Abstract
Cine cardiac magnetic resonance imaging (CMRI), the current gold standard for cardiac function analysis, provides images with high spatio-temporal resolution. Computing clinical cardiac parameters like ventricular blood-pool volumes, ejection fraction and myocardial mass from these high resolution images is an important step in cardiac disease diagnosis, therapy planning and monitoring cardiac health. An accurate segmentation of left ventricle blood-pool, myocardium and right ventricle blood-pool is crucial for computing these clinical cardiac parameters. U-Net inspired models are the current state-of-the-art for medical image segmentation. SegAN, a novel adversarial network architecture with multi-scale loss function, has shown superior segmentation performance over U-Net models with single-scale loss function. In this paper, we compare the performance of stand-alone U-Net models and U-Net models in SegAN framework for segmentation of left ventricle blood-pool, myocardium and right ventricle blood-pool from the 2017 ACDC segmentation challenge dataset. The mean Dice scores achieved by training U-Net models was on the order of 89.03%, 89.32% and 88.71% for left ventricle blood-pool, myocardium and right ventricle blood-pool, respectively. The mean Dice scores achieved by training the U-Net models in SegAN framework are 91.31%, 88.68% and 90.93% for left ventricle blood-pool, myocardium and right ventricle blood-pool, respectively.
Keywords: Image segmentation, cine cardiac MRI, convolutional neural network, deep learning
1. INTRODUCTION
Cardiac magnetic resonance imaging (CMRI), a non-invasive and non-ionizing radiation imaging modality, provides high resolution 3D images (parallel short axis slices stacked together) of the cardiac anatomy with superior soft tissue details. This makes CMRI the current gold standard for cardiac function analysis.1,2 The analysis of the ventricular structure and function is an important step in cardiac disease diagnosis, treatment and prognosis. Cardiac function indices like stroke volume, ejection fraction, cardiac output, myocardium thickness and strain analysis play a crucial role in predicting and planning therapy for diseases like myocardial infarction, ischemia, arrhythmogenic right ventricular cardiomyopathy, pulmonary hypertension, dilated and hypertrophic cardiomyopathy.2 The calculation of these cardiac function indices requires accurate delineation of the left ventricle endocardium, the left ventricle epicardium and the right ventricle endocardium. In order to avoid inter- and intra-expert variability that occurs in manual delineation, a robust automated segmentation method is necessary.
The automated segmentation of the cardiac chambers is challenging due to the fuzzy boundaries of the ventricular cavities, motion artifacts, banding artifacts, presence of trabeculae and papillary muscles, and shape variation across phases and pathologies. Prior to deep learning, a number of automated segmentation algorithms for segmentation of the cardiac chambers from CMR images have been proposed.2,3 These non-deep learning algorithms require considerable manual or semi-manual interactions (thresholding, clustering, region growing and edge-based methods3,4), fail to accurately delineate ventricles in basal and apical slices (graph based methods), and require extensive computational time (active shape models and active appearance models5,6). The inadequacies of these segmentation methods render them unsuitable for clinical applications.
In recent years, deep learning techniques have shown exceptional performance in image classification and segmentation. With the availability of large number of medical images for supervised training, fully convolutional networks (FCN) significantly improved the medical image segmentation performance. The introduction of U-Net by Ronneberger et al.,7 a fully convolutional network with a downsampling network that captures context information and an upsampling network that enables accurate localization of the annotated objects is currently the most popular method used for biomedical image segmentation. A majority of the medical image segmentation algorithms introduced in the past few years are variants of the U-Net model.
Generative adversarial networks (GAN)8 are a type of adversarial networks in which two neural networks compete against each other in a min-max game to generate new image which is as close as possible to the original training image. This inspired algorithms like cycle-GAN for automated segmentation of epithelial tissue from microscopic Drosophilia embryos images which outperformed the U-Net models.9 Xue et al.10 proposed SegAN, an end-to-end adversarial network architecture that achieved better Dice score than the U-Net models in the MICCAI BRATS (2013 and 2015) brain tumor segmentation challenge dataset.
In our previous work, we showed that the U-Net models, when combined with the SegAN architecture (Fig. 1), performs better in segmenting left ventricle blood-pool from CMR images than standalone U-Net training.11 Here, we combine a U-Net model7 and one of its variant, a modified U-Net (mod U-Net)10 with the SegAN adversarial architecture to segment the left ventricle (LV), myocardium (MC) and right ventricle (RV) simultaneously and show its effect on clinical cardiac parameters like stroke volume, ejection fraction and myocardial mass, using the image dataset made available through the 2017 Automatic Cardiac Diagnosis Challenge (ACDC).
