SUMMARY
Dietary restriction (DR) is the most robust means to extend lifespan and delay age-related diseases across species. An underlying assumption in the aging field is that DR enhances both lifespan and physical activity through similar mechanisms, but this has not been rigorously tested in different genetic backgrounds. Furthermore, nutrient response genes responsible for lifespan extension or age-related decline in functionality remain underexplored in natural populations. To address this, we measured nutrient-dependent changes in lifespan and age-related decline in climbing ability in the Drosophila Genetic Reference Panel fly strains. On average, DR extended lifespan and delayed decline in climbing ability, but there was a lack of correlation between these traits across individual strains, suggesting that distinct genetic factors modulate these traits independently and that genotype determines response to diet. Only 50% of strains showed positive response to DR for both lifespan and climbing ability, 14% showed a negative response for one trait but not both, and 35% showed no change in one or both traits. Through GWAS, we uncovered a number of genes previously not known to be diet-responsive nor to influence lifespan or climbing ability. We validated decima as a gene that alters lifespan and daedalus as one that influences age-related decline in climbing ability. We found that decima influences insulin-like peptide transcription in the GABA receptor neurons downstream of short neuropeptide F precursor (sNPF) signaling. Modulating these genes produced independent effects on lifespan and physical activity decline, which suggests that these age-related traits can be regulated through distinct mechanisms.
eTOC:
Wilson et al. show that genetic variation impacts lifespan and functional decline with age in response to dietary restriction. These two traits were not correlated. They validate daedalus in affecting physical activity under dietary restriction and that decima impacts lifespan under ad libitum conditions through insulin-like peptide signaling.
Introduction
Dietary restriction (DR), the reduction in total nutrients [1] or specific macromolecules [2, 3] without malnutrition, extends lifespan and slows aging in multiple species. Genes that impact diet response are of interest for their potential as anti-aging targets, but few such genes are known. Model organisms which have conserved pathways but shorter lifespan have been instrumental in understanding the basis of how diet impacts human aging. In D. melanogaster, varying the two components in the diet, yeast (source of protein and lipids) and sucrose, and not just total calories modulates metabolism, healthspan, and lifespan [4, 5]. Likewise, a low-protein, high carbohydrate diet extends healthspan in mammals [2]. The nutrient-response pathways target of rapamycin (TOR) [6], insulin-like signaling (ILS) [7], and sirtuins [6] mediate the effects of DR in many species. However, these and other DR response pathways have largely been found through candidate-based screens. It is unclear whether these pathways universally confer diet-dependent changes in natural populations [8]. There is evidence in humans that selective pressure on the response to nutrient availability may vary across populations, which results in natural genetic differences that influence obesity [9]. Consistent with this notion, recombinant inbred strains of mice differ in responses to caloric restriction [10, 11], but mechanisms behind this are unknown. Thus, there is a need for whole-genome-scale studies in natural organismal populations.
Measuring lifespan has been the gold standard to identify age-related mechanisms, but studies in worms [12-14] and mice [15] in select genetic backgrounds demonstrate that lifespan extension is not necessarily accompanied by increased functional ability later in life. Furthermore, studies in humans have investigated how the end of life correlates with mortality: although disability increases as individuals approach death , the rate and severity of decline varies by individual [16]. Thus, it is unclear whether lifespan and functional decline are affected by the same mechanisms. Walking speed in humans is an indicator of health and declines with age [16]. Similarly, flies' innate tendency to climb in their enclosure also declines with age and is the most widely used healthspan measure of functional ability [17], amongst others such as memory decline [18] and intestinal homeostasis [19]. Previous studies that measured functional ability have relied on interventions that are known to extend lifespan [20]. Thus, there is a need to examine the relationship between lifespan- and age-related decline in function in an unbiased manner in diverse genetic backgrounds.
Genome-wide association studies (GWAS) have become the standard to determine novel genetic influencers of longevity or health in humans [21] and models [22-24], yet it has not been used to determine how diet impacts these traits. To address this gap, we have used the genetic variation present in a wild fly population to dissect nutrient-responsive effects on lifespan and age-related climbing ability. We used the Drosophila Genetic Reference Panel (DGRP) [25], which have been elegantly used for GWAS of dozens of traits including lifespan on a single diet [22, 23]. We have previously utilized the DGRP to identify genes that influence body mass, triglyceride levels, and starvation resistance in response to dietary protein [26] and others have utilized them for similar studies in response to sugar [27]. This study is an unbiased examination of the relationship between physical activity decline with age and lifespan. Identifying genes related to these traits provides a better understanding of the architecture that optimizes lifespan and healthspan in response to diet.
Using the DGRP, we examined the diet-dependent changes in lifespan and climbing ability across life. We observed variation in diet-dependent changes to both climbing ability and lifespan across the DGRP lines. Using GWAS, we discovered novel genetic modulators of lifespan and climbing ability upon a specific form of DR. Our results failed to demonstrate any correlation between these traits across the strains. We validated CG34351, which we name “decima” (dcma), for ad libitum (AL)-dependent changes in lifespan. We also validated that CG33690, a previously uncharacterized gene that modulates diet-dependent changes in physical activity, and named it “daedalus” (dls). We further demonstrate that dcma modulates lifespan as a regulator of insulin-like peptide production downstream of sNPF signaling in the fly GABA receptor neurons. This study uncovers genetic mechanisms that independently determine lifespan or age-related climbing ability.
