Abstract
Background
Given the unceasing worldwide surge in COVID-19 cases, there is an imperative need to develop highly specific and sensitive serology assays to define exposure to Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2).
Methods
Pooled plasma samples from PCR positive COVID-19 patients were used to identify linear B-cell epitopes from a SARS-CoV-2 peptide library of spike (S), envelope (E), membrane (M), and nucleocapsid (N) structural proteins by peptide-based ELISA. Hit epitopes were further validated with 79 COVID-19 patients with different disease severity status, 13 seasonal human CoV, 20 recovered SARS patients and 22 healthy donors.
Findings
Four immunodominant epitopes, S14P5, S20P2, S21P2 and N4P5, were identified on the S and N viral proteins. IgG responses to all identified epitopes displayed a strong detection profile, with N4P5 achieving the highest level of specificity (100%) and sensitivity (>96%) against SARS-CoV-2. Furthermore, the magnitude of IgG responses to S14P5, S21P2 and N4P5 were strongly associated with disease severity.
Interpretation
IgG responses to the peptide epitopes can serve as useful indicators for the degree of immunopathology in COVID-19 patients, and function as higly specific and sensitive sero-immunosurveillance tools for recent or past SARS-CoV-2 infections. The flexibility of these epitopes to be used alone or in combination will allow for the development of improved point-of-care-tests (POCTs).
Funding
Biomedical Research Council (BMRC), the A*ccelerate GAP-funded project (ACCL/19-GAP064-R20H-H) from Agency of Science, Technology and Research (A*STAR), and National Medical Research Council (NMRC) COVID-19 Research fund (COVID19RF-001) and CCGSFPOR20002. ATR is supported by the Singapore International Graduate Award (SINGA), A*STAR.
Keywords: Epitopes, SARS-CoV-2, COVID-19, Patients, Biomarkers
Research in Context.
Evidence before this study
We searched PubMed on March 30, 2020, for studies that performed immunoassays on the detection of individuals with Coronavirus Disease 2019 (COVID-19). The keywords used were “COVID-19″, OR “SARS-CoV-2″, AND “immunoassays”, OR “serology”. While there were several reports on serological assays using SARS-CoV-2 spike protein as a whole or subunits, and nucleocapsid protein to identify COVID-19 patients, the immunodominant linear B-cell epitopes on these viral antigens have not been determined and validated on patient cohorts. Moreover, serological approaches on using specific linear B-cell sequences in combination to distinguish COVID-19 patients from other seasonal coronavirus or healthy controls, and as potential markers for severity-associated disease progression were not reported.
Added value of this study
In our study, 81 patients from the Singapore COVID-19 outbreak were screened for antibodies against linear B-cell peptide library encompassing the entire SARS-CoV-2 structural proteome. Our initial screening with pooled peptides (five peptides in one pool) and pooled patient samples validated the immunogenicity of coronavirus spike and nucleocapsid proteins. Further characterisation to identify immunodominant peptide sequences was subsequently demonstrated. Immunodominant epitopes were validated to be highly specific and sensitive in differentiating SARS-CoV-2 infections from other coronavirus infections. Moreover, several epitopes were found to be strongly associated with COVID-19 severity. In addition to guiding vaccine designs and assessment, determining specific linear B-cell epitopes will allow a more targeted immune-based approach than whole viral protein. Using these immunodominant peptides alone or in combination would allow for the development of a highly specific and sensitive immunoassay to detect individuals with recent or past exposure to SARS-CoV-2. Ideally, the implementation of peptides on point-of-care tests would be more specific and cost-effective than protein-based systems.
Implications of all the available evidence
Current immunoassays against SARS-CoV-2 utilise recombinant viral protein, which may be less specific and sensitive, thereby giving rise to issues of false positive results. We have highlighted the importance of using specific immunodominant B-cell linear epitopes as a targeted approach for identifying individuals with past or recent SARS-CoV-2 exposure.
Alt-text: Unlabelled box
1. Introduction
The ongoing pandemic caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) [1] has resulted in more than 4.5 million confirmed human infection cases in 213 countries, with a global mortality rate of almost 7% [2], which has led to partial or total confinement of various countries. This unprecedented crisis has generated major socio-economic disruptions worldwide, overwhelming healthcare systems in both developed and developing countries. While quantitative reverse transcriptase PCR (qRT-PCR) remains as a gold standard for the early detection of SARS-CoV-2-infected individuals, the incidence of false negative diagnosis due to insufficient viral genetic material at the point of detection has limited such molecular-based assays [3]. Although rapid tests to detect acute infections are crucial in light of the current pandemic, it is also necessary to develop efficient assays for the detection of exposure in order to successfully mitigate virus transmission. Thus, there is an imperative need to rapidly develop highly specific and sensitive serology-based approaches to confirm molecular diagnosis, and enable immunosurveillance to identify individuals with recent or past exposure to SARS-CoV-2.
To date, several studies and pre-prints have described serological assays to detect SARS-CoV-2-specific antibodies in patient blood specimens [3], [4], [5], [6], [7]. These immunoassays primarily use recombinant coronavirus proteins, spike (S), which is exposed on the surface of the viral particle, and nucleocapsid (N), which is highly expressed during infection [5, 8]. The wealth of findings from these studies has shed some light on the general humoral response upon SARS-CoV-2 infection, where anti-SARS-CoV-2-IgM and IgG are detectable as early as 3 and 4 days post-illness onset (pio), respectively [3, 7]. Given the seasonal circulation of several human coronaviruses (hCoV) including 229E, OC43, NL63 and HKU1, which are responsible for common cold [9], coupled with individuals who have prior exposure to Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) during the outbreak in 2003 [10], cross-reactivity amongst these genetically similar viruses is inevitable, which may lead to confounding detection outcomes when using recombinant proteins (sharing high homology) [11]. Furthermore, the use of recombinant proteins, in addition to high costs and storage constraints, may give rise to batch-to-batch variations, which would greatly affect the reproducibility of immunoassays [12]. Thus, it remains crucial to ascertain the specificity and sensitivity profiles for the development of efficient and precise immunoassays. Lastly, specific regions on the viral proteins recognised by virus-specific antibodies have yet to be further characterised, and this would be fundamental for a more targeted immune-based approach.
This study reports a proteome-wide linear B-cell epitope screen of SARS-CoV-2 S, envelope (E), membrane (M) and N virion proteins using plasma samples collected from COVID-19 patients during the SARS-CoV-2 outbreak in Singapore [13]. We present four immunodominant peptides: S14P5, S20P2, S21P2 and N4P5, located on the S and N proteins of SARS-CoV-2. We further propose the best peptide combination with the highest specificity and sensitivity for determination of SARS-CoV-2 exposure. Moreover, the magnitude of the humoral responses against these epitopes shows a strong positive association with increasing disease severity.
