Abstract
Ameloblastin (Ambn) has the potential to regulate cell-matrix adhesion through familiar cell-binding domains, but the proposed sequence motifs are not highly conserved across species. Here, we report that Ambn binds to ameloblast-like cell membranes through a highly evolutionary conserved amphipathic helix-forming (AH) motif encoded by exon 5. We applied high-resolution confocal microscopy to show colocalization of Ambn with ameloblast membrane surfaces in developing mouse incisors. Using a series of Ambn-derived peptides and Ambn variants, we showed that Ambn binds to cell membranes through a motif within the sequence encoded by exon 5. Using peptides derived from the N- or C-termini of this sequence, as well as Ambn variants that lacked or had a disrupted AH motif, we demonstrated that the AH motif located at the N-terminus of the sequence is involved in cell-Ambn adhesion. Sequence analysis revealed that this highly conserved AH motif is absent from other enamel matrix proteins, including amelogenin, enamelin, and amelotin. Collectively, these data suggest that Ambn binds to the cell surface membrane via a helix-forming motif and provide insight into the molecular mechanism and function of Ambn in enamel cell-matrix interaction.
Keywords: tooth, enamel biomineralization/formation, cell-matrix interactions, biophysics, biochemistry, extracellular matrix
Introduction
Enamel extracellular matrix (EECM) consists of a distinct set of molecules, including amelogenin (Amel), ameloblastin (Ambn), enamelin (Enam), amelotin (Amtn), and proteinases (Moradian-Oldak 2012). These proteins assemble to create a mineralizing 3-dimensional extracellular matrix (ECM) that eventually guides its own replacement by the mineral phase (Fincham et al. 1995; Smith 1998; Margolis et al. 2006; Lacruz et al. 2017).
Ambn, the second most abundant enamel matrix protein, is a proline-rich, intrinsically disordered macromolecule involved in cell-matrix adhesion, the construction of functional EECM, and enamel mineralization (Fukumoto et al. 2004; Zhang, Diekwisch, et al. 2011; Wald et al. 2017). Mutations such as deletion of AMBN exon 6, a homozygous splice-site mutation (c.532–1G>C), and a C-T point mutation causing a P357S mutation are all associated with human hypoplastic amelogenesis imperfecta (AI) (Poulter et al. 2014; Prasad et al. 2016; Lu et al. 2018). In mouse, deletion of Ambn exons 5 and 6 results in the detachment of ameloblasts, the loss of their polarized organization, and the replacement of enamel by a thin layer of dysplastic mineralized matrix (Fukumoto et al. 2004; Wazen et al. 2009).
Cell-matrix adhesion is typically mediated by integrin receptors, which can bind to a wide range of ECM proteins as well as to cell surface proteins and soluble factors (Mouw et al. 2014). In enamel, Ambn could mediate enamel cell-matrix adhesion through its integrin-binding motif (Cerny et al. 1996), heparin-binding motifs (Sonoda et al. 2009), or fibronectin-binding motif (Beyeler et al. 2010). However, our recent sequence conservation analysis from 47 species showed that these motifs are not highly conserved and have identical sequences in only a few species (Su, Kegulian, et al. 2019). We then identified a highly conserved amphipathic helix-forming (AH) motif in the sequence encoded by exon 5 of Ambn that binds to a synthetic lipid surface, implying that this motif could allow Ambn to directly bind ameloblast cell membrane.
Here, we build upon our recent in vitro findings on Ambn-liposome interactions and hypothesize that the highly conserved AH motif is involved in Ambn cell-matrix adhesion by binding to ameloblast-like cell membrane surfaces. We demonstrate that Ambn and cell membrane colocalize in developing mouse incisor enamel. In vitro, we used the LS8 (Chen et al. 1992) and ameloblast lineage cell (ALC) (Nakata et al. 2003) dental epithelial cell lines, which are well-characterized cell culture models representing ameloblasts, to study their affinity to recombinant exogenous Ambn protein, Ambn variants, and Ambn-derived peptides. We also used well-known cells with different origins, such as NIH3T3 fibroblasts and epithelial TCMK-1 cells, as controls to assess whether recombinant Ambn binds to these cells’ surfaces. We used a series of synthetic peptides and variants derived from Ambn in cell attachment and spreading assays to identify the motif that mediates the binding of Ambn to cell membranes. Ambn variants without the AH motif or with a disruption to AH motif were used to investigate the function of the AH motif in binding to cell membrane or a synthetic lipid membrane. Our findings provide insight into the molecular mechanism and function of Ambn in enamel cell-matrix interaction, which are in turn important for understanding the etiology of AI and developing new treatment strategies for enamel repair.
Materials and Methods
Protein Expression and Purification
Recombinant mouse Ambn proteins and their variants were expressed and purified following our published protocol, which is described briefly in the Appendix (Su, Bapat, et al. 2019), and characterized by mass spectroscopy (Appendix Fig. 1).
Peptide Synthesis
Peptides were synthesized and purified by Chempeptide Limited. Seven peptides (AB1, AB2, AB2N, AB2C, AB4, AB5, and AB6) were designed based on the amino acid sequence of mouse Ambn (Appendix Table 1).
Mouse Tissue Immunofluorescence-DiD Labeling and Imaging
The method was adapted from our published protocol (Bapat and Moradian-Oldak 2019) and is described in the Appendix.
Cell Culture
Mouse LS8 cells were obtained from Professor Malcolm Snead at the University of Southern California (Chen et al. 1992). Mouse ALCs were obtained from Professor Toshihiro Sugiyama at Akita University, Japan (Nakata et al. 2003). NIH3T3 (mouse fibroblast) and TCMK-1 (mouse kidney cells) were purchased from ATCC. All cells were cultured in high-glucose Dulbecco’s modified Eagle’s medium (DMEM) (Corning) containing 100 U/mL penicillin, 100 mg/mL streptomycin, and 10% heat-inactivated fetal bovine serum (Corning), in a 5.0% CO2 atmosphere at 37°C. For confocal microscopy, cells were cultured in an 8-well Nunc Lab-Tek II Chamber Slide System (Thermofisher).
