Xenophagy targets intracellular pathogens for destruction by the host autophagy pathway. Ubiquitin chains are conjugated to xenophagic targets and recruit multiple autophagy adaptors. The intracellular pathogen Legionella pneumophila resides in a vacuole that is ubiquitinated; however, this pathogen avoids xenophagic detection. Here, the mechanisms by which L. pneumophila can prevent the host xenophagy pathway from targeting the vacuole in which it resides were examined.
KEYWORDS: Legionella pneumophila, autophagy, bacterial effector proteins, type IV secretion
ABSTRACT
Xenophagy targets intracellular pathogens for destruction by the host autophagy pathway. Ubiquitin chains are conjugated to xenophagic targets and recruit multiple autophagy adaptors. The intracellular pathogen Legionella pneumophila resides in a vacuole that is ubiquitinated; however, this pathogen avoids xenophagic detection. Here, the mechanisms by which L. pneumophila can prevent the host xenophagy pathway from targeting the vacuole in which it resides were examined. Ubiquitin-labeled vacuoles containing L. pneumophila failed to recruit autophagy adaptors by a process that was independent of RavZ function. Coinfection studies were conducted using a strain of Listeria monocytogenes that served as a robust xenophagic target. Legionella pneumophila infection blocked xenophagic targeting of L. monocytogenes by a RavZ-dependent mechanism. Importantly, when coinfection studies were conducted with a RavZ-deficient strain of L. pneumophila, L. monocytogenes was targeted by the host xenophagy system but vacuoles containing L. pneumophila avoided targeting. Enhanced adaptor recruitment to the vacuole was observed by using a strain of L. pneumophila in which all of the effector proteins in the SidE family were deleted; however, this strain was still not targeted by the host autophagy pathway. Thus, there are at least two pathways by which L. pneumophila can disrupt xenophagic targeting of the vacuole in which it resides. One mechanism involves global disruption of the host autophagy machinery by the effector protein RavZ. A second cis-acting mechanism prevents the binding of autophagy adaptors to the ubiquitin-decorated surface of the L. pneumophila-containing vacuole.
INTRODUCTION
Legionella pneumophila is a Gram-negative intracellular pathogen that causes a severe form of pneumonia called Legionnaires’ disease (1, 2). Infection begins after L. pneumophila is phagocytosed by alveolar macrophages. Following uptake, this pathogen translocates over 300 effector proteins into the host cytosol through the specialized type IV secretion system (T4SS) named Dot/Icm (3–5). Effector proteins modulate intracellular transport to protect the L. pneumophila-containing vacuole (LCV) from lysosome-mediated degradation and remodel the vacuole into an endoplasmic reticulum (ER)-derived compartment that supports bacterial replication (6–8). These two hallmark events, bifurcation from the endocytic pathway and subversion of secretory traffic, require a functional Dot/Icm system (9). L. pneumophila must also couple extensive remodeling of the vacuole with protective mechanisms to counteract the host response against the modified phagosome (2). The autophagy pathway represents an important arm of the innate immune response that seeks to destroy pathogen-containing vacuoles (PCVs) and inhibit intracellular replication by a process called xenophagy (10–12).
The autophagy pathway was originally characterized as an adaptive response to starvation (13). Under nutrient-limiting conditions, double-membrane structures called phagophores localize to cytosolic cargo. As the phagophore membrane expands around the cargo and fuses, it is converted to a mature sequestering organelle called the autophagosome. The external membrane of the sealed autophagosome fuses with the lysosome to promote degradation of the inner cargo and liberate energy-rich substrates back into the cytosol to offset the effects of starvation (14). A hallmark of autophagosome formation is the lipidation and subsequent conjugation of Atg8 proteins such as LC3 (microtubule-associated protein 1A/1B-light chain 3) to the isolation membrane (15). Through the concerted action of autophagy-related genes (ATGs), lipidated LC3 is conjugated to both the inner and outer membranes of the maturing autophagosome (16). Importantly, LC3 is required for autophagosome expansion and fusion with lysosomes (14, 17). The autophagy pathway also functions as a bona fide arm of the innate and adaptive immune response by selectively targeting intracellular pathogens for degradation (18). Unlike bulk autophagy, selective autophagy targets specific substrates for degradation and requires molecular identifiers to coordinate this process.
Selective autophagy can be divided into three key events: cargo recognition, adaptor recruitment, and lysosome-mediated degradation. During infection, intracellular pathogens either modify or rupture the vacuole in which they reside, which can trigger a xenophagic response that marks these vacuoles for degradation. Ubiquitin labeling of xenophagic substrates is critical for targeting by the autophagy machinery by promoting localized cargo recognition (10). Ubiquitin E3 ligases attach ubiquitin to proteins on the pathogen-occupied vacuole or the surface of the pathogen once it is exposed to the cytosol of the host cell (12, 19–21). These ubiquitin-tagged substrates assemble into a “ubiquitin coat” that surrounds the pathogen to generate a signaling platform that stimulates the recruitment of autophagy adaptors. Autophagy adaptors contain conserved ubiquitin binding domains (UBD) and LC3-interacting regions (LIR) that bind Atg8 proteins attached to lipids on preautophagosomal membranes (22). Thus, autophagy adaptors serve as the molecular bridge that drives envelopment of the ubiquitin-tagged cargo into an autophagic vacuole, which enables the autophagy system to target intracellular pathogens for lysosomal degradation. The principal adaptor proteins that function in xenophagy include p62/SQSTM1, NDP52 (nuclear dot protein 52), NBR1 (neighbor of BRCA1 gene), and optineurin (23–25).
The adaptor protein p62 is one of the most extensively characterized mammalian autophagy receptors and has been demonstrated to participate in xenophagic targeting of Salmonella enterica (12), Listeria monocytogenes (26), Shigella flexneri (27), Streptococcus pyogenes (28), and Mycobacterium tuberculosis (29). Similar to p62, other members of the adaptor family, such as NDP52, play specialized roles in the removal of xenophagic substrates. For example, NDP52 is recruited to ruptured PCVs and cytosolic bacteria (25, 30). Similarly, optineurin localizes to ubiquitin-marked compartments and restricts bacterial replication by an autophagy-dependent mechanism (19, 23). Importantly, the autophagy regulatory protein TBK1 (TANK-binding kinase 1) phosphorylates these adaptor proteins to augment the autophagic response (30). Upon recruitment to xenophagic targets, these adaptors cooperate or act independently to restrict pathogen replication. Thus, many intracellular bacterial pathogens have evolved mechanisms to avoid ubiquitination and adaptor-mediated degradation.
It has been shown that L. pneumophila has the ability to avoid autophagy degradation (31). After bacterial uptake, the LCV accumulates ubiquitinated proteins (32). These ubiquitinated proteins coalesce into a ubiquitin coat that surrounds the LCV surface and remains associated with the vacuole throughout the infection. Importantly, a previous study revealed that both K48 and K63 ubiquitin linkages, which have been shown to activate a xenophagic response, compose the LCV ubiquitin coat (33). The intense enrichment of autophagy-activating ubiquitinated proteins at the LCV surface should result in targeting of the LCV by the host autophagy pathway; however, L. pneumophila delivers an effector protein called RavZ into host cells that disrupts the autophagy system. RavZ is a cysteine protease that irreversibly deconjugates lipidated Atg8 proteins from maturing autophagosomes by cleaving the peptide bond before the conserved glycine residue at the C terminus of all Atg8 family members (34). This cleavage event generates an Atg8 protein that can no longer be conjugated to the expanding phagophore membrane (34, 35). Another L. pneumophila effector named LpSPL consumes host sphingolipids, which reduces autophagosome biogenesis and dampens basal autophagy levels (36). A RavZ-deficient strain of L. pneumophila creates a vacuole that is decorated with ubiquitin but still avoids targeting by the host autophagy pathway (34). Similarly, mutants deficient in both LpSPL and RavZ are not targeted by autophagy (36). Thus, it is likely that L. pneumophila has additional effectors that can prevent xenophagy from targeting the vacuole in which it resides. Adaptor proteins are required to link “marked” cargo with the autophagy pathway, yet it is unclear whether ubiquitin-marked LCVs ever recruit these critical receptors. To better understand the mechanism by which L. pneumophila avoids autophagic targeting, this study focused on recruitment of autophagy adaptors to the LCV.
RESULTS
The xenophagy adaptor p62 is excluded from the LCV.