2. Materials and methods
2.1. Ethical approvals
The study design and protocols for COVID-19, recovered SARS and seasonal hCoV patient cohorts were evaluated by National Healthcare Group (NHG) Domain Specific Review Board (DSRB) and approved under study numbers 2012/00917, 2020/00091 and 2020/00076, respectively. Healthy donor samples were collected under study numbers 2017/2806 and NUS IRB 04-140. Residual serum sample from National Healthy Survey (NHS) collected in 2010 under study number 006/2010 was used as a positive control in peptide-based ELISA [14]. Written informed consent was obtained from participants in accordance with the Declaration of Helsinki for Human Research.
2.2. Patients and sample collection
2.2.1. COVID-19 patients
Eighty-one patients who tested PCR-positive for SARS-CoV-2 in nasopharyngeal swab samples and admitted to a public hospital in Singapore were recruited into the study from January to March 2020 [13]. Demographic data, clinical and laboratory parameters, and clinical severity during the hospitalisation period were retrieved from patient records (Table 1). Patients were classified into three groups based on clinical severity: mild (no pneumonia on chest radiographs (CXR) at baseline and during hospital admission; clinical severity 0), moderate (pneumonia on CXR without hypoxia; clinical severity 1), and severe (pneumonia on CXR with hypoxia (desaturation to ≤94%); clinical severity 2). Whole blood of patients was collected into BD Vacutainer® CPT™ tubes (BD Biosciences, #362753), and centrifuged at 1700 g for 20 min to obtain plasma fractions. Plasma samples were categorised according to three timepoints: median 5 days post-illness onset (pio), median 10–14 days pio, and median 23 days pio.
Table 1.
Demographic and clinical data of COVID-19 patients.
| Patients (n = 81) | |
|---|---|
| Demographics | |
| Age, years | 45 (13) |
| Sex | |
| Male | 48 (59.3%) |
| Female | 33 (40.7%) |
| Ethnicity | |
| Chinese | 68 (84.0%) |
| Others | 13 (16.0%) |
| Comorbidities | 28 (34.6%) |
| Diabetes | 7 (8.6%) |
| Hypertension | 15 (18.5%) |
| Others | 11 (13.6%) |
| Vital signs at admission | |
| Temperature, °C | 37.7 (0.9) |
| Heart rate, beats/min | 91.4 (16.6) |
| Respiratory rate, per min | 18.4 (1.9) |
| Diastolic blood pressure, mmHg | 97.5 (2.4) |
| Systolic blood pressure, mmHg | 132.2 (18.5) |
| Oxygen saturation, % | 77.8 (15.2) |
| Laboratory findings | |
| Haemoglobin, g/dL | 13.8 (1.6) |
| Haematocrit, % | 40.8 (4.6) |
| Platelets, x 109/L | 194.8 (69.8) |
| White blood cells, x 109/L | 5.3 (3.0) |
| Lymphocytes, x 109/L | 1.2 (0.6) |
| Neutrophils, x 109/L | 4.3 (7.7) |
| Monocytes, x 109/L | 0.6 (1.1) |
| C-reactive protein (CRP), mg/L | 37.4 (55.7) |
| Creatinine, μmol/L | 75.0 (45.3) |
| Lactate dehydrogenase (LDH), U/L | 514.3 (298.4) |
| Alanine aminotransferase (ALT), U/L | 34.6 (28.1) |
| Clinical outcome (clinical severity; group) | |
| No pneumonia (0; mild) | 34 (42.0%) |
| Pneumonia, without hypoxia (1; moderate) | 28 (34.5%) |
| Pneumonia, with hypoxia (2; severe) | 19 (23.5%) |
Data represented as Mean (S•D) or n (%). COVID-19: Coronavirus Disease-19.
2.2.2. Recovered SARS and seasonal hCoV patients
A total of 20 individuals previously diagnosed with SARS-CoV during the outbreak in 2003 [15] were contacted and enrolled. Plasma fractions from recovered SARS individuals were isolated as described above. Archived paired samples from hCoV patients collected between 2012 and 2013 were also used in this study. This included pre- and post-infected samples from seven alpha-CoV (229E/NL63) and six beta-CoV (OC43) infections confirmed using the SeeGene RV12 respiratory multiplex kit [16]. Demographic data were retrieved from patient records (Supplementary Table 1).
2.3. Design of linear peptide libraries
SARS-CoV-2 and SARS-CoV full-length peptide sequences spanning the spike (S), envelope (E), membrane (M) and nucleocapsid (N) were obtained from NCBI GenBank accession numbers MN908947.3 and NC_004718.3, respectively. Biotinylated linear peptides of 18-mer overlapping sequences of 10 residues were synthesised (Mimotopes) and reconstituted in DMSO (Sigma-Aldrich, #D2650). An initial screen was conducted using pooled peptide sets of five to eight peptides per pool.
2.4. Plasma inactivation and peptide-based ELISA
Plasma fractions were treated with Triton™ X-100 (ThermoFisher Scientific, #28314) to a final concentration of 1% for 2 h at room temperature (RT) for virus inactivation [17]. Epitope screening was performed using a peptide-based ELISA as previously described [18], [19], [20], [21]. Briefly, Maxisorp flat-bottom 96-well plates (ThermoFisher Scientific, #442404) were coated overnight at 4 °C with 1:2000 dilution of NeutrAvidin protein (1 mg/ml) (ThermoFisher Scientific, #31050) in PBS. Plates were blocked with a 0.01% Polyvinyl Alcohol (PVA) (Sigma-Aldrich, #341584) solution in 0.1% PBST (blocking buffer) before addition of pooled or single biotinylated peptides (1:2000 dilution in 0.1% PBST), and inactivated plasma samples (1:1000 dilution in 0.1% PBST). Goat anti-human IgM-HRP (Jackson ImmunoResearch, #109-035-043, RRID: AB_2337581) or goat anti-human IgG-HRP (Jackson ImmunoResearch, #109-035-088, RRID: AB_2337584) diluted in blocking buffer was used for detection of peptide-bound antibodies. For development, tetramethylbenzidine (TMB) substrate (Sigma-Aldrich, #T8665) was added to the plates and the reaction was stopped using 0.16 M sulfuric acid (Merck, #1.00731.1000). Absorbance measurements were done using two wavelengths (450 nm and 690 nm) on an Infinite M200 plate reader (Tecan, firmware V_2.02_11/06). In all steps, plates were incubated at RT for 1 h on a rotating shaker and washed twice with 0.1% PBST in between steps.
