linear size alone of 0.797 in comparison to 0.846 and 0.848 for feature selection protocols including and excluding size features, respectively [51]. Several models or radiomics classifiers allowed distinguishing benign from malignant lesions accurately with varying but acceptable AUC values (0.842–0.851). These values are lower than those of expert breast radiologists (AUC of 0.959), suggesting that the adjunct value of radiomics in differentiating malignant from benign lesion needs to be better evaluated [44]. Bickelhaupt et al. [23] used unenhanced MRI sequences in a multicentric prospective study to evaluate a radiomics model of suspicious breast lesions (BI-RADS 4 and 5) extracted from breast tissue, and reported promising results. Using radiological X-ray based techniques, a multicentric and prospective study applied a radiomics approach to digital breast tomosynthesis (DBT) examinations from women with dense breast [13,45] and was the first study to explore radiomics in this clinical context. In this study, twenty patients with negative standard mammography and a DBT detected histology-proven breast cancer were enrolled as well as a control group of 20 patients of similar age and breast density with negative DBT. From 104 radiomics features extracted, 3 (skewness, entropy, and 90 percentile) were found to differ significantly between the two groups [41]. These preliminary results are encouraging, suggesting that a radiomics analysis of DBT images could have potential utility in cancer detection. It has been reported that, using more sophisticated computational methods including deep-learning and machine learning algorithms, an AI system achieved a cancer detection accuracy comparable to an average breast radiologist in a retrospective setting [10]. In this study, nine multi-reader, multi-case and multi-vendor study datasets previously used for different research purposes in seven countries were collected. The radiological exams of digital mammography were verified by histopathological analysis or follow-up, yielding a total of 2652 exams (containing 653 malignant cases) and interpretations by 101 radiologists for a total of 28 296 independent interpretations. The AI system analyzed these exams yielding an AUC of 0.840 (95% confidence interval [CI] = 0.820 to 0.860) whereas the averaged AUC of the radiologists was 0.814 (95% CI = 0.787 to 0.841) (difference 95% CI = −0.003 to 0.055). The AI system had an AUC higher than 61.4% of the radiologists [10] so clearly a good proportion of radiologists still outperformed the AI model. The authors of the study stated that the performance and impact of such a system in a screening setting need further investigation [10]; nonetheless the results seem promising and could impact on diagnostic imaging of the breast as well as screening pending further studies to determine generalizability. Another recent study tested the feasibility of AI algorithms to reduce the breast cancer screening reading workload by automatically identifying normal digital mammography exams [46]. This study hypothesized that there is potential to use AI to automatically reduce the breast cancer screening reading workload by excluding exams with a low likelihood of cancer and that the exclusion of exams with the lowest likelihood of cancer in screening might not change radiologists’ breast cancer detection performance [46]. This area is worthy of further research including the acceptability of such a strategy in real-world screening settings.
6. Radiomics in breast cancer prognostication
The concept that by using radiomics it should be possible to obtain supplemental data that are not identified by human eyes [47], is particularly useful in breast cancer prognostication. Indeed, maximal information from standard of care images are extracted using radiomics to predict several features of breast cancer such as: lymph node status, prognosis and even treatment response [13]. Although some researchers have claimed that standard mammography can be used to implement AI tools to predict BC development in advance, we have to consider that the wide application of radiomics in clinical practice is only in early development, and much remains exploratory at this stage. Researchers created a deep-learning model that can predict from a mammogram if a patient is likely to develop breast cancer as much as five years in advance with an accuracy reported to be better than the Tyrer-Cuzick model (version 8) [48]. That AI model was trained on mammograms with known outcomes from over 60 000 patients; the model presumably learned the subtle patterns in breast tissue that are precursors to malignant tumors. In addition, patients with non-dense breasts and model-assessed high risk had 3.9 times the cancer incidence of patients with dense breasts and model-assessed low risk [48] suggesting good discrimination of future breast cancer risk. Breast cancer prediction models based on radiomics to predict BC development are only a small part of the entire radiomics arsenal in BC prognostication. For example two different studies found that a radiomics model is able to predict sentinel