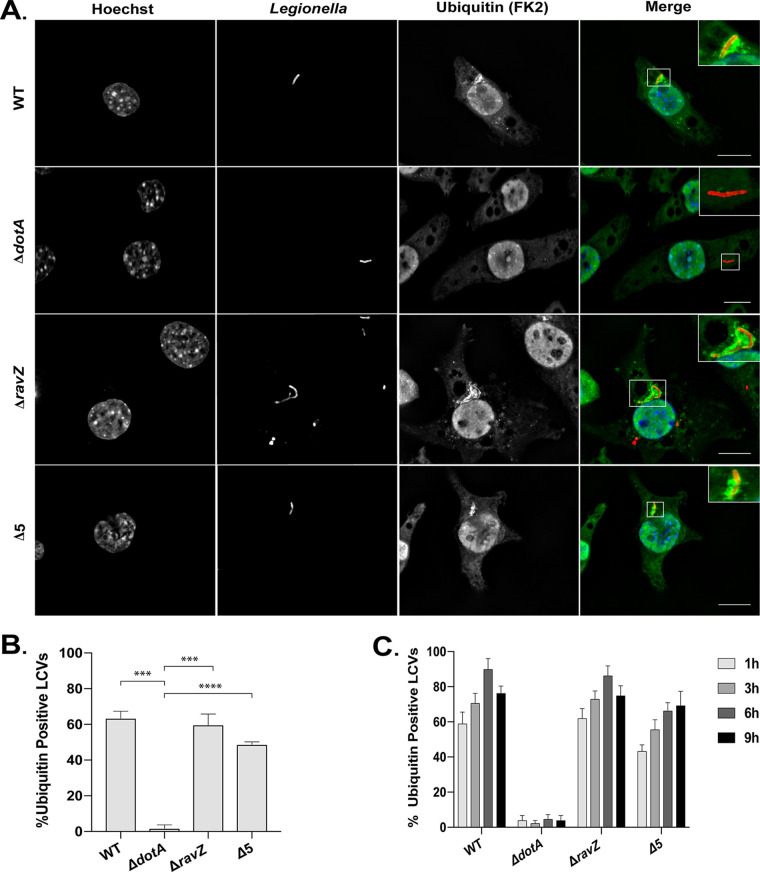
As described previously (32), vacuoles containing L. pneumophila stained positive for ubiquitin at both early and late time points of infection (Fig. 1), with the majority of vacuoles displaying an intense ubiquitin signature as early as 1 h after infection (Fig. 1B). Vacuoles containing ΔdotA mutants did not display this ubiquitin staining, which indicates that acquisition of a ubiquitin coat on the vacuole membrane requires a functional Dot/Icm secretion system. Importantly, vacuoles containing the pentuple mutant of L. pneumophila, a strain that has five large chromosomal deletions that eliminate 71 effector proteins (37), displayed robust ubiquitin recruitment similar to that of vacuoles containing the wild-type (WT) strain (Fig. 1C). Thus, these 71 effectors are not essential for ubiquitination of the vacuole.
FIG 1.
L. pneumophila-containing vacuoles (LCVs) display a ubiquitin signature. (A) Representative deconvolved fluorescence micrographs of J77A.1 macrophage-like cells at 1 h after infection with the indicated L. pneumophila strains. Merged panels show staining for L. pneumophila (red), ubiquitin (green), and Hoechst (blue). The ubiquitin FK2 antibody detects both mono- and polyubiquitinated proteins. Bars, 7.5 μm. (B) Quantification of ubiquitin colocalization to vacuoles at 1 h p.i. (C) Quantification of ubiquitin colocalization to vacuoles at each time point over a 9-h time course. Colocalization data are representative of 3 independent experiments. For each independent experiment, approximately 100 vacuoles were scored in triplicate, for a total of 300 vacuoles per time point. The unpaired t test was used to analyze the differences between samples. ****, P < 0.0001; ***, P < 0.001.
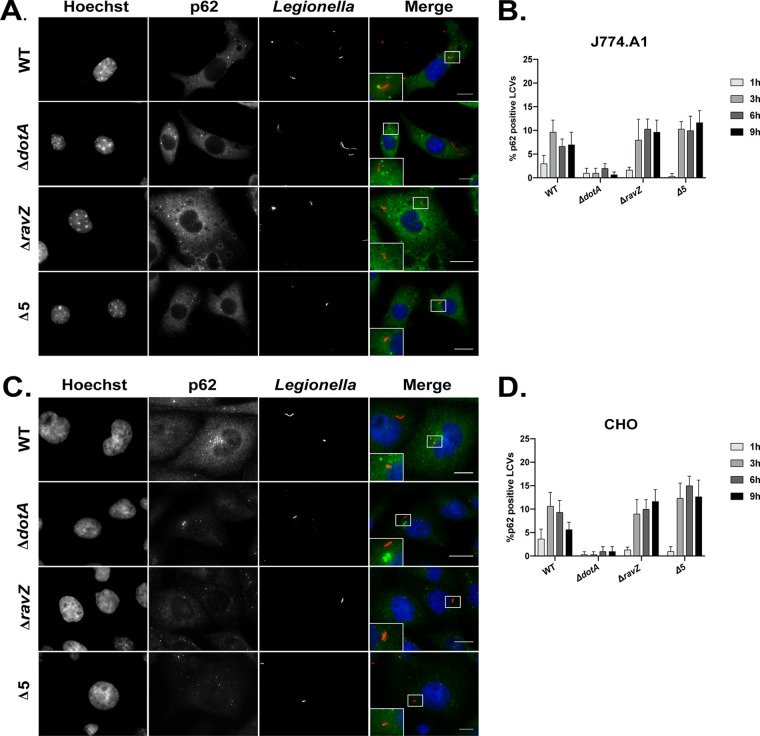
Previous data also indicated that the ubiquitin coat on the LCV is composed of both K48 and K63 linkages (33). Both K48 and K63 ubiquitin chains have been shown to stimulate the recruitment of p62, an autophagy adaptor recruited to most xenophagic substrates. Immunofluorescence (IF) microscopy was used to determine if p62 was recruited to these ubiquitin-tagged vacuoles. Despite the intense enrichment of ubiquitin on the majority of vacuoles containing L. pneumophila, very few compartments stained positive for p62 (Fig. 2). Similar results were obtained for vacuoles containing either the ΔravZ mutant or the pentuple mutant, which indicates that the 71 different effector proteins deleted in the pentuple mutant were not essential for blocking p62 recruitment to the LCV. Localization of p62 to the vacuole, although rare, was observed more frequently with L. pneumophila that had a functional Dot/Icm system than with a mutant defective in Dot/Icm function, which was expected given that ubiquitination of the vacuole requires Dot/Icm function. The defect in p62 recruitment was observed in both J774.A1 macrophage-like cells and Chinese hamster ovary (CHO) cells (Fig. 2B and D). These data indicate that the ubiquitin chains on the surface of the LCV are not efficiently recognized by p62.
FIG 2.
LCVs do not recruit the autophagy adaptor p62/SQSTM1. (A and C) Representative fluorescence micrographs of J774.A1 cells (A) and CHO cells (C) at 3 h after infection with WT L. pneumophila (Lp01), the ΔdotA mutant, the ΔravZ mutant, and the pentuple mutant (Δ5). Merged panels show staining for L. pneumophila (red), p62/SQSTM1 (green), and Hoechst (blue). Bars, 7.5 μm. (B and D) Data measuring colocalization of p62/SQSMT1 to vacuoles containing the indicated L. pneumophila strains in J774.A1 cells and CHO cells, respectively. Recruitment was measured over a 9-h time course at the indicated intervals. All colocalization data are a representative of 3 independent experiments. For each independent experiment, approximately 100 vacuoles were scored in triplicate, for a total of 300 vacuoles per time point.
Multiple xenophagy adaptors are excluded from the LCV.
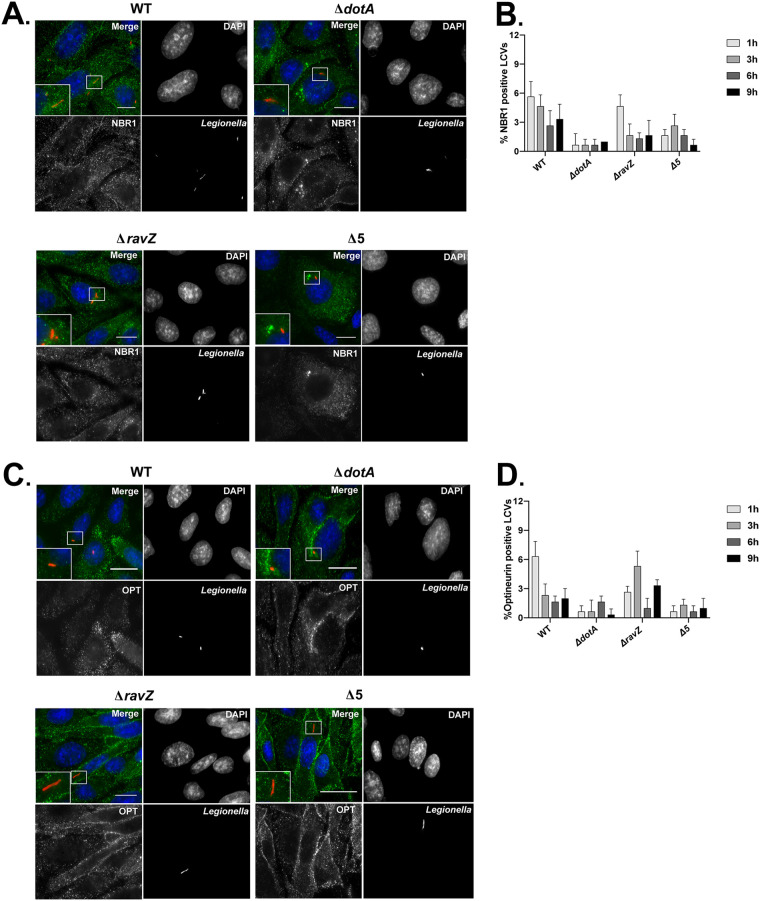
In addition to p62, the adaptor proteins NBR1, optineurin, and NDP52 have all been shown to recognize xenophagic substrates and deliver them to the autophagy pathway (18). Colocalization experiments were used to determine if the exclusion of p62 from the LCV extended to the other members of the autophagy adaptor family. Similar to what was observed for p62, colocalization of NBR1 to the LCV was rare for vacuoles containing either the parental strain of L. pneumophila, the isogenic ΔravZ mutant, or the pentuple mutant (Fig. 3A and B). Optineurin was also excluded from these LCVs (Fig. 3C and D).
FIG 3.
NBR1 and optineurin (OPT) autophagy adaptor proteins do not colocalize with LCVs. (A and C) Representative fluorescence micrographs of CHO cells at 3 h after infection with WT L. pneumophila (Lp01), the ΔdotA mutant, the ΔravZ mutant, and the pentuple mutant (Δ5). Merged panels show staining for L. pneumophila (red), NBR1 or optineurin (green), and DAPI (blue). Bars, 7.5 μm. (B and D) Colocalization of NBR1 or optineurin to L. pneumophila vacuoles in CHO cells. Recruitment was measured over a 9-h time course at the indicated intervals. Colocalization data represent the averages from 3 independent experiments. Approximately 100 vacuoles were scored in triplicate, for a total of 300 vacuoles per time point.