2.5. Sequence alignment and computational modelling
Sequence alignment was conducted for each structural protein of SARS-CoV-2 and SARS-CoV S, E, M and N using Clustal Omega (EMBL-EBI, version 1.2.4). Structural data of immunodominant regions on SARS-CoV-2 S and N proteins were retrieved from PDB 6VSB and 6YVO and were modelled in PyMol (Schrodinger, version 2.2.0).
2.6. Data and statistical analyses
Optical density (OD) values of samples were normalised to a positive control to account for plate-to-plate variations, and background signals were subtracted. In the first screening phase of the peptide pools, a cut-off value of mean + 3SD of healthy control samples was used to single out individual peptides detected by COVID-19 patients. For subsequent analyses with individual peptides, receiver operating characteristic (ROC) curves for peptides to differentiate between SARS-CoV-2 infections and others were performed, and the areas under the curve (AUCs) were calculated for each peptide (supplementary methods). The best threshold for each ROC curve was determined as the maximum of the Youden's J statistic [22] with a constraint that the threshold has to be a positive value (supplementary methods). For peptide combination analyses, logistic regression models were used to model the combinatory effects of two, three or four peptides’ OD readings towards the prediction of SARS-CoV-2 infection from others as a binary outcome. The logistic regression model fitted values were then used for ROC analysis to identify the optimal thresholds (via Youden's J statistic) as well as AUCs (supplementary methods). Data analyses were done using GraphPad Prism (GraphPad Software, version 8.0.0). Kruskal–Wallis tests and post hoc tests using Dunn's multiple comparison tests were used to identify significant differences. Correlation analysis was carried out using Spearman's rank correlation. Absolute rho values greater than 0.3 are considered to be a fair correlation [23]. P-values less than 0.05 are considered significant.
2.7. Data availability
Data can be obtained upon request to the corresponding author.
3. Results
3.1. Immunodominant linear B-cell epitopes on SARS-CoV-2 S and N proteome
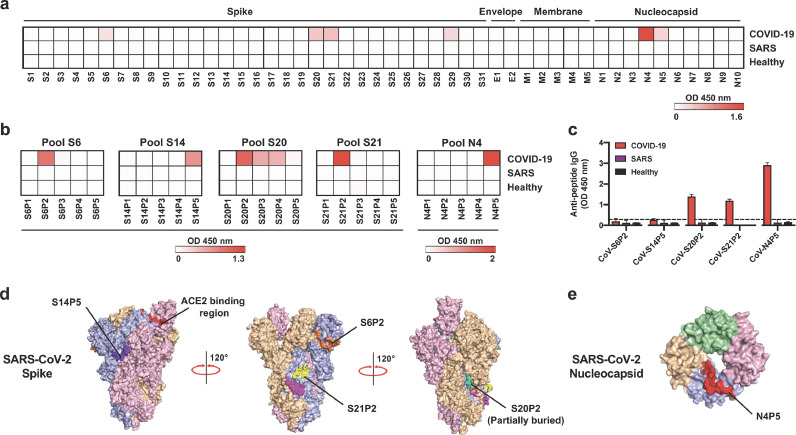
In order to identify potential linear B-cell epitopes, pooled plasma samples from 18 COVID-19 patients of the Singapore outbreak [13] (Table 1) collected during the convalescent phase of median 14 days pio were screened by peptide-based ELISA for IgG reactivity against a pooled peptide library of five peptides, encompassing the entire SARS-CoV-2 structural proteome (Fig. 1a). Freshly drawn plasma samples of 20 recovered SARS individuals and 22 healthy donors were also assessed in parallel (Fig. 1a). While there was no specific recognition on the E and M peptide pools by the COVID-19 patient samples (Fig. 1a), several hits were identified in the S and N proteins, based on the cut-off value using mean + 3SD of healthy controls. Positive pools S6, S14, S20, S21 from S glycoprotein, and N4 from N protein, were chosen for further characterisation of the specific peptide recognised (Fig. 1a).
Fig. 1.
Identification of IgG-specific linear B-cell epitopes on spike (S), envelope (E), membrane (M), and nucleocapsid (N) SARS-CoV-2 proteins with COVID-19 patient plasma. (a–c) Plasma samples of COVID-19 patients collected at timepoint of median 14 days post-illness onset (pio, n = 18), recovered SARS individuals (n = 20) and healthy donors (n = 22) were pooled, respectively. (a) Pooled plasma samples (1:1000 dilution) were tested in a peptide-based ELISA of peptide pools covering SARS-CoV-2 S, E, M, and N proteins for IgG. Each pool consists of 5–8 peptides, with an overlapping sequence of 10 amino acids. OD results are presented in a heatmap, with white and red colours denoting low and high OD values, respectively. The cut-off for positive pools was derived from mean + 3SD of pooled healthy donors. (b) Individual peptides of the positive peptide pools in (a) were screened with pooled plasma samples (1:1000 dilution) to determine the IgG hit peptides. OD results are presented in a heatmap, with white and red colours denoting low and high OD values, respectively. (c) Peptide-binding response of pooled plasma samples (1:1000 dilution) against the corresponding hit peptides on SARS-CoV, with bar graphs presented as mean ± SEM, and dotted line denoting mean + 3SD of pooled healthy donors. All data in (a–c) are of two independent experiments. (d,e) Diagrams showing the localisation of SARS-CoV-2 specific IgG peptides S6P2, S14P5, S20P2, S21P2, and N4P5 on (d) SARS-CoV-2 S (PDB: 6VSB), and (e) N (PDB: 6YVO) protein. Each monomer is denoted as either pink, blue, orange or green. All peptides are annotated on the blue monomer.
Within each pool of five overlapping peptides, further detailed screening on each individual peptide allowed the identification of distinct peptides highly recognised by IgGs present in the plasma samples of COVID-19 patients. In addition to two reported peptide epitopes S14P5 and S21P2 [21], peptide S6P2 from pool S6, peptide S20P2 from pool S20, and peptide N4P5 from pool N4 were dominant hits from their respective pools (Fig. 1b, Table 2 and Supplementary Table 2). It is also noteworthy that IgG levels of recovered SARS and healthy individuals in all of the assays were very low or negligible, highlighting that COVID-19 patients recognised these peptides with high specificity (Fig. 1a and b).
Table 2.