Fluorescein Isothiocyanate Labeling of Ambn, AmbnΔ5, AmbnΔ6, Transferrin, Amel, Lysozyme, and AB2
The lyophilized proteins and peptide were dissolved in pH 10.0, 100 mM NaHCO3 buffer. Fresh fluorescein isothiocyanate (FITC) solution was prepared in dimethyl sulfoxide (DMSO) and then added to the protein or peptide solution to obtain a FITC/protein molar ratio of 100:1. The mixture was kept in the dark and shaken gently at 37°C for 4 h. The unreacted FITC was removed by HiTrap desalting columns (GE Healthcare). Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) confirmed that Ambn did not degrade during labeling (Appendix Fig. 2).
Cell Membrane Binding Assay
The cells were cultured in DMEM containing 10.0 µM 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine perchlorate (DiD; Thermofisher) overnight, then incubated in DMEM containing 5.0 µM FITC-labeled protein or 20 µM AB2-FITC for 15 min, washed gently with phosphate-buffered saline (PBS) buffer 3 times, fixed with 4.0% paraformaldehyde for 20 min, washed with PBS buffer 3 times, kept in ProLong Diamond Antifade Mountant with DAPI (Thermofisher), and covered with glass cover-slips. All samples were imaged using a Leica TCS SP8 confocal microscope with an oil immersion objective HCX PL APO CS × 60 (NA 1.4). Detection of FITC and DiD was performed at 498 to 560 nm (excitation 488 nm) and at 643 to 750 nm (excitation 633 nm), respectively. Three-dimensional images were reconstructed using 163 optical sections (0.1 µm step) for ALCs with Ambn-FITC or 17 optical sections (0.3 µm step) for ALCs with AB2-FITC. Colocalization analysis was performed using Leica Application Suite X (version 1.8.1.13759).
Cell Attachment and Spreading Assays
The assays were conducted following published protocols and as described in the Appendix (Humphries 2001; Sonoda et al. 2009).
Unilamellar Lipid Vesicle Preparation and Biophysical Assays
Biophysical assays were conducted using large unilamellar vesicles (LUVs), mimicking the membrane domain involved in epithelial cell-ECM adhesion (Márquez et al. 2008), following published protocols and as described in the Appendix (Su, Kegulian, et al. 2019).
Data Analysis
Statistical analysis was carried out using 1-way analysis of variance (ANOVA). Significance was accepted when P < 0.05. All data were representative of at least 3 independent experiments.
Results
Ambn Adheres to Ameloblast Cells
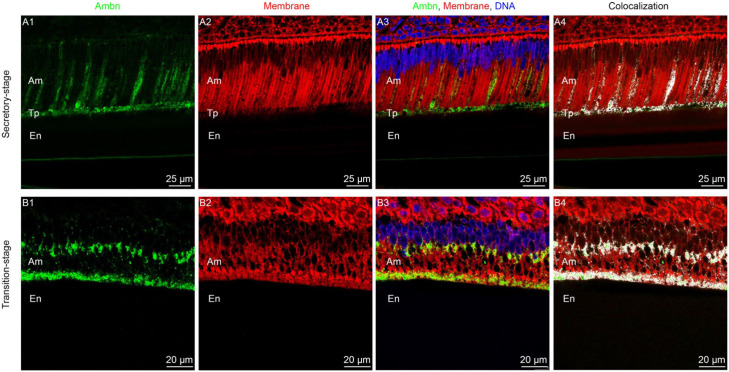
To show colocalization of Ambn with the ameloblast membrane, in vivo immunofluorescence together with DiD staining of enamel sections from P8 mouse incisors was performed by modifying a recently published protocol (Bapat and Moradian-Oldak 2019) (Fig. 1 and Appendix Fig. 3). In situ immunofluorescence of incisor sections showed that Ambn (green) was expressed in both secretory and transition stages, as well as localized at the interface between ameloblasts and enamel matrix (Fig. 1A1, B1). Colocalization analysis further showed that Ambn colocalized with membrane lipid (white dots in Fig. 1A4, B4).
Figure 1.
Ameloblastin (Ambn) colocalizes with cell membrane in vivo. (A1–A3) Confocal images of secretory-stage ameloblasts in a mouse mandibular incisor (P8). (A4) Colocalization between Ambn and cell membrane highlighted in white. (B1–B3) Confocal images of transition-stage ameloblasts in a mouse mandibular incisor (P8). (B4) Colocalization between Ambn and cell membrane highlighted in white. Am, ameloblast; En, enamel; Tp, Tomes’ processes. Blue: DAPI-stained nucleus. Green: Ambn immunostained with anti-Ambn primary antibody M-300 and anti-rabbit secondary antibody conjugated with Alexa 488. Red: 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine perchlorate (DiD)–stained membrane.