lymph node or axillary metastases [49,50], which may have clinical utility since axillary lymph node status is still a mandatory variable in the diagnostic and prognostic evaluation of BC patients. Dong et al. [55] found that radiomics features extracted from diffusion weighted imaging (DWI) sequences, which are considered stable sequences, showed high correlation with sentinel lymph node metastases [49]. Further validation of these results is still needed, but it seems possible that radiomics could help in clinical decision-making potentially avoiding invasive procedures to the axilla. Another application is related to the Ki67 labeling index, which is routinely used as a prognostic marker in breast cancer and aims to estimate both cell proliferation and therapeutic response [13]. Recent studies have examined the possibility to predict the proliferation marker Ki67 expression through a radiomics approach. One study used a semiautomatic segmentation of DCE-MRI images, extracting radiomics features (morphological, grey-scale statistic, and texture features) on 377 women diagnosed with invasive breast cancer and found that quantitative radiomics imaging features of breast tumor extracted from these data are associated with breast cancer Ki67 expression [51]. Differentiation form low-Ki67 and high-Ki67 expression using naive Bayes classification method achieved the best performance yielding an AUC of 0.773, 0.757 for overall accuracy, 0.777 for sensitivity and 0.769 for specificity [51]. Another prospective study based on DBT acquired in 70 women diagnosed with invasive breast cancer, 40 patients with low Ki-67 expression (Ki-67 proliferation index < 14%) and 30 patients with high Ki-67 expression (Ki-67 proliferation index ≥ 14%), found that a combination of five features (sphericity, autocorrelation, interquartile range, robust mean absolute deviation, and short run high grey level emphasis) yielded AUC of up to 0.698 to differentiate low- and high Ki67 expression, and that thirty-four radiomics features were significantly (p ≤ 0.001) correlated with Ki-67 [52]. Despite the above-described encouraging results, future larger studies are needed to further evaluate these preliminary findings and to find to what extent radiomics and AI approaches can be integrated in clinical practice in a useful and reliable strategy [53,54]. A recent study based on ultrasound images found the a radiomics approach demonstrated a strong correlation between receptor status and BC subtypes (P < 0.05; area under the curve, 0.760) and that the appearance of hormone receptor-positive cancer and human epidermal growth factor receptor 2–negative cancer on ultrasound scans differs from that of triple-negative cancer [55].
7. Current limitations of radiomics
Promising results of radiomics approaches are still not widely available in daily clinical practice. A quick PubMed search for radiomics, imaging biomarkers or radiogenomics reveals well over 4000 articles. However, surprisingly, given this amount of published research, outside of academic literature there is no widespread clinical application or clinically-based evaluation of these technologies [56]. Several issues reduce the application of the proposed radiomics approaches in clinical practice: the lack of knowledge of its basic concepts among radiologists, limited availability of efficient and standardized or reproducible systems of feature extraction, and limited data sharing for external validation. In addition, the majority of radiomics studies are mostly preliminary with a retrospective design, a relatively small sample size, and often with questionable or uncertain repeatability assessment [57,58]. Larger, high-quality prospective studies are needed to validate such preliminary results. Reproducibility of methods in radiomics research is crucial and should be extensively assessed. Indeed, according to a recent study, the overall scientific quality and reporting of radiomics studies is insufficient, especially regarding feature reproducibility, analysis of clinical utility, and open science categories [57]. Using the TRIPOD statement improvements are possible in stating study objective, blind assessment of outcome, sample size, and missing data categories [57].
8. Conclusion
Without a crystal ball it cannot be said whether further advances in AI might one day replace radiologists or other roles in diagnostics currently performed by humans, but certainly AI will play a role in radiology, one that is unfolding rapidly at present. Furthermore, an important strength of radiomics analysis is that it is a non-invasive approach to characterize the tumor directly from clinical medical images. Therefore focus on better quality research studies with potential to influence treatment, patient outcome, and social impact should be encouraged [19]. Possibly in the next decade, Radiomics will be used to speed-up workflow and reduce the number of invasive procedure.
Funding
None.
Ethical approval
Not required.
Declaration of competing interest
Nothing to declare; N. Houssami declares research support from the National Breast Cancer Foundation Australia.
