The intracellular bacterial pathogen Salmonella is able to evade the immune system and persist within the host. In some cases, these persistent infections are asymptomatic for long periods and represent a significant public health hazard because the hosts are potential chronic carriers, yet the mechanisms that control persistence are incompletely understood. Using a mouse model of chronic typhoid fever combined with major histocompatibility complex (MHC) class II tetramers to interrogate endogenous, Salmonella-specific CD4+ helper T cells, we show that certain host microenvironments may favorably contribute to a pathogen’s ability to persist in vivo.
KEYWORDS: CD4 T cell, Salmonella, immune regulation, liver immunology, macrophages
ABSTRACT
The intracellular bacterial pathogen Salmonella is able to evade the immune system and persist within the host. In some cases, these persistent infections are asymptomatic for long periods and represent a significant public health hazard because the hosts are potential chronic carriers, yet the mechanisms that control persistence are incompletely understood. Using a mouse model of chronic typhoid fever combined with major histocompatibility complex (MHC) class II tetramers to interrogate endogenous, Salmonella-specific CD4+ helper T cells, we show that certain host microenvironments may favorably contribute to a pathogen’s ability to persist in vivo. We demonstrate that the environment in the hepatobiliary system may contribute to the persistence of Salmonella enterica subsp. enterica serovar Typhimurium through liver-resident immunoregulatory CD4+ helper T cells, alternatively activated macrophages, and impaired bactericidal activity. This contrasts with lymphoid organs, such as the spleen and mesenteric lymph nodes, where these same cells appear to have a greater capacity for bacterial killing, which may contribute to control of bacteria in these organs. We also found that, following an extended period of infection of more than 2 years, the liver appeared to be the only site that harbored Salmonella bacteria. This work establishes a potential role for nonlymphoid organ immunity in regulating chronic bacterial infections and provides further evidence for the hepatobiliary system as the site of chronic Salmonella infection.
INTRODUCTION
Typhoid fever is caused by the intracellular bacterium Salmonella enterica subsp. enterica serovar Typhi, with an estimated global burden of over 27 million cases annually and a case mortality of about 1% (1). For reasons not fully understood, it is estimated that 5% of typhoid patients fail to clear the infection within a year, progressing instead to a chronic carrier state. These individuals are usually asymptomatic, and survival of the bacterium is likely to involve successful localization to a permissive niche within the host. In humans, it is believed that this niche is primarily within macrophages of the hepatobiliary system and gallstones in the gallbladder (2). This observation is also supported by data collected from S. Typhi-positive cadavers showing S. Typhi present in 85.7% of liver tissues (3). The potential maintenance of Salmonella infection in the hepatobiliary system is perhaps not surprising, as the liver is an immune-tolerogenic organ that can be transplanted without immunosuppressive drugs, whereas other organs would be rejected; this phenomenon may be coordinated by CD4+ regulatory T (Treg) cells expressing the Fas ligand protein (4, 5). It has been suggested that anti-inflammatory mediators, such as interleukin 10 (IL-10), are responsible for restricting unnecessary inflammation in the liver and may contribute to persistence of certain infections, such as those with hepatitis virus (6–10). The typhoid carrier state is difficult to identify and treat because affected individuals are normally asymptomatic, and antibiotic treatment is often ineffective, possibly leading to increased bacterial shedding in the feces. Therefore, a better understanding of the physiological mechanisms that define the carrier state is needed, as these individuals remain a reservoir of Salmonella transmission. Furthermore, increased comprehension of the role of the immune response against bacteria during this persistent state has the potential to contribute to novel treatment and clearance of chronic infection, breaking this cycle of transmission.
Engagement of the adaptive immune response is critical for Salmonella clearance during persistent infection. In particular, CD4+ T cells seem to be required for preventing the increase in bacterial burden and mortality (11–13). The protective role of CD4+ T cells against Salmonella infection is attributed to the production of inflammatory cytokines, such as gamma interferon (IFN-γ) and tumor necrosis factor alpha (TNF-α) (14–19). Furthermore, studies using mice that lack Th1 cells due to a deficiency in the canonical Th1 transcription factor T-bet have confirmed that these CD4+ T cells are necessary to resolve a primary Salmonella infection (20). The balance of cytokines can potentially dictate the outcome of Salmonella infection, in which the antimicrobial cytokine IFN-γ activates macrophages to control intracellular replication (21, 22), while the anti-inflammatory cytokine IL-10 decreases the bactericidal activity of macrophages (23).
Based on the fact that CD4+ T cells are important mediators during Salmonella infection, and because the hepatobiliary system is a site of bacterial persistence, we hypothesized that Salmonella persistence may be affected by the anatomical location of the bacteria and the accompanying Salmonella-specific CD4+ T cells. We tested this hypothesis by comparing Salmonella-specific CD4+ T-cell responses between lymphoid organs and the liver to assess how these responses may affect infection outcome. Furthermore, we analyzed the influence the adaptive immune response at these infection sites may have on macrophage bactericidal activity, as well as on macrophage polarization. We show that during persistent infection, Salmonella-specific liver CD4+ T cells produce the immunosuppressive cytokine IL-10 and may contribute to bacterial persistence. The anatomical T-cell differences are reflected in differential macrophage polarization between the liver and lymphoid tissues. Finally, we show that, at extremely late time points after infection, the liver continues to harbor Salmonella bacteria. These findings suggest that the liver provides a protected niche for Salmonella persistence that is supported by the liver’s immunosuppressive environment.
RESULTS
Salmonella-specific CD4+ T cells are maintained throughout chronic infection and are important for controlling lymphoid bacterial burdens.
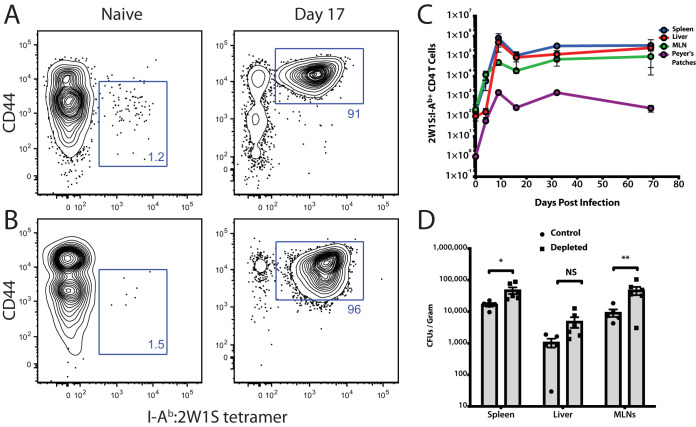
Previous studies in the field have analyzed CD4+ T cell responses in lymphoid organs during chronic Salmonella infection; however, the role of the liver during persistent infection in regulating these responses is less clear. This is particularly true in Salmonella-resistant mouse strains, such as 129/SvJ mice, which possess a functional natural resistance-associated macrophage protein 1 (NRAMP-1) gene (known as Slc11a1). In contrast, other common strains, such as C57BL/6, have a single G169D substitution in NRAMP1, leading to a nonfunctional protein, which renders these strains highly susceptible to Salmonella infection (24, 25). The current study sought to determine the host immune response against a chronic Salmonella infection in the livers of resistant mice, paying particular attention to endogenous, pathogen-specific CD4+ T cells. To date, most chronic liver immune responses have been studied in the context of viral hepatitis. There is currently a lack of knowledge about liver-specific immunity with bacterial infections, especially Salmonella. In order to define the role of CD4+ immune responses to chronic Salmonella infection in the liver, we utilized a strain of Salmonella enterica subsp. enterica serovar Typhimurium genetically tagged on the C terminus of the bacterial outer membrane porin C (OmpC) with 2W1S (EAWGALANWAVDSA), a well-known CD4+ T-cell epitope (here referred to as ST OmpC-2W1S). This strain has been used in other studies to visualize endogenous anti-2W1S CD4+ T cell responses utilizing major histocompatibility complex (MHC) class II tetramers (26–30). This engineered strain allowed us to analyze the Salmonella-specific T-cell response in multiple anatomic sites throughout the course of infection in resistant mice. In an uninfected mouse, there are approximately 100 naive 2W1S-specific CD4+ T cells in the liver (Fig. 1A). Upon oral infection with ST OmpC2W1S, liver Salmonella-specific CD4 T cells exhibit a rapid expansion in numbers similar to that seen in the lymphoid tissues. These Salmonella-specific CD4+ T cells then underwent a 70% reduction, on average, across all tissues, with numbers stabilizing as mice entered the chronic phase of infection, where they were maintained throughout (Fig. 1A).
FIG 1.
OmpC–2W1S-specific CD4+ T cells remain elevated throughout persistent Salmonella infection and are required to control bacterial burden. (A and B) Representative flow cytometry plots are shown for I-Ab:2W1S tetramer staining for naive T cells and for T cells 17 days postinfection. (A) Spleen and (B) shows liver 2W1S-specific T cells. Numbers are the percentage of cells in each gate. (C) Total Salmonella-specific CD4+ T cells (2W1S:I-Ab+) were enumerated from each organ and are shown at the indicated times postinfection. (D) Mice chronically infected with S. Typhimurium for 44 days were antibody depleted of CD4+ T cells for 12 days before bacterial burdens were assessed. Bacterial burdens were determined in each organ after CD4+ T-cell depletion. Statistical differences were determined using two-way analysis of variance (ANOVA) and a Sidak’s multiple-comparison test. *, P < 0.05; **, P < 0.01. Representative of 2 independent experiments for time course and 4 independent experiments for depletion.
Other groups have shown CD4+ T cells to be important mediators of anti-Salmonella immunity (11–13). To confirm this in our model, we allowed the infection to persist for approximately 6 weeks, at which time CD4+ T cells were depleted for 12 days using an anti-CD4 antibody (12). Mice lacking CD4+ T cells during chronic infection showed increased bacterial burdens in the lymphoid tissues compared to T cell-intact mice (Fig. 1B). Although not significant, the livers of CD4-depleted mice also demonstrated an expansion in bacterial burdens compared to T cell-intact mice.