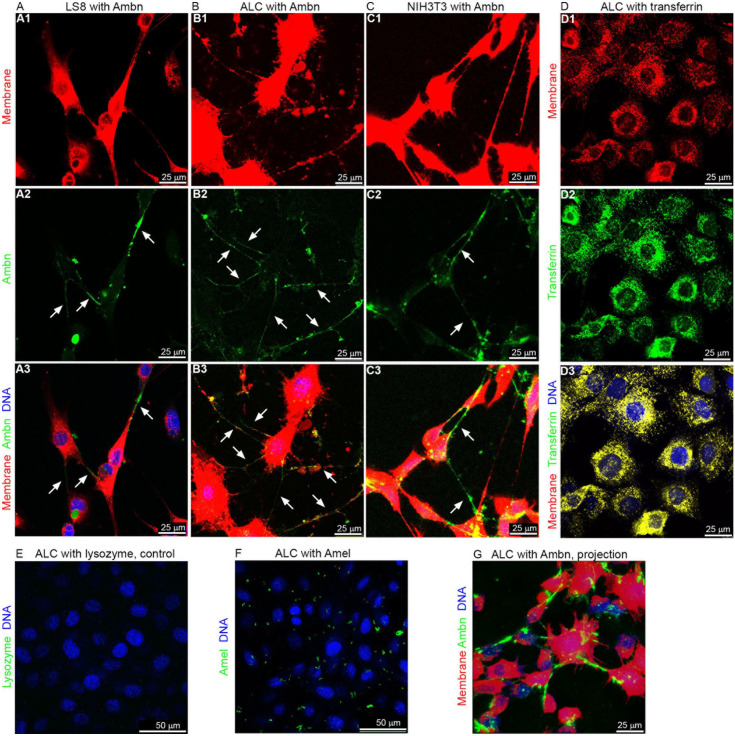
To investigate Ambn-cell adhesion, ameloblast-like cells (LS8 and ALC) and fibroblasts (NIH3T3) were incubated with FITC-labeled recombinant Ambn. Confocal images showed that wild-type Ambn (green) colocalized with membrane (red) on both ameloblast-like cells and fibroblasts, particularly on their pseudopodia (Fig. 2A–C), suggesting that Ambn adheres to the cell membrane surface and the adhesion may not be specific to ameloblasts. The adhesion to ameloblast-like cells was clearly visible at higher cell density (Appendix Fig. 4). In contrast, transferrin (positive control, green), whose receptor is known to be expressed on ameloblasts (McKee et al. 1987), bound evenly to the cells, covering the entire cell surface (Fig. 2D). Lysozyme (negative control, green) did not show any significant binding to ALCs (Fig. 2E). Amel (green), the most abundant enamel matrix protein, showed nonspecific adhesion to ALCs (Fig. 2F). Three-dimensional imaging (Appendix Video 1) and maximum intensity projection of z-stack (Fig. 2G) confocal images demonstrated Ambn-cell adhesion more clearly.
Figure 2.
Ameloblastin (Ambn) adheres to LS8, ameloblast lineage cell (ALC), and NIH3T3 cells. (A–C) Confocal images of LS8, ALC, and NIH3T3 cells incubated with 5 µM Ambn–fluorescein isothiocyanate (FITC). The brightness of red was increased for better visibility of the pseudopodia. (D) Confocal images of ALCs incubated with 5 µM transferrin-FITC as positive control. Yellow color in D3 indicates co-localization of green (transferrin) and red (cell membrane). (E, F) Confocal images of ALCs incubated with 5 µM lysozyme-FITC or Amel-FITC. (G) Maximum intensity projection of the z-stack confocal images of ALCs incubated with 5 µM Ambn-FITC (see Appendix Video 1). Red: DiD-stained membrane. Green: FITC-labeled proteins. Blue: DAPI-stained nucleus.
Ambn-Derived Peptide AB2 Inhibits Ambn-Cell Adhesion
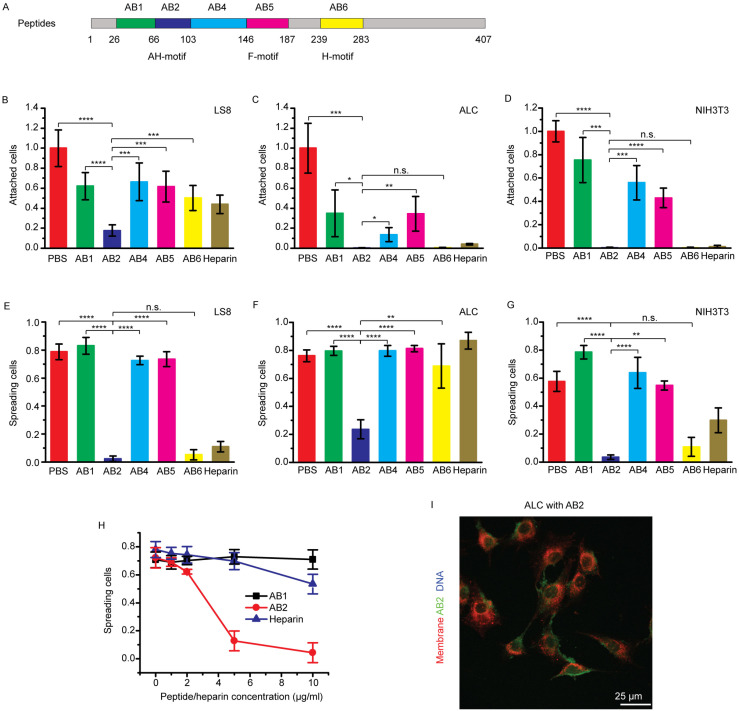
To identify the motif responsible for Ambn-cell adhesion, competitive cell attachment and spreading assays were conducted using Ambn-derived peptides and heparin. Peptide AB2 is the sequence encoded by exon 5 of Ambn and contains the AH motif (Fig. 3A and Appendix Table 1). AB5 and AB6 contain the reported fibronectin- and heparin-binding motifs, respectively (Sonoda et al. 2009; Beyeler et al. 2010). These peptides and heparin were expected to inhibit Ambn-cell adhesion by competing with the AH, fibronectin-binding, or heparin-binding motifs of the recombinant full-length Ambn coated on the dishes. AB1 and AB4 were negative controls.
Figure 3.
Ameloblastin (Ambn)–derived peptides and heparin inhibit Ambn-cell adhesion. (A) Schematic drawing of peptides (AB1, AB2, AB4, AB5, and AB6) derived from wild-type (WT) mouse Ambn. F-motif indicates that peptide AB5 contains the reported fibronectin-binding motif (VPIMDFADPQF). H-motif indicates that peptide AB6 contains one of the reported heparin-binding motifs (VTKG). (B–D) Inhibition of LS8, ameloblast lineage cell (ALC), and NIH3T3 cell attachment on a 5-µg/mL Ambn-coated plate by 10 µg/mL Ambn-derived peptides or heparin. (E–G) Inhibition of LS8, ALC, and NIH3T3 cell spreading on a 5-µg/mL Ambn-coated plate by 10 µg/mL Ambn-derived peptides or heparin. (H) Inhibition of ALC cell spreading on a 5-µg/mL Ambn-coated plate by different concentrations of AB1 (negative control), AB2, and heparin. (I) Maximum intensity projection of the z-stack confocal images of ALCs incubated with 20 µM AB2–fluorescein isothiocyanate (FITC). Red: DiD-stained membrane. Green: FITC-labeled AB2. Statistical analysis was conducted with 1-way analysis of variance with 3 technical replicates. n.s., not significant. *P < 0.05. **P < 0.01. ***P < 0.001. ****P < 0.0001.