References
- 1.Ferlay J., Colombet M., Soerjomataram I., Dyba T., Randi G., Bettio M. Cancer incidence and mortality patterns in Europe: estimates for 40 countries and 25 major cancers in 2018. Eur J Cancer. 2018;103:356–387. doi: 10.1016/j.ejca.2018.07.005. [DOI] [PubMed] [Google Scholar]
- 2.Carioli G., Malvezzi M., Rodriguez T., Bertuccio P., Negri E., La Vecchia C. Trends and predictions to 2020 in breast cancer mortality in Europe. Breast (Edinburgh, Scotland) 2017;36:89–95. doi: 10.1016/j.breast.2017.06.003. [DOI] [PubMed] [Google Scholar]
- 3.Cedolini C., Bertozzi S., Londero A.P., Bernardi S., Seriau L., Concina S. Type of breast cancer diagnosis, screening, and survival. Clin Breast Canc. 2014;14:235–240. doi: 10.1016/j.clbc.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 4.Tagliafico A., Houssami N., Calabrese M. Digit Breast Tomosynthesis: Pract Approach. 2016 doi: 10.1007/978-3-319-28631-0. [DOI] [Google Scholar]
- 5.Sardanelli F., Boetes C., Borisch B., Decker T., Federico M., Gilbert F.J. Magnetic resonance imaging of the breast: recommendations from the EUSOMA working group. Eur J Cancer. 2010;46:1296–1316. doi: 10.1016/j.ejca.2010.02.015. [DOI] [PubMed] [Google Scholar]
- 6.van den Ende C., Oordt-Speets A.M., Vroling H., van Agt H.M.E. Benefits and harms of breast cancer screening with mammography in women aged 40-49 years: a systematic review. Int J Cancer. 2017;141:1295–1306. doi: 10.1002/ijc.30794. [DOI] [PubMed] [Google Scholar]
- 7.Phi X.-A., Tagliafico A., Houssami N., Greuter M.J.W., de Bock G.H. Digital breast tomosynthesis for breast cancer screening and diagnosis in women with dense breasts - a systematic review and meta-analysis. BMC Canc. 2018:18. doi: 10.1186/s12885-018-4263-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sprague B.L., Arao R.F., Miglioretti D.L., Henderson L.M., Buist D.S.M., Onega T. National performance benchmarks for modern diagnostic digital mammography: update from the breast cancer surveillance consortium. Radiology. 2017;283:59–69. doi: 10.1148/radiol.2017161519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bi W.L., Hosny A., Schabath M.B., Giger M.L., Birkbak N.J., Mehrtash A. Artificial intelligence in cancer imaging: clinical challenges and applications. CA A Cancer J Clin. 2019;69:127–157. doi: 10.3322/caac.21552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodriguez-Ruiz A., Lång K., Gubern-Merida A., Broeders M., Gennaro G., Clauser P. Stand-alone artificial intelligence for breast cancer detection in mammography: comparison with 101 radiologists. J Natl Cancer Inst. 2019 doi: 10.1093/jnci/djy222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodríguez-Ruiz A., Krupinski E., Mordang J.-J., Schilling K., Heywang-Köbrunner S.H., Sechopoulos I. Detection of breast cancer with mammography: effect of an artificial intelligence support system. Radiology. 2019;290:305–314. doi: 10.1148/radiol.2018181371. [DOI] [PubMed] [Google Scholar]
- 12.European Society of Radiology (ESR) Medical imaging in personalised medicine: a white paper of the research committee of the European Society of Radiology (ESR) Insights Imag. 2015;6:141–155. doi: 10.1007/s13244-015-0394-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crivelli P., Ledda R.E., Parascandolo N., Fara A., Soro D., Conti M. A new challenge for radiologists: radiomics in breast cancer. BioMed Res Int. 2018;2018:1–10. doi: 10.1155/2018/6120703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bi W.L., Hosny A., Schabath M.B., Giger M.L., Birkbak N.J., Mehrtash A. Artificial intelligence in cancer imaging: clinical challenges and applications. CA A Cancer J Clin. 2019;69 doi: 10.3322/caac.21552. caac.21552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Valdora F., Houssami N., Rossi F., Calabrese M., Tagliafico A.S. Rapid review: radiomics and breast cancer. Breast Canc Res Treat. 2018;169:217–229. doi: 10.1007/s10549-018-4675-4. [DOI] [PubMed] [Google Scholar]