Lymphoid Salmonella-specific CD4+/− T cells protect against infection and produce IFN-γ, while liver T cells exacerbate infection and produce both IL-10 and IFN-γ.
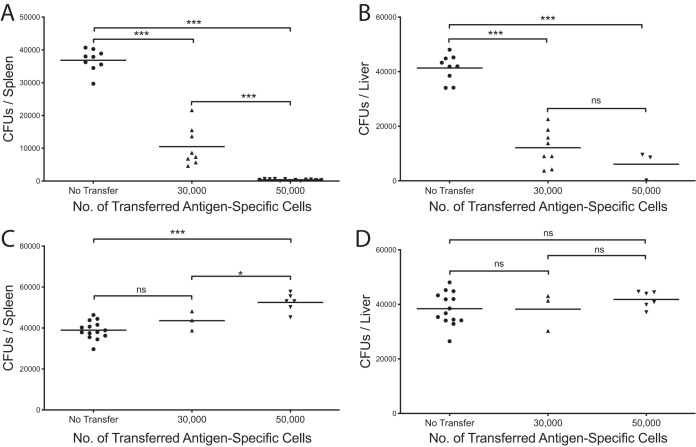
In order to evaluate potential differences between lymphoid- and nonlymphoid-derived Salmonella-specific CD4+ T cells, we employed an adoptive transfer strategy to assess whether these cells could protect mice from subsequent challenge. 2W1S-specific CD4+ T cells were harvested and enriched from either the lymphoid tissues (spleen and mesenteric lymph nodes [MLNs]) or the livers of chronically infected mice (≥40 days postinfection). Cell numbers were matched, and cells were transferred intravenously into naive animals. Mice were challenged 24 h later with ST OmpC-2W1S, and bacterial burdens were assessed 3 weeks after infection. Mice receiving CD4+ T cells from lymphoid tissues were significantly protected during the challenge, demonstrating reduced bacterial burdens in the spleen, liver, and MLNs (Fig. 2A and B). The conferred protection also showed a cellular dose-dependent response. Surprisingly, CD4+ T cells transferred from the livers of chronically infected mice failed to protect mice during the challenge and instead, at higher transfer numbers, exacerbated the infection (Fig. 2C and D). These disparities led us to next assess potential functional differences between these cells.
FIG 2.
Spleen Salmonella-specific helper T cells contribute to protection, while liver T cells contribute to exacerbation of subsequent infections. Salmonella-specific CD4+ T cells were transferred from spleens (A and B) or livers (C and D) of chronically infected mice into naive mice 24 h before bacterial challenge at the indicated numbers. Spleens (A and C) and livers (B and D) were harvested 22 days postinfection, and bacterial burdens in each organ were determined. One-way ANOVA with Tukey’s post hoc analysis was used to determine statistical significance between groups. *, P < 0.05; **, P < 0.01; ***, P < 0.001. From 2 or 3 combined independent experiments. Representative of more than 5 independent experiments.
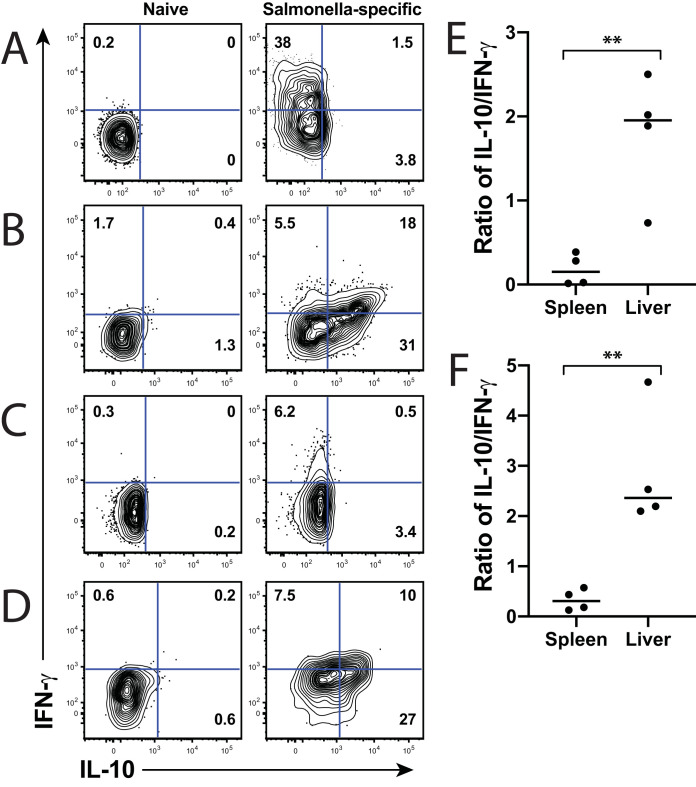
To evaluate functional differences between lymphoid and liver-derived Salmonella-specific CD4+ T cells, we initially assessed their cytokine production. Multiple studies have demonstrated that IFN-γ is a crucial cytokine for Salmonella clearance (14–16). Since liver CD4+ T cells exacerbated infection, we predicted that liver cells produced the immunosuppressive cytokine interleukin 10. As expected, Salmonella-specific CD4+ T cells predominantly produced IFN-γ during both the acute and chronic phases of infection in the lymphoid organs (Fig. 3A and C), while these same cells in the liver produced both IFN-γ and IL-10 (Fig. 3B and D). To best delineate the cytokine differences between lymphoid and liver CD4+ T cells, we normalized cytokine production by comparing the ratio of IL-10 to IFN-γ in each tissue. Higher IL-10 production would result in a ratio greater than 1.0, whereas more IFN-γ production would lead to a ratio less than 1.0, with 0.0 representing a cell that produces only IFN-γ. In both the acute (Fig. 3E) and chronic (Fig. 3F) phases of infection, the liver demonstrated a significantly higher ratio of IL-10 to IFN-γ, while the splenic T cells almost entirely produced IFN-γ. These data demonstrate that Salmonella-specific T cells from each organ are differently regulated to assume an immunosuppressive (liver) or a bactericidal (spleen) phenotype. The different phenotypes imply that the T cells from each organ may contribute to clearance in the spleen or lymph nodes and to persistence in the liver.
FIG 3.
Salmonella-specific lymphoid T cells produce predominantly IFN-γ, while liver T cells produce both IFN-γ and IL-10. (A and C) Spleens and (B and D) livers were harvested from (A and B) acutely or (C and D) chronically infected mice and stimulated with phorbol 12-myristate 13-acetate (PMA)/ionomycin for 2 h, then for an additional 4 h in the presence of brefeldin A. Cells were permeabilized, and intracellular cytokine staining was performed to measure IFN-γ and IL-10 production. Cytokine gates were set on naive (CD44low) CD4+ T cells, as shown in the left column for panels A to D. Representative flow plots are shown for each time point and tissue. Numbers in gates indicate percentage of cells positive for each cytokine. For (E) acute and (F) chronic times, the ratio of IL-10 to IFN-γ was calculated. A ratio of <1 indicates greater IFN-γ, while a ratio of >1 indicates greater IL-10. Unpaired Student’s t test was used to determine statistical significance between groups; **, P < 0.01. Combined from 2 experiments out of a total of 4 independent experiments.
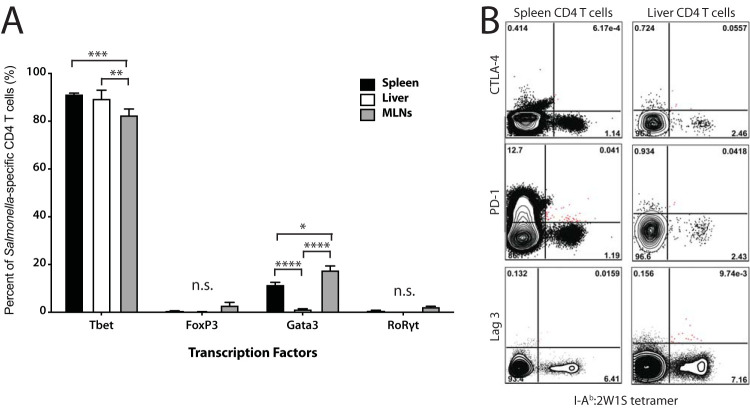
Because Salmonella-specific CD4+ T cells from the liver appeared to contribute to the exacerbation of infection and produced IL-10, we hypothesized that they may be FoxP3+ Tregs, which have been shown to play an important role during the acute phase of infection by delaying T-cell effector responses, particularly through their expression of CTLA-4; however, they seem to play less of a role during chronic infection (12). Therefore, we analyzed the T-cell transcription factor profile of Salmonella-specific CD4+ T cells during chronic infection in the lymphoid tissues and the liver. Surprisingly, the majority of Salmonella-specific CD4+ T cells in both organs were not FoxP3+ but expressed the Th1 transcription factor T-bet (Fig. 4A). Interestingly, lymphoid Salmonella-specific CD4 T cells expressed the canonical Th2 transcription factor, Gata3, at significantly higher rates than those in the liver, perhaps indicating that some of these cells were potentially driving humoral immune responses in the spleen and lymph nodes. We next assessed whether liver Salmonella-specific T cells exhibited any other common T-cell suppression markers. Both the lymphoid tissue and liver CD4+ T cells were uniformly negative for the traditional suppression makers CTLA-4, PD-1, and Lag3 (Fig. 4B), indicating that these cells were not classical Tregs.
FIG 4.
Salmonella-specific T cells from lymphoid organs and liver predominately express the transcription factor, T-bet, and do not express inhibitory markers. (A) Organs were harvested from infected mice infected for 40 days, and cells were stained with 2W1S-IAb-APC tetramer and surface markers. Cells were then fixed/permeabilized for transcription factor staining. Percentages of Salmonella-specific cells are shown for each transcription factor. (B) Spleens and livers were harvested from mice infected for 63 days, and cells were stained with 2W1S-IAb-APC tetramer. Cells were flow stained for each inhibitory molecule shown. Gates indicate the percentages of cell positive for each marker. Two-way ANOVA with Tukey’s post hoc analysis was used to determine statistical significance between groups. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. In panel A, N = 4 from two combined independent experiments. Panel B is representative of two independent experiments.