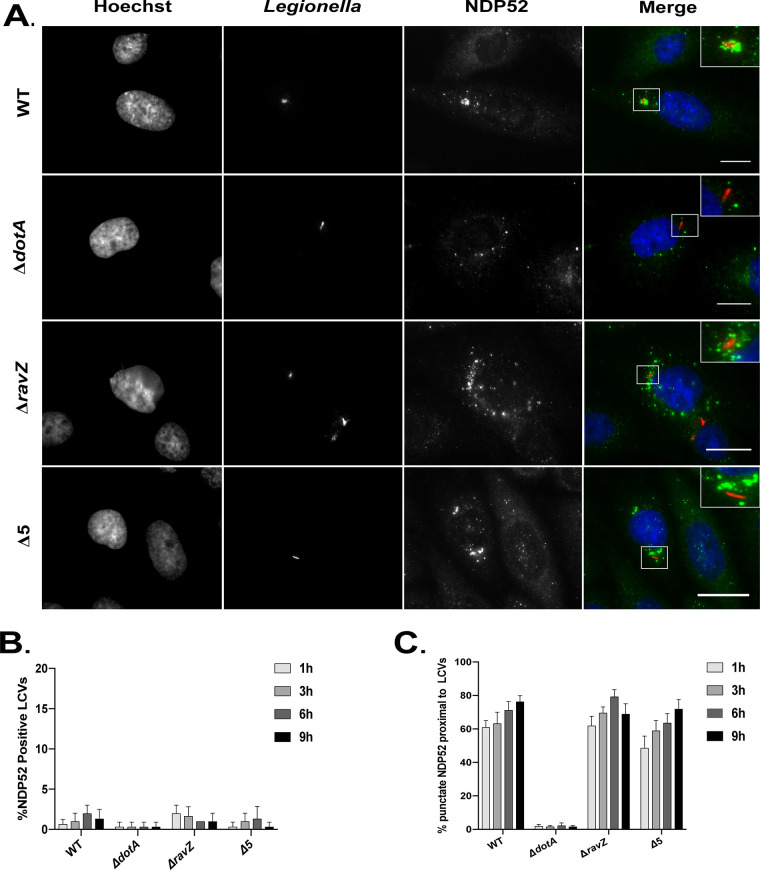
Staining for NDP52 revealed a pattern of punctate staining in proximity to the vacuole, but NDP52 did not appear to specifically colocalize with LCVs (Fig. 4). At 1 h postinfection, approximately 60% of vacuoles containing L. pneumophila had NDP52-positive puncta near the perimeter of the vacuole, but specific vacuolar colocalization was not observed (Fig. 4B and C). This punctate distribution of NDP52 was also observed near vacuoles containing the ΔravZ mutant and the pentuple mutant but not for vacuoles containing the ΔdotA mutant (Fig. 4C), which suggests that the recruitment of NDP52 to targets adjacent to the LCV is enhanced by Dot/Icm-mediated activities. These data indicate that peripheral recruitment of NDP52 could be triggered by an effector-dependent modification to host or bacterial proteins in proximity to the vacuole.
FIG 4.
NDP52 localization to the LCV. (A) Representative fluorescence micrographs of CHO cells 3 h after infection with WT L. pneumophila (Lp01), the ΔdotA mutant, the ΔravZ mutant, and the pentuple mutant (Δ5). Merged panels and insets show staining for Legionella (red), NDP52 (green), and Hoechst (blue). (B) Colocalization of NDP52 to vacuoles containing L. pneumophila in CHO cells. Recruitment was measured over a 9-h time course at the indicated intervals. (C) Colocalization of NDP52 puncta near vacuoles containing the indicated L. pneumophila strains in CHO cells. Recruitment was measured over a 9-h time course at the indicated intervals. Positive recruitment was defined as localization of more than 7 distinct NDP52-positive puncta near the vacuole. Bars, 7.5 μm.
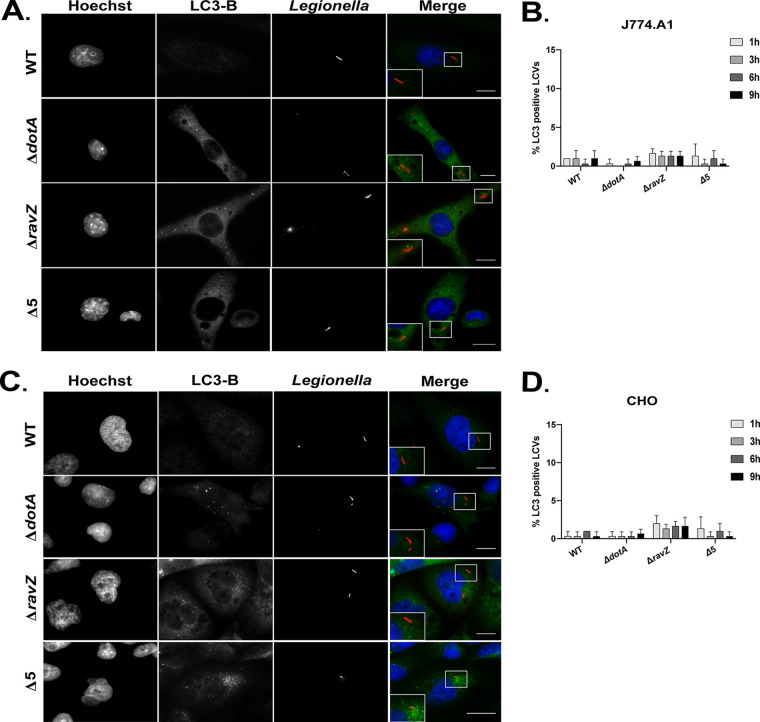
The recruitment of autophagosomes to L. pneumophila vacuoles was also investigated (Fig. 5). As expected, the lack of adaptor localization was consistent with the lack of autophagosome recruitment to LCVs. The percentage of LCVs that colocalized with LC3B in both macrophage and nonmacrophage cell lines was below 5% (Fig. 5B and D).
FIG 5.
Vacuoles containing L. pneumophila do not recruit LC3B. (A and C) Representative fluorescence micrographs of J774.A1 cells (A) and CHO cells (C) 3 h after infection with WT L. pneumophila (Lp01), the ΔdotA mutant, the ΔravZ mutant, and the pentuple mutant (Δ5). Merged panels show staining for L. pneumophila (red), LC3B (green), and DAPI (blue). Bars, 7.5 μm. (B and D) Colocalization of LC3B to vacuoles containing the indicated L. pneumophila strains in J774.A1 cells and CHO cells, respectively. Recruitment was measured over a 9-h time course at the indicated intervals. All colocalization data are representative of 3 independent experiments. For each independent experiment, approximately 100 vacuoles were scored in triplicate, for a total of 300 vacuoles per time point.
Phosphoribosyl-ubiquitination contributes to the exclusion of p62 on the LCV.
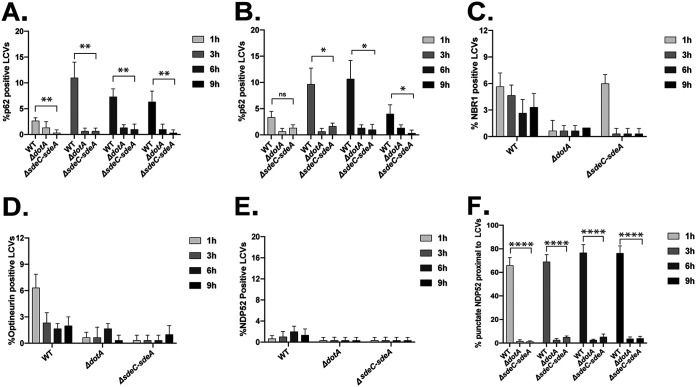
The SidE family of effectors catalyze ubiquitination of proteins on the LCV through an unconventional mechanism (38). These effectors contain a mono-ADP-ribosyltransferase (mART) domain that generates an ADP-ribosylated ubiquitin intermediate that mediates transfer of phosphoribosyl-ubiquitin to proteins on the LCV, which generates a noncanonical ubiquitin linkage that may not be recognized efficiently by autophagy adaptors (38, 39). There are four SidE family effector proteins in the L. pneumophila Philadelphia 1 strain that function as phosphoribosyl-ubiquitinating proteins. The eponymous SidE protein is encoded at a locus unlinked from those of the other three members of the family (SdeA, SdeB, and SdeC), which are encoded together in a contiguous region of the chromosome called the sde locus. Effector proteins within the sde locus (SdeA, SdeB, and SdeC) mediate phosphoribosyl-ubiquitination of the host ER-associated protein reticulon 4 at the LCV surface (40). This raised the possibility that the exclusion of p62 from the LCV may result from the enrichment of phosphoribosyl-ubiquitinated reticulon 4 on this organelle. To test this hypothesis, J774.A1 macrophage-like cells were infected with a sde locus mutant of L. pneumophila that has a chromosomal deletion removing the genes sdeA, sdeB, sdeC, lpg2154, and sidJ [Δ(sdeC-sdeA)] (40). No significant increase in p62 localization was observed on vacuoles containing the sde locus mutant [Δ(sdeC-sdeA)] compared to either the parental strain of L. pneumophila or the ΔdotA mutant (Fig. 6). Importantly, vacuoles containing the sde locus mutant displayed reduced recruitment of p62 compared to vacuoles containing the parental strain in both macrophage and nonmacrophage cell lines (Fig. 6A and B). Thus, the effectors within the sde locus are not essential for the exclusion of p62 from the LCV.