- 16.Tagliafico A.S., Valdora F., Mariscotti G., Durando M., Nori J., La Forgia D. An exploratory radiomics analysis on digital breast tomosynthesis in women with mammographically negative dense breasts. Breast. 2018;40:92–96. doi: 10.1016/j.breast.2018.04.016. [DOI] [PubMed] [Google Scholar]
- 17.Lambin P., Leijenaar R.T.H., Deist T.M., Peerlings J., de Jong E.E.C., van Timmeren J. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14:749–762. doi: 10.1038/nrclinonc.2017.141. [DOI] [PubMed] [Google Scholar]
- 18.Valdora F., Houssami N., Rossi F., Calabrese M., Tagliafico A.S. Rapid review: radiomics and breast cancer. Breast Canc Res Treat. 2018;169:217–229. doi: 10.1007/s10549-018-4675-4. [DOI] [PubMed] [Google Scholar]
- 19.Tagliafico A.S., Wilson D., Sconfienza L.M. European society of musculoskeletal radiology (ESSR) research committee. Encouraging MSK imaging research towards clinical impact is a necessity: opinion paper of the european society of musculoskeletal radiology (ESSR) Eur Radiol. 2019;29:3410–3413. doi: 10.1007/s00330-019-06218-4. [DOI] [PubMed] [Google Scholar]
- 20.Fedorov A., Beichel R., Kalpathy-Cramer J., Finet J., Fillion-Robin J.-C., Pujol S. 3D slicer as an image computing platform for the quantitative imaging network. Magn Reson Imag. 2012;30:1323–1341. doi: 10.1016/j.mri.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu Y., Guo Y., Wang Y., Yu J., Li J., Zhou S. Automatic tumor segmentation in breast ultrasound images using a dilated fully convolutional network combined with an active contour model. Med Phys. 2019;46:215–228. doi: 10.1002/mp.13268. [DOI] [PubMed] [Google Scholar]
- 22.Zhou Y., He L., Huang Y., Chen S., Wu P., Ye W. CT-based radiomics signature: a potential biomarker for preoperative prediction of early recurrence in hepatocellular carcinoma. Abdom Radiol (New York) 2017;42:1695–1704. doi: 10.1007/s00261-017-1072-0. [DOI] [PubMed] [Google Scholar]
- 23.Bickelhaupt S., Paech D., Kickingereder P., Steudle F., Lederer W., Daniel H. Prediction of malignancy by a radiomic signature from contrast agent-free diffusion MRI in suspicious breast lesions found on screening mammography. J Magn Reson Imaging. 2017;46:604–616. doi: 10.1002/jmri.25606. [DOI] [PubMed] [Google Scholar]
- 25.Vallières M., Kay-Rivest E., Perrin L.J., Liem X., Furstoss C., Aerts H.J.W.L. Radiomics strategies for risk assessment of tumour failure in head-and-neck cancer. Sci Rep. 2017;7:10117. doi: 10.1038/s41598-017-10371-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X., Mao K., Wang L., Yang P., Lu D., He P. An appraisal of lung nodules automatic classification algorithms for CT images. Sensors (Basel, Switzerland) 2019;19:194. doi: 10.3390/s19010194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jain A K M.N.M., Data F.P.J. clustering: a review. ACM Comput Surv. 1999;31:264. [Google Scholar]
- 28.Ismail S.Z.S., K-Means-Type Algorithms M.A. A generalized convergence theorem and characterization of local optimality. IEEE Trans. Pattern Anal. Mach. Intell. 1998;PAMI. 1984;6:81–87. doi: 10.1109/TPAMI. 4767478. [DOI] [PubMed] [Google Scholar]
- 29.Xie X.L., Beni G. A validity measure for fuzzy clustering. IEEE Trans Pattern Anal Mach Intell. 1991;13:841–846. doi: 10.1109/34.85677. [DOI] [Google Scholar]
- 30.Isensee F., Kickingereder P., Wick W., Bendszus M., Maier-Hein K.H. Brain tumor segmentation and radiomics survival prediction: contribution to the BRATS 2017 challenge. Lect Notes Comput Sci. 2018:287–297. doi: 10.1007/978-3-319-75238-9_25. (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics) 10670 LNCS. [DOI] [Google Scholar]