IL-10 prevents bacterial clearance during the acute phase of infection.
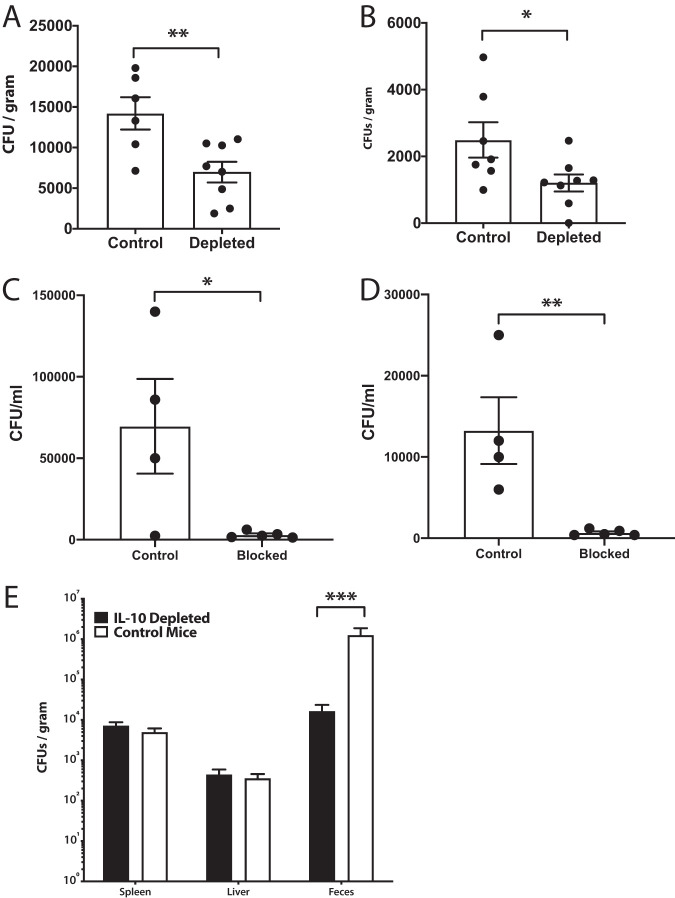
Since liver Salmonella-specific T cells produce large amounts of IL-10, we sought to determine the role for IL-10 in our infection model. Initially, we acutely infected mice orally for 2 weeks with wild-type (WT) Salmonella and subsequently neutralized IL-10 over the next 3 weeks. Mice in which IL-10 was neutralized in this fashion had decreased bacterial burdens in their systemic tissues compared to those in isotype control antibody-treated animals (Fig. 5A and B). To further confirm the importance of IL-10 during the acute phase of infection and whether this was important even earlier than 2 weeks after infection, mice were injected with an anti-IL-10 receptor (IL-10R) antibody 1 day prior to Salmonella infection, and then, 8 days after infection with WT bacteria, spleens and livers were assessed for bacterial burdens. Similarly to what was observed with IL-10 cytokine neutralization, burdens were also reduced in the spleen and liver in mice when the IL-10R was acutely blocked demonstrating that binding of IL-10 to its receptor is essential for action and that blocking IL-10 activity prior to infection accelerates bacterial clearance (Fig. 5C and D). Interestingly, when mice were depleted at 40 days after the initial infection (chronic phase) by neutralizing IL-10 over the next 3 weeks, bacterial burdens did not decrease in the systemic compartments as they did early in the infection; however, decreased bacterial loads were evident in the feces of depleted mice, suggesting that IL-10 contributes to increased bacterial shedding during later periods of infection (Fig. 5E).
FIG 5.
IL-10 prevents Salmonella clearance during the acute phase of persistent infection but not during the chronic phase. (A and B) Mice infected with WT Salmonella for 14 days (acute) were intraperitoneally (i.p.) administered anti-IL-10 neutralizing antibody continuously every 3 days for 21 days (total of 35 days), at which time organs were harvested and plated for bacterial burdens. (A) Spleens and (B) livers of cytokine-blocked mice or controls are shown. In a separate experiment, mice were injected with an anti-IL-10R blocking antibody 1 day prior to infection with WT Salmonella and bacterial burdens were assessed 8 days (acute) postinfection in (C) spleen and (D) liver. (E) Mice were first infected with WT Salmonella for 40 days (chronic) at which time anti-IL-10 neutralizing antibody was administered i.p. every 3 days for 21 days (total of 61 days), at which time organs were harvested and plated for bacterial burdens. An unpaired Student’s t test was used to determine statistical significance between groups (A to D) or a two-way ANOVA and Sidak’s multiple-comparison test were used (E). *, P < 0.05; **, P < 0.01; ***, P < 0.001. Representative of 2 or 3 independent repeats for each experiment.
Liver T cells cannot control intracellular Salmonella replication, and liver macrophages display an alternatively activated phenotype.
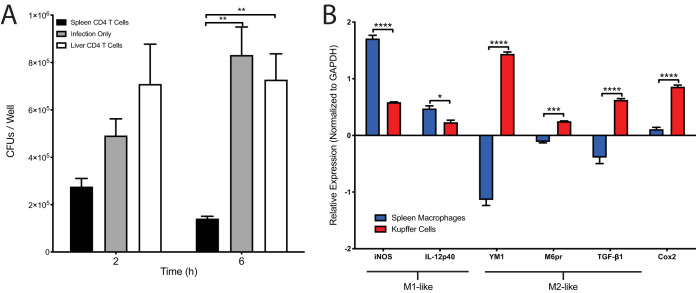
To test the ability of Salmonella-specific CD4+ T cells to aid in the clearance of bacteria from macrophages, we adapted an in vitro coculture method developed by Karen Elkins (31). Bulk CD4 T cells, likely containing multiple Salmonella-specific CD4+ T cells, were isolated from either the lymphoid tissues or the livers of chronically infected mice using a CD4 T-cell enrichment and cultured in the upper chamber of a transwell, while a macrophage cell line was cultured in the lower chamber. The T cells were then stimulated with anti-CD3/CD28 microbeads for 24 h, removed, and then the macrophages were infected with Salmonella, where intracellular bacterial replication and cellular survival was assessed using a gentamicin protection assay (32). Macrophages incubated with T cells from lymphoid tissues controlled intracellular bacterial replication, while those from the liver did not (Fig. 6A), demonstrating an impaired ability of liver CD4+ T cells to control intracellular replication in macrophages, the principal Salmonella reservoir. While we cannot rule out a possible contribution of CD4 T cells with specificities other than Salmonella, it is likely that many of the enriched T cells are Salmonella-specific based on our and others’ previous work (33–35). It is clear that there were important differences in the activity of CD4 T cells between the spleen and liver, highlighting that these organs appear to differentially imprint distinct phenotypes on resident T cells.
FIG 6.
Liver CD4+ T cells are unable to control intracellular bacterial replication in macrophages, and liver macrophages have an M2-like phenotype during chronic Salmonella infection. (A) Liver or spleen CD4+ T cells from chronically infected mice (139 days postinfection) were purified and cocultured with Salmonella-infected Raw 264.7 macrophages. Infection only refers to RAW264.7 macrophages that were infected with Salmonella in the absence of any T cells. Macrophage bacterial burdens were assessed at the indicated time points. One-way ANOVA with Tukey’s post hoc analysis was used to determine statistical significance between groups. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (B) RNA was extracted from purified splenic or liver macrophages from mice 111 days postinfection. Real-time quantitative PCR (RT-qPCR) results were analyzed using the threshold cycle (ΔΔCT) method, normalizing samples to GAPDH and comparing relative expression levels to those in naive, uninfected splenic or liver macrophages. Two-way ANOVA with Sidak’s multiple-comparison test was used to determine statistical significance between groups. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001. Representative of 4 independent experiments (A) and 3 independent experiments (B).
The above results led us to hypothesize that the CD4+ T cells from the liver imparted an altered phenotype on Kupffer cells (liver-resident macrophages) compared to that of macrophages isolated from the lymphoid tissues. It has been shown that Salmonella can exploit a replicative niche in alternatively activated macrophages (also called M2 macrophages) (36). To test this, we enriched macrophages from the spleen or liver using expression of the macrophage marker F4-80, and, using real-time quantitative PCR (RT-qPCR), assessed gene expression in each macrophage type and assessed the fold change of each infected macrophage compared to naive, uninfected control macrophages from each organ (threshold cycle [2−ΔΔCT] method). Comparing markers associated with either an M1 or M2 phenotype, we found that splenic macrophages exhibited a more M1-like phenotype, with increased expression of inducible nitric oxide synthase (iNOS) and production of the Th1-skewing cytokine IL-12p40 compared to that in Kupffer cells (Fig. 6B). Conversely, Kupffer cells isolated from chronically infected livers showed a more M2-like phenotype expressing the M2-associated genes YM-1, M6Pr, and TGF-b1. Interestingly, Kupffer cells also showed an increased expression of COX2, which has been shown to be associated with increased prostaglandin E2 (PGE2) levels and increased bacterial burdens (37).
Viable Salmonella is culturable from the livers of long-term chronically infected mice.
It has been shown that mice infected with Salmonella for a year showed bacteria in the livers, spleens, and MLNs, with livers showing signs of induced microgranulomas (14). To determine if Salmonella established a survival niche in the livers of mice infected for an even longer period, we permitted mice to remain infected for nearly 2 years. Mice were euthanized at various time points (all mice were ≥687 days postinfection), and we assessed various organs for the presence of Salmonella. At this late time point, we were only able to culture viable Salmonella bacteria from the livers of these infected mice (Fig. 7A). To ensure that we were culturing bona fide Salmonella bacteria, we confirmed all colonies via PCR (Fig. 7B). Additionally, we created frozen sections from select animal tissue and used immunofluorescence to search for Salmonella bacilli. We were only able to find a plausible bacillus in the liver from chronically infected mice (Fig. 7C), demonstrating that the liver is a potential long-term niche for bacterial survival in mice, although we cannot rule out the possibility that there may be viable bacteria in the spleen or lymph nodes that are below the limit of detection.