FIG 6.
Vacuoles containing the sde locus mutant [Δ(sdeC-sdeA)] suppress adaptor recruitment. J774.A1 and CHO cells were infected with WT L. pneumophila, the ΔdotA mutant, and the Δsde locus mutant [Δ(sdeC-sdeA)]. The recruitment of autophagy adaptors was evaluated by fluorescence microscopy. (A and B) Colocalization of p62/SQSTM1 to vacuoles containing L. pneumophila strains in J774.A1 cells (A) or CHO cells (B). (C and D) CHO cells were infected with the strains indicated, and colocalization of NBR1 and optineurin was measured over a 9-h infection time course. (E) CHO cells were infected with the indicated strains, and colocalization of NDP52 to vacuoles was measured over a 9-h time course. (F) Colocalization of NDP52-positive puncta near vacuoles containing L. pneumophila strains in CHO cells. Positive recruitment was defined as proximal localization of more than 7 NDP52-positive puncta near the vacuole. The unpaired t test was used to analyze the differences between samples. ****, P < 0.0001; **, P < 0.01; *, P < 0.05; ns, not significant.
To determine if the remaining xenophagy adaptors are excluded from vacuoles containing the sde locus mutant, colocalization studies were performed. Similar to what was observed with p62, recruitment of NBR1 and optineurin was rarely detected on vacuoles (Fig. 6C and D). Additionally, most vacuoles containing the sde locus mutant were NDP52 negative (Fig. 6E). Interestingly, peripheral NDP52 staining, which was observed around vacuoles containing L. pneumophila, was not apparent around vacuoles containing the sde locus mutant (Fig. 6F), which suggests that phosphoribosyl-ubiquitination of reticulon 4 molecules may provide a signal that recruits NDP52 to membranes in close proximity to the LCV.
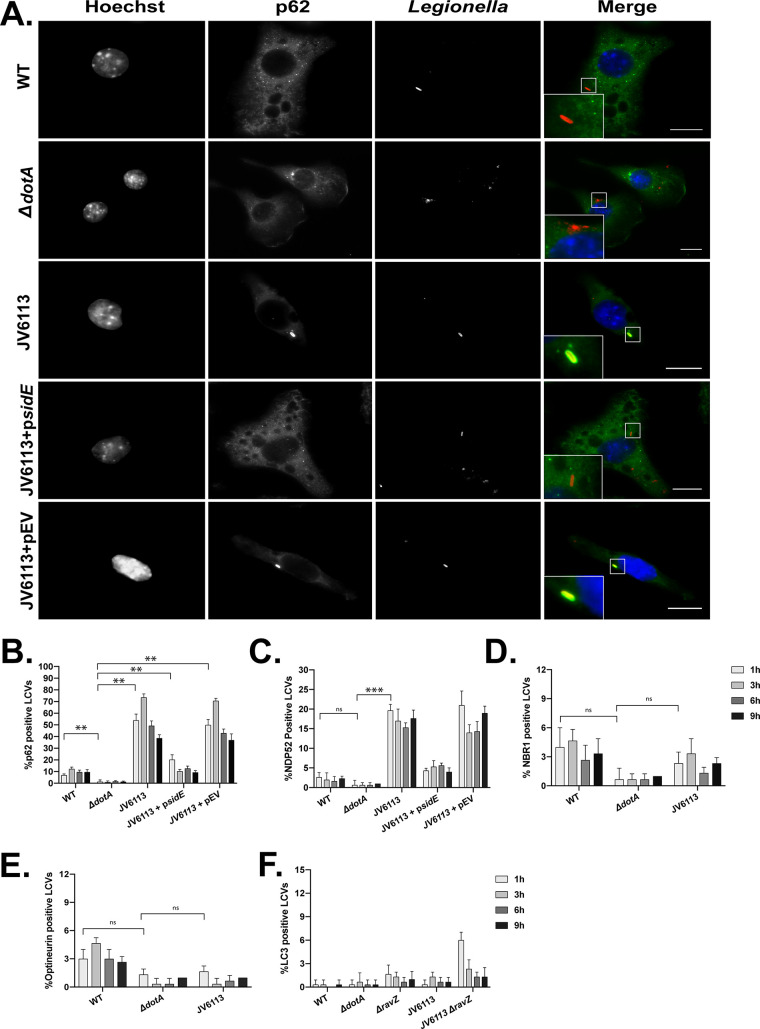
The sde locus mutant still encodes the effector protein SidE, which does not appear to mediate phosphoribosyl-ubiquitination of reticulon 4 but likely modifies other proteins on the LCV. Thus, the L. pneumophila strain JV6113, which is an isogenic mutant deficient in all of the proteins in the SidE family (41), was examined for p62 recruitment to determine if the complete absence of phosphoribosyl-ubiquitination activity had an effect on adaptor recruitment to the LCV (Fig. 7). The recruitment of p62 to vacuoles containing the mutant deficient in all SidE family members was measured over the first 9 h of infection. After 1 h of infection, roughly 50% of vacuoles containing the SidE family mutant stained positive for p62 (Fig. 7A and B). Approximately 75% of vacuoles containing the SidE family mutant stained positive for p62 at 3 h, and then p62 staining became less frequent at 6 h and 9 h after infection. The ability of the SidE family mutant to suppress p62 recruitment to the LCV was restored when a plasmid encoding the sidE gene was introduced in trans (Fig. 7A and B; JV6113 + psidE). Recruitment of the adaptor protein NDP52 was also more frequent on vacuoles containing the SidE family mutant, although the majority of vacuoles remained negative for NDP52 (Fig. 7C). Vacuoles containing the SidE family mutant did not display enhanced recruitment of the adaptor NBR1 or optineurin (Fig. 7D and E). These data suggest that phosphoribosyl-ubiquitination by SidE family members can interfere with p62 recruitment to the LCV but that additional mechanisms likely suppress the localization of other autophagy adaptors.
FIG 7.
Vacuoles containing a sidE family mutant have a defect in suppressing p62 recruitment but evade xenophagy. (A) Representative fluorescence micrographs of J774.A1 cells infected for 1 h with WT L. pneumophila, the ΔdotA mutant, the sidE family deletion mutant (JV6113), the complemented sidE family mutant (JV6113 + psidE), and the empty vector control (JV6113 + pEV). Merged panels show staining for L. pneumophila (red), p62 (green), and Hoechst (blue). Bars, 7.5 μm. (B) Colocalization of p62 to vacuoles containing the indicated L. pneumophila strains in J774.A1 cells. (C) Colocalization of NDP52 to the indicated L. pneumophila strains in CHO cells. (D) Colocalization of NBR1 to the indicated L. pneumophila strains in CHO cells. (E) Colocalization of optineurin to the indicated L. pneumophila strains in CHO cells. (F) Colocalization of LC3B to the indicated L. pneumophila strains in J774.A1 cells. For all colocalization studies, recruitment was measured over a 9-h time course at the indicated intervals. All colocalization data are representative of 3 independent experiments. For each independent experiment, approximately 100 vacuoles were scored in triplicate, for a total of 300 vacuoles per time point. The unpaired t test was used to analyze the differences between samples. ***, P < 0.001; **, P < 0.01; ns, not significant.
The localization of p62 to vacuoles containing the SidE family mutant raised the question of whether these p62 molecules at the LCV were sufficient to induce a xenophagic response. To address this question, the ravZ gene was deleted in the SidE family mutant (JV6113 ΔravZ), and the recruitment of LC3B to vacuoles containing this mutant was measured by fluorescence microscopy (Fig. 7F). There was no significant increase in LC3B recruitment to vacuoles containing the SidE family mutant (JV6113 ΔravZ) compared to vacuoles containing an isogenic ravZ mutant (Fig. 7F). Importantly, there was no significant increase in LC3B localization observed at 3 h postinfection, at which time roughly 75% of vacuoles containing the SidE family mutant were positive for p62. Thus, the increase in p62 recruitment to vacuoles that contain mutants unable to mediate phosphoribosyl-ubiquitination is not sufficient to promote xenophagic targeting of the LCV.
RavZ translocation results in a global defect in host xenophagy.