Results
Genetic variation in diet-dependent changes in lifespan
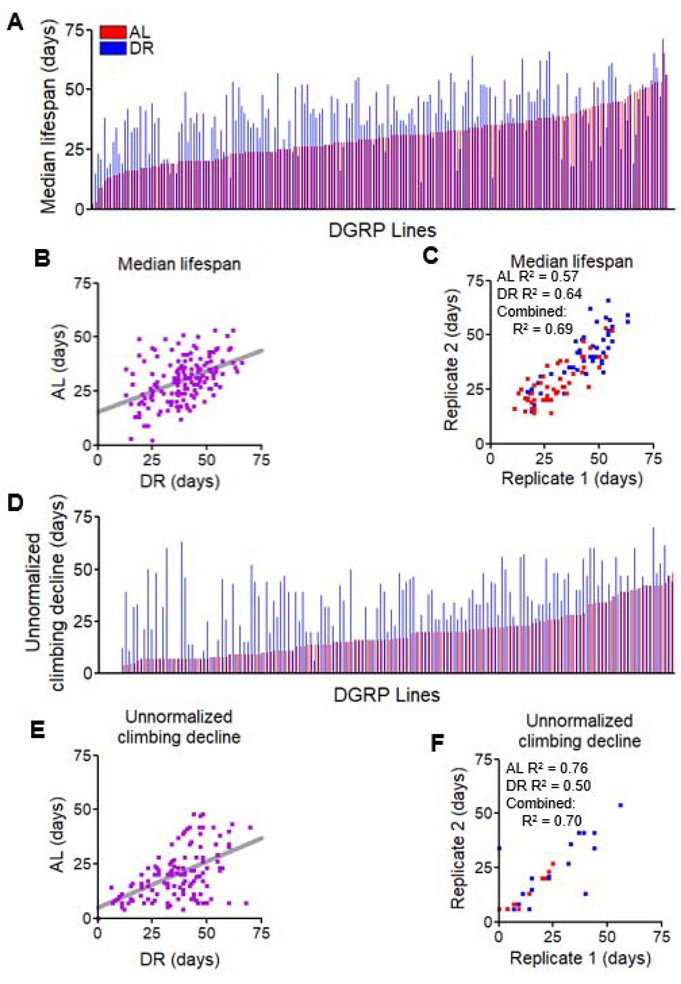
To determine the effects of genetic variation under DR-mediated changes in lifespan, we reared ~200 mated females in eight vials from 161 DGRP lines under AL conditions or a DR protocol previously demonstrated to consistently extend lifespan [5, 28]. The DR diet contained 0.5% yeast and the AL diet contained 5% yeast, while the sugar and cornmeal content was the same, as described previously [3, 26]. We observed a broad range of diet-dependent changes in lifespan across strains, ranging from a 65% reduction to a 1250% increase in median lifespan from AL to DR (Figure 1A, Data S1). 83% of lines survived longer under DR than on AL (Figure 1B). Of the longer-lived DGRP strains on AL (>35 days median lifespan), less than 46% received additional longevity benefits by DR, whereas 82% of shorter-lived strains (<35 days median lifespan) had median lifespan extended by DR. We repeated lifespan measurements for 52 strains and saw reproducible median lifespans (Figure 1C, R2= 0.57 for AL, R2 = 0.64 for DR). As with prior reports [10], we observed that DR extends lifespan in most, but not all, strains. We also observed that DR was less effective in extending the lifespan of strains that were already relatively long-lived, consistent with previous studies demonstrating that long-lived strains may not benefit from lifespan extension methods [29].
Figure 1. Genotype influences variation in lifespan, climbing ability, and response to DR across the DGRP lines.
(A) Median lifespan of 161 DGRP lines in ascending order under AL diet (red). Adjacent lines in blue represent the same strain raised under DR diet. (B) Comparison of median lifespan under AL of each strain with its DR counterpart. Same data as in A, displayed as a scatterplot. Grey bar represents best-fit trendline. (C) Comparison of median lifespan values across biological replicates of 52 DGRP lines under AL (red) and DR (blue). (D) The age (in days) at which fewer than 20% of the surviving population can climb in the allotted time. Data are arranged in ascending order by the phenotype under AL diet (red) with adjacent lines representing the same strain under DR (blue). (E) Comparison of each strain’s climbing data between AL and DR diets. Grey line represents best-fit trendline. (F) Comparison of biological replicates for 25 tested DGRP lines for the day at which less than 20% of surviving flies are able to climb under AL (red) and DR (blue). N = 50-200 flies per strain per diet. Data used found in Data S1 and Data S2. See also Figure S1.
Age-related decline in negative geotaxis varies by genotype and diet
We were also interested in determining genetic components that influence functional health. Because walking speed is used as a marker of health in humans [16], we utilized flies’ natural tendency to climb as a representative measure of functional health. Age-related climbing ability is also one of the most commonly used measures of healthspan in D. melanogaster [17, 30]. We recorded the percentage of flies that climbed the wall of an empty vial. Data was collected for 156 DGRP strains once per week throughout their life [17] (detailed in Methods, Data S2). As an index of physical activity decline, we analyzed the day at which a strain fell below 50% of its initial climbing ability. Some strains fell below this threshold within the first week after being placed under DR or AL, while others maintained climbing capacity for longer than 60 days (Figure S1C). DR generally improved climbing ability and delayed the age-related decline in climbing ability in 69% of all tested lines; 25% of lines showed no difference between the two diets (Figure S1D).
We also analyzed age-related changes in the absolute percentage of flies that were capable of climbing. For this, we recorded the day at which the percentage of flies able to climb fell under 20% (Figure 1D, Figure S1B). DR extended this parameter in 87% of lines, while 12% of the lines were unaffected by diet (Figure 1E). For both climbing measures, we re-tested 17 lines and found our recorded values to be reproducible (Figure 1F, Figure S1E, R2 = 0.62 for AL 50% decline and 0.63 for DR, R2 = 0.76 for AL day below 20% climbing and 0.50 for DR). Together, these results indicate that DR generally improves climbing ability, but the degree to which it is beneficial varies by genotype.