Figure 1.
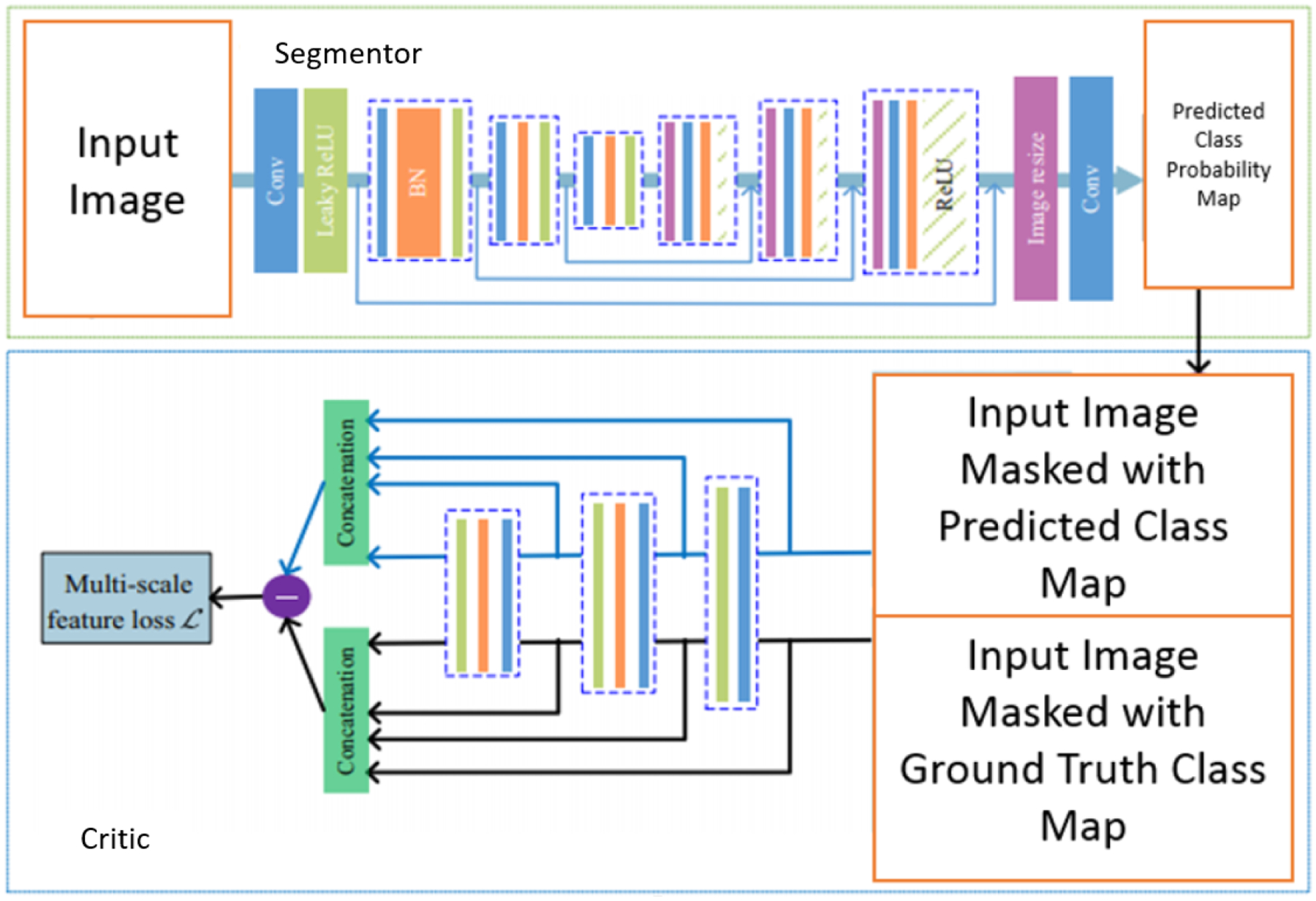
SegAN Architecture10
2. METHODOLOGY
2.1. SegAN Architecture
The conventional GAN consists of two convolutional neural networks - a generator G and a discriminator D. The generator G aims to produce an image that is similar to the image from the training dataset and the discriminator D aims to differentiate between the real image and the fake image generated by generator G.8 This generative adversarial network inspired Xue et al. to propose an adversarial architecture SegAN for segmentation of biomedical images. The SegAN adversarial architecture consists of two networks - segmentor and critic. The segmentor network can be any encoder-decoder type fully convolutional neural network that produces a predicted class probability map from a raw image input. The critic network is the encoder part of the segmentor network. It requires two inputs - a raw input image masked by ground truth and a raw input image masked by predicted class probability map produced by the segmentor network (See Fig. 1). The features are extracted from multiple layers of the critic network and concatenated to calculate the L1 loss function.10 This multi-resolution approach to feature extraction enables the SegAN model to learn the dissimilarities between the segmentor network predicted segmentation maps and the ground truth across multiple layers of the critic network.