Peptide sequences of putative immunodominant SARS-CoV-2 IgG linear B-cell epitopes.
| SARS-CoV-2 | SARS-CoV | Identity with SARS-CoV-2 (%) | |||||
|---|---|---|---|---|---|---|---|
| Peptide | Starting aa | Ending aa | Polypeptide sequence | Starting aa | Ending aa | Polypeptide sequence | |
| Spike (S) glycoprotein | |||||||
| S6P2 | 209 | 226 | PINLVRDLPQGFSALEPL | 201 | 218 | QPIDVVRDLPSGFNTLKP | 64.71 |
| S14P5 | 553 | 570 | TESNKKFLPFQQFGRDIA | 537 | 554 | VLTPSSKRFQPFQQFGRD | 75.00 |
| S20P2 | 769 | 786 | GIAVEQDKNTQEVFAQVK | 753 | 770 | AAEQDRNTREVFAQVKQM | 81.25 |
| S21P2 | 809 | 826 | PSKPSKRSFIEDLLFNKV | 793 | 810 | KPTKRSFIEDLLFNKVTL | 93.75 |
| Nucleocapsid (N) phosphoprotein | |||||||
| N4P5 | 153 | 170 | NNAAIVLQLPQGTTLPKG | 153 | 170 | NNNAATVLQLPQGTTLPK | 94.12 |
Sequences were obtained from GenBank (accession number: MN908947.3 and NC_004718.3), National Centre for Biotechnology Information (NCBI); aa: amino acid, SARS-CoV: Severe Acute Respiratory Syndrome Coronavirus, SARS-CoV-2: Severe Acute Respiratory Syndrome Coronavirus 2.
Given the high level of homology between SARS-CoV-2 and the closely-related SARS-CoV [8, 24], pooled COVID-19 patient plasma samples were also screened against the corresponding hit regions on SARS-CoV S and N proteins to assess for potential cross-reactivity (Fig. 1c and Table 2). Interestingly, while COVID-19 patient plasma samples were able to recognise SARS-CoV versions of peptides S20P2 and N4P5, there was no binding to S6P2, alluding to the identification of a potential SARS-CoV-2 specific epitope (Fig. 1c). The localisation of all the putative epitopes on the respective viral proteins is shown in Fig. 1d and e. All epitopes are located on the surface of the viral protein, except for S20P2, which is partially buried in the S protein 3D structure (Fig. 1d).
3.2. SARS-CoV-2 epitopes can serve as highly specific and sensitive tools
Anti-SARS-CoV-2 antibody levels differ with infection phases, with IgM being detected as early as 3 days pio [25]. To investigate whether IgM epitopes could be identified with the same approach, pooled patient plasma samples collected during the early acute phase of median 5 days pio were screened with samples taken at median 14 days pio (Supplementary Fig. 1a). Two positive pools, S20 from S glycoprotein, and N10 from N protein, were detected albeit at a lower recognition level at median 5 days pio, as compared to samples taken during median 14 days pio (Supplementary Fig. 1a). These peptide pools were further characterised to determine the specific regions. Peptides S20P4 and N10P1 were dominant hits in their respective pools (Supplementary Fig. 1b and supplementary Table 2). Interestingly, COVID-19 patient plasma samples were able to recognise both corresponding hit regions on SARS-CoV S and N proteins, suggesting that these could be SARS cross-reactive epitopes for IgM detection (Supplementary Fig. 1b). Peptide S20P4 is localised on the surface of the S viral protein (Supplementary Fig. 1d). However, when validated with a larger number of patient samples, the putative epitopes were only recognised by a minority of the COVID-19 patients, indicating that this approach is inefficient for IgM detection (Supplementary Fig. 1E and supplementary Table 3).
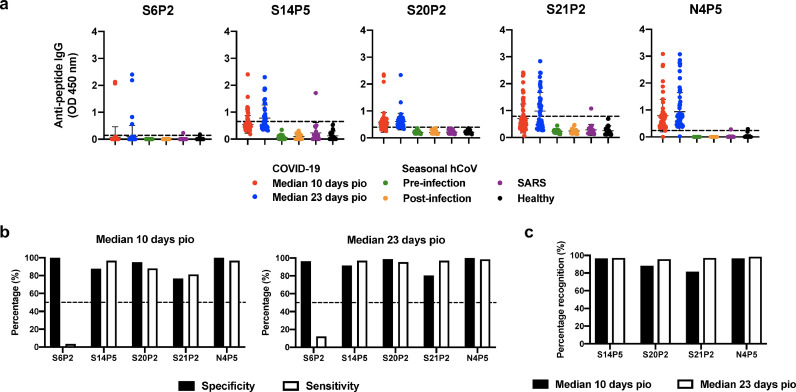
To further determine the longitudinal IgG profiling in a larger cohort of COVID-19 patients (n = 79), individual plasma samples collected at median 10 days pio (n = 59) and median 23 days pio (n = 66) were assessed and compared. All the four putative epitopes, S14P5, S20P2, S21P2, and N4P5, except S6P2, were distinctly recognised by plasma samples from most COVID-19 patients (Fig. 2a). Strikingly, there was no recognition to all epitopes by samples obtained from patients infected with seasonal hCoV (baseline samples from patients before infection were used as controls), and healthy donors (Fig. 2a). However, cross-reactivity was observed for epitopes S14P5 and S21P2 from one out of 20 recovered SARS individuals (Fig. 2a).
Fig. 2.
Specificity and sensitivity of putative IgG epitopes to detect anti-SARS-CoV-2 antibodies from COVID-19 patients. (a) Plasma samples from COVID-19 patients collected at timepoints of median 10 days post-illness onset (pio, n = 59) and median 23 days pio (n = 66), sera from seasonal hCoV-infected patients (n = 13) either pre- or post-infection, and plasma samples from recovered SARS patients (n = 20) and healthy donors (n = 22) were screened at 1:1000 dilution against five IgG-specific putative epitopes: S6P2, S14P5, S20P2, S21P2, and N4P5. Data are shown as mean ± SD of two independent experiments, with dotted lines indicating mean + 3SD of healthy donors. (b) Percentage specificity and sensitivity of putative epitopes to detect SARS-CoV-2-specific antibodies in plasma samples from COVID-19 patients during the timepoints of median 10 days pio (left panel) and median 23 days pio (right panel). Specificity and sensitivity values were derived from a threshold determined by the maximisation of the Youden's J statistic with a constraint that the threshold value has to be positive. (c) Percentage recognition of epitopes during the timepoints of median 10 days pio (black bars) and median 23 days pio (white bars).
In order to define an optimal cut-off value for each peptide, we used the Youden J statistical method, which determines the sensitivity and specificity profiles of the assay in one single test [22] (Methods, supplementary methods). This will guide the development of diagnostic tests and estimate the probability of an informed decision. Epitopes S20P2 and N4P5 exhibited specificity and sensitivity profiles of about 90% (Fig. 2b, Table 3 and Supplementary Fig. 3). Both S14P5 and S21P2 have a moderate specificity and sensitivity level of >80%, while S6P2 has a poor ability to correctly detect COVID-19 patients with a sensitivity level of <10% (Fig. 2b, Table 3 and Supplementary Fig. 3). S14P5 and N4P5 were consistently recognised by COVID-19 patients at a frequency of >97%, whereas the frequency of recognising S20P2 and S21P2 ranged between 81% and 88% at median 10 days pio (Fig. 2c). However, at median 23 days pio, the percentage recognition for all four epitopes is >95%. Interestingly, when the various peptide combinations of S14P5, S20P2, S21P2 and N4P5 were predicted using bioinformatics tools (supplementary methods), the specificity and sensitivity of detection could approach 100% at median 10 days (Table 4).