Lack of correlation between lifespan and age-related activity decline
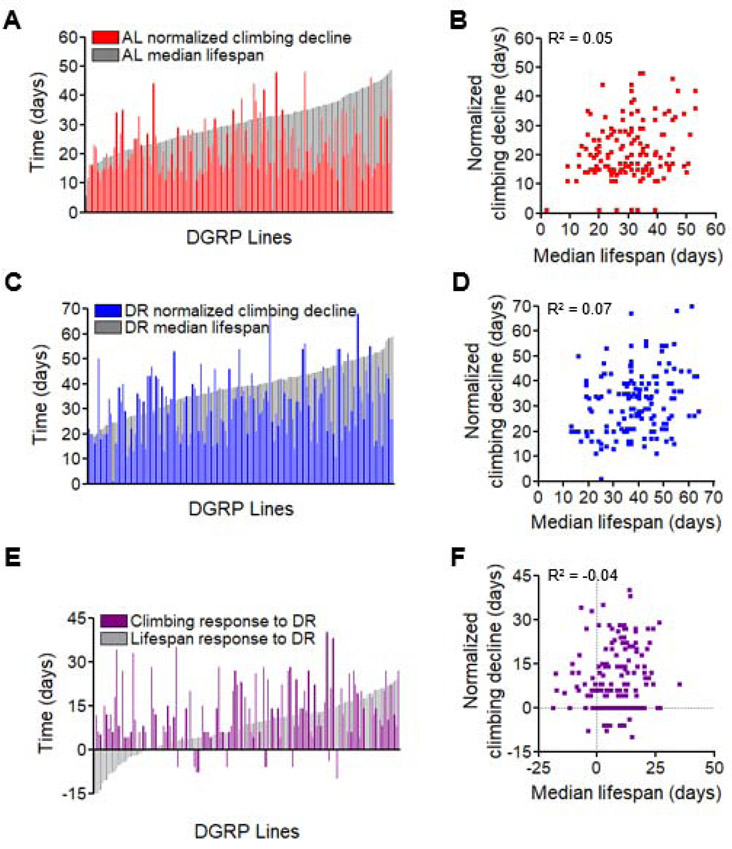
To better understand the relationship between our lifespan and climbing measures, we compared median lifespan and decline in age-related climbing ability across the DGRP strains. We found no evidence of correlation between median lifespan and climbing decline on AL across individual strains (Figure 2A-B, R2 = 0.05), nor on DR (Figure 2C-D, R2 = 0.07). Similarly, we found no correlation between median lifespan and the day at which less than 20% of the population climbed (Figure S2A-D, AL R2 = −0.06, DR R2 = −0.01). We also examined responsiveness of each DGRP strain to DR for either increase in lifespan or delayed climbing decline. 50% of strains showed a >3 day improvement in both lifespan and climbing decline in response to DR. 14% showed opposing phenotypes, either with reduced climbing ability and increased lifespan under DR or vice-versa. The remaining 36% showed no change in either or both phenotypes (Figure 2E). We observed no evidence of correlation between change in climbing ability and change in median lifespan in response to DR (Figure 2F, R2 = −0.04). We compared the period of life in which flies were below 50% of their maximal climbing ability to their lifespan. The longer-lived half of all strains under both diets spent more time being inactive than the shorter-lived half of strains, but DR improved the active period of life on average (Figure S2E). Additional measures associated with climbing ability also failed to correlate with lifespan on either diet (Figure S2F-H). Similarly, lifespan also failed to correlate with starvation resistance, triglyceride levels, and body mass, which were previously measured [26] (Figures S2I-K), indicating that these metabolic health phenotypes known to associate with lifespan are also not indicative of length of lifespan across a panel of wild strains. In summary, though on average DR increased median lifespan and slowed age-related decline in physical activity, our data fail to support the idea that extended lifespan is accompanied by improved climbing ability across DGRP strains.
Figure 2. Genotype and diet differentially influence lifespan and climbing decline.
(A-D) Comparison of each tested strain’s day below 50% of maximal climbing proportion with median lifespan, on (A-B) AL or (C-D) DR. Each bar in A and C represents a DGRP strain, ordered by median lifespan on each diet. Colored bars represent climbing half-life and grey bars represent median lifespan. (B and D) Scatter plots depicting climbing ability compared to median lifespan on the (B) AL diet or (D) DR. Each dot represents a single DGRP strain. (E) Comparison of DR responsiveness with regards to median lifespan (grey bars) and time above 50% initial climbing ability (purple bars). (F) Scatter plot depicting response to DR of each tested DGRP line with regards to median lifespan and amount of time above 50% initial climbing ability. Each dot represents a single DGRP strain. N = 50-200 flies per strain per diet. Data used found in Data S1 and Data S2. See also Figure S2.
Genome-wide association analysis
Next, we determined the genes whose expression influenced lifespan and age-related physical activity decline. We performed GWAS using a linear regression model with terms for genotype, diet, and the interaction between genotype and diet (called “Interaction” terms), as described in the Methods. We identified a list of candidate loci with a minor allele frequency ≥25% with statistical signals of ≤10% FDR based on permutation analysis (Table 1, Methods). Effect size was also calculated for each locus (Data S4). We conducted a preliminary RNAi screen with all candidate genes to determine how altered gene expression could impact longevity or climbing ability. We induced whole-body RNAi against four candidate genes indicated from GWAS, two of which showed marginal lifespan effects (Figure S3A-D, Table S1). Of the candidates tested, we narrowed our focus to two genes relevant to diet-dependent climbing decline or lifespan: CG33690 (which we name “daedalus” with the symbol dls) and CG34351 (which we name “decima” with the symbol dcma).
Table 1. Lifespan and climbing gene candidates identified by genome-wide association.
Effect size data shown in Data S4. See also Figure S3A-D.