- 31.Ferreira Junior J.R., Koenigkam-Santos M., Cipriano F.E.G., Fabro A.T. Azevedo-Marques PM de. Radiomics-based features for pattern recognition of lung cancer histopathology and metastases. Comput Methods Progr Biomed. 2018;159:23–30. doi: 10.1016/J.CMPB.2018.02.015. [DOI] [PubMed] [Google Scholar]
- 32.Zhang B., He X., Ouyang F., Gu D., Dong Y., Zhang L. Radiomic machine-learning classifiers for prognostic biomarkers of advanced nasopharyngeal carcinoma. Cancer Lett. 2017;403:21–27. doi: 10.1016/j.canlet.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 33.Le E.P.V., Wang Y., Huang Y., Hickman S., Gilbert F.J. Artificial intelligence in breast imaging. Clin Radiol. 2019;74:357–366. doi: 10.1016/j.crad.2019.02.006. [DOI] [PubMed] [Google Scholar]
- 34.Bertero M. Regularization methods for linear inverse problems. Inverse Problems. In: Talenti G., Berlin H, editors. Lecture notes in mathematics. Springer; Berlin, Heidelberg: 1986. p. 1225. [DOI] [Google Scholar]
- 35.Benvenuto F., Piana M., Campi C., Massone A.M. A hybrid supervised/unsupervised machine learning approach to solar flare prediction. Astrophys J. 2018;853:90. doi: 10.3847/1538-4357/aaa23c. [DOI] [Google Scholar]
- 36.Lambin P., Rios-Velazquez E., Leijenaar R., Carvalho S., van Stiphout R.G.P.M., Granton P. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer (Oxford, England?: 1990) 2012;48:441–446. doi: 10.1016/j.ejca.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coroller T.P., Grossmann P., Hou Y., Rios Velazquez E., Leijenaar R.T.H., Hermann G. CT-based radiomic signature predicts distant metastasis in lung adenocarcinoma. Radiotherapy and Oncology . J Eur Soc Ther Radiol Oncol. 2015;114:345–350. doi: 10.1016/j.radonc.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huynh B.Q., Li H., Giger M.L. Digital mammographic tumor classification using transfer learning from deep convolutional neural networks. J Med Imaging. 2016:3. doi: 10.1117/1.JMI.3.3.034501. 034501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lao J., Chen Y., Li Z.-C., Li Q., Zhang J., Liu J. A deep learning-based radiomics model for prediction of survival in glioblastoma multiforme. Sci Rep. 2017;7:10353. doi: 10.1038/s41598-017-10649-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tagliafico A.S., Bignotti B., Rossi F., Signori A., Sormani M.P., Vadora F. Diagnostic performance of contrast-enhanced spectral mammography: systematic review and meta-analysis. Breast. 2016:28. doi: 10.1016/j.breast.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 41.Tagliafico A.S., Valdora F., Mariscotti G., Durando M., Nori J., La Forgia D. An exploratory radiomics analysis on digital breast tomosynthesis in women with mammographically negative dense breasts. Breast. 2018:40. doi: 10.1016/j.breast.2018.04.016. [DOI] [PubMed] [Google Scholar]
- 42.Parekh V.S., Jacobs M.A. Integrated radiomic framework for breast cancer and tumor biology using advanced machine learning and multiparametric MRI. NPJ Breast Cancer. 2017;3:43. doi: 10.1038/s41523-017-0045-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whitney H.M., Taylor N.S., Drukker K., Edwards A.V., Papaioannou J., Schacht D. Additive benefit of radiomics over size alone in the distinction between benign lesions and luminal a cancers on a large clinical breast MRI dataset. Acad Radiol. 2019;26:202–209. doi: 10.1016/j.acra.2018.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Crivelli P., Ledda R.E., Parascandolo N., Fara A., Soro D., Conti M. A new challenge for radiologists: radiomics in breast cancer. BioMed Res Int. 2018;2018:1–10. doi: 10.1155/2018/6120703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tagliafico A.S., Valdora F., Mariscotti G., Durando M., Nori J., La Forgia D. An exploratory radiomics analysis on digital breast tomosynthesis in women with mammographically negative dense breasts. Breast (Edinburgh, Scotland) 2018;40:92–96. doi: 10.1016/j.breast.2018.04.016. [DOI] [PubMed] [Google Scholar]