Table 3.
Receiver operating characteristic (ROC) profiles of IgG-specific SARS-CoV-2 linear B-cell epitopes.
| Peptide | Threshold | Specificity (%) | Sensitivity (%) | AUC | Wilcoxon Padj value |
|---|---|---|---|---|---|
| Median 10 days pio | |||||
| S6P2 | 1.153 | 100.00 | 3.39 | 0.847 | <0.0001 |
| S14P5 | 0.288 | 87.81 | 96.61 | 0.915 | <0.0001 |
| S20P2 | 0.328 | 95.12 | 88.14 | 0.974 | <0.0001 |
| S21P2 | 0.275 | 76.83 | 81.36 | 0.864 | <0.0001 |
| N4P5 | 0.313 | 100.00 | 96.61 | 0.994 | <0.0001 |
| Median 23 days pio | |||||
| S6P2 | 0.022 | 96.34 | 12.12 | 0.895 | <0.0001 |
| S14P5 | 0.354 | 91.46 | 96.97 | 0.962 | <0.0001 |
| S20P2 | 0.360 | 98.78 | 95.46 | 0.993 | <0.0001 |
| S21P2 | 0.291 | 80.49 | 96.97 | 0.943 | <0.0001 |
| N4P5 | 0.335 | 100.00 | 98.49 | 0.999 | <0.0001 |
pio: post-illness onset, AUC: area under curve, SARS-CoV-2: Severe Acute Respiratory Syndrome Coronavirus 2.
Table 4.
Receiver operating characteristic (ROC) profiles of IgG-specific SARS-CoV-2 linear B-cell epitopes used in combination.
| Number of peptides in combination | Peptide combination | Threshold | Specificity (%) | Sensitivity (%) | AUC |
|---|---|---|---|---|---|
| Median 10 days pio | |||||
| 4 | S14P5 + S20P2 + S21P2 + N4P5 | 0.500 | 100.00 | 100.00 | 1.000 |
| 3 | S14P5 + S20P2 + N4P5 | 0.500 | 100.00 | 100.00 | 1.000 |
| 3 | S20P2 + S21P2 + N4P5 | 0.500 | 100.00 | 100.00 | 1.000 |
| 3 | S14P5 + S21P2 + N4P5 | 0.804 | 100.00 | 98.30 | 0.997 |
| 3 | S14P5 + S20P2 + S21P2 | 0.414 | 92.70 | 91.50 | 0.981 |
| 2 | S20P2 + N4P5 | 0.269 | 98.80 | 100.00 | 1.000 |
| 2 | S14P5 + N4P5 | 0.803 | 100.00 | 98.30 | 0.997 |
| 2 | S21P2 + N4P5 | 0.481 | 98.80 | 98.30 | 0.992 |
| 2 | S20P2 + S21P2 | 0.407 | 92.70 | 91.50 | 0.982 |
| 2 | S14P5 + S20P2 | 0.503 | 93.90 | 88.10 | 0.975 |
| 2 | S14P5 + S21P2 | 0.373 | 84.10 | 96.60 | 0.929 |
| Median 23 days pio | |||||
| 4 | S14P5 + S20P2 + S21P2 + N4P5 | 0.500 | 100.00 | 100.00 | 1.000 |
| 3 | S14P5 + S20P2 + N4P5 | 0.500 | 100.00 | 100.00 | 1.000 |
| 3 | S20P2 + S21P2 + N4P5 | 0.500 | 100.00 | 100.00 | 1.000 |
| 3 | S14P5 + S21P2 + N4P5 | 0.794 | 100.00 | 98.50 | 0.999 |
| 3 | S14P5 + S20P2 + S21P2 | 0.353 | 98.80 | 100.00 | 0.998 |
| 2 | S20P2 + N4P5 | 0.500 | 100.00 | 100.00 | 1.000 |
| 2 | S14P5 + N4P5 | 0.820 | 100.00 | 98.50 | 0.999 |
| 2 | S21P2 + N4P5 | 0.792 | 100.00 | 98.50 | 0.999 |
| 2 | S20P2 + S21P2 | 0.307 | 97.60 | 97.00 | 0.998 |
| 2 | S14P5 + S20P2 | 0.278 | 97.60 | 100.00 | 0.997 |
| 2 | S14P5 + S21P2 | 0.485 | 92.70 | 97.00 | 0.971 |
pio: post-illness onset, AUC: area under curve, SARS-CoV-2: Severe Acute Respiratory Syndrome Coronavirus 2.
3.3. Epitope-specific IgG levels associate with disease severity
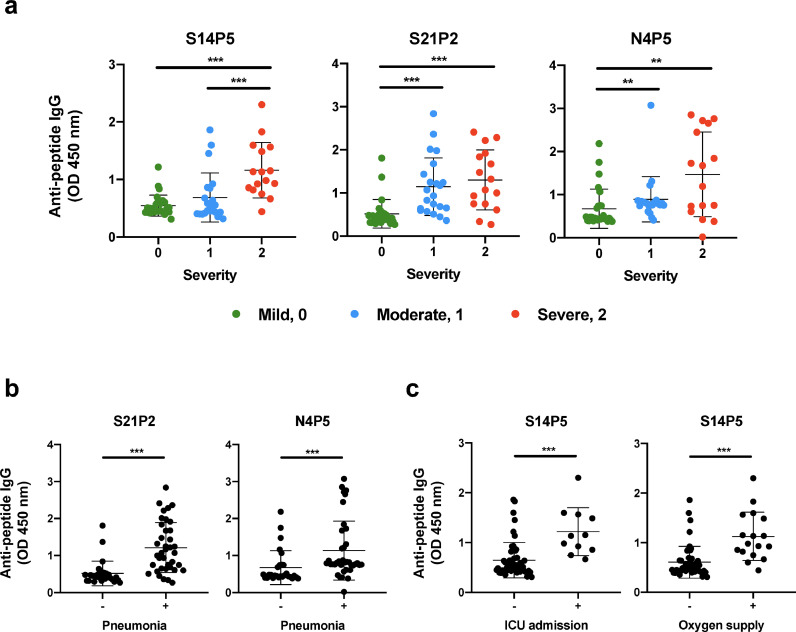
SARS-CoV-2 infected patients manifest different disease outcomes that can be categorised into three groups: mild (no pneumonia, clinical severity 0), moderate (pneumonia with no hypoxia, clinical severity 1), or severe (pneumonia with hypoxia, clinical severity 2) [26]. To elucidate if anti-viral IgG responses at different days pio were associated with disease severity, antibody responses against the four highly recognised epitopes were analysed based on disease severity (green denotes mild, blue denotes moderate, and red denotes severe) (Supplementary Fig. 2a and Fig. 3a). Surprisingly, only IgG responses at median 23 days pio against epitopes S14P5, S21P2 and N4P5 were higher in severe COVID-19 patients than in the mild and moderate groups (Supplementary Fig. 2a and Fig. 3a). It was also observed that IgG responses to epitopes S21P2 and N4P5 were associated with pneumonia at median 23 days pio (Fig. 3b). In more severe cases with patients requiring admission into ICU or oxygen supplementation, IgG levels to S14P5 at median 23 days pio were also significantly higher (Fig. 3c). Correlation analyses of these epitopes with other clinical parameters of severity showed positive association with C-reactive protein (CRP), lactate dehydrogenase (LDH) and lymphopenia (Supplementary Fig. 2b–d).