FIG 7.
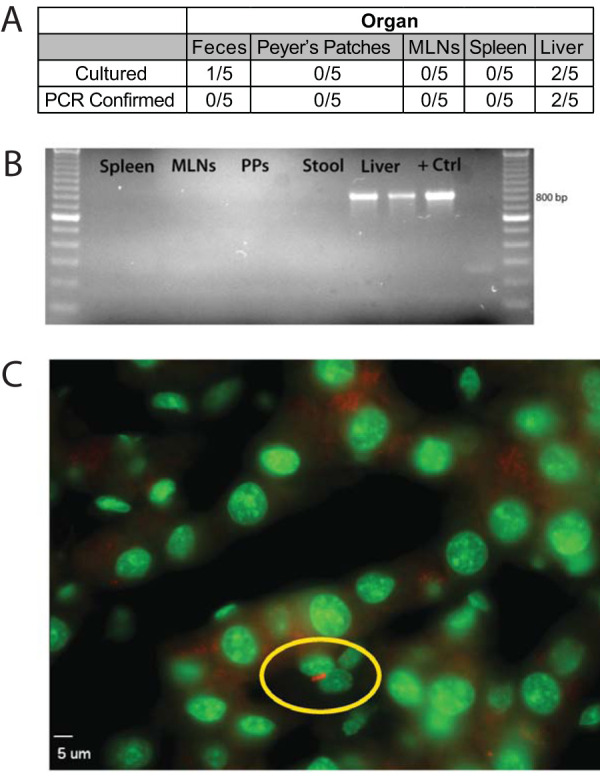
Salmonella can be isolated from late chronically infected mouse livers. (A) Organs were harvested from late chronically infected mice (≥648 days postinfection) with S. Typhimurium. Organs were homogenized in 0.1% Triton X-100, then plated on LB agar plates containing 50 μg/ml streptomycin, and colonies were quantified. (B) Cultured colonies were verified by PCR by amplifying an 832-bp region specific for a conserved region of the ompC gene and a kanamycin (Kan) resistance cassette used for bacterial selection. (C) Liver sections were fixed in 4% paraformaldehyde, permeabilized, and stained with an anti-Salmonella antibody and Sytox green for nuclear counterstaining. Images were acquired at ×630 magnification using a Zeiss fluorescence microscope. Representative of 2 independent experiments.
DISCUSSION
In the present study, we showed that CD4+ T cells are important for maintaining immunity against chronic Salmonella infection. However, different immune responses occurred at different anatomical sites during infection, in that lymphoid T cells were phenotypically antibacterial and liver T cells were immunosuppressive. The Salmonella-specific CD4+ T cells in the spleen and MLNs appeared to represent canonical Th1 cells, marked by the transcription factor T-bet and production of the proinflammatory cytokine IFN-γ. Our data suggest that lymphoid Th1 cells provide a protective role during chronic Salmonella infection through the production of inflammatory cytokines that classically activate bactericidal activity in macrophages. This may also explain why we were unable to recover viable bacteria from the lymphoid organs during the late, chronic stages of infection. In contrast to the immune responses noted in the lymphoid organs, we observed a distinctly different response in the liver. Hepatic Salmonella-specific CD4+ T cells produced large amounts of the anti-inflammatory cytokine IL-10 and, when adoptively transferred into mice, led to amplified bacterial burdens in the lymphoid organs. Notably, these liver Salmonella-specific CD4+ T cells were not the classical, thymus-derived FoxP3+ regulatory T cells. In fact, they exhibit a transcription factor and cytokine profile most similar to that of type-1 regulatory T cells (Tr1), which have been shown to produce large amounts of IL-10, lack FoxP3 expression, and express T-bet (38–41). Furthermore, it has been shown that Tr1 cells can be generated in the liver in response to inflammation (8, 42). We speculate that these antigen-specific Tr1-like cells arise in the liver in response to Salmonella infection, possibly to prevent immunopathology. In doing so, however, these Tr1-like cells may inadvertently provide Salmonella with a permissive niche for persistence. In support of this, we were only able to cultivate bacteria from the livers of very late chronically infected mice (≥650 days). This theory agrees with other liver infection models that show that immune suppression induced in the liver allows pathogen persistence (43–47). However, while those studies analyzed viral pathogens, we believe that this is the first instance of the liver providing a permissive niche for a bacterial pathogen. It should be noted that our late chronic infection data conflict with some previous studies showing the MLNs are the chronic sites of Salmonella persistence (14, 48, 49). It is possible that these discrepancies are due to differences in experimental design, use of a different mouse model (discussed below), use of different Salmonella strains, or perhaps because of the late times we are assessing after infection.
It is important to discuss how different models of Salmonella infection might potentially lead to differing and, in some cases, opposing results. In the susceptible C57B/6 model of infection, mice either completely clear attenuated Salmonella or succumb to wild-type Salmonella, in which case bacterial burdens rapidly become overwhelming (50). In neither case does long-term persistence of infection occur nor, in the case of attenuated infection, is bacterial fecal shedding observed. Furthermore, in the C57B/6 infection model, a protective Th1 CD4 T-cell response predominates in all organs, including the liver (51). This is distinctly different from the Salmonella infection model described here in the highly resistant 129/Sv × C57B/6 F1 mouse model that expresses a functional NRAMP-1 gene product and which can harbor virulent wild-type bacteria in systemic organs such as the spleen, liver, and lymph nodes for weeks to months. Rarely in this model do mice surrender to infection unless the infectious dose is exceptionally high. Each model contributes uniquely to further understanding how the mammalian immune system responds to Salmonella infection. In the case of the persistent F1 model used here, it allows for an understanding of why the immune response is unable to completely eliminate an active, systemic infection even when potent immunity is induced. This helps mimic the situation in a small percentage of typhoid fever cases where individuals can become lifelong, asymptomatic carriers of the bacteria (2, 52). This feature is important because the causative agent of typhoid fever, Salmonella enterica serovar Typhi is host specific and does not infect wild-type mice, so the persistent F1 mouse model can act as a suitable surrogate to replicate this persistent infection state (53–56). The C57BL/6 model can serve several major functions. The first is that it is well suited to modeling acute infection with attenuated bacteria that are cleared within a month. In contrast to the persistent F1 model, this provides a window into the early innate and adaptive immune response that is effective at actually clearing the infection as opposed to being permissive of persistence. The second is that, because the C57BL/6 model is susceptible to lethal infection with wild-type Salmonella infection, it can serve as an excellent surrogate for studying vaccine-mediated protection against a highly virulent infection. In this way, a variety of vaccine formulations against Salmonella infection can be tested as to whether they demonstrate preclinical efficacy (50, 57). Lastly, the C57BL/6 model allows for the use of a variety of genetic knockout mouse strains that are not available when using other mouse strains. It is possible, and even likely, that our observations regarding how liver and lymphoid CD4 T cells behave differently are due to the use of the resistant F1 model and that these differences bear some responsibility for the persistence of bacteria. We expect that if we were to perform similar experiments in the susceptible C57BL/6 model, then these organ-specific differences would diminish or even disappear, as has been shown in other recent studies (51). These model-specific differences are why it will be important to perform studies in both models, covering a broad range of responses, to help our understanding of how Salmonella can subvert immunity and how the immune system can recognize and prevent or clear infection.
It has been well established that Salmonella resides within macrophages during infection (14, 58, 59), yet how T cells and macrophages from distinct organs interact and the effect this has on bacterial survival in the persistent F1 infection mouse model has not been previously determined. We showed that lymphoid CD4+ T cells were potent activators of macrophage bactericidal activity, possibly through the activity of IFN-γ. These findings are in agreement with those of other studies in the field, showing that IFN-γ is important for the control of intracellular bacterial replication in macrophages (15, 22, 60–62). Conversely, we found that CD4+ T cells from persistently infected livers were unable to control bacterial replication. IL-10 can alternatively activate and induce M2-like macrophages, a phenotype we ascribe to the liver macrophage subset, potentially leading to increased bacterial numbers (23, 63–65). Notably, while we didn’t observe control of intracellular bacteria, neither did we find that intracellular bacterial burdens increased. One possible explanation for this is that the liver T cells were simultaneously producing IL-10 and IFN-γ. Perhaps these opposing signals from liver CD4+ T cells prevented an increase in bacterial loads compared to that in macrophages not stimulated with T cells. Alternatively, it is possible that the bacterial burdens have reached a maximum within the cell, and, when the cell eventually dies, released bacteria are exposed to extracellular antibiotics and die, preventing them from being counted at higher numbers. It is known that Salmonella bacteria survive in M2-like macrophages and can replicate due to the altered metabolism of these cells (66). It would be intriguing in future work to determine whether liver CD4+ T cells regulate the metabolic profile of undifferentiated macrophages or if they can shift M1-like macrophages metabolically toward an M2-like profile.