| Phenotype | Gene | Marker | Effect/ Location |
p value | FDR | Type | Description |
|---|---|---|---|---|---|---|---|
| 75% Survival | 3L:12,638,741 | 2.50E-05 | 0% | ||||
| CR32111 | 3L:12,638,748 | 3’ UTR | 4.16E-05 | 3% | Interaction | nc transcript | |
| 3L:12,638,743 | 4.50E-05 | 3% | |||||
| CG43203 | 3R:15,179,932 | ns | 3.39E-05 | 3% | Interaction | Unknown function | |
| CG34351 | 2L:4,707,945 | Intron | 8.34E-05 | 8% | Interaction | Neuronal G-protein regulator | |
| CG8312 | 3R:5,443,281 | Intron | 1.04E-04 | 10% | Interaction | PHD protein | |
| Median LS | CG5888 | 2L:16,447,864 | Intron | 4.67E-10 | 10% | Genotype | Leucine-rich repeat protein |
| CG5888 | 2L:16,447,727 | Intron | 2.02E-10 | 0% | Interaction | Leucine-rich repeat protein | |
| 3L:12,638,741 | 4.81E-05 | 0% | |||||
| CR32111 | 3L:12,638,748 | 3’ UTR | 5.80E-05 | 0% | Interaction | nc transcript | |
| 3L:12,638,743 | 8.13E-05 | 3% | |||||
| CG31221 | 3R:15,265,527 | Intron | 7.62E-05 | 3% | Interaction | Low-density lipoprotein receptor | |
| CR45580 | 3R:722,994 | nc region | 1.44E-04 | 10% | Interaction | nc transcript | |
| 50% climbing decline | CR43930 | 3L:20,907,785 | Upstream | 1.50E-04 | 0% | Interaction | Unknown function |
| Day below 20% climbing | CG33690 | 3L:16,195,836 | Downstream | 3.42E-05 | 3% | Interaction | Unknown function |
| 3L:16,195,839 | 3.54E-05 | 3% | |||||
| 3L:16,195,821 | 4.36E-05 | 3% | |||||
| 3L:16,195,833 | 5.07E-05 | 3% | |||||
| 3L:16,195,854 | 5.97E-05 | 4% | |||||
| 3L:16,195,865 | 1.43E-04 | 5% | |||||
| 3L:16,195,864 | 1.81E-04 | 9% |
LS = lifespan; ns = non-synonymous; nc = non-coding; genotype, interaction, and case control terms detailed in Methods
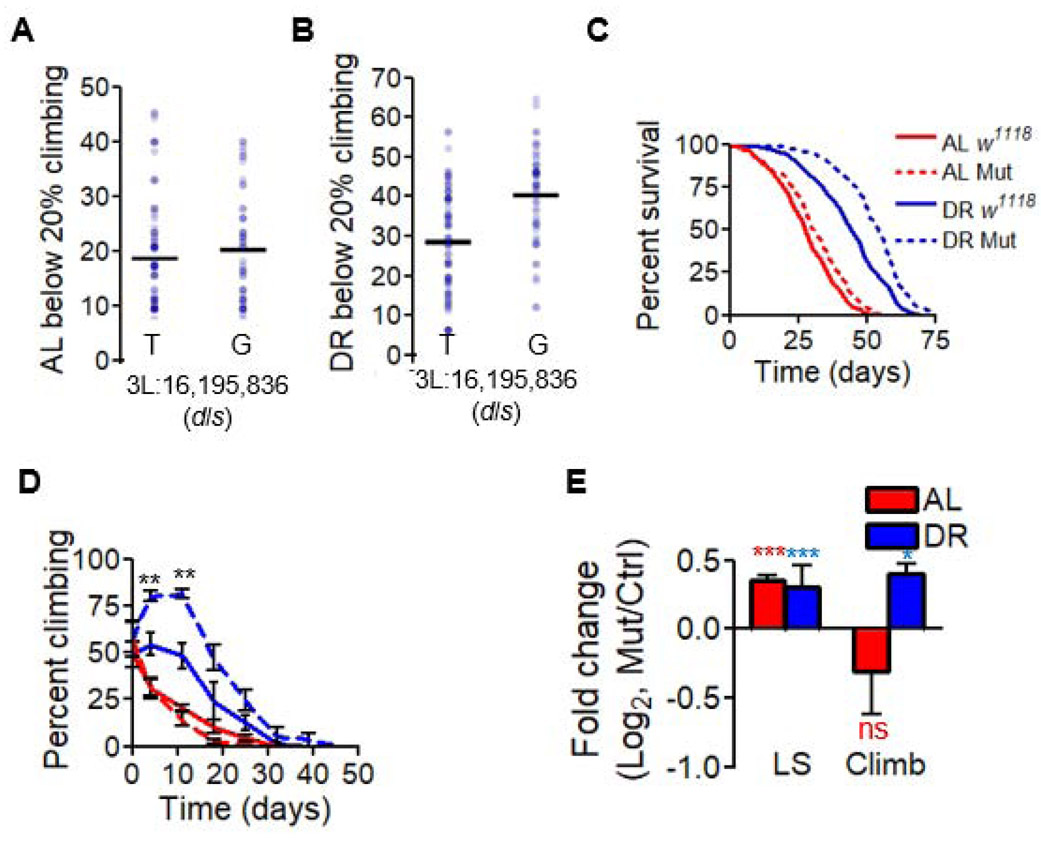
The most significant locus associated with climbing modulation (Table 1) was mapped to position 16,195,836 on chromosome 3L, downstream of dls (Figure 3A-B). Since expression of dls was altered between DGRP strains with the two different alleles (Figure S3L), we used a Minos element insertion line for dls for our validation experiments to determine how knockout of dls influenced climbing ability. We observed a ~90% reduction in mRNA expression under DR in this mutant line (Figure S4A-B). Compared to w1118 background control, mutants had a 19% and an 8% increase in median lifespan under DR and AL, respectively (Figure 3C). However, when outcrossed into Canton-S, these flies showed a 7% reduction in median lifespan under DR and a 19% reduction under AL (Figure S3E). Climbing ability consistently increased in mutant dls flies under DR, regardless of strain background; no significant changes in climbing were observed under AL (Figure 3D, Figure S3F, Data S3). In total, while the longevity effects of this mutation depended on strain background, climbing was significantly increased only in flies under DR (Figure 3E, Figure S3G). dls mutants under DR also had increased 24-hour spontaneous activity (see Methods, Figure S4E-F), which had previously also been found to have a weak correlation with lifespan [31]. Because CG33690 influences activity under DR, we propose the name daedalus (dls) for this gene, after the mythological Greek inventor who created wings for his son, Icarus, and himself to escape incarceration. Together, these data support the conclusion that lifespan and age-related climbing ability are regulated by distinct mechanisms, and thus can be uncoupled.
Figure 3. daedalus modulates DR-specific climbing ability.