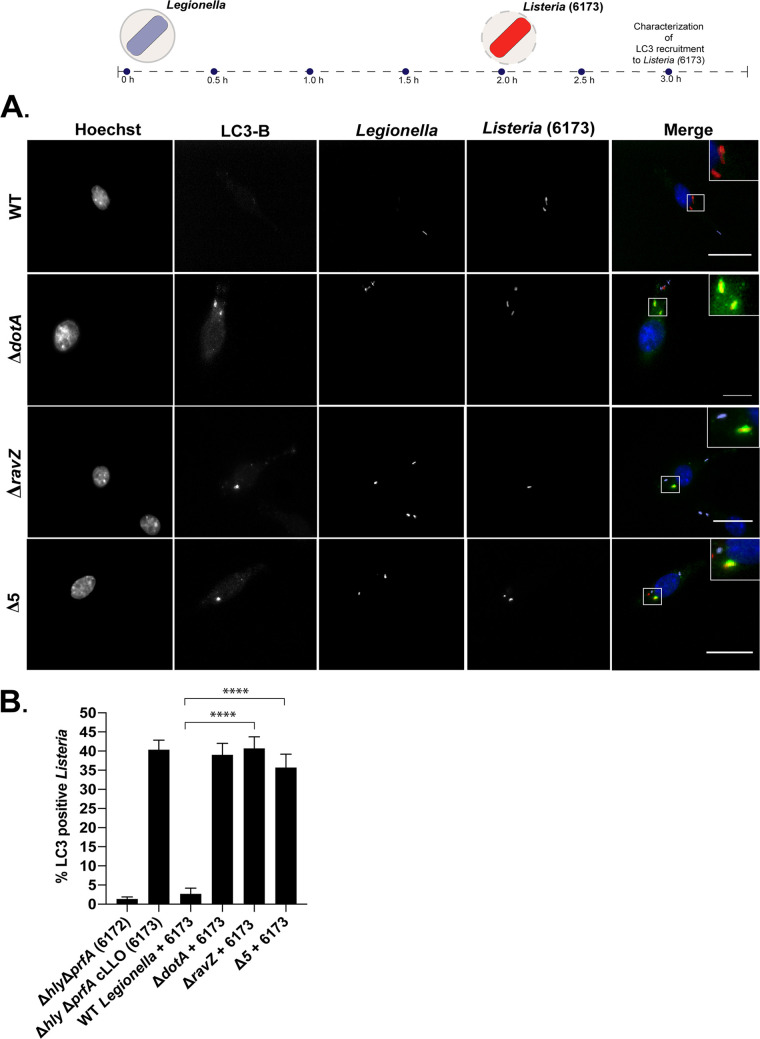
Previous studies have demonstrated that LC3-positive autophagic structures are depleted in cells infected with L. pneumophila that produce RavZ (34), which suggests that RavZ may have trans-acting effects that globally disrupt autophagy. To determine if RavZ translocated by L. pneumophila during infection globally interferes with xenophagic targeting of intracellular pathogens, host cells containing L. pneumophila were coinfected with a Listeria monocytogenes mutant that is efficiently targeted for destruction by xenophagy (26). This mutant strain of L. monocytogenes does not express most virulence proteins due to chromosomal deletions that eliminate genes encoding multiple phospholipases and the transcriptional activator protein PrfA, but it constitutively produces the major pore-forming protein listeriolysin O (LLO), which mediates escape from the phagosome after macrophage uptake (26). Thus, this Listeria strain (Δhly ΔprfA cLLO) is targeted by a robust xenophagic response following rupture of the vacuole membrane.
Single-infection experiments demonstrated that there was no recruitment of LC3B to LCVs containing either the parental strain of L. pneumophila, the ΔravZ mutant strain, or the pentuple mutant. In contrast, robust LC3B recruitment was observed around L. monocytogenes within the first 2 h of infection (Fig. 8). In cells coinfected with the parental strain of L. pneumophila and L. monocytogenes, there was a dramatic defect in LC3B recruitment around L. monocytogenes. This inhibition of LC3B recruitment was mediated by RavZ, as LC3B recruitment to L. monocytogenes was restored in cells coinfected with either the ΔravZ mutant or the pentuple mutant that is also deficient in ravZ (Fig. 8B). Thus, RavZ translocated during infection by L. pneumophila has the ability to globally suppress the xenophagy pathway from targeting intracellular pathogens.
FIG 8.
L. pneumophila organisms producing RavZ protect L. monocytogenes from xenophagic targeting. (A) Diagram showing recruitment of LC3B to L. monocytogenes 6173 in coinfected cells and representative fluorescence micrographs of J774.A1 cells coinfected with L. monocytogenes 6173 and L. pneumophila. Merged panels show staining for L. pneumophila (purple), L. monocytogenes (red), LC3B (green), and Hoechst (blue). Bars, 7.5 μm. (B) Colocalization of LC3B to L. monocytogenes strains in J774.A1 cells; recruitment was measured 1 h after L. monocytogenes invaded cells that had been infected with the indicated L. pneumophila strains. For each independent experiment, approximately 100 bacteria were scored in triplicate, for a total of 300 bacteria per time point. The unpaired t test was used to analyze the differences between samples. ****, P < 0.0001.
Legionella pneumophila inhibits recruitment of autophagy adaptors to the vacuole by a cis-acting mechanism that is RavZ independent.
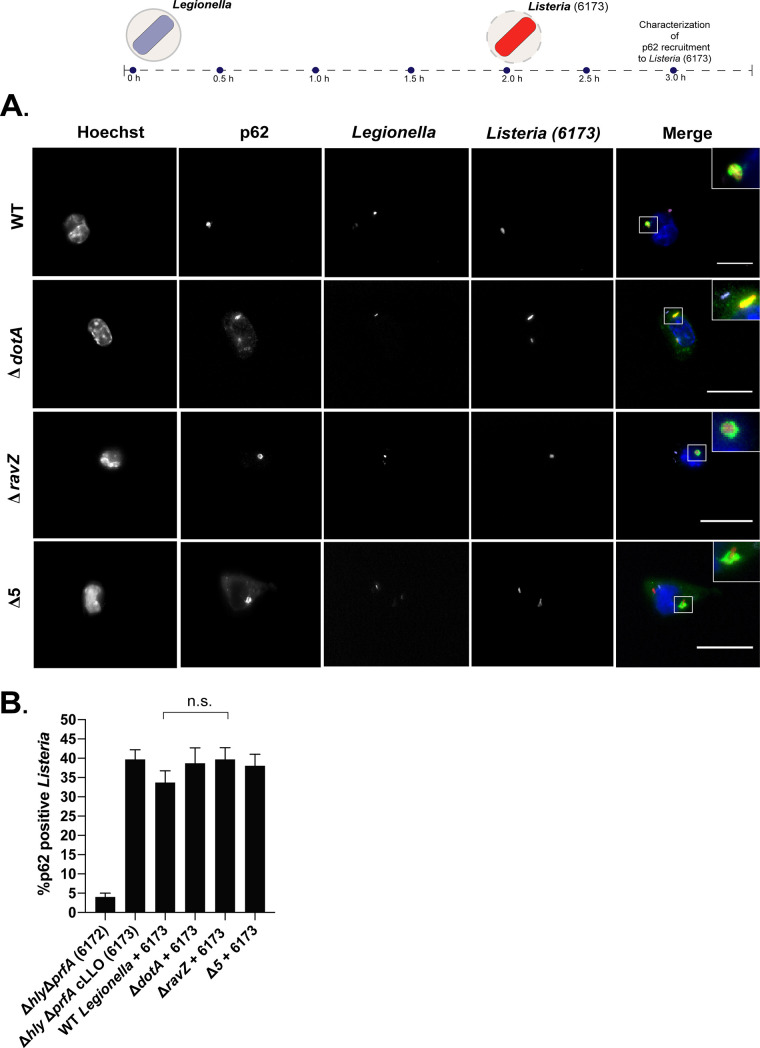
The observation that LC3B localizes to L. monocytogenes in host cells infected with a ΔravZ mutant of L. pneumophila suggested that adaptors remain functional to bind to xenophagic targets in these cells. To test this hypothesis, the recruitment of p62 to L. monocytogenes was examined in coinfected cells. These experiments revealed that p62 was recruited to L. monocytogenes in macrophages that were first infected with the parental strain of L. pneumophila producing RavZ (Fig. 9). Similar levels of p62 localization to L. monocytogenes were observed in host cells coinfected with the ΔravZ mutant and those with the pentuple mutant, which indicated that p62 recruitment to L. monocytogenes was independent of LC3B localization (Fig. 9B). Importantly, in macrophages that displayed robust localization of p62 to L. monocytogenes, there remained no evidence of intense p62 recruitment to the vacuoles containing L. pneumophila, which is consistent with what was observed during single-infection assays (Fig. 2). These data indicate that the ubiquitinated vacuole containing L. pneumophila is protected from recognition by p62 but that L. pneumophila infection does not protect other xenophagic targets from being recognized by p62 and degraded by the autophagy pathway. Thus, L. pneumophila has evolved a cis-acting mechanism to restrict p62 binding to ubiquitin-tagged LCVs.
FIG 9.
L. pneumophila infection does not disrupt recruitment of autophagy adaptors to L. monocytogenes. (A) Diagram showing recruitment of p62 to L. monocytogenes 6173 in cells coinfected with L. pneumophila and representative fluorescence micrographs of J774.A1 cells coinfected with L. monocytogenes 6173 and the indicated L. pneumophila strains. Merged panels show staining for L. pneumophila (purple), L. monocytogenes (red), p62 (green), and Hoechst (blue). Bars, 7.5 μm. (B) Colocalization of p62 to L. monocytogenes strains was measured 1 h after L. monocytogenes invaded cells infected with L. pneumophila. For each independent experiment, approximately 100 bacteria were scored in triplicate, for a total of 300 bacteria per time point. The unpaired t test was used to analyze the differences between samples. n.s., not significant.
DISCUSSION
This study has uncovered new insights about L. pneumophila autophagy avoidance through the use of adaptor localization assays and a coinfection system. These data show that xenophagy receptors are not enriched on ubiquitinated LCVs (Fig. 1 to 4). Given that adaptor recruitment is required for xenophagy, these data indicate that L. pneumophila can avoid xenophagic targeting at the stage between ubiquitin signaling and adaptor localization. Pathogens that are targeted by xenophagy acquire an activating signal such as ubiquitin, which drives the recruitment of adaptor proteins. Unlike other pathogen-occupied vacuoles, the LCV is unique in that it is not recognized by adaptor proteins that link ubiquitinated cargo with the xenophagy pathway. To date, most studies have focused on the lack of LC3B recruitment to L. pneumophila vacuoles; however, LC3B localization is a relatively late step in xenophagic targeting (14). Data here revealed that xenophagic avoidance by L. pneumophila occurred at multiple stages in the xenophagy pathway. The effector RavZ inhibited xenophagy globally in infected cells. Importantly, these data revealed that even in the absence of RavZ, L. pneumophila can suppress the xenophagic response at a stage between the ubiquitin labeling of the LCV and the binding of xenophagic adaptors to the ubiquitin-coated vacuole. Suppression of adaptor recruitment likely represents a mechanism by which L. pneumophila strains that do not encode RavZ are able to avoid xenophagic targeting.