Fig. 3.
Association of anti-SARS-CoV-2 antibody response of COVID-19 patients with clinical severity during SARS-CoV-2 infection. Antibody profiles of COVID-19 patients at timepoint of median 23 days post-illness onset (pio, n = 66) against significant IgG peptides under different clinical measures of (a) disease severity, (b) pneumonia, and (c) intensive care unit (ICU) admission and oxygen supply requirement. Samples in (a) were classified based on severity with green denoting mild (clinical severity 0; n = 28), blue denoting moderate (clinical severity 1; n = 22), and red denoting severe (clinical severity 2; n = 16). Data are presented as mean ± SD of two independent experiments. Negative and positive observations in (b-c) are denoted as - and +, respectively. Statistical analysis was carried out using Kruskal-Wallis tests, followed by post hoc Dunn's multiple comparison tests. (**P<0•01, ***P<0•001).
4. Discussion
The development of sensitive and specific immunoassays that can determine SARS-CoV-2 exposure, and serve as sero-epidemiological surveillance tools would greatly help to control virus transmission. In this study, four immunodominant IgG-specific SARS-CoV-2 linear epitopes, S14P5, S20P2, S21P2 and N4P5, located on the S and N viral proteins were described. The S and N antigens are immunodominant amongst coronavirus antigens, and are commonly used in the development of serological assays against coronaviruses [5, 8, 27]. Sequences of these epitopes are well conserved across multiple SARS-CoV-2 strains [21] (Supplementary Table 4), with no detectable cross-reactivity against seasonal hCoV and recovered SARS individuals, despite a high level of homology with the corresponding regions on SARS-CoV (Table 2) [11]. Moreover, sequences of several identified epitopes overlap either completely or partially with reported SARS-CoV B-cell linear epitopes at regions QQFGRD in S14P5, FIEDLLFNKVTLADAGF in S21P2, and QLPQGTTLPKG in N4P5 [28]. Corroborating a recent study, no response was also detected against the SARS-CoV S protein by recovered SARS patients who were infected 17 years ago [21], suggesting that antibodies against SARS-CoV linear epitopes are short-lived [29]. As there is currently no active SARS-CoV transmission, it remains elusive whether antibodies against SARS-CoV would react with these immunodominant SARS-CoV-2 epitopes and generate false positive responses. We demonstrated in this report that the best performing linear B-cell epitopes, S20P2 and N4P5, could correctly discriminate SARS-CoV-2 from seasonal coronavirus infection in patients, with a specificity and sensitivity levels of >90% for single peptide detection.
Interestingly, simulation on the combinatorial use of two or more peptides can approach 100% sensitivity and specificity for the detection of COVID-19 patients. This is an important aspect for the design of point-of-care tests (POCTs) where the combination of two peptides (i.e. S20P2 plus N4P5) could allow for rapid IgG detection with a high level of specificity and sensitivity. This remains as one of the current limitation in the development of POCTs based on single SARS-CoV-2 proteins. Furthermore, the use of recombinant proteins may lead to false positive results and hamper specificity of the immune-based assays due to high level of cross-reactivities between SARS-CoV-2 and other human coronaviruses such as the closely-related SARS-CoV [11, 24, 30]. For instance, in a hypothetical scenario, a test that is 90% specific and 100% sensitive is used to screen a population of 10,000 persons without COVID-19 and 100 persons with COVID-19. In this scenario, 1100 persons will test positive, of which 1000 are false positives, creating a larger burden for follow-up diagnostic assays and potentially unnecessary quarantine measure.
The prevalence of human coronaviruses differ geographically and in different populations [9, 31]. As a result, this will influence the specificity and/or sensitivity of the immune-based assays due to the different cut-off values taken from various healthy controls around the world. However, the use of specific epitopes reported in this study may overcome the various specificity and sensitivity issues. Moreover, the use of high purity peptide-based immunoassays can overcome batch-to-batch inconsistencies resulting from variations in protein refolding and storage stability. Although the samples screened in this study were collected at median 10 days pio (with hospital admission of median 5 days pio), the value of developing POCTs with these immunodominant IgG epitopes would still be useful in identifying undiagnosed asymptomatic carriers. In the event that SARS-CoV-2 transmission becomes seasonal, this rapid testing will allow medical workers to quickly decide the best course of action and provide the critically-ill patients with the best chance of survival.
The wide spectrum of COVID-19 clinical severity ranges from asymptomatic, to a mild upper respiratory tract infection, to viral pneumonia, and to death from respiratory failure or associated complications [8, 25, 32, 33]. Our study revealed that IgG levels against epitopes S14P5, S21P2 and N4P5 in COVID-19 patients were associated with disease severity, confirming recent studies that reported an association of antibody levels against the S protein with disease severity [3, 5, 7, 26]. Moreover, within the severe group, high antibody levels against these linear B-cell epitopes were also detected by patients who either developed pneumonia or were admitted into the ICU. SARS-CoV-2 is just one of the many pathogens that can cause severe pneumonia. Ideally, screening an independent cohort with individuals experiencing severe pneumonia against these IgG epitopes, followed by validation with pathogen testing and other clinical data would be useful to confirm or exclude the diagnosis of COVID-19.