(A and B) Plot of the day at which fewer than 20% of flies climb in tested DGRP lines, split by genotype at the most significant locus downstream of dls on (A) AL or (B) DR. The day at which 20% or fewer surviving flies are able to climb is represented by blue dots, black bars represent mean values across all tested strains with a given genotype and diet. Significance for diet interaction p < 4E-5, FDR = 3%. (C-E) The effect of Minos element insertion on (C) lifespan and (D) climbing ability over the course of life in a w1118 genetic background, and (E) the log2 difference median lifespan and unnormalized climbing decline between mutant and controls. AL shown in red, DR in blue. Significant differences between mutant and controls are indicated by *. * = p < 0.05, ** = p < 0.005, *** = p < 0.0005. nc = no change, ns = not significant. p values shown in Data S3. N = 200 flies per condition for each mutant experiment. Data in (C-E) show collective results from three biological replicates. Error bars represent SD between replicates. Data used found in Data S3. See also Figure S3E-G, L, Figure S4A-B, E-F.
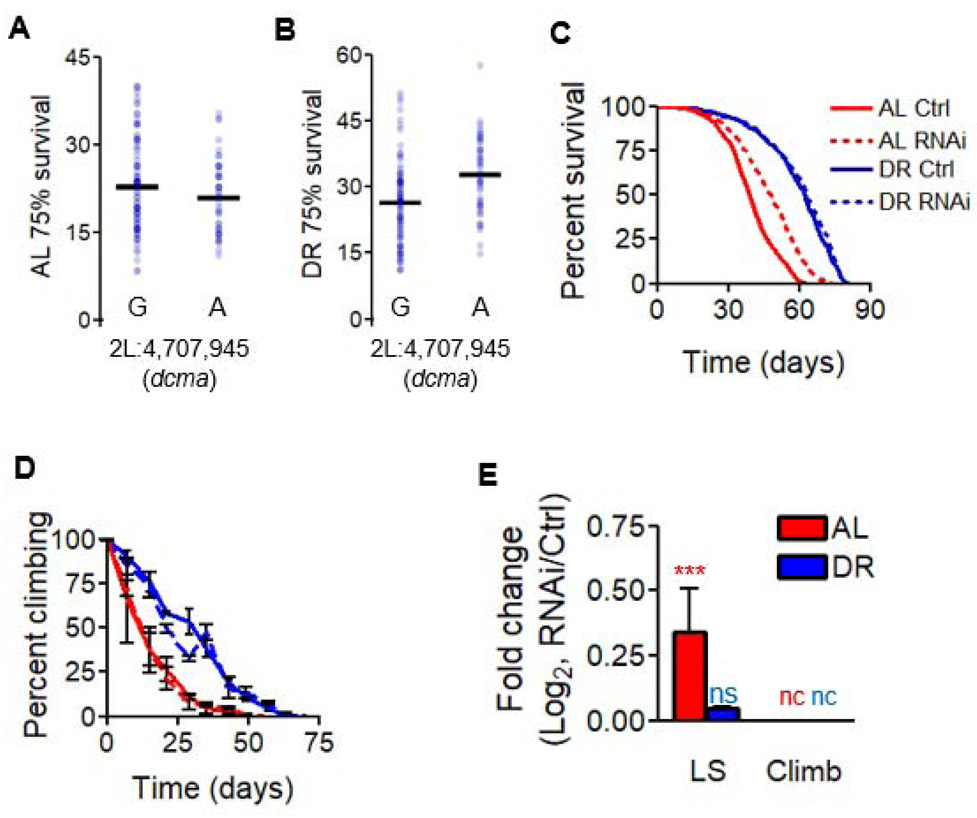
Through our interaction GWAS for longevity, we identified an intronic variant in CG34351 associated with the day at which less than 75% of a strain’s population was still alive (Figure 4A-B). CG34351 is homologous to the human Regulator of G-Protein Signaling 7 Binding Protein (RGS7BP), and was previously noted in a screen for wing size modulators [32], but is uncharacterized with respect to longevity and diet. We identified that expression of CG34351 was altered by genetic variation (Figure S3K), and thus examined the role of dcma expression in lifespan modulation. From publicly available expression data [33], we found that dcma is expressed neuronally. We used the pan-neuronal driver Elav-GS-Gal4 to examine diet-dependent changes in longevity and climbing ability in adulthood. We observed a 20% median lifespan increase under AL conditions over control flies with one dcmaRNAi transgenic line (v30160, Data S4, Figure 4C) and a 29% median lifespan increase under AL with another line (v30163, Figure S3H). We propose the name decima (dcma) for CG34351 after the Roman goddess of fate, who used threads to measure one's length of life. In parallel, we tested the effect of dcmaRNAi on climbing ability but there was no significant (Figure 4D, Figure S3I, Data S3). Knockdown efficiency of RNAi strains was confirmed by qRT-PCR (Figure S4C-D). Overall, we saw increased longevity under AL with dcma RNAi in neurons but did not see a change in age-related climbing ability (Figure 4E, Figure S3J). Combined with our data from dls, these results further reiterate a genetic uncoupling of lifespan and climbing.
Figure 4. decima modulates longevity in a diet-dependent manner.
(A and B) ALignment of all 161 DGRP lines according to genotype at a particular locus in dcma and according to the day at which ≤ 75% of flies in a strain remain alive on (A) AL or (B) DR. Strains’ median lifespans are represented by blue dots, black bars represent mean values across all tested strains with a given genotype and diet. Significance for diet interaction p < 9E-5, FDR = 8%. (C-E) The effect of neuron-specific RNAi of dcma using the v30160 transgenic line in modulating (C) lifespan and (D) climbing ability over the course of life, with (E) log2 fold-change between RNAi and control for both median lifespan and unnormalized climbing decline values. AL shown in red, DR in blue. Significant differences between RNAi and controls are indicated by *. * = p < 0.05, ** = p < 0.005, *** = p < 0.0005, determined by unpaired t test. p values shown in Data S3. nc = no change, ns = not significant. N = 200 flies per condition for each RNAi experiment. Data in (C-E) show collective results from three biological replicates. Error bars represent SD between replicates. See also Figure S3H-K, Figure S4C-D. RU486 controls shown in Figure S5.