A coinfection system was used to determine if the lack of adaptor recruitment to LCVs was specific to vacuoles containing L. pneumophila or due to global suppression of adaptor function. To date, p62/SQSTM1 is the only member of the adaptor family to be implicated in several antibacterial processes. Recruitment of p62 to vacuoles containing L. pneumophila encoding a functional Dot/Icm system was observed infrequently, even though most of these vacuoles displayed an intense ubiquitin signature, which indicated that L. pneumophila has the capacity to suppress p62 recognition of the ubiquitin molecules associated with the vacuole. When recruitment of p62 and LC3B to Listeria monocytogenes was investigated under coinfection conditions, two observations were made that further defined the processes that enable L. pneumophila to avoid xenophagic targeting. First, in coinfected cells, translocation of RavZ was found to block LC3B recruitment to L. monocytogenes (Fig. 8). This demonstrates that RavZ translocated at physiological levels will globally disrupt autophagic processes induced during L. pneumophila infection and that this includes xenophagy. Second, coinfection experiments revealed that p62 recruitment to L. monocytogenes was not blocked in cells infected with L. pneumophila (Fig. 9). At the molecular level, this reveals that host ubiquitin conjugation systems have the ability to detect pathogen-occupied vacuoles containing either L. pneumophila or L. monocytogenes. However, L. pneumophila utilizes a cis-acting mechanism to prevent xenophagic adaptors from being recruited to the ubiquitin-positive vacuole in which it resides and not from other xenophagic targets in the cell, such as L. monocytogenes. Thus, this cis-acting mechanism can protect L. pneumophila from a xenophagic response without altering the host cells’ ability to target other invading pathogens.
This study also revealed an atypical pattern of NDP52 distribution that was observed along the perimeter of the LCV (Fig. 4 and 6). Importantly, this peripheral distribution of NDP52 was dependent on the phosphoribosyl-ubiquitination activity of the Sde proteins. A recent study reported that NDP52 is required to engage protein complexes that initiate the early stages of autophagosome formation, such as FIP200 and SINTBAD/NAP1 (42). Thus, one possible explanation is that this distinct pattern of NDP52 localization could indicate sites of localized autophagosome nucleation near the vacuole in which L. pneumophila resides. Because the punctate distribution of NDP52 around the vacuole was significantly reduced during infection by a mutant that does not produce the Sde proteins, it is possible that phosphoribosyl-ubiquitination of reticulon 4 by the Sde proteins drives the recruitment of NDP52 to membranes near the vacuole. In this scenario, the effectors that act in cis on the LCV membrane would prevent the pathogen-occupied organelle from being recognized by adaptor proteins. Further experimentation is required to determine what drives this atypical distribution of NDP52 near the vacuole during infection and whether phosphoribosyl-ubiquitination of reticulon 4 is required.
Many intracellular pathogens have evolved mechanisms to coopt the host ubiquitin system and modulate ubiquitin signaling to dampen or suppress cell-intrinsic immune responses (43–45). Because L. pneumophila encodes a number of effectors that either mimic or hijack the host ubiquitin system (46), it is possible that cis-acting effectors may have novel activities that modify the ubiquitin coat to disrupt efficient recruitment of adaptors to the vacuole. The SidE family of effectors were of particular interest because they are capable of introducing an atypical phosphoribosyl-ubiquitin linkage onto proteins associated with the LCV (38, 40) and have also been proposed to poison the host ubiquitin system in infected cells by modifying all available ubiquitin monomers to phosphoribosyl-ubiquitin (39). Thus, it was important to determine if mutants deficient in these effectors would occupy vacuoles that were capable of recruiting xenophagic adaptors.
These data showed that a mutant deficient in the three Sde proteins retained the ability to suppress the recruitment of p62 to the ubiquitinated LCV (Fig. 6A and B). Unexpectedly, when the SidE protein was deleted in addition to these Sde proteins, there was robust recruitment of p62 to the vacuoles containing this SidE family mutant (Fig. 7A and B). Importantly, the percentage of vacuoles containing the SidE family mutant that were p62 positive was similar to the percentage of vacuoles that stained positive for ubiquitin (Fig. 1). Thus, phosphoribosyl-ubiquitination of proteins by members of the SidE family of effectors plays a role in preventing the accumulation of p62 on the LCV. Although the mechanism by which this occurs is unknown, these coinfection data indicate that the SidE family of proteins act locally on the LCV and do not globally poison the ubiquitin system during infection, as L. monocytogenes was efficiently targeted for xenophagic recognition in cells that were infected with L. pneumophila producing the SidE family of proteins (Fig. 8).
The SidE family of effectors were important for exclusion of p62 from the LCV, but these vacuoles were still not efficiently recognized by other adaptors that participate in xenophagy (Fig. 7C to E). Additionally, the accumulation of p62 on vacuoles containing the SidE family mutant was not sufficient to promote the recruitment of autophagic membranes to the LCV (Fig. 7F). This suggests that L. pneumophila has evolved additional mechanisms to avoid xenophagic capture. Additional studies are needed both to understand how phosphoribosyl-ubiquitination by the SidE family of effectors can prevent p62 accumulation at the LCV and to identify other cis-acting effector proteins that participate in xenophagic avoidance.
MATERIALS AND METHODS
Bacterial strains and cell lines.
Legionella pneumophila serogroup 1 (Lp01) strains used in this study (Table 1) were cultured on supplemented charcoal yeast extract (CYE) plates at 37°C as described previously (47, 48). CYE medium is composed of 1% yeast extract, 1% N-(2-acetamido)-2-aminoethanesulfonic acid (ACES; pH 6.9), 3.3 mM l-cysteine, 0.33 mM Fe(NO3)3, 1.5% Bacto agar, and 0.2% activated charcoal. When necessary, CYE was supplemented with streptomycin (100 μg/ml) or thymidine (100 μg/ml). For coinfection experiments, mono-DsRed-expressing Legionella strains were used (49) and were cultured in the presence of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) to activate DsRed expression and chloramphenicol (100 μg/ml). Listeria monocytogenes strains were generously provided by Dan Portnoy (University of California, Berkeley). L. monocytogenes strains were cultured in brain heart infusion (BHI) medium at 30°C overnight to reach post-exponential phase as previously described (26). Infections were performed by diluting overnight samples in fresh BHI, until an optical density at 600 nm (OD600) between 0.9 and 1.2 was reached. Samples were then subjected to centrifugation to remove the growth medium, and the pellet was rinsed in phosphate-buffered saline (PBS) prior to infection. All experiments described in this study included the use of J774.A1 mouse macrophages (ATCC) and Chinese hamster ovary (CHO) cells (ATCC). Both J774.A1 and CHO cells were cultured in RPMI medium supplemented with 10% fetal bovine serum (FBS) and maintained at 37°C and 5% CO2.
TABLE 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Description | Reference(s) |
|---|---|---|
| Lp01 (WT) | Legionella pneumophila Philadelphia 1; rpsL hsdR | 4 |
| ΔdotA mutant | Lp01, avirulent mutant defective in effector secretion; ΔdotA | 4 |
| ΔravZ mutant | Lp01, ΔravZ | 34 |
| Δ5 mutant | Lp02, pentuple mutant from which 18.5% of genome has been genetically removed, which includes approx 31% of Dot/Icm substrates; Δ(lpg1603-lpg1686) Δ(lpg1104-lpg1128) Δ(lpg1136-lpg1169) Δ(lpg1933-lpg1999) Δ(lpg2369-lpg2465) Δ(lpg2508-lpg2573) | 37 |
| sde locus mutant | Lp02, sdeC-sdeA deletion; Lux+ Δ(sdeC-sdeA) [Δ(lpg2153-lpg2157)] Kanr PahpC::lux; KK034 | 40 |
| JV6113 | Lp02, sidE family deletion; ΔsidE ΔsdeC ΔsdeB-A [Δlpg0234 Δlpg2153 Δ(lpg2156-lpg2157)] | 40, 41 |
| JV6113+pEV | Lp02, sidE family deletion with empty vector plasmid; ΔsidE ΔsdeC ΔsdeB-A [Δlpg0234, Δlpg2153 Δ(lpg2156-lpg2157)] + pJB1806 | This study |
| JV6113+psidE | Lp02, sidE family deletion with sidE (lpg0234) complementation plasmid; ΔsidE ΔsdeC ΔsdeB-A [Δlpg0234 Δlpg2153 Δ(lpg2156-lpg2157)] + pJB1806:lpg0234(sidE) | |
| JV6113 ΔravZ | Lp02, sidE family deletion with ravZ deletion; ΔsidE ΔsdeC ΔsdeB-A [Δlpg0234 Δlpg2153 Δ(lpg2156-lpg2157)] + ΔravZ | This study |
| 6172 | Listeria monocytogenes, deficient in expression of PrfA-regulated virulence factors; Δhly ΔprfA, with the empty integrated vector pHpPL3 | 26 |
| 6173 | Listeria monocytogenes, deficient in expression of PrfA-regulated virulence factors; lyses phagosome through ectopic and constitutive expression of cLLO (hly); Δhly ΔprfA+cLLO pHpPL3-hly | 26 |
| psidE | sidE complementation plasmid; pJB1806:lpg0234 (sidE) | This study |
| ravZ deletion construct | Construct generated to genetically remove ravZ from recipient strains; λpir; pSRS47s harboring flanking genetic region | 34 |
Molecular cloning, plasmid construction, and deletion experiments.