The relative numbers of attached LS8, ALC, and NIH3T3 cells decreased in the presence of all Ambn-derived peptides and heparin. A significant decrease in cell adhesion was particularly noted in the presence of AB2 (P < 0.00001, P < 0.001, P < 0.00001), AB6 (P < 0.05, P < 0.001, P < 0.00001), and heparin (P < 0.01, P < 0.001, P < 0.00001) (Fig. 3B–D), suggesting that all peptides inhibited cell attachment to some extent, and AB2, AB6, and heparin inhibited cell attachment more significantly. Note that the 3 P values in parentheses correspond to LS8, ALC, and NIH3T3, respectively.
Many molecules can mediate attachment of cells in a nonphysiological manner. In contrast, only a very few molecules are able to mediate spreading, and spreading assays are more sensitive to inhibitors (Humphries 2001). For this reason, cell-spreading assays were conducted to confirm whether the peptides and heparin are directly involved in the cell adhesion process. The results showed that the percentage of LS8 cells with spread morphology decreased from 78.8% ± 5.5% to 2.2% ± 1.9%, 5.2% ± 3.6%, and 11.0% ± 3.8% in the presence of AB2, AB6, and heparin, respectively, but did not decrease to a similar extent in the presence of other peptides (Fig. 3E). The percentage of ALCs with spread morphology decreased from 76.1% ± 4.3% to 23.6% ± 6.8% in the presence of AB2, while the percentages did not decrease to a similar extent in the presence of other peptides or even heparin (Fig. 3F). A significant inhibitory effect of AB2 was also found with NIH3T3 fibroblasts and TCMK-1 mouse kidney epithelial cells (Fig. 3G and Appendix Fig. 5). Remarkably, AB2 inhibited ALC cell spreading more effectively than heparin (Fig. 3H). Confocal images showed that AB2 (green) presented on the ALC cell surface (Fig. 3I and Appendix Video 2). These results, together with the observation that AB2 inhibits cell attachment, strongly suggested that, in addition to the heparin-binding VTKG motif, a domain within the AB2 sequence is involved in Ambn-cell adhesion.
Removal of Ambn Exon 5 and 6 Sequences Affects Ambn-Cell Adhesion
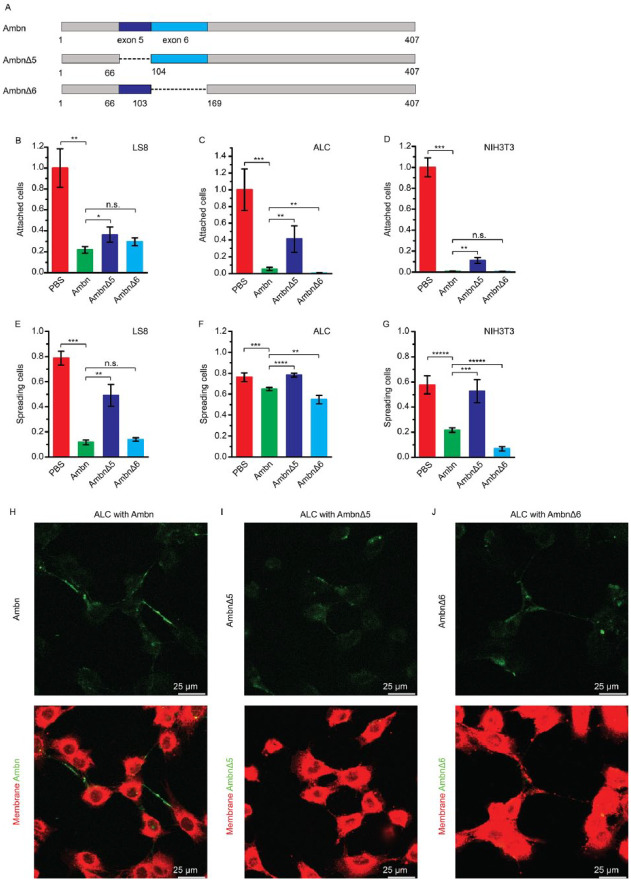
To confirm whether the sequence encoded by exon 5 (AB2) is involved in Ambn-cell interactions, AmbnΔ5 and AmbnΔ6 (Fig. 4A) were tested as competitors in the cell attachment and spreading assays. Cell attachment assay showed that the relative percentages of attached LS8, ALC, and NIH3T3 cells were 21.9% ± 3.1%, 5.4% ± 1.9%, and 0.6% ± 0.3%, respectively, in the presence of Ambn and 29.6% ± 3.7%, 0.5% ± 0.3%, and 0.4% ± 0.4%, respectively, in the presence of AmbnΔ6 (Fig. 4B–D), suggesting that both Ambn and AmbnΔ6 inhibited the cell attachment. In contrast, in the presence of AmbnΔ5, the relative percentages of attached LS8, ALC, and NIH3T3 cells were higher (36.4% ± 7.2%, 41.2% ± 15.8%, and 11.0% ± 2.8%, respectively). Notably, because of the lack of the binding domain, AmbnΔ5 did not inhibit the attachment of the 3 cell lines on Ambn-coated plates as effectively as Ambn or AmbnΔ6.
Figure 4.