decima regulates insulin-like peptide production
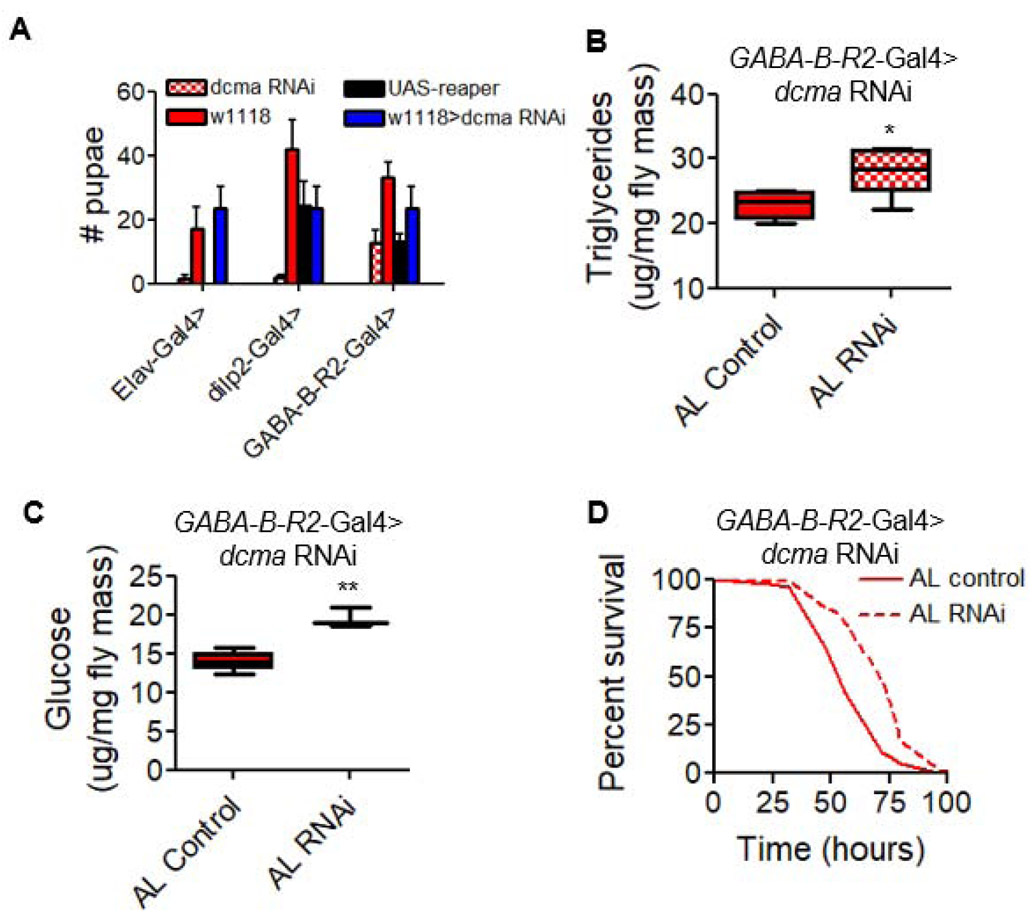
Next, we determined how neuronal dcmaRNAi extends lifespan. We used pan-neuronal knockdown of dcma using Elav-GS and measured body mass, triglyceride levels, and starvation resistance, but found no significant effects (Figure S6A-C, Data S3). To induce a stronger knockdown of dcma, we used the constitutively active neuronal Gal4 driver Elav-Gal4, which also induced RNAi in larval development. This inhibited development beyond the larval stage, as the majority of flies were unable to reach pupation and adulthood (Figure 5A, left), suggesting a disruption of growth signaling [34]. Thus, we tested the influence of dcma on the production of Drosophila insulin-like peptides (dilps). With dcmaRNAi in the dilp-producing cells (IPCs), we found a similar lack of development among larvae (Figure 5A, middle). To identify more specifically which cells may be impacting this phenotype, we used a panel of neuronal subtype Gal4 drivers. dcmaRNAi in the GABA receptor neurons using GABA-B-R2-Gal4 induced a similar inhibition of development (Figure 5A, right) but provided enough adult flies for testing adult phenotypes. Inhibition of the ILS pathway in flies influences body composition [35]. We observed that dcma inhibition in the GABA-B-R2 cells elevates levels of triglycerides (Figure 5B) and glucose (Figure 5C). These flies also survived in starvation conditions longer than controls after rearing under AL (Figure 5D), as has been observed for mutants that inhibit the ILS pathway [36]. Elevated body mass, triglyceride levels, and glucose levels were also observed with dilp2-driven Gal4 (Figure S6E-G).
Figure 5. decima knockdown inhibits development, promotes elevated triglyceride and glucose levels, and improves starvation resistance.
(A) Number of larvae reaching pupariation from dcmaRNAi driven pan-neuronally (left), in IPCs (middle), and in GABA receptor cells (right). N = 60 embryos per condition. (B) Whole-body triglyceride levels of flies with GABA-B-R2-Gal4-driven dcmaRNAi (checkered boxes) versus controls with no RNAi (solid boxes). N = 15 flies per condition. (C) Whole-body glucose levels in flies with GABA-B-R2-Gal4-driven dcmaRNAi. N = 15 flies per condition. (D) Starvation resistance of flies with GABA-B-R2-Gal4-driven dcmaRNAi (dotted line) versus control (solid line). N = 100 flies per condition. RNAi represented by checkered bars and control conditions in solid colors. Significant differences between RNAi and controls are indicated by *. * = p < 0.05, ** = p < 0.005, *** = p < 0.0005, determined by unpaired t test. p values shown in Data S3. nc = no change, ns = not significant. All experiments show collective results of three biological replicates. Error bars represent SD between replicates. See also Figure S6.