For complementation experiments, lpg0234 was amplified from Lp01 genomic DNA using the primer set TM1F (GACTCTAGAGGATCCCCGGGTACCGAGCTCGAATTCgtgctgatttttaagtcac) and TM1R (GAGCGGATAACAATTTCACACAGGAAACAGAATTCttacaatcttaagagagttc). These primers were used to generate the complementation plasmid. The sequence- and ligation-independent cloning (SLIC) method (50) was used to insert lpg0243 into the EcoRI site of pJB1806. The resulting plasmid (psidE) was then transformed into electrocompetent L. pneumophila, which was grown on selective medium and subsequently sequenced to confirm the genotype of the final product (Keck DNA Sequencing Facility, Yale University). To delete ravZ (lpg1683) from the chromosome, a deletion construct expressing flanking genetic regions (pSRS47s::ravZ) (34) was used. Allelic exchange was performed as previously described (51), and sucrose-resistant, kanamycin-sensitive colonies were screened by PCR to validate proper gene deletion. In addition, phenotypic analysis was performed to confirm removal of ravZ by qualitatively assessing the presence of LC3B puncta in infected macrophages by immunofluorescence microscopy.
Single-infection and coinfection experiments.
Single-infection experiments with Legionella strains were performed at a multiplicity of infection (MOI) of 25; under certain conditions, thymidine (100 μg ml−1) was added to the infection. Single-infection experiments with L. monocytogenes strains were performed at a MOI of 1, and at 30 min postinfection (p.i.), cells were rinsed in PBS to remove extracellular bacteria and resuspended in RPMI supplemented with gentamicin (50 μg ml−1) as described previously (26). For coinfection experiments, J774.A1 cells were first infected with DsRed-expressing Legionella for 2 h at an MOI of 50, and IPTG was added at a final concentration of 1 mM to induce the expression of DsRed. After 2 h, L. monocytogenes strains (OD600, 0.9 to 1.2) were added to Legionella-infected cells at an MOI of 1. Coinfected samples were incubated at 37°C for an additional 30 min to allow uptake. To remove extracellular bacteria, the coinfected cells were rinsed in PBS and the medium was replaced with RPMI supplemented with gentamicin (50 μg ml−1), and the coinfection was allowed to proceed for an additional 60 min.
Indirect immunofluorescence and image analysis.
J774.A1 and CHO cells were plated in RPMI supplemented with 10% FBS at a concentration of 1.5 × 105 cells per well on coverslips treated with 0.02 mM poly-l-lysine (Sigma-Aldrich). Legionella pneumophila and L. monocytogenes samples were prepared as described above for both single-infection and coinfection experiments. For LC3B and NBR1 localization assays, coverslips were fixed and permeabilized with cold methanol on ice for 60 s. For detection of endogenous p62/SQSTM1, NDP52, and optineurin, samples were fixed with 8% paraformaldehyde (PFA) on ice for 10 min and subsequently treated with 0.4% Triton (Sigma-Aldrich). Ubiquitinated proteins were detected by fixing samples in 4% PFA and permeabilizing with cold methanol. Following fixation, coverslips were thoroughly rinsed in PBS (Gibco) and blocked for 1 h in 2% bovine serum albumin (BSA) (suspended in PBS) at room temperature. Then coverslips were incubated in each respective primary antibody diluted in blocking solution for 1 h, rinsed in PBS, and incubated with secondary antibodies for 30 min. Next, coverslips were rinsed in PBS and mounted onto glass slides using Prolong Gold antifade reagent (Life Technologies).
Primary antibodies and dilutions used were 1:5,000 rabbit anti-Legionella (Abcam), 1:4,000 rabbit anti-Listeria (Abcam), 1:200 mouse anti-LC3B (2G6; Nanotools), 1:300 mouse anti-SQSTM1/p62 (BD Biosciences), 1:300 mouse anti-NBR1 (BD Biosciences), 1:300 mouse antioptineurin (BD Biosciences), 1:300 mouse anti-NDP52 (Abnova), and 1:100 fluorescein-labeled mono- and polyubiquitinated conjugates (Enzo Life Sciences). The following secondary antibodies were used: Alexa Fluor 488-conjugated goat anti-mouse immunoglobulin, Alexa Fluor 568-conjugated goat anti-rabbit immunoglobulin, and Alexa Fluor 647-conjugated goat anti-rabbit immunoglobulin. All secondary antibodies were obtained from Life Technologies and used at a final concentration of 1:2,000. Hoechst (Abcam) or DAPI (4′,6-diamidino-2-phenylindole; Abcam) was added to secondary-antibody solutions at a final concentration of 1:10,000. The samples were then imaged on a Nikon Eclipse TE2000-S inverted fluorescence microscope with a 100×/1.4 numerical aperture lens objective and a Photometrics CoolSNAP EZ camera. SlideBook (version 6.2) software was used for image acquisition and analysis, and ImageJ (52) was used for final analysis of all images. For deconvolved images, 4-μm z-projections were deconvolved with SlideBook software.
Colocalization quantification and statistical analysis.
For single-infection experiments, the colocalization of endogenous host proteins to the LCV was quantified by scoring 100 infected cells. Average colocalization was quantified from three technical replicates for each time point, for a total of 300 samples for statistical analysis, and three independent iterations were performed. For NDP52 punctate recruitment, positive recruitment was defined as proximal localization of more than 7 NDP52-positive puncta near the vacuole, whereas NDP52 recruitment was defined by conventional standards, which qualitatively measure colocalization between endogenous NDP52 and bacterial surface markers (e.g., Legionella-containing vacuole). For coinfection assays, coinfected cells were defined as cells containing no more than 3 bacteria per cell. Coinfected cells were then scored for recruitment of autophagy proteins (p62 or LC3-B) to L. monocytogenes (strains 6172 and 6173); 100 coinfected cells were scored in triplicate for a total of 300 cells. Three independent iterations were performed for both experimental setups. Data from quantitative indirect immunofluorescence were analyzed using Prism GraphPad 8. Data are presented as means ± standard deviations, and the statistical significance was analyzed by Student's t test, as indicated in the figure legends. A P value of ≤0.05 was considered to indicate statistical significance. The unpaired t test was used to analyze the differences between samples.
ACKNOWLEDGMENTS
We thank Dan Portnoy and Gabriel Mitchell for generously providing all Listeria strains used in this study. We are also grateful to Ralph Isberg for providing both the Δ(sdeC-sdeA) locus strain and the JV6113 strain. We thank all members of the Roy lab for their insight, especially Sandhya Ganesan. We also thank Jorge Galan, Barbara Kazmierczak, and Hesper Rego for their constructive feedback.
This work was supported by NIH grant R37AI041699 (C.R.R.) and a Howard Hughes Medical Institute (HHMI) Gilliam predoctoral fellowship (T.O.O.).
REFERENCES
- 1.Fraser DW, Tsai TR, Orenstein W, Parkin WE, Beecham HJ, Sharrar RG, Harris J, Mallison GF, Martin SM, McDade JE, Shepard CC, Brachman PS. 1977. Legionnaires’ disease: description of an epidemic of pneumonia. N Engl J Med 297:1189–1197. doi: 10.1056/NEJM197712012972201. [DOI] [PubMed] [Google Scholar]
- 2.Hubber A, Roy CR. 2010. Modulation of host cell function by Legionella pneumophila type IV effectors. Annu Rev Cell Dev Biol 26:261–283. doi: 10.1146/annurev-cellbio-100109-104034. [DOI] [PubMed] [Google Scholar]
- 3.Horwitz MA. 1983. Formation of a novel phagosome by the Legionnaires’ disease bacterium (Legionella pneumophila) in human monocytes. J Exp Med 158:1319–1331. doi: 10.1084/jem.158.4.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berger KH, Isberg RR. 1993. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol Microbiol 7:7–19. doi: 10.1111/j.1365-2958.1993.tb01092.x. [DOI] [PubMed] [Google Scholar]
- 5.Horwitz MA. 1987. Characterization of avirulent mutant Legionella pneumophila that survive but do not multiply within human monocytes. J Exp Med 166:1310–1328. doi: 10.1084/jem.166.5.1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kagan JC, Roy CR. 2002. Legionella phagosomes intercept vesicular traffic from endoplasmic reticulum exit sites. Nat Cell Biol 4:945–954. doi: 10.1038/ncb883. [DOI] [PubMed] [Google Scholar]
- 7.Swanson MS, Isberg RR. 1995. Association of Legionella pneumophila with the macrophage endoplasmic reticulum. Infect Immun 63:3609–3620. doi: 10.1128/IAI.63.9.3609-3620.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Isaac DT, Isberg R. 2014. Master manipulators: an update on Legionella pneumophila Icm/Dot translocated substrates and their host targets. Future Microbiol 9:343–359. doi: 10.2217/fmb.13.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vogel JP, Andrews HL, Wong SK, Isberg RR. 1998. Conjugative transfer by the virulence system of Legionella pneumophila. Science 279:873–876. doi: 10.1126/science.279.5352.873. [DOI] [PubMed] [Google Scholar]
- 10.Randow F. 2011. How cells deploy ubiquitin and autophagy to defend their cytosol from bacterial invasion. Autophagy 7:304–309. doi: 10.4161/auto.7.3.14539. [DOI] [PubMed] [Google Scholar]
- 11.Zheng YT, Shahnazari S, Brech A, Lamark T, Johansen T, Brumell JH. 2009. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J Immunol 183:5909–5916. doi: 10.4049/jimmunol.0900441. [DOI] [PubMed] [Google Scholar]
- 12.Birmingham CL, Smith AC, Bakowski MA, Yoshimori T, Brumell JH. 2006. Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole. J Biol Chem 281:11374–11383. doi: 10.1074/jbc.M509157200. [DOI] [PubMed] [Google Scholar]