- 46.Rodriguez-Ruiz A., Lång K., Gubern-Merida A., Teuwen J., Broeders M., Gennaro G. Can we reduce the workload of mammographic screening by automatic identification of normal exams with artificial intelligence? A feasibility study. Eur Radiol. 2019 doi: 10.1007/s00330-019-06186-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gillies R.J., Kinahan P.E., Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278:563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yala A., Lehman C., Schuster T., Portnoi T., Barzilay R. A deep learning mammography-based model for improved breast cancer risk prediction. Radiology. 2019;292:60–66. doi: 10.1148/radiol.2019182716. [DOI] [PubMed] [Google Scholar]
- 49.Dong Y., Feng Q., Yang W., Lu Z., Deng C., Zhang L. Preoperative prediction of sentinel lymph node metastasis in breast cancer based on radiomics of T2-weighted fat-suppression and diffusion-weighted MRI. Eur Radiol. 2018;28:582–591. doi: 10.1007/s00330-017-5005-7. [DOI] [PubMed] [Google Scholar]
- 50.Han L., Zhu Y., Liu Z., Yu T., He C., Jiang W. Radiomic nomogram for prediction of axillary lymph node metastasis in breast cancer. Eur Radiol. 2019;29:3820–3829. doi: 10.1007/s00330-018-5981-2. [DOI] [PubMed] [Google Scholar]
- 51.Ma W., Ji Y., Qi L., Guo X., Jian X., Liu P. Breast cancer Ki67 expression prediction by DCE-MRI radiomics features. Clin Radiol. 2018;73:909. doi: 10.1016/j.crad.2018.05.027. e1-909.e5. [DOI] [PubMed] [Google Scholar]
- 52.Tagliafico A., Bignotti B., Rossi F., Matos J., Calabrese M., Valdora F. Breast cancer Ki-67 expression prediction by digital breast tomosynthesis radiomics features. Eur Radiol Exp. 2019 doi: 10.1186/s41747-019-0117-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Houssami N., Kirkpatrick-Jones G., Noguchi N., Lee C.I. Artificial Intelligence (AI) for the early detection of breast cancer: a scoping review to assess AI’s potential in breast screening practice. Expert Rev Med Devices. 2019;16:351–362. doi: 10.1080/17434440.2019.1610387. [DOI] [PubMed] [Google Scholar]
- 54.Houssami N., Lee C.I., Buist D.S.M., Tao D. Artificial intelligence for breast cancer screening: opportunity or hype? Breast. 2017;36:31–33. doi: 10.1016/j.breast.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 55.Guo Y., Hu Y., Qiao M., Wang Y., Yu J., Li J. Radiomics analysis on ultrasound for prediction of biologic behavior in breast invasive ductal carcinoma. Clin Breast Canc. 2018 doi: 10.1016/j.clbc.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 56.Daniel Pinto dos Santos Why radiomics research does not translate to clinical practice: evaluation of literature using RQS and TRIPOD. https://ai.myesr.org/publications/why-radiomics-research-does-not-translate-to-clinical-practice-evaluation-of-literature-using-rqs-and-tripod/ n.d.
- 57.Park J.E., Kim D., Kim H.S., Park S.Y., Kim J.Y., Cho S.J. Quality of science and reporting of radiomics in oncologic studies: room for improvement according to radiomics quality score and TRIPOD statement. Eur Radiol. 2019 doi: 10.1007/s00330-019-06360-z. [DOI] [PubMed] [Google Scholar]
- 58.Tagliafico A.S., Bignotti B., Rossi F., Valdora F., Martinoli C. AY;JH]]]] ∖’’Æ‘]∖Local recurrence of soft tissue sarcoma . Radiomic Analysis. 2019 doi: 10.2478/raon-2019-0041. [DOI] [PMC free article] [PubMed] [Google Scholar]