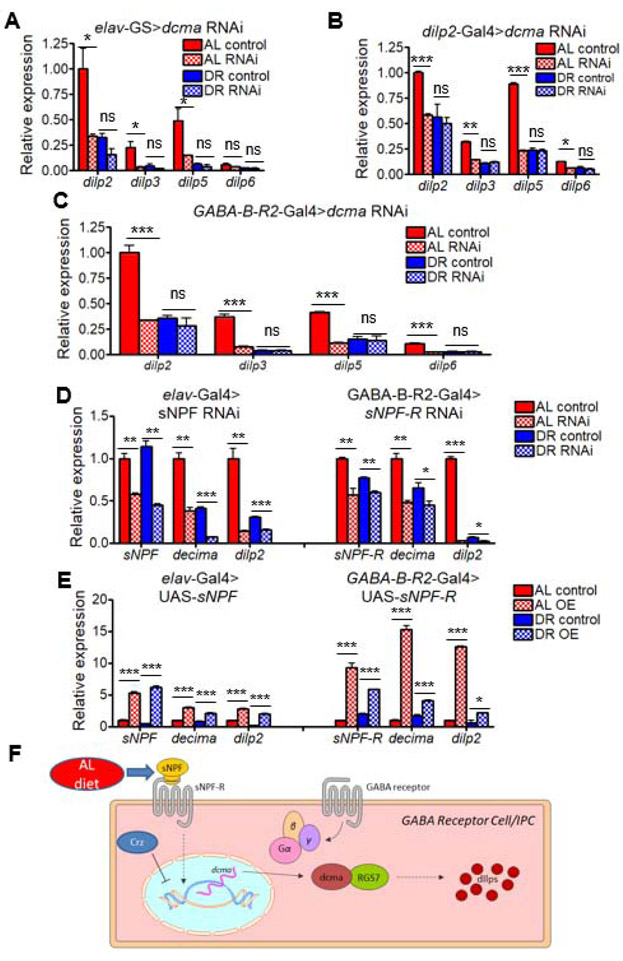
We next measured dilp mRNA levels in heads with dcmaRNAi. Using Elav-GS, we found that mRNA expression of the three neuronal dilps (dilp2, dilp3, dilp5) was significantly reduced in the heads of flies raised under AL but not DR conditions (Figure 6A); expression of the other dilps was very low and did not show significant reduction by dcmaRNAi (Figure S7A, Data S3). We also tested expression of dilps using constitutively active dilp2-Gal4 driven dcmaRNAi to determine whether dcma regulation of dilps occurs in a cell-autonomous manner. Consistent with Elav-GS-driven dcmaRNAi, we observed a significant reduction in the expression of dilp2, dilp3, and dilp5 (Figure 6B, Data S3). In addition, a reduction in dilp6 expression was observed and the non-neuronally expressed dilps showed either low levels of expression or no significant effects (Figure S7B, Data S3). dcmaRNAi in GABA-B-R2 cells also demonstrated a significant reduction in all neuronal dilps under AL only (Figure 6C) as well as non-significant effects on the expression of non-neuronal dilps (Figure S7C, Data S3). As RGS7BP is known to interact with the protein Regulator of G-protein Signaling 7 (RGS7) to regulate downstream signaling [37], we induced pan-neuronal RGS7RNAi, which also caused reduced dilp2 expression, the most prominently expressed neuronal dilp (Figure S7D). To test whether dcma-mediated dilp repression was required for lifespan extension, we simultaneously induced dcmaRNAi and overexpressed dilp2. dilp2 overexpression prevented the lifespan extension seen with dcmaRNAi alone when expressed in IPCs (Figure S7F-H) or pan-neuronally (Figure S7I-K).
Figure 6. decima regulates insulin-like peptide production in GABA receptor neurons through sNPF signaling.
(A-C) Levels of neuronally-expressed dilps in heads of flies with dcmaRNAi driven (A) pan-neuronally under control of RU486, (B) in dilp2-producing cells, and (C) in GABA receptor neurons. (D) Relative expression of dcma and dilp2 with pan-neuronal RNAi of sNPF (left) or GABA Receptor cell-specific sNPF-RRNAi (right) in heads of flies relative to rp49 housekeeping gene. (E) Relative expression of dcma and dilp2 with pan-neuronal overexpression of sNPF (left) or GABA Receptor cell-specific sNPF-R overexpression (right) in heads of flies relative to rp49 housekeeping gene. AL shown in red, DR in blue. RNAi represented by checkered bars and control conditions in solid bars. N = 50 fly heads per condition. Significant differences between RNAi and controls are indicated by *. * = p < 0.05, ** = p < 0.005, *** = p < 0.0005, determined by unpaired t test. p values shown in Data S3. nc = no change, ns = not significant. N = 200 flies per condition for each RNAi experiment. All experiments show collective results of three biological replicates. Error bars represent SD between replicates. (F) Model for decima’s regulation of dilp production. dcma transcription is upregulated through dietary and sNPF signals, and in turn then regulates the transcription of Drosophila insulin-like peptides in the GABA receptor neurons/insulin-producing cells. See also Figure S7.
To test whether known dilp-modulating neuropeptides [38] operate upstream of dcma, we examined dcma expression after knocking down or overexpressing these neuropeptides. Pan-neuronal knockdown of short neuropeptide F precursor (sNPF) or GABA-B-R2-Gal4-driven RNAi of sNPF receptor (sNPF-R) resulted in reduced dcma and dilp2 expression (Figure 6D). Overexpression of sNPF or sNPF-R elevated the expression of dcma and dilp2, especially under AL (Figure 6E). Pan-neuronal CorazoninRNAi (Crz) elevated expression of dcma and dilp2 under AL, but RNAi of Tachykinin (Tk), Octopamine receptor in mushroom bodies (Oamb) and 5-hydroxytryptamine (serotonin) receptor 1A (5-HT1A) did not alter expression of dcma and dilp2 in the same manner (Figure S7E). sNPF is implicated in modulating lifespan through dilp signaling [39], and our results suggest that dcma is involved in this process.
Because GABA receptor neurons are known to be IPCs [40], we sought to determine if dcma’s regulation of dilps was through upd2 signaling, a previously described diet-responsive protein, albeit independently of protein consumption [40]. upd2 overexpression did not change dcma expression, and dcmaRNAi did not change upd2 expression (Figure S7L-N). These data suggest that dcma’s regulation of dilps occurs independently of upd2. Together, our results demonstrate a novel role for dcma in the regulation of dilp expression and lifespan (Figure 6F).