In this resistant mouse model, IL-10 appears to play a major role early after infection in allowing the systemic persistence of Salmonella in both the spleens and livers of infected mice. When we blocked IL-10 from exerting its function, either by neutralizing the cytokine itself or by blocking the IL-10 receptor, we found that bacterial burdens were significantly reduced in spleens and livers. Notably, this systemic effect was lost when IL-10 was blocked during the chronic phase of the infection; however, fecal shedding of bacteria was significantly increased. There are several potential explanations for these potentially contradictory findings about the role of IL-10 during the acute versus chronic phase of infection. One possibility is that IL-10 expression is essential for the initial systemic establishment of infection, particularly in the liver, but is dispensable later in the infection. Perhaps during the later stages of infection, more bacteria in the spleens and liver are capable of entering an intracellular persister state in macrophages, as described by the Holden group (67). It is possible that these metabolically inactive bacteria are not affected when IL-10 is neutralized and that the immune response is presumably shifted back toward a more Th1 phenotype. Another possibility is that there is greater shuttling of bacteria between the gallbladder, another essential organ in the hepatobiliary system (30), and liver during the late phase of infection and that IL-10 blockade allows bacterial escape into the bile and eventually into the feces, potentially reinfecting via the small intestine and thereby maintaining high numbers of systemic bacteria. The concept that the hepatobiliary system displays a distinct immune profile that is differentially affected by the function of IL-10 is further supported by our recent study showing a shift toward a type 2 T-cell and macrophage immune response in the gallbladder. It is possible that the liver delivers immunoregulatory cells to the gallbladder, where they are also permissive of bacterial persistence at this site. The combination of the liver and gallbladder allowing for persistence of Salmonella would explain the mechanism for fecal shedding of bacteria from infected people and mice. Combined, these findings highlight that targeting IL-10 blockade may be a potential therapeutic target for reducing fecal bacterial shedding in human typhoid patients.
Taken together, we propose that in our mouse model of chronic Salmonella Typhimurium infection, a balance is reached between pathogen persistence and immunopathology, and that this balance is largely contributed to by opposing proinflammatory signals, such as IFN-γ, and by anti-inflammatory signals, such as IL-10, produced by Salmonella-specific CD4+ T cells in different anatomical locations. This balance seems to be most important in the liver, a crucial organ for host homeostasis, whereas the lymphoid organs can tolerate a certain degree of inflammation (68, 69). Some evidence from chronic typhoid human patients suggests the hepatobiliary system might be a site of bacterial persistence (3, 70, 71). Our group has shown this to also be the case in mice (72–74). The current study further bolsters the idea that the hepatobiliary system is a site of Salmonella persistence and also adds to the concept that the liver acts as an immunosuppressive organ during infection. Future studies will assess the potential interplay between the infected host organs, particularly how the liver and gallbladder may share immune cells. While we did not evaluate in the current study whether T cells in each organ were the recently described T resident memory cells (Trm), it is possible that these cells do not migrate between organs and are permanent resident cells (75). There is recent evidence for a noncirculating pool of Salmonella-specific memory Th1 CD4+ T cells in the liver in response to attenuated infection (51). It is possible that the cells we observe in the chronic virulent infection model are similarly resident but that, instead of assuming the Th1 phenotype observed in that study, the persistent exposure to infection induces a more Tr1-like phenotype. More studies are needed to address this possibility.
MATERIALS AND METHODS
Ethics statement.
This study was carried out in accordance with recommendations from the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. Tulane University is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC). All experimental procedures involving animals were approved and performed in compliance with the guidelines established by Tulane University School of Medicine’s Institutional Animal Care and Use Committee.
Bacterial strains, epitope tagging, and growth conditions.
Salmonella enterica subsp. enterica serovar Typhimurium strain SL1344 (a gift from Marc Jenkins, University of Minnesota) or strain 14028 (ATCC) were used for infections as indicated. SL1344 Salmonella was tagged chromosomally with the 2W1S peptide (EAWGALANWAVDSA) as previously described, using the lambda Red recombinase system (26, 76).
Mice, infections, and bacterial burdens.
C57BL/6J and 129X1/SvJ mice were purchased from Jackson Laboratory (Bar Harbor, ME) and bred to generate naturally resistant NRAMP1(SLC11A1)+/− F1 hybrid mice (C57BL/6J × 129X1/SvJ F1 mice), which were maintained under specific-pathogen-free conditions in the vivarium at Tulane University School of Medicine. Prior to infection, mice were deprived of food for approximately 18 h and then infected at 6 to 10 weeks of age orally with 5 × 107 CFU of S. Typhimurium SL1344 in 100 μl of phosphate-buffered saline (PBS). Chronic infection was defined as infection at ≥40 days postinfection. At designated times postinfection, mice were euthanized by CO2 asphyxiation, and spleens, mesenteric lymph nodes (MLNs), and livers were harvested for determination of bacterial burdens. Organs were homogenized in sterile 0.1% Triton X-100 and plated on LB agar containing 100 μg/ml streptomycin for enumeration of viable CFU. The data were expressed as the mean CFU per tissue or per gram of tissue.
Generation of single-cell preparations from infected mice.
Spleens and MLNs were made in single-cell suspensions by homogenizing the organs over a 100-μm nylon mesh filter in cold sorter buffer (1× phosphate-buffered saline, 2% newborn calf serum, and 0.1% sodium azide). Hepatocytes were removed from livers by sedimenting the suspension at 50 × g for 2 min. Supernatant was collected and spun at 500 × g to pellet the remaining cells. Cells were then underlayered with a 15% OptiPrep gradient (Sigma-Aldrich) prepared with Hanks’ balanced salt solution (HBSS) and spun at 750 × g for 20 min at room temperature.
MHC-II tetramer staining, magnetically activated cell sorting, and flow cytometry.
Single-cell preparations were resuspended in 200 μl with FcBlock. Cells were enriched as previously described (77) by staining with 10 μM 2W1S:I-Ab MHC-II tetramer, conjugated to allophycocyanin (APC), incubated at room temperature in the dark for 1 h, then washed, stained with anti-APC magnetic beads (Miltenyi), and passed over an LS column on a quadroMACS magnet. Eluted cells were stained with the following anti-mouse antibodies: CD19, F4/80, CD11c, CD11b (eFluor 450), CD3ε (fluorescein isothiocyanate [FITC]), CD4 (BV605), CD8a (BV510), and CD44 (PerCP-Cy5.5). Cells were collected on an LSRFortessa instrument (Beckton-Dickson). Data were analyzed using FlowJo software (TreeStar, Ashland, OR).
In vivo CD4 depletion.
Antibodies used for depletion were purchased from Bio X Cell (West Lebanon, NH). For CD4+ T-cell depletions, purified anti-mouse CD4 (clone GK1.5) was diluted in sterile saline, and mice were injected intraperitoneally (i.p.) with 750 μg of antibody in a 200-μl volume every 3 days for 12 days, starting at 44 days postinfection. Depletion of CD4+ T cells was confirmed by flow cytometry of peripheral blood. After 2 weeks of depletion, mice were sacrificed, and the bacterial burdens in the MLNs, spleens, and livers were determined as described above.
In vitro intracellular cytokine staining.
Single-cell suspensions from organs were stimulated in vitro with phorbol 12-myristate 13-acetate (PMA) 50 ng/ml and ionomycin (Iono) 100 ng/ml (Sigma-Aldrich) in complete RPMI at 37°C. After 2 h of stimulation, 10 μg/ml brefeldin A (Sigma-Aldrich) was added for an additional 4 h. Cells were then fixed and permeabilized using the BD Cytofix/Cytoperm kit according to the manufacturer’s instructions, then stained overnight for intracellular cytokines. Cytokine-positive gates were set on naive, CD44low T cells, which do not produce cytokines, from each organ. Other studies have shown this to be an accurate gating strategy for measuring intracellular cytokines (27).
Adoptive transfer of Salmonella-specific CD4+/− T cells.
Absolute cell numbers of antigen-specific CD4+ T cells were determined, and tetramer-enriched cells were transferred into anesthetized mice via retroorbital injection. Mice were infected 24 h later as described above.
Transcription factor staining.
Spleens, MLNs, and livers were harvested from infected mice, and cells were made into single-cell preparations, as described above. Cells were stained with 2W1S-IAb-APC and enriched for antigen-specific cells, as described above. Cells were washed and blocked with FcBlock and subsequently stained with anti-mouse surface antibodies. Cells were then fixed and permeabilized using the Foxp3 fixation/permeabilization buffer kit (eBioscience) according to the manufacturer’s instructions, then stained overnight for nuclear transcription factors. The next morning, samples were washed twice with 1× permeabilization wash buffer and resuspended in sorter buffer for fluorescence-activated cell sorter (FACS) analysis.
In vivo depletion of IL-10 and IL-10R blocking.
For acute-phase IL-10 depletion studies, mice were infected with WT Salmonella Typhimurium SL1344 as described above. The infection was allowed to persist for 14 days before cytokine depletion. IL-10 was depleted by administering 200 μg of rat monoclonal antibody against IL-10 (clone JES5-2A5; Bio X Cell) intraperitoneally in 200 μl PBS every 3 days for 21 days. An isotype was administered in the same fashion and served as a negative control (clone HRPN; Bio X Cell). To replicate the conditions of an IL-10 knockout mouse for certain experiments, 129X1/SvJ mice were injected with 1 mg of anti-IL-10R (Bio X Cell, West Lebanon, NH) or with 1× PBS 1 day before intraperitoneal injection with S. Typhimurium strain 14028.
Macrophage invasion assay and CD4+ T cell coculture.
Raw264.7 macrophages were cultured in complete Dulbecco’s modified Eagle’s medium (DMEM) and seeded at 5 × 105 cells per well in 24-well plates overnight before coculture assays commenced. CD4+ T cells were isolated from the lymphoid organs (spleen and MLNs) or livers of mice chronically infected with Salmonella Typhimurium using the CD4+ T cell isolation kit II (Miltenyi) following the manufacturer’s instructions. An aliquot of purified CD4+ T cells was analyzed and counted by flow cytometry, as described above. CD4+ T cells (8 × 104) were added to 0.4-μm transwell inserts (Greiner Bio-One) with a 1:1 ratio of anti-CD3/anti-CD28 Dynabeads (Invitrogen). CD4+ T cells were stimulated for 36 to 48 h to activate/polarize macrophages before a gentamicin protection assay was performed. To perform the gentamicin protection assay, antibiotics were removed from the macrophages, and monolayers were rinsed in PBS, then cultured in complete DMEM (cDMEM) without antibiotics. Macrophages were infected with wild-type SL1344 Salmonella Typhimurium at a multiplicity of infection (MOI) of 1:1 and spun onto the macrophage monolayers at 450 × g to synchronize the infection. Macrophages were infected for 1 h at 37°C, before extracellular bacteria were killed by the addition of 100 μg/ml gentamicin for 1 h at 37°C. Monolayers were then washed with PBS to remove dead bacteria and cDMEM medium with 10 μg/ml gentamicin was added for the remainder of the assay. At the indicated times postinfection, macrophages were lysed for 10 min at room temperature in lysis buffer (150 mM NaCl, 0.1% Triton X-100, 50 mM Tris-HCl, and 1× SigmaFast protease inhibitor cocktail), to liberate intracellular Salmonella bacteria. Serial dilutions were spot plated onto LB plates containing 50 μg/ml streptomycin and incubated at 37°C for 24 h, and bacterial colonies were counted and back calculated to determine the CFU/well.