- 13.Mizushima N. 2007. Autophagy: process and function. Genes Dev 21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 14.He C, Klionsky DJ. 2009. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet 43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. 2004. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci 117:2805–2812. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- 16.Mizushima N, Yoshimori T, Ohsumi Y. 2011. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol 27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 17.Alemu EA, Lamark T, Torgersen KM, Birgisdottir AB, Larsen KB, Jain A, Olsvik H, Overvatn A, Kirkin V, Johansen T. 2012. ATG8 family proteins act as scaffolds for assembly of the ULK complex: sequence requirements for LC3-interacting region (LIR) motifs. J Biol Chem 287:39275–39290. doi: 10.1074/jbc.M112.378109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kohler LJ, Roy CR. 2017. Autophagic targeting and avoidance in intracellular bacterial infections. Curr Opin Microbiol 35:36–41. doi: 10.1016/j.mib.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noad J, von der Malsburg A, Pathe C, Michel MA, Komander D, Randow F. 2017. LUBAC-synthesized linear ubiquitin chains restrict cytosol-invading bacteria by activating autophagy and NF-kappaB. Nat Microbiol 2:17063. doi: 10.1038/nmicrobiol.2017.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Franco LH, Nair VR, Scharn CR, Xavier RJ, Torrealba JR, Shiloh MU, Levine B. 2017. The ubiquitin ligase Smurf1 functions in selective autophagy of Mycobacterium tuberculosis and anti-tuberculous host defense. Cell Host Microbe 21:59–72. doi: 10.1016/j.chom.2016.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitchell G, Cheng MI, Chen C, Nguyen BN, Whiteley AT, Kianian S, Cox JS, Green DR, McDonald KL, Portnoy DA. 2018. Listeria monocytogenes triggers noncanonical autophagy upon phagocytosis, but avoids subsequent growth-restricting xenophagy. Proc Natl Acad Sci U S A 115:E210–E217. doi: 10.1073/pnas.1716055115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johansen T, Lamark T. 2011. Selective autophagy mediated by autophagic adapter proteins. Autophagy 7:279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, Richter B, Korac J, Waidmann O, Choudhary C, Dotsch V, Bumann D, Dikic I. 2011. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 333:228–233. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamark T, Kirkin V, Dikic I, Johansen T. 2009. NBR1 and p62 as cargo receptors for selective autophagy of ubiquitinated targets. Cell Cycle 8:1986–1990. doi: 10.4161/cc.8.13.8892. [DOI] [PubMed] [Google Scholar]
- 25.Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. 2009. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol 10:1215–1221. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- 26.Mitchell G, Ge L, Huang Q, Chen C, Kianian S, Roberts MF, Schekman R, Portnoy DA. 2015. Avoidance of autophagy mediated by PlcA or ActA is required for Listeria monocytogenes growth in macrophages. Infect Immun 83:2175–2184. doi: 10.1128/IAI.00110-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mostowy S, Sancho-Shimizu V, Hamon MA, Simeone R, Brosch R, Johansen T, Cossart P. 2011. p62 and NDP52 proteins target intracytosolic Shigella and Listeria to different autophagy pathways. J Biol Chem 286:26987–26995. doi: 10.1074/jbc.M111.223610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barnett TC, Liebl D, Seymour LM, Gillen CM, Lim JY, Larock CN, Davies MR, Schulz BL, Nizet V, Teasdale RD, Walker MJ. 2013. The globally disseminated M1T1 clone of group A Streptococcus evades autophagy for intracellular replication. Cell Host Microbe 14:675–682. doi: 10.1016/j.chom.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang R, Varela M, Vallentgoed W, Forn-Cuni G, van der Vaart M, Meijer AH. 2019. The selective autophagy receptors optineurin and p62 are both required for zebrafish host resistance to mycobacterial infection. PLoS Pathog 15:e1007329. doi: 10.1371/journal.ppat.1007329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thurston TL, Boyle KB, Allen M, Ravenhill BJ, Karpiyevich M, Bloor S, Kaul A, Noad J, Foeglein A, Matthews SA, Komander D, Bycroft M, Randow F. 2016. Recruitment of TBK1 to cytosol-invading Salmonella induces WIPI2-dependent antibacterial autophagy. EMBO J 35:1779–1792. doi: 10.15252/embj.201694491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sherwood RK, Roy CR. 2013. A Rab-centric perspective of bacterial pathogen-occupied vacuoles. Cell Host Microbe 14:256–268. doi: 10.1016/j.chom.2013.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dorer MS, Kirton D, Bader JS, Isberg RR. 2006. RNA interference analysis of Legionella in Drosophila cells: exploitation of early secretory apparatus dynamics. PLoS Pathog 2:e34. doi: 10.1371/journal.ppat.0020034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ivanov SS, Roy CR. 2009. Modulation of ubiquitin dynamics and suppression of DALIS formation by the Legionella pneumophila Dot/Icm system. Cell Microbiol 11:261–278. doi: 10.1111/j.1462-5822.2008.01251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choy A, Dancourt J, Mugo B, O'Connor TJ, Isberg RR, Melia TJ, Roy CR. 2012. The Legionella effector RavZ inhibits host autophagy through irreversible Atg8 deconjugation. Science 338:1072–1076. doi: 10.1126/science.1227026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horenkamp FA, Kauffman KJ, Kohler LJ, Sherwood RK, Krueger KP, Shteyn V, Roy CR, Melia TJ, Reinisch KM. 2015. The Legionella anti-autophagy effector RavZ targets the autophagosome via PI3P- and curvature-sensing motifs. Dev Cell 34:569–576. doi: 10.1016/j.devcel.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rolando M, Escoll P, Buchrieser C. 2016. Legionella pneumophila restrains autophagy by modulating the host’s sphingolipid metabolism. Autophagy 12:1053–1054. doi: 10.1080/15548627.2016.1166325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Connor TJ, Adepoju Y, Boyd D, Isberg RR. 2011. Minimization of the Legionella pneumophila genome reveals chromosomal regions involved in host range expansion. Proc Natl Acad Sci U S A 108:14733–14740. doi: 10.1073/pnas.1111678108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qiu J, Sheedlo MJ, Yu K, Tan Y, Nakayasu ES, Das C, Liu X, Luo ZQ. 2016. Ubiquitination independent of E1 and E2 enzymes by bacterial effectors. Nature 533:120–124. doi: 10.1038/nature17657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhogaraju S, Kalayil S, Liu Y, Bonn F, Colby T, Matic I, Dikic I. 2016. Phosphoribosylation of ubiquitin promotes serine ubiquitination and impairs conventional ubiquitination. Cell 167:1636–1649.e1613. doi: 10.1016/j.cell.2016.11.019. [DOI] [PubMed] [Google Scholar]
- 40.Kotewicz KM, Ramabhadran V, Sjoblom N, Vogel JP, Haenssler E, Zhang M, Behringer J, Scheck RA, Isberg RR. 2017. A single Legionella effector catalyzes a multistep ubiquitination pathway to rearrange tubular endoplasmic reticulum for replication. Cell Host Microbe 21:169–181. doi: 10.1016/j.chom.2016.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jeong KC, Sexton JA, Vogel JP. 2015. Spatiotemporal regulation of a Legionella pneumophila T4SS substrate by the metaeffector SidJ. PLoS Pathog 11:e1004695. doi: 10.1371/journal.ppat.1004695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ravenhill BJ, Boyle KB, von Muhlinen N, Ellison CJ, Masson GR, Otten EG, Foeglein A, Williams R, Randow F. 2019. The cargo receptor NDP52 initiates selective autophagy by recruiting the ULK complex to cytosol-invading bacteria. Mol Cell 74:320–329.e6. doi: 10.1016/j.molcel.2019.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mesquita FS, Thomas M, Sachse M, Santos AJ, Figueira R, Holden DW. 2012. The Salmonella deubiquitinase SseL inhibits selective autophagy of cytosolic aggregates. PLoS Pathog 8:e1002743. doi: 10.1371/journal.ppat.1002743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kamanova J, Sun H, Lara-Tejero M, Galán JE. 2016. The Salmonella effector protein SopA modulates innate immune responses by targeting TRIM E3 ligase family members. PLoS Pathog 12:e1005552. doi: 10.1371/journal.ppat.1005552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Jong MF, Liu Z, Chen D, Alto NM. 2016. Shigella flexneri suppresses NF-κB activation by inhibiting linear ubiquitin chain ligation. Nat Microbiol 1:16084. doi: 10.1038/nmicrobiol.2016.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hubber A, Kubori T, Nagai H. 2013. Modulation of the ubiquitination machinery by Legionella. Curr Top Microbiol Immunol 376:227–247. doi: 10.1007/82_2013_343. [DOI] [PubMed] [Google Scholar]
- 47.Feeley JC, Gibson RJ, Gorman GW, Langford NC, Rasheed JK, Mackel DC, Baine WB. 1979. Charcoal-yeast extract agar: primary isolation medium for Legionella pneumophila. J Clin Microbiol 10:437–441. doi: 10.1128/JCM.10.4.437-441.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saito A, Rolfe RD, Edelstein PH, Finegold SM. 1981. Comparison of liquid growth media for Legionella pneumophila. J Clin Microbiol 14:623–627. doi: 10.1128/JCM.14.6.623-627.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ivanov SS, Charron G, Hang HC, Roy CR. 2010. Lipidation by the host prenyltransferase machinery facilitates membrane localization of Legionella pneumophila effector proteins. J Biol Chem 285:34686–34698. doi: 10.1074/jbc.M110.170746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li MZ, Elledge SJ. 2012. SLIC: a method for sequence- and ligation-independent cloning. Methods Mol Biol 852:51–59. doi: 10.1007/978-1-61779-564-0_5. [DOI] [PubMed] [Google Scholar]
- 51.Nagai H, Roy CR. 2001. The DotA protein from Legionella pneumophila is secreted by a novel process that requires the Dot/Icm transporter. EMBO J 20:5962–5970. doi: 10.1093/emboj/20.21.5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]