Antibody levels against these epitopes also correlated with clinical laboratory parameters such as CRP and LDH levels, which are elevated in severe COVID-19 patients [34, 35]. High IgG levels against epitope N4P5 correlated with lymphopenia, a phenomenon found in COVID-19 patients [36]. One plausible hypothesis for these associations is that severe patients had a higher viral load during the early infection phase that resulted in higher antibody responses. While a causality link between antibody response and enhanced severity remains to be determined, it is also possible that high specific-antibody levels may contribute to an enhanced lung inflammation in COVID-19 patients. Such an observation was reported in SARS-CoV-infected monkeys treated with an anti-S protein monoclonal antibody [37]. In addition, there could also be a higher level of non-neutralising antibody responses in severe patients that may lead to a detrimental consequence such as antibody dependant enhancement, which was demonstrated in vitro using SARS-CoV patients serum samples [38]. Nevertheless, these putative epitopes can serve as useful indicators on the degree of immunopathology in COVID-19 patients, and validation on a larger cohort remains necessary.
Altogether, in view of the recent studies demonstrating the presence of mutation and deletions in SARS-CoV-2, the identification of multiple epitopes for the detection of SARS-CoV-2 infection allows for constant refinement of the peptide sequences to be used in serological assays [39, 40]. An advantage of the epitopes reported in this study is the low rate of potential mutations, with a highest mutation rate of 1•35% for epitope S21P2 (16 SNP in 236 sequences out of 17,462 deposited complete genomes), and below 0•19% for all other peptides (Supplementary Table 4). An immediate application of these peptide epitopes could be developed as paper-based lateral flow assays [25]. Ideally, the implementation of peptides on POCTs would be easier and cost effective than protein-based systems. However, it is important to note that serological POCTs should not replace RT-PCR especially in the diagnosis of an early acute SARS-CoV-2 infection due to the delayed antibody response [25, 41, 42]. Nonetheless, the identification of IgG-specific epitopes on the S and N viral proteins expands the tool box needed for sero-epidemiological studies and vaccine assessment.
Funding sources
Biomedical Research Council (BMRC), the A*ccelerate GAP-funded project (ACCL/19-GAP064-R20H-H) from Agency of Science, Technology and Research (A*STAR), and National Medical Research Council (NMRC) COVID-19 Research fund (COVID19RF-001) and CCGSFPOR20002. ATR is supported by the Singapore International Graduate Award (SINGA), A*STAR. The funding sources had no role in the study design; collection, analysis, and interpretation of data; in the writing of the report; and in the decision to submit the paper for publication.
Declaration of Competing Interest
SNA, CYPL, GC, CMP, LR and LFPN have filed a technology disclosure on the identified peptides with the patent application number 10202002981P. All other authors declare no conflicts.
Acknowledgements
The authors would like to thank the study participants who donated their blood samples to this project, and the healthcare workers caring for COVID-19 patients. The authors also wish to thank Ding Ying and the Singapore Infectious Disease Clinical Research Network (SCRN) team for their help in patient recruitment, and Chai Siaw Ching and National Centre for Infectious Diseases Research Office team for their help in coordinating the recovered SARS and seasonal hCoV patients’ samples and data. The authors wish to thank Dr Danielle Anderson and her team at Duke-NUS for their technical assistance, and also to Dr Anis Larbi at the Singapore Immunology Network (SIgN), A*STAR for providing the healthy donor samples.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2020.102911.
Appendix. Supplementary materials
References
- 1.Cohen J., Normile D. New SARS-like virus in China triggers alarm. Science. 2020;367(6475):234. doi: 10.1126/science.367.6475.234. [DOI] [PubMed] [Google Scholar]
- 2.WHO. Coronavirus disease (COVID-2019) situation reports. 2020. Available from:https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/.
- 3.Long Q-x, Deng H-j, Chen J., Hu J., Liu B-z, Liao P. Antibody responses to SARS-CoV-2 in COVID-19 patients: the perspective application of serological tests in clinical practice. medRxiv. 2020;26(6):845–848. [Google Scholar]
- 4.Amanat F., Nguyen T., Chromikova V., Strohmeier S., Stadlbauer D., Javier A. A serological assay to detect SARS-CoV-2 seroconversion in humans. medRxiv. 2020;26:1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okba N.M.A., Muller M.A., Li W., Wang C., GeurtsvanKessel C.H., Corman V.M. SARS-CoV-2 specific antibody responses in COVID-19 patients. Emerg Infect Dis. 2020;26(7) doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiao D.A.T., Gao D.C., Zhang D.S. Profile of Specific Antibodies to SARS-CoV-2: the First Report. J Infect. 2020;81(1):147–178. doi: 10.1016/j.jinf.2020.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang B., Zhou X., Zhu C., Feng F., Qiu Y., Feng J. Immune phenotyping based on neutrophil-to-lymphocyte ratio and IgG predicts disease severity and outcome for patients with COVID-19. medRxiv. 2020 doi: 10.3389/fmolb.2020.00157. [Preprint]2020.03.12.20035048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahmed S.F., Quadeer A.A., McKay M.R. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses. 2020;12(3):254. doi: 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaunt E.R., Hardie A., Claas E.C.J., Simmonds P., Templeton K.E. Epidemiology and clinical presentations of the four human coronaviruses 229E, HKU1, NL63, and OC43 detected over 3 years using a novel multiplex real-time PCR method. J Clin Microbiol. 2010;48(8):2940. doi: 10.1128/JCM.00636-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parashar U.D., Anderson L.J. Severe acute respiratory syndrome: review and lessons of the 2003 outbreak. Int J Epidemiol. 2004;33(4):628–634. doi: 10.1093/ije/dyh198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Che X.Y., Qiu L.W., Liao Z.Y., Wang Y.D., Wen K., Pan Y.X. Antigenic cross-reactivity between severe acute respiratory syndrome-associated coronavirus and human coronaviruses 229E and OC43. J Infect Dis. 2005;191(12):2033–2037. doi: 10.1086/430355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoelzle K., Grimm J., Ritzmann M., Heinritzi K., Torgerson P., Hamburger A. Use of recombinant antigens to detect antibodies against Mycoplasma suis, with correlation of serological results to hematological findings. Clin Vaccine Immunol. 2007;14(12):1616–1622. doi: 10.1128/CVI.00345-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pung R., Chiew C.J., Young B.E., Chin S., Chen M.I., Clapham H.E. Investigation of three clusters of COVID-19 in Singapore: implications for surveillance and response measures. Lancet. 2020;395(10229):1039–1046. doi: 10.1016/S0140-6736(20)30528-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ang L.W., Kam Y.W., Lin C., Krishnan P.U., Tay J., Ng L.C. Seroprevalence of antibodies against chikungunya virus in Singapore resident adult population. PLoS Negl Trop Dis. 2017;11(12) doi: 10.1371/journal.pntd.0006163. e0006163-e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leong H.N., Earnest A., Lim H.H., Chin C.F., Tan C., Puhaindran M.E. SARS in Singapore–predictors of disease severity. Ann Acad Med Singap. 2006;35(5):326–331. [PubMed] [Google Scholar]