The L1 loss function is given by -
| (1) |
where θS and θC are the parameters of segmentor network and critic network, respectively and N is the number of training images. fC(xn ∘ S(xn)) and fC(xn ∘ yn) are the features extracted from ground truth masked input image and segmentor predicted class probability map masked input image, respectively. The mean absolute error lmae is given by -
| (2) |
where L represents the number of layers in the critic network.10
The segmentor and critic networks are trained in an alternating fashion, with the segmentor network aiming to minimize the multi-scale L1 loss function and the critic network aiming to maximize the the multi-scale L1 loss function, resembling a min-max game.
2.2. Experiments and Implementation Details
The focus of our experiment is to compare the results of a stand-alone 2D U-Net architecture with a SegAN architecture. In our experiments, we train the U-Net7 model by back propagation using cross entropy loss as cost function and compare its results with SegAN architecture, where, the U-Net model is used as the segmentor network and the downsampling part of this U-Net model is used as the critic network (SegAN + U-Net). The SegAN architecture is trained using multi-scale L1 loss as cost function. We perform our experiments on two stand-alone U-Net models - an original U-Net7 and a modified U-Net (mod U-Net).10 In the modified U-Net, the downsampling part consists of convolutional layers with kernel size 4×4 and stride 2, while the upsampling part consists of convolutional layers with kernel size 3×3 and stride 1. The segmentation results obtained using this modified U-Net are compared with its SegAN integration, i.e., the modified U-Net model is used as the segmentor network and the downsampling part of the modified U-Net model is used as the critic network (SegAN + mod U-Net).
For stand-alone 2D U-Net model training, the images are resized to 224×224 and fed into the network in batches of 10. The U-Net model is trained using Adam optimizer with a learning rate of 0.0001 for 100 epochs.
In SegAN architecture, the input to the segmentor network is a 224×224×1 resized CMR image and the output is a 224×224×3 predicted class probability map, where the three layers correspond to the three different segmented classes - left ventricle blood-pool (LV), myocardium (MC) and right ventricle blood-pool (RV). Here, we train three different critic networks, one for each label class. The segmentor network and the three critic networks are trained using the average loss computed from the three different critic networks. Since we are training four networks (one segmentor network and three critic networks) per epoch, the images to SegAN network are fed in batches of two to avoid memory allocation issues. The SegAN network is trained using Adam optimizer with a learning rate of 0.00001 for 50 epochs. All the experiments were performed on a machine equipped with NVIDIA RTX 2080 Ti GPU with 11GB of memory.
2.3. Dataset
The Automated Cardiac Diagnosis Challenge (ACDC) dataset was released during the MICCAI 2017 conference in conjunction with the STACOM workshop. The dataset consists of short axis cine cardiac MR images for 100 subjects evenly distributed into five subgroups - normal, previous myocardial infarctions, dilated cardiomyopathy, hypertrophic cardiomyopathy and abnormal right ventricle. The image dataset corresponding to each subject consists of two image volumes, one at end-diastole and one at end-systole, with each containing 8–12 slices, leading to a total of 1, 902 images.
In our experiments, we divide the ACDC dataset into 80% training data and 20% validation data with five non-overlapping folds for cross validation. During training, we augment the dataset using translation, rotation, gamma correction and flipping operations.