Deletion of ameloblastin (Ambn) exon 5 or 6 sequences affects Ambn-cell adhesion. (A) Schematic drawing of Ambn variants (AmbnΔ5 and AmbnΔ6) derived from wild-type (WT) mouse Ambn. (B–D) Inhibition of LS8, ameloblast lineage cell (ALC), and NIH3T3 cell attachment on a 5-µg/mL Ambn-coated plate by 10 µg/mL Ambn, AmbnΔ5, and AmbnΔ6. (E–G) Inhibition of LS8, ALC, and NIH3T3 cell spreading on a 5-µg/mL Ambn-coated plate by 10 µg/mL Ambn, AmbnΔ5, and AmbnΔ6. (H–J) Confocal images of ALCs incubated with 5 µM Ambn–fluorescein isothiocyanate (FITC), AmbnΔ5-FITC, or AmbnΔ6-FITC. Red: DiD-stained membrane. Green: FITC-labeled Ambn or variants. Statistical analysis was conducted with 1-way analysis of variance with 3 technical replicates. n.s., not significant. *P < 0.05. **P < 0.01. ***P < 0.001. ****P < 0.0001.
Cell-spreading assay showed that the percentages of LS8, ALC, and NIH3T3 cells with spread morphology were 16.7% ± 9.0%, 65.0% ± 1.5%, and 21.7% ± 1.9%, respectively, when treated with Ambn and 14.0% ± 1.3%, 54.9% ± 4.1%, and 6.8% ± 1.7%, respectively, with AmbnΔ6. The percentages were lower than PBS control, suggesting that Ambn and Ambn Δ6 inhibited the cell spreading. In contrast, the percentages were higher (49.1% ± 8.6%, 72.1% ± 3.4%, and 52.7% ± 9.2%, respectively) with AmbnΔ5 (Fig. 4E–G). AmbnΔ5 did not inhibit cell spreading on Ambn-coated plates as effectively as Ambn or AmbnΔ6.
Confocal images of ALCs with FITC-labeled proteins further showed that the deletion of exon 5 or exon 6 sequences significantly altered the pattern of Ambn-cell adhesion (Fig. 4F–H). These 3 independent experiments suggested that the sequence encoded by exon 5 (AB2) is involved in Ambn-cell adhesion.
Ambn Adheres to Cells via the Highly Conserved AH-Forming Motif
Recently, we showed that the N-terminal of the sequence encoded by exon 5 of Ambn exhibits a highly conserved AH-forming motif (N-terminal 18 amino acids of AB2 used in this study) (Su, Kegulian, et al. 2019). A comparative analysis of the human, mouse, and pig amino acid sequences of Amel, Enam, and Ambn in the present study showed that only Ambn has potential AH motifs in different isoforms of the protein and across the 3 species (Appendix Tables 2 and 3).
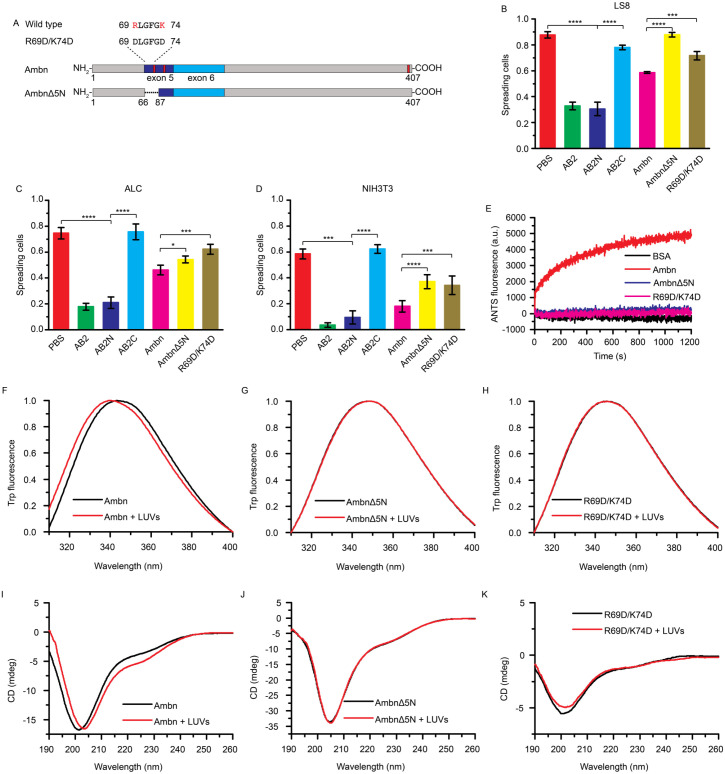
To confirm the role of the Ambn-cell adhesion motif, 2 AB2-derived peptides and 2 Ambn variants were designed (Fig. 5A and Appendix Table 1). Cell-spreading assays with LS8, ALC, NIH3T3, and TMCK-1 cells showed that AB2N, representing the AH motif at the N-terminus of AB2, exhibited similar cell-spreading percentages to those of AB2, while AB2C, representing the C-terminus of AB2, did not (Fig. 5B–D and Appendix Fig. 5). AmbnΔ5N without the AH motif and AmbnR69D/K74D with a disrupted AH motif exhibited higher cell-spreading ratios in comparison to that of full-length Ambn, suggesting their weaker inhibitory effects. Confocal images of ALCs with FITC-labeled proteins showed that without the AH motif (AmbnΔ5N) or with an inactive AH motif (AmbnR69D/K74D), Ambn-cell adhesion was more diffuse (Appendix Fig. 6). Therefore, we confirmed that AB2N, the AH motif at the N-terminus of AB2, plays a role in Ambn-cell adhesion.
Figure 5.
The amphipathic helix-forming motif is vital for the adhesion of ameloblastin (Ambn) to cells and large unilamellar vesicles (LUVs). (A) Schematic drawing of Ambn variants (AmbnΔ5N and AmbnR69D/K74D) disrupting amphipathic helix-forming motif. (B–D) Inhibition of LS8, ameloblast lineage cell (ALC), or NIH3T3 cell spreading on a 5-µg/mL Ambn-coated plate by 10 µg/mL AB2N, AB2C, AmbnΔ5N, or AmbnR69D/K74D. (E) Membrane leakage of 300 µM LUVs in the presence of 1 µM Ambn, AmbnΔ5N, or R69D/K74D and bovine serum albumin (BSA) as negative control. (F–H) Normalized Trp fluorescence spectra of 2.0 µM Ambn, AmbnΔ5N, or R69D/K74D with 300 µM LUVs. (I–K) Circular dichroism (CD) spectra of 2.0 µM Ambn, AmbnΔ5N, or R69D/K74D with 300 µM LUVs. All spectra are representative of at least 3 replicates. Statistical analysis was conducted by 1-way analysis of variance test with 3 technical replicates. *P < 0.05. ***P < 0.001. ****P < 0.0001.