Real-time quantitative PCR.
Spleens and livers were harvested from mice chronically infected with Salmonella Typhimurium and single-cell preparations were made, as described above. Before magnetically activated cell sorting (MACS), crude liver nonparenchymal cells (NPCs) were first isolated by mixing the single-cell suspensions into an 11.7% OptiPrep solution and sandwiching the suspension between two density layers, an underlayer of 17.6% (wt/vol) OptiPrep solution and a top layer of HBSS. The discontinuous gradients were centrifuged at 1,400 × g for 17 min at 4°C. The NPCs were harvested from the interface and further purified by MACS. Briefly, both the splenic cell preparations and crude NPCs were blocked with FcBlock, before incubation with a biotinylated F4-80 antibody (eBioscience). Streptavidin microbeads (Miltenyi) were then added to the preparations, and F4-80+ cells were positively selected by passing cells through a LS column attached to a quadroMACS separator (Miltenyi). RNA was then immediately purified from the F4-80+ cells using the RNeasy microkit (Qiagen) following the manufacturer’s directions. RNA purity and concentration were determined on a NanoDrop 2000 spectrophotometer (Thermo Scientific). RNA samples were stored at −80°C until used for RT-qPCR. cDNA was made by adding 100 ng of pure RNA to the iScript cDNA synthesis kit (Bio-Rad), according to the manufacturer’s instructions. Controls lacking reverse transcriptase were included to ensure primer specificity. After reverse transcription of the mRNA, a 20-μl reaction mixture containing 200 μM each primer (see Table 1 for primer sequences) and 1 μl of cDNA was added to the iQ SYBR green Supermix (Bio-Rad) following the manufacturer’s recommendations. RT-qPCR was performed on a CFX Connect real-time PCR machine (Bio-Rad). The results were analyzed using the ΔΔCT method, normalizing all samples to GAPDH and comparing relative expression levels to those in uninfected macrophages.
TABLE 1.
Primer sequences used in this study
| Gene | PrimerBank identifier | Amplicon size (bp) | Sequence fora
: |
|
|---|---|---|---|---|
| Fwd primer (5′→3′) | Rev primer (5′→3′) | |||
| GAPDH | 6679937a1 | 123 | AGGTCGGTGTGAACGGATTTG | TGTAGACCATGTAGTTGAGGTCA |
| NOS2 | 6754872a1 | 127 | GTTCTCAGCCCAACAATACAAGA | GTGGACGGGTCGATGTCAC |
| Il12b | 6680397a1 | 123 | TGGTTTGCCATCGTTTTGCTG | ACAGGTGAGGTTCACTGTTTCT |
| Chi3l3 | 6753416a1 | 197 | CAGGTCTGGCAATTCTTCTGAA | GTCTTGCTCATGTGTGTAAGTGA |
| M6PR | 14916479a1 | 127 | TGCTGGAGGACTGAACTGTTA | GAGCCACCTCGTTCTTTGACT |
| TGFB1 | 6755775a1 | 133 | CTCCCGTGGCTTCTAGTGC | GCCTTAGTTTGGACAGGATCTG |
| PTGS | 118130137c3 | 124 | TGCACTATGGTTACAAAAGCTGG | TCAGGAAGCTCCTTATTTCCCTT |
Fwd, forward; Rev, reverse.
Salmonella-specific PCR.
To ensure that the colonies grown on LB plates containing 50 μg/ml streptomycin from organs of late chronically infected mice were Salmonella Typhimurium, any colonies observed were verified by PCR for the presence of the 2W1S genomic insert. Colonies were plucked from LB plates and aseptically transferred to 10 μl of sterile PCR-grade water. A 5-μl aliquot of the suspension was used as the template DNA for the PCR. Primers were designed to amplify an 833-bp product specific for a conserved region of the ompC gene (5′-GATGTCGGCGCGACTTACTACTTC-3′) and an inserted kanamycin-resistance cassette (5′-CTCGTCCTGCAGTTCATTCAGGGC-3′) used for bacterial selection. Purified DNA from the SL1344 OmpC-2W1S strain of Salmonella Typhimurium used to originally infect the mice was used as a positive control. To rule out the possibility of reagents being contaminated with Salmonella DNA, a no-DNA template control (water only) was run with each experiment. Platinum Taq (Invitrogen) was used according to the manufacturer’s instructions; however, a 2-min denaturation step at 94°C was added to allow lysis of the bacterial cells. The primer annealing temperature was set at 60°C.
Fluorescent immunohistochemistry.
Fluorescent immunostaining was performed on frozen liver sections embedded in optimal cutting temperature (OCT) medium from mice chronically infected with Salmonella Typhimurium. Tissues were fixed for 15 min with 4% paraformaldehyde. Tissues were washed three times with PBS, then incubated with 0.05% Triton X-100 (Sigma-Aldrich) in PBS for 20 min to permeabilize the cells. Tissues were blocked with 10% normal goat serum (NGS) (Invitrogen) for 1 h to block nonspecific binding. Sections were incubated with a polyclonal primary antibody (Rabbit anti-Salmonella [O+H antigens], 1:1,000, MyBioSource) in PBS and 1% NGS for 1 h at room temperature. Tissues were washed three times with PBS containing 0.1% Tween 20 (PBST, Sigma-Aldrich). Tissues were then incubated with secondary antibody (DyLight 549-conjugated goat anti-rabbit [H&L], 1:5,000; Rockland, Inc.) in PBST for 1 h at room temperature in the dark, followed by three washes with PBST. To counterstain the cell nuclei, tissues were stained with Sytox green (300 nM in water) for 2 min, then washed extensively with PBST. Slides were mounted using ProLong Gold antifade mounting medium (Invitrogen) and allowed to cure for 48 h before imaging. Images were acquired with a Zeiss Axioplan II microscope (Carl Zeiss, Thornwood, NY), using a 63× objective (numerical aperture [NA] = 1.4). Series of horizontal optical sections (0.3 μm each) were collected and subsequently deconvolved using SlideBook 6.0 software (Intelligent Imaging Innovations, Denver, CO). Postcollection processing of the images was performed using Volocity 6.3 software (Perkin-Elmer).
Statistical analysis.
Statistical differences between data sets were assessed using Prism (GraphPad) software. Outliers were removed using the Grubbs outlier test.
ACKNOWLEDGMENTS
We thank the McLachlan lab for reading and editing of the manuscript.
This study was supported in part by National Institutes of Health grants R01 AI116917 (to J.S.G.) and R01 AI103343 (to J.B.M.)