3. RESULTS AND DISCUSSION
In Table 1, we summarize the segmentation performance of U-Net models with and without SegAN integration. We can observe that the SegAN architecture, when integrated with both the variants of U-Net models, achieves better Dice score for left ventricle blood-pool and right ventricle blood-pool segmentation in both end diastole and end systole phases. However, in case of left ventricle myocardium segmentation, the Dice scores achieved in the stand-alone U-Net models training are better or similar to the U-Net models trained in SegAN framework. These segmentation results are in compliance with the results of the brain tumor segmentation in BRATS 2015 dataset.10 The SegAN architecture obtained better Dice score in segmenting the whole brain tumor region, but had some drawbacks in segmenting the tumor core and Gd-enhanced tumor core. This is attributed to the fact that the SegAN architecture extracts features from multiple layers of the critic network and the segmentation of regions of smaller areas, like the left ventricle myocardium, may require more concentration at pixel-level features. Therefore, U-Net model with cross entropy loss (pixel-level loss) could have better segmentation performance than SegAN architecture with multi-scale L1 loss for segmentation of the left ventricle myocardium. Fig. 2 shows examples of the segmented left ventricle blood-pool, myocardium and right ventricle blood-pool in mid, apical and basal slices for the two variants of U-Net models, with and without SegAN integration.
Table 1.
Segmentation evaluation, mean Dice score (std-dev) for end diastole (ED) and end systole (ES) for left ventricle blood-pool (LV), myocardium (MC) and right ventricle blood-pool (RV) segmentation in the 2017 ACDC dataset. Statistical significance (T-test) of the results of SegAN architecture compared against U-Net models are represented by * for p < 0.05 and ** for p < 0.005. The best Dice scores achieved are labeled in bold.
| LV Dice (ED) (%) | LV Dice (ES)(%) | MC Dice (ED) (%) | MC Dice (ES)(%) | RV Dice (ED) (%) | RV Dice (ES)(%) | |
|---|---|---|---|---|---|---|
| U-Net | 89.04 (1.97) | 88.65 (2.03) | 90.62 (2.72) | 88.17 (3.21) | 88.09 (2.05) | 88.80 1.92 |
| SegAN + U-Net | 91.69 (1.49)** | 90.29 (1.61)** | 88.19 (4.21) | 88.42 (5.93) | 91.55 (4.19) | 90.06 (1.74)** |
| mod U-Net | 90.14 (1.78) | 88.29 (1.55) | 91.61 (2.14) | 88.89 (2.32)** | 90.55 (4.41) | 87.41 (4.78) |
| SegAN + mod U-Net | 92.19 (1.67)* | 91.09 (2.08)* | 91.18 (3.50) | 87.95 (7.97) | 92.06 (3.41) | 90.05 (2.02)* |
Figure 2.

Examples of segmentation of the left ventricle blood-pool (blue), myocardium (green) and right ventricle blood-pool (red) in mid, apical and basal slices (top to bottom)
In our previous work, we showed that U-Net model (and two of its variants) integrated in SegAN framework improves the segmentation of left ventricle blood-pool from CMR images.11 In this paper, we investigate the viability of SegAN framework for multi-class segmentation of left ventricles, myocardium and right ventricles by evaluating the clinical cardiac parameters like LV stroke volume, LV ejection fraction, RV stroke volume, RV ejection fraction and myocardial mass. In Table 2, we show the correlation coefficient of these clinical cardiac parameters calculated using the segmentation results obtained from the above mentioned U-Net models and its SegAN integrated counterparts with the clinical cardiac parameters calculated using the ground truth. The correlation coefficient values of ventricular stroke volume and the ejection fraction computed from the segmentation results of SegAN framework are higher than the correlation coefficient values of ventricular stroke volume and the ejection fraction computed from the U-Net models. However, the correlation coefficient values of myocardial mass computed from the segmentation results of SegAN framework are lower than the correlation coefficient values of myocardial mass computed from the U-Net models. These observations are in agreement with the Dice score results shown in Table 1, where the segmentation of the myocardial tissue using SegAN integration is not superior to the segmentation using stand-alone U-Net models, i.e., the myocardial mass estimates are affected since they are directly related to uncertainties present in the myocardium segmentation.
Table 2.
Evaluation of clinical indices - LV stroke volume (SV) correlation coefficient, LV ejection fraction (EF) correlation coefficient, myocardium mass correlation coefficient, RV stroke volume correlation coefficient and RV ejection fraction correlation coefficient.
| LV SV Correlation | LV EF Correlation | MC Mass Correlation | RV SV Correlation | RV EF Correlation | |
|---|---|---|---|---|---|
| U-Net | 0.923 | 0.904 | 0.955 | 0.899 | 0.841 |
| SegAN + U-Net | 0.941 | 0.918 | 0.931 | 0.944 | 0.901 |
| mod U-Net | 0.937 | 0.957 | 0.956 | 0.908 | 0.882 |
| SegAN + mod U-Net | 0.974 | 0.963 | 0.937 | 0.935 | 0.921 |
The major drawback of the SegAN architecture in multi-class segmentation is the computational time and the memory required to train one segmentor network and three critic networks simultaneously. The computational time required for one epoch for an U-Net model is around 225 seconds, whereas the U-Net model in SegAN framework requires around 900 seconds (for multi-class segmentation).