Ambn Adheres to LUVs via the Highly Conserved AH-Forming Motif
Ambn-LUVs interaction mimics the Ambn-membrane interaction and eliminates the potential interference of fibronectin- and/or heparin-binding motifs found on ameloblast-like cells. Membrane leakage assays showed that the 8-aminonaphthalene-1,3,6-trisulfonic acid (ANTS) fluorescence intensity increased when Ambn was added to the LUVs, whereas there was no increase in the presence of AmbnΔ5N, AmbnR69D/K74D, or bovine serum albumin (BSA; negative control) (Fig. 5E), implying that Ambn cannot disrupt LUVs without an active AH motif.
Intrinsic fluorescence spectra of AmbnΔ5N and Ambn-R69D/K74D in the presence of LUVs did not exhibit any blue or red shift, indicating a lack of tertiary structural changes with LUVs. In contrast, Ambn had a significant blue shift in the presence LUVs, indicating a strong tertiary structural change with LUVs (Fig. 5F–H). Circular dichroism (CD) spectra of AmbnΔ5N and AmbnR69D/K74D had no significant changes in the presence of LUVs. In contrast, Ambn had deeper and more pronounced peaks at 222 nm in the presence of LUVs, indicating a coil-helix transition, which is a typical behavior of AH motifs (Fig. 5I–K). These results suggested that AH motif is vital for Ambn-LUV interaction.
Discussion
Despite extensive studies over the past few decades, specific receptors that recognize EECM proteins have not yet been fully identified on ameloblast cells. We have recently identified a membrane-binding motif within Ambn that has the potential to form an amphipathic α-helix (AH) when binding to lipid bilayers (Su, Kegulian, et al. 2019). We located this motif within a sequence encoded by exon 5 of Ambn (69RLGFGKALNSLWLHGLLP86). An amphipathic α-helix is defined as a helix with polar and nonpolar residues on opposite sides along its long axis. It is a common motif encountered in various proteins and peptides, and it can act as a membrane anchor, deform lipid membranes, or recognize membrane curvature (Drin and Antonny 2010; Bornholdt et al. 2013).
Here, we showed that the evolutionarily conserved AH motif is vital for the adhesion of Ambn to ameloblast-like LS8 and ALC cells, TCMK-1 kidney epithelial cells, and NIH3T3 fibroblasts. We specifically showed that AB2 and AB2N, the sequence encoded by exon 5 of Ambn and the AH motif, inhibited LS8, ALC, NIH3T3, and TCMK-1 cells’ adhesion to Ambn-coated plates. AmbnΔ5, AmbnΔ5N, and AmbnR69D/K74D, which lack the active AH motif, did not inhibit cells’ adhesion to Ambn-coated plates as effectively as wild-type Ambn or AmbnΔ6. The Ambn variants also lost their ability to effectively bind to cell membranes and LUVs.
The reported integrin-binding (Cerny et al. 1996), heparin-binding (Sonoda et al. 2009), and fibronectin-binding (Beyeler et al. 2010) motifs present in Ambn, together with our present data, suggest that Ambn-cell adhesion in vivo might be the result of a cooperative function of the AH motif and other receptor-binding motifs. Interestingly, in all cells studied, particularly the ALC and TCMK-1 epithelial cells, the effect of AB2 (AH motif) was found to be much stronger in inhibiting cell adhesion when compared to the effect of heparin. We note that amelogenin, enamelin, and amelotin do not contain such AH motifs in their sequences, supporting the notion that Ambn has a unique role in cell-matrix adhesion (Fukumoto et al. 2004).
Consistent with our in vitro data, in mutant mouse models in which exons 5 and 6 of Ambn are deleted, ameloblasts detach from the ECM and enamel formation is severely disturbed (Fukumoto et al. 2004). In another mouse model with 3 mutations in the N-terminus of exon 5 (Y/F-x-x-Y/L/F-x-Y/F motif), enamel appears with disordered hydroxyapatite crystallites (Wald et al. 2017).
Like Amel, another intrinsically disordered protein, Ambn has the tendency to self-assemble and the potential to interact with many other targets, including cell membrane, other enamel proteins, and minerals (Murakami et al. 1997; Mazumder et al. 2014; Su et al. 2016). Ambn may tether enamel matrix to the cell surfaces of ameloblasts through its interaction and coassembly with Amel. Ambn interacts with Amel via its N-terminal sequence in vitro (Ravindranath et al. 2004; Su et al. 2016; Wald et al. 2017) and coassembles with Amel in the matrix during the secretory stage of enamel formation, when nucleation of enamel crystallites occurs (Mazumder et al. 2014; Mazumder et al. 2016). Ambn and Amel are processed simultaneously in the Golgi complex and cosecreted by the secretory granules in Tomes’ processes (Zalzal et al. 2008), implying that the interaction between Ambn and Amel may occur before secretion. This notion is supported by previous in vivo studies of Amel X−/−/Ambn−/− double-knockout mice that showed additional enamel defects beyond those of Amel X−/− or Ambn−/− mice (Hatakeyama et al. 2009).
The AH motif may also be involved in Ambn protein assembly and therefore serve a dual function. Indeed, these 2 functions may be related, since self-assembly may be a prerequisite for Ambn-lipid interaction. As reported recently, triple mutations in the N-terminus of exon 5 affect the self-assembly of Ambn and result in abnormal enamel (Wald et al. 2017). Ambn has proline-rich regions located in the C-terminus of the sequence encoded by exon 5 and in the sequence encoded by exon 6 (Delsuc et al. 2015). We found that when the sequence encoded by exon 6 was deleted, the pattern of Ambn-cell adhesion was altered. This may be because the proline-rich region, which may be involved in protein assembly (Wald et al. 2013), was disrupted, interrupting Ambn self-assembly. This is consistent with a recent report that deletion of human AMBN exon 6 is associated with amelogenesis imperfecta (Poulter et al. 2014).