REFERENCES
- 1.Buckle GC, Walker CLF, Black RE. 2012. Typhoid fever and paratyphoid fever: systematic review to estimate global morbidity and mortality for 2010. J Glob Health 2:010401. doi: 10.7189/jogh.01.010401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gunn JS, Marshall JM, Baker S, Dongol S, Charles RC, Ryan ET. 2014. Salmonella chronic carriage: epidemiology, diagnosis, and gallbladder persistence. Trends Microbiol 22:648–655. doi: 10.1016/j.tim.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nath G, Singh YK, Maurya P, Gulati AK, Srivastava RC, Tripathi SK. 2010. Does Salmonella Typhi primarily reside in the liver of chronic typhoid carriers? J Infect Dev Ctries 4:259–261. doi: 10.3855/jidc.820. [DOI] [PubMed] [Google Scholar]
- 4.Watanabe T, Katsukura H, Shirai Y, Yamori M, Nishi T, Chiba T, Kita T, Wakatsuki Y. 2003. A liver tolerates a portal antigen by generating CD11c+ cells, which select Fas ligand+ Th2 cells via apoptosis. Hepatology 38:403–412. doi: 10.1053/jhep.2003.50343. [DOI] [PubMed] [Google Scholar]
- 5.Watanabe T, Yoshida M, Shirai Y, Yamori M, Yagita H, Itoh T, Chiba T, Kita T, Wakatsuki Y. 2002. Administration of an antigen at a high dose generates regulatory CD4+ T cells expressing CD95 ligand and secreting IL-4 in the liver. J Immunol 168:2188–2199. doi: 10.4049/jimmunol.168.5.2188. [DOI] [PubMed] [Google Scholar]
- 6.Wohlleber D, Knolle PA. 2012. The liver as an immune-privileged site, p 93–106. In Stein-Streilein J. (ed), Infection, immune homeostasis and immune privilege. Springer Basel, Basel, Switzerland. [Google Scholar]
- 7.Blackburn SD, Wherry EJ. 2007. IL-10, T cell exhaustion and viral persistence. Trends Microbiol 15:143–146. doi: 10.1016/j.tim.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 8.Xu L, Yin W, Sun R, Wei H, Tian Z. 2013. Liver type I regulatory T cells suppress germinal center formation in HBV-tolerant mice. Proc National Acad Sci 110:16993–16998. doi: 10.1073/pnas.1306437110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peppa D, Micco L, Javaid A, Kennedy PTF, Schurich A, Dunn C, Pallant C, Ellis G, Khanna P, Dusheiko G, Gilson RJ, Maini MK. 2010. Blockade of immunosuppressive cytokines restores NK cell antiviral function in chronic hepatitis B virus infection. PLoS Pathog 6:e1001227. doi: 10.1371/journal.ppat.1001227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miroux C, Vausselin T, Delhem N. 2010. Regulatory T cells in HBV and HCV liver diseases: implication of regulatory T lymphocytes in the control of immune response. Expert Opin Biol Ther 10:1563–1572. doi: 10.1517/14712598.2010.529125. [DOI] [PubMed] [Google Scholar]
- 11.van Diepen A, van de Gevel JS, Koudijs MM, Ossendorp F, Beekhuizen H, Janssen R, van Dissel JT. 2005. Gamma irradiation or CD4+-T-cell depletion causes reactivation of latent Salmonella enterica serovar Typhimurium infection in C3H/HeN mice. Infect Immun 73:2857–2862. doi: 10.1128/IAI.73.5.2857-2862.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johanns TM, Ertelt JM, Rowe JH, Way SS. 2010. Regulatory T cell suppressive potency dictates the balance between bacterial proliferation and clearance during persistent Salmonella infection. PLoS Pathog 6:e1001043. doi: 10.1371/journal.ppat.1001043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nauciel C. 1990. Role of CD4+ T cells and T-independent mechanisms in acquired resistance to Salmonella typhimurium infection. J Immunology 145:1265–1269. [PubMed] [Google Scholar]
- 14.Monack DM, Bouley DM, Falkow S. 2004. Salmonella Typhimurium persists within macrophages in the mesenteric lymph nodes of chronically infected Nramp1+/+ mice and can be reactivated by IFNγ neutralization. J Exp Medicine 199:231–241. doi: 10.1084/jem.20031319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nauciel C, Espinasse-Maes F. 1992. Role of gamma interferon and tumor necrosis factor alpha in resistance to Salmonella typhimurium infection. Infect Immun 60:450–454. doi: 10.1128/IAI.60.2.450-454.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pie S, Matsiota-Bernard P, Truffa-Bachi P, Nauciel C. 1996. Gamma interferon and interleukin-10 gene expression in innately susceptible and resistant mice during the early phase of Salmonella Typhimurium infection. Infection and immunity 64:849–854. doi: 10.1128/IAI.64.3.849-854.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mittrücker H-W, Köhler A, Kaufmann S. 2002. Characterization of the murine T-lymphocyte response to Salmonella enterica serovar Typhimurium infection. Infect Immun 70:199–203. doi: 10.1128/IAI.70.1.199-203.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mizuno Y, Takada H, Nomura A, Jin C-H, Hattori H, Ihara K, Aoki T, Eguchi K, Hara T. 2003. Th1 and Th1-inducing cytokines in Salmonella infection. Clin Exp Immunol 131:111–117. doi: 10.1046/j.1365-2249.2003.02060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stoycheva M, Murdjeva M. 2005. Serum levels of interferon-gamma, interleukin-12, tumour necrosis factor-alpha, and interleukin-10, and bacterial clearance in patients with gastroenteric Salmonella infection. Scand J Infect Dis 37:11–14. doi: 10.1080/00365540410026068. [DOI] [PubMed] [Google Scholar]
- 20.Ravindran R, Foley J, Stoklasek T, Glimcher LH, McSorley SJ. 2005. Expression of T-bet by CD4 T cells is essential for resistance to Salmonella infection. J Immunol 175:4603–4610. doi: 10.4049/jimmunol.175.7.4603. [DOI] [PubMed] [Google Scholar]
- 21.Kagaya K, Watanabe K, Fukazawa Y. 1989. Capacity of recombinant gamma interferon to activate macrophages for Salmonella-killing activity. Infect Immun 57:609–615. doi: 10.1128/IAI.57.2.609-615.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herbst S, Schaible UE, Schneider BE. 2011. Interferon gamma activated macrophages kill mycobacteria by nitric oxide induced apoptosis. PLoS One 6:e19105. doi: 10.1371/journal.pone.0019105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee K-S, Jeong E-S, Heo S-H, Seo J-H, Jeong D-G, Choi Y-K. 2011. IL-10 suppresses bactericidal response of macrophages against Salmonella Typhimurium. J Microbiol 49:1050–1053. doi: 10.1007/s12275-011-1043-z. [DOI] [PubMed] [Google Scholar]
- 24.Govoni G, Gros P. 1998. Macrophage NRAMP1 and its role in resistance to microbial infections. Inflamm Res 47:277–284. doi: 10.1007/s000110050330. [DOI] [PubMed] [Google Scholar]
- 25.Eckmann L, Fierer J, Kagnoff MF. 1996. Genetically resistant (Ityr) and susceptible (Itys) congenic mouse strains show similar cytokine responses following infection with Salmonella dublin. J Immunol 156:2894–2900. [PubMed] [Google Scholar]
- 26.Nelson RW, McLachlan JB, Kurtz JR, Jenkins MK. 2013. CD4+ T cell persistence and function after infection are maintained by low-level peptide:MHC class II presentation. J Immunol 190:2828–2834. doi: 10.4049/jimmunol.1202183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McLachlan JB, Catron DM, Moon JJ, Jenkins MK. 2009. Dendritic cell antigen presentation drives simultaneous cytokine production by effector and regulatory T cells in inflamed skin. Immunity 30:277–288. doi: 10.1016/j.immuni.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moon JJ, Chu HH, Pepper M, McSorley SJ, Jameson SC, Kedl RM, Jenkins MK. 2007. Naive CD4+ T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity 27:203–213. doi: 10.1016/j.immuni.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frederick DR, Goggins JA, Sabbagh LM, Freytag LC, Clements JD, McLachlan JB. 2018. Adjuvant selection regulates gut migration and phenotypic diversity of antigen-specific CD4+ T cells following parenteral immunization. Mucosal Immunol 11:549–561. doi: 10.1038/mi.2017.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.González JF, Kurtz J, Bauer DL, Hitt R, Fitch J, Wetzel A, Perle KL, White P, McLachlan J, Gunn JS. 2019. Establishment of chronic typhoid infection in a mouse carriage model involves a type 2 immune shift and T and B cell recruitment to the gallbladder. mBio 10:e02262-19. doi: 10.1128/mBio.02262-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elkins KL, Cowley SC, Conlan JW. 2011. Measurement of macrophage-mediated killing of intracellular bacteria, including Francisella and Mycobacteria. Curr Prot Immunol Chapter 14:Unit 14.25. [DOI] [PubMed] [Google Scholar]
- 32.Loetscher Y, Wieser A, Lengefeld J, Kaiser P, Schubert S, Heikenwalder M, Hardt W-D, Stecher B. 2012. Salmonella transiently reside in luminal neutrophils in the inflamed gut. PLoS One 7:e34812. doi: 10.1371/journal.pone.0034812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Srinivasan A, Foley J, McSorley SJ. 2004. Massive number of antigen-specific CD4 T cells during vaccination with live attenuated Salmonella causes interclonal competition. J Immunol 172:6884–6893. doi: 10.4049/jimmunol.172.11.6884. [DOI] [PubMed] [Google Scholar]
- 34.Lee S-J, McLachlan JB, Kurtz JR, Fan D, Winter SE, Bäumler AJ, Jenkins MK, McSorley SJ. 2012. Temporal expression of bacterial proteins instructs host CD4 T cell expansion and Th17 development. PLoS Pathog 8:e1002499. doi: 10.1371/journal.ppat.1002499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barat S, Willer Y, Rizos K, Claudi B, Mazé A, Schemmer AK, Kirchhoff D, Schmidt A, Burton N, Bumann D. 2012. Immunity to intracellular Salmonella depends on surface-associated antigens. PLoS Pathog 8:e1002966. doi: 10.1371/journal.ppat.1002966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eisele NA, Ruby T, Jacobson A, Manzanillo PS, Cox JS, Lam L, Mukundan L, Chawla A, Monack DM. 2013. Salmonella require the fatty acid regulator PPARδ for the establishment of a metabolic environment essential for long-term persistence. Cell Host Microbe 14:171–182. doi: 10.1016/j.chom.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Woolard MD, Wilson JE, Hensley LL, Jania LA, Kawula TH, Drake JR, Frelinger JA. 2007. Francisella tularensis-infected macrophages release prostaglandin E 2 that blocks T cell proliferation and promotes a Th2-like response. J Immunol 178:2065–2074. doi: 10.4049/jimmunol.178.4.2065. [DOI] [PubMed] [Google Scholar]
- 38.Fujio K, Okamura T, Yamamoto K. 2010. The family of IL-10-secreting CD4+ T cells. Adv Immunol 105:99–130. doi: 10.1016/S0065-2776(10)05004-2. [DOI] [PubMed] [Google Scholar]