4. CONCLUSION AND FUTURE WORK
In this paper, we demonstrate the use of an adversarial architecture, SegAN with multi-scale L1 loss function, to segment the left ventricle blood-pool, myocardium and right ventricle blood-pool from cine cardiac MR images. This multi-scale L1 loss function captures features at multiple levels - pixel-level, superpixel-level and patches-level. Our experiments reveal that this integration of U-Net models in the SegAN framework leads to significant improvement of left ventricle blood-pool and right ventricle blood-pool segmentation in the 2017 ACDC segmentation challenge dataset. The adversarial nature of the architecture and the multi-resolution approach enables the SegAN model to accurately segment the above mentioned heart chambers, which in turn, enables accurate computation of critical clinical parameters like ventricular stroke volumes and ejection fraction.
We observed that the segmentation result of left ventricle myocardium did not improve with the SegAN integration. An alternative solution to this could be an integration of a weighted cross entropy loss as cost function along with the multi-scale L1 loss function.
ACKNOWLEDGMENTS
Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award No. R35GM128877 and by the Office of Advanced Cyber-infrastructure of the National Science Foundation under Award No. 1808530.
REFERENCES
- [1].La AG, Claessen G, de Bruaene Van A, Pattyn N, Van JC, Gewillig M, Bogaert J, Dymarkowski S, Claus P, and Heidbuchel H, “Cardiac MRI: a new gold standard for ventricular volume quantification during high-intensity exercise.,” Circulation. Cardiovascular imaging 6(2), 329–338 (2013). [DOI] [PubMed] [Google Scholar]
- [2].Peng P, Lekadir K, Gooya A, Shao L, Petersen SE, and Frangi AF, “A review of heart chamber segmentation for structural and functional analysis using cardiac magnetic resonance imaging,” Magnetic Resonance Materials in Physics, Biology and Medicine 29(2), 155–195 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Petitjean C and Dacher J-N, “A review of segmentation methods in short axis cardiac MR images,” Medical image analysis 15(2), 169–184 (2011). [DOI] [PubMed] [Google Scholar]
- [4].Alattar MA, Osman NF, and Fahmy AS, “Myocardial segmentation using constrained multi-seeded region growing,” in [International Conference Image Analysis and Recognition], 89–98, Springer; (2010). [Google Scholar]
- [5].Santiago C, Nascimento JC, and Marques JS, “A new ASM framework for left ventricle segmentation exploring slice variability in cardiac MRI volumes,” Neural Computing and Applications 28(9), 2489–2500 (2017). [Google Scholar]
- [6].Tzimiropoulos G and Pantic M, “Optimization problems for fast AAM fitting in-the-wild,” in [Proceedings of the IEEE international conference on computer vision], 593–600 (2013). [Google Scholar]
- [7].Ronneberger O, Fischer P, and Brox T, “U-Net: Convolutional networks for biomedical image segmentation,” in [International Conference on Medical image computing and computer-assisted intervention], 234–241, Springer; (2015). [Google Scholar]
- [8].Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, Courville A, and Bengio Y, “Generative adversarial nets,” in [Advances in neural information processing systems], 2672–2680 (2014). [Google Scholar]
- [9].Haering M, Grosshans J, Wolf F, and Eule S, “Automated segmentation of epithelial tissue using cycle-consistent generative adversarial networks,” bioRxiv, 311373 (2018). [Google Scholar]
- [10].Xue Y, Xu T, Zhang H, Long LR, and Huang X, “Segan: Adversarial network with multi-scale L1 loss for medical image segmentation,” Neuroinformatics 16(3–4), 383–392 (2018). [DOI] [PubMed] [Google Scholar]
- [11].Upendra RR, Dangi S, and Linte CA, “An adversarial network architecture using 2d U-Net models for segmentation of left ventricle from cine cardiac MRI,” in [International Conference on Functional Imaging and Modeling of the Heart], Lecture Notes in Computer Science. vol. 11504 pp. 415–424, Springer; (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]