Observations in different knockout animal models have provided evidence that Ambn may be associated with Tomes’ process formation (Wazen et al. 2009; Bartlett et al. 2011; Zhang, Zhang, et al. 2011). One proposed mechanism was based on the report that Ambn upregulates the expression level of RhoA (Zhang, Zhang, et al. 2011), which in turn regulates the polymerization of actin to produce pseudopodium-like Tomes’ processes (Nobes and Hall 1995). Interestingly, when exon 5 and exon 6 of Ambn are deleted in mice, Tomes’ processes do not develop (Wazen et al. 2009). In light of our present in vitro and cell culture data and these previous reports, we propose that during the early stages of enamel formation, Ambn is likely to bind to the cell membrane via the AH motif encoded by exon 5 and might be involved in regulation of the Tomes’ processes. Whether Ambn interacts with the cell membrane directly or via specific receptors is not known and is the subject of further investigation.
Author Contributions
J. Su, contributed to conception, design, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript; R.A. Bapat, G. Visakan, contributed to data acquisition and analysis, critically revised the manuscript; J. Moradian-Oldak, contributed to conception, design, data analysis and interpretation, drafted and critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Supplemental Material
Supplemental material, DS_10.1177_0022034520918521 for An Evolutionarily Conserved Helix Mediates Ameloblastin-Cell Interaction by J. Su, R.A. Bapat, G. Visakan and J. Moradian-Oldak in Journal of Dental Research
Acknowledgments
We thank Prof. Ralf Langen and Prof. Michael Paine for helpful discussions and valuable suggestions, Dr. Malcolm Snead for providing the LS8 cells, Mrs. Hong Shi and Dr. Shuhui Geng for technical assistance with cell culture experiments, Dr. Shuxing Li (NanoBiophysics Center, USC) for assistance with CD and fluorescence spectra, and Mr. Chris Martinez for assistance with counting the cells.
Footnotes
A supplemental appendix to this article is available online.
This project was funded by National Institutes of Health–National Institute of Dental and Craniofacial Research grants R01DE013414, R01DE027632, and DE020099 to J. Moradian-Oldak.
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- Bapat RA, Moradian-Oldak J. 2019. Immunohistochemical co-localization of amelogenin and ameloblastin in developing enamel matrix. In: Papagerakis P, editor. Odontogenesis: methods and protocols. New York (NY): Springer; p. 219–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett JD, Yamakoshi Y, Simmer JP, Nanci A, Smith CE. 2011. MMP20 cleaves E-cadherin and influences ameloblast development. Cells Tissues Organs. 194(2–4):222–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyeler M, Schild C, Lutz R, Chiquet M, Trueb B. 2010. Identification of a fibronectin interaction site in the extracellular matrix protein ameloblastin. Exp Cell Res. 316(7):1202–1212. [DOI] [PubMed] [Google Scholar]
- Bornholdt ZA, Noda T, Abelson DM, Halfmann P, Wood MR, Kawaoka Y, Saphire EO. 2013. Structural rearrangement of Ebola virus VP40 begets multiple functions in the virus life cycle. Cell. 154(4):763–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerny R, Slaby I, Hammarstrom L, Wurtz T. 1996. A novel gene expressed in rat ameloblasts codes for proteins with cell binding domains. J Bone Miner Res. 11(7):883–891. [DOI] [PubMed] [Google Scholar]
- Chen LS, Couwenhoven RI, Hsu D, Luo W, Snead ML. 1992. Maintenance of amelogenin gene expression by transformed epithelial cells of mouse enamel organ. Arch Oral Biol. 37(10):771–778. [DOI] [PubMed] [Google Scholar]
- Delsuc F, Gasse B, Sire JY. 2015. Evolutionary analysis of selective constraints identifies ameloblastin (AMBN) as a potential candidate for amelogenesis imperfecta. BMC Evol Biol. 15:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drin G, Antonny B. 2010. Amphipathic helices and membrane curvature. FEBS Lett. 584(9):1840–1847. [DOI] [PubMed] [Google Scholar]
- Fincham AG, Moradian-Oldak J, Diekwisch TG, Lyaruu DM, Wright JT, Bringas P, Jr, Slavkin HC. 1995. Evidence for amelogenin “nanospheres” as functional components of secretory-stage enamel matrix. J Struct Biol. 115(1):50–59. [DOI] [PubMed] [Google Scholar]
- Fukumoto S, Kiba T, Hall B, Iehara N, Nakamura T, Longenecker G, Krebsbach PH, Nanci A, Kulkarni AB, Yamada Y. 2004. Ameloblastin is a cell adhesion molecule required for maintaining the differentiation state of ameloblasts. J Cell Biol. 167(5):973–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama J, Fukumoto S, Nakamura T, Haruyama N, Suzuki S, Hatakeyama Y, Shum L, Gibson C, Yamada Y, Kulkarni A. 2009. Synergistic roles of amelogenin and ameloblastin. J Dent Res. 88(4):318–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries MJ. 2001. Cell adhesion assays. Mol Biotechnol. 18(1):57–61. [DOI] [PubMed] [Google Scholar]
- Lacruz RS, Habelitz S, Wright JT, Paine ML. 2017. Dental enamel formation and implications for oral health and disease. Physiol Rev. 97(3):939–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T, Li M, Xu X, Xiong J, Huang C, Zhang X, Hu A, Peng L, Cai D, Zhang L, et al. 2018. Whole exome sequencing identifies an AMBN missense mutation causing severe autosomal-dominant amelogenesis imperfecta and dentin disorders. Int J Oral Sci. 10(3):26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis H, Beniash E, Fowler C. 2006. Role of macromolecular assembly of enamel matrix proteins in enamel formation. J Dent Res. 85(9):775–793. [DOI] [PubMed] [Google Scholar]