- 16.Jiang L., Lee V.J., Cui L., Lin R., Tan C.L., Tan L.W. Detection of viral respiratory pathogens in mild and severe acute respiratory infections in Singapore. Sci Rep. 2017;7:42963. doi: 10.1038/srep42963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Darnell M.E.R., Taylor D.R. Evaluation of inactivation methods for severe acute respiratory syndrome coronavirus in noncellular blood products. Transfusion. 2006;46(10):1770–1777. doi: 10.1111/j.1537-2995.2006.00976.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kam Y.-.W., Lum F.-.M., Teo T.-.H., Lee W.W.L., Simarmata D., Harjanto S. Early neutralizing IgG response to Chikungunya virus in infected patients targets a dominant linear epitope on the E2 glycoprotein. EMBO Mol Med. 2012;4(4):330–343. doi: 10.1002/emmm.201200213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kam Y.W., Lee C.Y., Teo T.H., Howland S.W., Amrun S.N., Lum F.M. Cross-reactive dengue human monoclonal antibody prevents severe pathologies and death from Zika virus infections. JCI Insight. 2017;2(8):e92428. doi: 10.1172/jci.insight.92428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kam Y.W., Leite J.A., Amrun S.N., Lum F.M., Yee W.X., Bakar F.A. ZIKV-specific NS1 epitopes as serological markers of acute Zika virus infection. J Infect Dis. 2019;220(2):203–212. doi: 10.1093/infdis/jiz092. [DOI] [PubMed] [Google Scholar]
- 21.Poh C.M., Carissimo G., Wang B., Amrun S.N., Lee C.Y.-.P., Chee R.S.-.L. Two linear epitopes on the SARS-CoV-2 spike protein that elicit neutralising antibodies in COVID-19 patients. Nat Commun. 2020;11(1):2806. doi: 10.1038/s41467-020-16638-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Youden W.J. Index for rating diagnostic tests. Cancer. 1950;3(1):32–35. doi: 10.1002/1097-0142(1950)3:1<32::aid-cncr2820030106>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 23.Chan Y.H. Biostatistics 104: correlational analysis. Singap Med J. 2003;44(12):614–619. [PubMed] [Google Scholar]
- 24.Lv H., Wu N.C., Tsang O.T.-.Y., Yuan M., Perera R.A.P.M., Leung W.S. Cross-reactive antibody response between SARS-CoV-2 and SARS-CoV infections. bioRxiv. 2020 doi: 10.1016/j.celrep.2020.107725. [Preprint]2020.03.15.993097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee C.Y.-.P., Lin R.T.P., Renia L., Ng L.F.P. Serological approaches for COVID-19: epidemiologic perspective on surveillance and control. Front Immunol. 2020;11:879. doi: 10.3389/fimmu.2020.00879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wong J.E.L., Leo Y.S., Tan C.C. COVID-19 in Singapore—current experience: critical global issues that require attention and action. JAMA. 2020;323(13):1243–1244. doi: 10.1001/jama.2020.2467. [DOI] [PubMed] [Google Scholar]
- 27.Lin Y., Shen X., Yang R.F., Li Y.X., Ji Y.Y., He Y.Y. Identification of an epitope of SARS-coronavirus nucleocapsid protein. Cell Res. 2003;13(3):141–145. doi: 10.1038/sj.cr.7290158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ahmed S.F., Quadeer A.A., McKay M.R. COVIDep: a web-based platform for real-time reporting of vaccine target recommendations for SARS-CoV-2. Nat Protoc. 2020;15:2141–2142. doi: 10.1038/s41596-020-0358-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang F., Quan Y., Xin Z.T., Wrammert J., Ma M.J., Lv H. Lack of peripheral memory B cell responses in recovered patients with severe acute respiratory syndrome: a six-year follow-up study. J Immunol. 2011;186(12):7264–7268. doi: 10.4049/jimmunol.0903490. [DOI] [PubMed] [Google Scholar]
- 30.Ou X., Liu Y., Lei X., Li P., Mi D., Ren L. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020;11(1):1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van der Hoek L. Human coronaviruses: what do they cause? Antivir Ther. 2007;12(4 Pt B):651–658. [PubMed] [Google Scholar]
- 32.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao Y., Li T., Han M., Li X., Wu D., Xu Y. Diagnostic utility of clinical laboratory data determinations for patients with the severe COVID-19. J Med Virol. 2020;92(7):791–796. doi: 10.1002/jmv.25770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mardani R., Ahmadi Vasmehjani A., Zali F., Gholami A., Mousavi Nasab S.D., Kaghazian H. Laboratory parameters in detection of COVID-19 patients with positive RT-PCR; a diagnostic accuracy study. Arch Acad Emerg Med. 2020;8(1) e43-e. [PMC free article] [PubMed] [Google Scholar]
- 36.Fan B.E., Chong V.C.L., Chan S.S.W., Lim G.H., Lim K.G.E., Tan G.B. Hematologic parameters in patients with COVID-19 infection. Am J Hematol. 2020;95(6):E131–E134. doi: 10.1002/ajh.25774. [DOI] [PubMed] [Google Scholar]
- 37.Liu L., Wei Q., Lin Q., Fang J., Wang H., Kwok H. Anti-spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight. 2019;4(4):e123158. doi: 10.1172/jci.insight.123158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang S.-.F., Tseng S.-.P., Yen C.-.H., Yang J.-.Y., Tsao C.-.H., Shen C.-.W. Antibody-dependent SARS coronavirus infection is mediated by antibodies against spike proteins. Biochem Biophys Res Commun. 2014;451(2):208–214. doi: 10.1016/j.bbrc.2014.07.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang C., Liu Z., Chen Z., Huang X., Xu M., He T. The establishment of reference sequence for SARS-CoV-2 and variation analysis. J Med Virol. 2020;92(6):667–674. doi: 10.1002/jmv.25762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Phan T. Genetic diversity and evolution of SARS-CoV-2. Infect Genet Evol. 2020;81 doi: 10.1016/j.meegid.2020.104260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nicholson L.B. The immune system. Essays Biochem. 2016;60(3):275–301. doi: 10.1042/EBC20160017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shen L., Wang C., Zhao J., Tang X., Shen Y., Lu M. Delayed specific IgM antibody responses observed among COVID-19 patients with severe progression. Emerg Microbes Infect. 2020;9(1):1096–1101. doi: 10.1080/22221751.2020.1766382. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data can be obtained upon request to the corresponding author.