- 39.Roncarolo MG, Gregori S, Battaglia M, Bacchetta R, Fleischhauer K, Levings MK. 2006. Interleukin-10-secreting type 1 regulatory T cells in rodents and humans. Immunol Rev 212:28–50. doi: 10.1111/j.0105-2896.2006.00420.x. [DOI] [PubMed] [Google Scholar]
- 40.Roncarolo MG, Bacchetta R, Bordignon C, Narula S, Levings MK. 2001. Type 1 T regulatory cells. Immunol Rev 182:68–79. doi: 10.1034/j.1600-065X.2001.1820105.x. [DOI] [PubMed] [Google Scholar]
- 41.Groux H, O’Garra A, Bigler M, Rouleau M, Antonenko S, de Vries JE, Roncarolo MG. 1997. A CD4+ T-cell subset inhibits antigen-specific T-cell responses and prevents colitis. Nature 389:737–742. doi: 10.1038/39614. [DOI] [PubMed] [Google Scholar]
- 42.Erhardt A, Biburger M, Papadopoulos T, Tiegs G. 2007. IL-10, regulatory T cells, and Kupffer cells mediate tolerance in concanavalin A-induced liver injury in mice. Hepatology 45:475–485. doi: 10.1002/hep.21498. [DOI] [PubMed] [Google Scholar]
- 43.Mellor AL, Munn DH. 2008. Creating immune privilege: active local suppression that benefits friends, but protects foes. Nat Rev Immunol 8:74–80. doi: 10.1038/nri2233. [DOI] [PubMed] [Google Scholar]
- 44.Carambia A, Herkel J. 2010. CD4 T cells in hepatic immune tolerance. J Autoimmun 34:23–28. doi: 10.1016/j.jaut.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 45.Erhardt A, Tiegs G. 2010. Tolerance induction in response to liver inflammation. Dig Dis 28:86–92. doi: 10.1159/000282069. [DOI] [PubMed] [Google Scholar]
- 46.Donaghy L, Cabillic F, Corlu A, Rostan O, Toutirais O, Guguen-Guillouzo C, Guiguen C, Gangneux J-P. 2010. Immunostimulatory properties of dendritic cells after Leishmania donovani infection using an in vitro model of liver microenvironment. PLoS Negl Trop Dis 4:e703. doi: 10.1371/journal.pntd.0000703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kruse N, Neumann K, Schrage A, Derkow K, Schott E, Erben U, Kühl A, Loddenkemper C, Zeitz M, Hamann A, Klugewitz K. 2009. Priming of CD4+ T cells by liver sinusoidal endothelial cells induces CD25low forkhead box protein 3− regulatory T cells suppressing autoimmune hepatitis. Hepatology Baltim Md 50:1904–1913. doi: 10.1002/hep.23191. [DOI] [PubMed] [Google Scholar]
- 48.Voedisch S, Koenecke C, David S, Herbrand H, Förster R, Rhen M, Pabst O. 2009. Mesenteric lymph nodes confine dendritic cell-mediated dissemination of Salmonella enterica serovar Typhimurium and limit systemic disease in mice. Infect ImmunIAI 77:3170–3180. doi: 10.1128/IAI.00272-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Griffin AJ, Li L-X, Voedisch S, Pabst O, McSorley SJ. 2011. Dissemination of persistent intestinal bacteria via the mesenteric lymph nodes causes typhoid relapse. Infect Immun 79:1479–1488. doi: 10.1128/IAI.01033-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kurtz JR, Petersen HE, Frederick DR, Morici LA, McLachlan JB. 2014. Vaccination with a single CD4 T cell peptide epitope from a Salmonella type III-secreted effector protein provides protection against lethal infection. Infect Immun 82:2424–2433. doi: 10.1128/IAI.00052-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Benoun JM, Peres NG, Wang N, Pham OH, Rudisill VL, Fogassy ZN, Whitney PG, Fernandez-Ruiz D, Gebhardt T, Pham Q-M, Puddington L, Bedoui S, Strugnell RA, McSorley SJ. 2018. Optimal protection against Salmonellainfection requires noncirculating memory. Proc Natl Acad Sci U S A 115:10416–10421. doi: 10.1073/pnas.1808339115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Levine MM, Black RE, Lanata C. 1982. Precise estimation of the numbers of chronic carriers of Salmonella Typhi in Santiago, Chile, an endemic area. J Infect Dis 146:724–726. doi: 10.1093/infdis/146.6.724. [DOI] [PubMed] [Google Scholar]
- 53.Song J, Willinger T, Rongvaux A, Eynon EE, Stevens S, Manz MG, Flavell RA, Galán JE. 2010. A mouse model for the human pathogen Salmonella Typhi. Cell Host Microbe 8:369–376. doi: 10.1016/j.chom.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Song J, Wilhelm CL, Wangdi T, Maira-Litran T, Lee S-J, Raetz M, Sturge CR, Mirpuri J, Pei J, Grishin NV, McSorley SJ, Gewirtz AT, Bäumler AJ, Pier GB, Galán JE, Yarovinsky F. 2016. Absence of TLR11 in mice does not confer susceptibility to Salmonella Typhi. Cell 164:827–828. doi: 10.1016/j.cell.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mathur R, Oh H, Zhang D, Park S-G, Seo J, Koblansky A, Hayden MS, Ghosh S. 2012. A mouse model of Salmonella Typhi infection. Cell 151:590–602. doi: 10.1016/j.cell.2012.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mathur R, Zeng W, Hayden MS, Ghosh S. 2016. Mice lacking TLR11 exhibit variable Salmonella Typhi susceptibility. Cell 164:829–830. doi: 10.1016/j.cell.2016.02.020. [DOI] [PubMed] [Google Scholar]
- 57.Lee S-J, Benoun J, Sheridan BS, Fogassy Z, Pham O, Pham Q-M, Puddington L, McSorley SJ. 2017. Dual immunization with SseB/flagellin provides enhanced protection against Salmonella infection mediated by circulating memory cells. J Immunol 199:1353–1361. doi: 10.4049/jimmunol.1601357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Richter-Dahlfors A, Buchan AM, Finlay BB. 1997. Murine salmonellosis studied by confocal microscopy: Salmonella Typhimurium resides intracellularly inside macrophages and exerts a cytotoxic effect on phagocytes in vivo. J Exp Med 186:569–580. doi: 10.1084/jem.186.4.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nix RN, Altschuler SE, Henson PM, Detweiler CS. 2007. Hemophagocytic macrophages harbor Salmonella enterica during persistent infection. PLoS Pathog 3:e193. doi: 10.1371/journal.ppat.0030193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cowley SC, Elkins KL. 2003. Multiple T cell subsets control Francisella tularensis LVS intracellular growth without stimulation through macrophage interferon γ receptors. J Exp Medicine 198:379–389. doi: 10.1084/jem.20030687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Buchmeier NA, Schreiber RD. 1985. Requirement of endogenous interferon-gamma production for resolution of Listeria monocytogenes infection. Proc National Acad Sci 82:7404–7408. doi: 10.1073/pnas.82.21.7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Santanirand P, Harley VS, Dance DA, Drasar BS, Bancroft GJ. 1999. Obligatory role of gamma interferon for host survival in a murine model of infection with Burkholderia pseudomallei. Infect Immun 67:3593–3600. doi: 10.1128/IAI.67.7.3593-3600.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Katakura T, Miyazaki M, Kobayashi M, Herndon DN, Suzuki F. 2004. CCL17 and IL-10 as effectors that enable alternatively activated macrophages to inhibit the generation of classically activated macrophages. J Immunol 172:1407–1413. doi: 10.4049/jimmunol.172.3.1407. [DOI] [PubMed] [Google Scholar]
- 64.Koscsó B, Csóka B, Kókai E, Németh ZH, Pacher P, Virág L, Leibovich SJ, Haskó G. 2013. Adenosine augments IL-10-induced STAT3 signaling in M2c macrophages. J Leukoc Biol 94:1309–1315. doi: 10.1189/jlb.0113043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tomioka H, Tatano Y, Maw WW, Sano C, Kanehiro Y, Shimizu T. 2012. Characteristics of suppressor macrophages induced by mycobacterial and protozoal infections in relation to alternatively activated M2 macrophages. Clin Dev Immunol 2012:635451. doi: 10.1155/2012/635451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Behnsen J, Perez-Lopez A, Nuccio S-P, Raffatellu M. 2015. Exploiting host immunity: the Salmonella paradigm. Trends Immunol 36:112–120. doi: 10.1016/j.it.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Helaine S, Cheverton AM, Watson KG, Faure LM, Matthews SA, Holden DW. 2014. Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science 343:204–208. doi: 10.1126/science.1244705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nathan C. 2002. Points of control in inflammation. Nature 420:846–852. doi: 10.1038/nature01320. [DOI] [PubMed] [Google Scholar]
- 69.Bellingan GJ, Caldwell H, Howie SE, Dransfield I, Haslett C. 1996. In vivo fate of the inflammatory macrophage during the resolution of inflammation: inflammatory macrophages do not die locally, but emigrate to the draining lymph nodes. J Immunol 157:2577–2585. [PubMed] [Google Scholar]
- 70.Nath G, Pratap CB, Patel SK, Gulati AK, Tripathi SK. 2011. Isolation of Salmonella Typhi from apparently healthy liver. Infect Genet Evol 11:2103–2105. doi: 10.1016/j.meegid.2011.08.029. [DOI] [PubMed] [Google Scholar]
- 71.Nath G, Mauryal P, Gulati AK, Singh TB, Srivastava R, Kumar K, Tripathi SK. 2010. Comparison of Vi serology and nested PCR in diagnosis of chronic typhoid carriers in two different study populations in typhoid endemic area of India. Southeast Asian J Trop Med Public Health 41:636–640. [PubMed] [Google Scholar]
- 72.Gonzalez-Escobedo G, Marshall JM, Gunn JS. 2011. Chronic and acute infection of the gall bladder by Salmonella Typhi: understanding the carrier state. Nat Rev Microbiol 9:9–14. doi: 10.1038/nrmicro2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Crawford RW, Rosales-Reyes R, Ramírez-Aguilar MDLL, Chapa-Azuela O, Alpuche-Aranda C, Gunn JS. 2010. Gallstones play a significant role in Salmonella spp. gallbladder colonization and carriage. Proc National Acad Sci 107:4353–4358. doi: 10.1073/pnas.1000862107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gonzalez-Escobedo G, Gunn JS. 2013. Gallbladder epithelium as a niche for chronic Salmonella carriage. Infect Immun 81:2920–2930. doi: 10.1128/IAI.00258-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gebhardt T, Palendira U, Tscharke DC, Bedoui S. 2018. Tissue-resident memory T cells in tissue homeostasis, persistent infection, and cancer surveillance. Immunol Rev 283:54–76. doi: 10.1111/imr.12650. [DOI] [PubMed] [Google Scholar]
- 76.Uzzau S, Figueroa-Bossi N, Rubino S, Bossi L. 2001. Epitope tagging of chromosomal genes in Salmonella. Proc Natl Acad Sci U S A 98:15264–15269. doi: 10.1073/pnas.261348198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Moon JJ, Chu HH, Hataye J, Pagán AJ, Pepper M, McLachlan JB, Zell T, Jenkins MK. 2009. Tracking epitope-specific T cells. Nat Protoc 4:565–581. doi: 10.1038/nprot.2009.9. [DOI] [PMC free article] [PubMed] [Google Scholar]