- Márquez MG, Nieto FL, Fernández-Tome MC, Favale NO, Sterin-Speziale N. 2008. Membrane lipid composition plays a central role in the maintenance of epithelial cell adhesion to the extracellular matrix. Lipids. 43(4):343–352. [DOI] [PubMed] [Google Scholar]
- Mazumder P, Prajapati S, Bapat R, Moradian-Oldak J. 2016. Amelogenin-ameloblastin spatial interaction around maturing enamel rods. J Dent Res. 95(9):1042–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumder P, Prajapati S, Lokappa SB, Gallon V, Moradian-Oldak J. 2014. Analysis of co-assembly and co-localization of ameloblastin and amelogenin. Front Physiol. 5:274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee MD, Zerounian C, Martineau-Doizé B, Warshawsky H. 1987. Specific binding sites for transferrin on ameloblasts of the enamel maturation zone in the rat incisor. Anat Rec. 218(2):123–127. [DOI] [PubMed] [Google Scholar]
- Moradian-Oldak J. 2012. Protein-mediated enamel mineralization. Front Biosci. 17:1996–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouw JK, Ou G, Weaver VM. 2014. Extracellular matrix assembly: a multiscale deconstruction. Nat Rev Mol Cell Biol. 15(12):771–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami C, Dohi N, Fukae M, Tanabe T, Yamakoshi Y, Wakida K, Satoda T, Takahashi O, Shimizu M, Ryu O, et al. 1997. Immunochemical and immunohistochemical study of the 27- and 29-kDa calcium-binding proteins and related proteins in the porcine tooth germ. Histochem Cell Biol. 107(6):485–494. [DOI] [PubMed] [Google Scholar]
- Nakata A, Kameda T, Nagai H, Ikegami K, Duan Y, Terada K, Sugiyama T. 2003. Establishment and characterization of a spontaneously immortalized mouse ameloblast-lineage cell line. Biochem Biophys Res Commun. 308(4):834–839. [DOI] [PubMed] [Google Scholar]
- Nobes CD, Hall A. 1995. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 81(1):53–62. [DOI] [PubMed] [Google Scholar]
- Poulter JA, Murillo G, Brookes SJ, Smith CE, Parry DA, Silva S, Kirkham J, Inglehearn CF, Mighell AJ. 2014. Deletion of ameloblastin exon 6 is associated with amelogenesis imperfecta. Hum Mol Genet. 23(20):5317–5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad MK, Geoffroy V, Vicaire S, Jost B, Dumas M, Le Gras S, Switala M, Gasse B, Laugel-Haushalter V, Paschaki M, et al. 2016. A targeted next-generation sequencing assay for the molecular diagnosis of genetic disorders with orodental involvement. J Med Genet. 53(2):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravindranath HH, Chen LS, Zeichner-David M, Ishima R, Ravindranath RM. 2004. Interaction between the enamel matrix proteins amelogenin and ameloblastin. Biochem Biophys Res Commun. 323(3):1075–1083. [DOI] [PubMed] [Google Scholar]
- Smith C. 1998. Cellular and chemical events during enamel maturation. Crit Rev Oral Biol Med. 9(2):128–161. [DOI] [PubMed] [Google Scholar]
- Sonoda A, Iwamoto T, Nakamura T, Fukumoto E, Yoshizaki K, Yamada A, Arakaki M, Harada H, Nonaka K, Nakamura S, et al. 2009. Critical role of heparin binding domains of ameloblastin for dental epithelium cell adhesion and ameloblastoma proliferation. J Biol Chem. 284(40):27176–27184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J, Bapat RA, Moradian-Oldak J. 2019. The expression and purification of recombinant mouse ameloblastin in E. coli. In: Papagerakis P, editor. Odontogenesis: methods and protocols. New York (NY): Springer; p. 229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J, Chandrababu KB, Moradian-Oldak J. 2016. Ameloblastin peptide encoded by exon 5 interacts with amelogenin N-terminus. Biochem Biophys Rep. 7:26–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J, Kegulian NC, Bapat RA, Moradian-Oldak J. 2019. Ameloblastin binds to phospholipid bilayers via a helix-forming motif within the sequence encoded by exon 5. ACS Omega. 4(2):4405–4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wald T, Osickova A, Sulc M, Benada O, Semeradtova A, Rezabkova L, Veverka V, Bednarova L, Maly J, Macek P, et al. 2013. Intrinsically disordered enamel matrix protein ameloblastin forms ribbon-like supramolecular structures via an N-terminal segment encoded by exon 5. J Biol Chem. 288(31):22333–22345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wald T, Spoutil F, Osickova A, Prochazkova M, Benada O, Kasparek P, Bumba L, Klein OD, Sedlacek R, Sebo P, et al. 2017. Intrinsically disordered proteins drive enamel formation via an evolutionarily conserved self-assembly motif. Proc Natl Acad Sci USA. 114(9):E1641–E1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wazen RM, Moffatt P, Zalzal SF, Yamada Y, Nanci A. 2009. A mouse model expressing a truncated form of ameloblastin exhibits dental and junctional epithelium defects. Matrix Biol. 28(5):292–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalzal SF, Smith CE, Nanci A. 2008. Ameloblastin and amelogenin share a common secretory pathway and are co-secreted during enamel formation. Matrix Biol. 27(4):352–359. [DOI] [PubMed] [Google Scholar]
- Zhang X, Diekwisch TG, Luan X. 2011. Structure and function of ameloblastin as an extracellular matrix protein: adhesion, calcium binding, and CD63 interaction in human and mouse. Eur J Oral Sci. 119(Suppl 1):270–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang X, Lu X, Atsawasuwan P, Luan X. 2011. Ameloblastin regulates cell attachment and proliferation through RhoA and p27. Eur J Oral Sci. 119(Suppl 1):280–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, DS_10.1177_0022034520918521 for An Evolutionarily Conserved Helix Mediates Ameloblastin-Cell Interaction by J. Su, R.A. Bapat, G. Visakan and J. Moradian-Oldak in Journal of Dental Research