Discussion
Dietary restriction remains the most robust method for lifespan and healthspan extension. Despite this, enthusiasm for DR has been tempered by the observation that organisms of different genotypes do not always gain these extensions [11, 26], with some genotypes even showing reduced lifespan [10] or worsened health [41] under DR. Thus, identifying mechanisms that mediate differential health and longevity effects under DR may provide novel targets and a better understanding of mechanisms by which it extends lifespan. Further, the observation that some strains perform better than others under our form of DR suggest that our DR may not be optimal for these particular strains and may be too extreme, as has previously been suggested [28], and that understanding nutritional geometry is necessary to understand how diet influences aging [42]. The genetics impacting this will provide a better understanding for optimizing diets for maximal lifespan and healthspan responses.
Here, we have utilized the DGRP to perform the first GWAS for longevity and traits relating to healthspan using two different dietary compositions. We utilized a form of dietary restriction that has repeatedly extended lifespan [4, 19], consisting of reduced dietary yeast. Our data demonstrates that about 50% of strains showed positive response to DR for both lifespan and healthspan, while 14% showed a negative response in one trait but not both. 35% showed no change in one or both traits. Only one strain out of 160 showed a negative response for both lifespan and healthspan. Our findings suggest that even robust lifespan-extending methods are not ubiquitously beneficial, and it is likely that different dietary compositions are more optimal for different genetic backgrounds. Our work details some genes which influence lifespan and period of life spent physically functional, but it is worth noting that many DGRP strains did not live long or were largely inactive during their lives. This allowed for more phenotypic diversity in our tested strains, but might also demonstrate the ills of inbreeding. Thus, more analyses using other collections and with more complex organisms continue to be useful for future analyses relating to our study. It is also important to note that in our study we used female flies, as we have previously shown that females show a larger change in physiology and lifespan in response to DR, and this difference is suggested to be due to females showing a stronger preference for dietary yeast, presumably due to its effect on egg production [43]. We found that DR generally extended median lifespan and climbing ability, but a number of strains showed no effect or negative effects, consistent with previous findings where nutrient manipulations failed to induce universal effects across mouse strains of different genotypes [10, 11]. Our work identifies how natural genetic variation influences climbing ability and lifespan across wild Drosophila strains.
Analysis of the DGRP strains showed no evidence of correlation between climbing ability and lifespan. Climbing ability remains the most common method for assessing overall functional fly health [17]. While others have suggested that physical activity should be presented in the context of maximal functionality rather than absolute activity [44], we observed no correlation with lifespan in either of these parameters (Figure 2, Figure S2). We also saw that DR affected lifespan and the active period of life differentially across the strains. Despite the prevailing notion in the field that lifespan-extending interventions also improve functionality, our data argue that these traits are uncoupled. ALthough DR is the most robust means for lifespan and healthspan extension [20], our data suggest that genotype significantly influences the extent and type of benefits that DR provides. Thus, in some backgrounds, DR will only optimize for specific traits, and a different diet might be more beneficial for improving another trait. In studies of human longevity, it is suggested that a healthy lifestyle can increase the functional period of life without altering lifespan by compressing morbidity [45]. Additionally, through GWAS across all tested lines, we have identified a number of genes unique to either altering longevity or activity. Though climbing ability is not an end-all measure for overall health, our results suggest that the link between lifespan and various healthspan measures needs to be examined in an unbiased manner to determine the mechanisms which impact them.
The connection between lifespan and healthspan has been an important topic of debate. It is conceivable that these traits do not always go hand-in-hand, as physically fit individuals can suffer from cognitive decline whereas others can live with crippling disorders for decades, but the genetics of traits relating to fitness as it relates to lifespan had previously not been studied. Recent studies have suggested either that lifespan and healthspan, as measured by activity, are uncoupled [12, 13, 15, 24] or that they are positively correlated [44]. One study investigated the age-related decline in activity and response to heat or oxidative stress in long-lived C. elegans mutants to determine the healthspan and gerospan of these worms [14]. Surprisingly, this work demonstrated that long-lived strains such as daf-2 mutants, which have reduced ILS activity, and eat-2 mutants, which have reduced dietary consumption, had a shortened period of activity relative to other worm strains. Another study, however, found that when movement of the same strain of daf-2 mutants was normalized to their maximal velocity, this period was not shorter [44]. It is worth noting that our work here also identifies that a regulator of the ILS pathway, dcma, is an effector of lifespan but not activity. This was observed even when normalizing to the maximal climbing ability of individual strains Another C. elegans study investigated the individual variation in animals by tracking the activity of individual worms within an isogenic population throughout life [12]. They found that the inactive period of life is generally longer in long-lived worms, suggesting that living longer does not equate to being healthier longer even in isogenic animals. Consistent with this, research in mice has shown that variation in grip strength and metabolic measures such as fat levels do not correlate with length of life [15]. Our comparisons between metabolic traits and lifespan also failed to correlate, suggesting that these health-related phenotypes are also not necessarily linked to length of lifespan. A study in primates has suggested that a specific regime of calorie restriction was capable of improving health measures like triglyceride levels and disease onset but did not extend lifespan [46], though another primate study showed that both lifespan and measures of healthspan are improved with DR [1]. These two studies used different nutrient compositions but are nevertheless consistent with the idea that lifespan and healthspan can be uncoupled. Taking these studies into consideration, it is conceivable that lifespan is uncoupled from healthspan as lifespan is likely to be affected by the weakest link that leads to changes in mortality and thus may not reflect the underlying rate of aging or a particular healthspan trait. Unlike previous reports, which have focused on candidate-based targets, we provide the first instance of a direct comparison of lifespan and a measure of health across ~150 wild strains with naturally arising genetic variation. Understanding the genetics that modulate morbidity, particularly in response to diet, allows for more targeted approaches to maximizing healthspan.
Our GWAS analysis enabled us to successfully pinpoint loci that are significantly associated with lifespan or climbing modulation. From these candidates, we demonstrate that dcma interacts with the ILS pathway. We noticed the absence of genetic loci in genes that take part in well-studied diet-responsive pathways involved in longevity or health, such as those in the TOR and sirtuin pathways [5]. One reason for this could be that polymorphisms that alter the function of these genes would likely be lethal or inhibit development to adulthood, and thus may not be well-represented in adult wild isolates like the DGRP. A second possibility is that some of the genes we have identified have not yet been shown to influence well-established longevity pathways. A third possibility is that there are many genes that influence lifespan phenotypes, and genes in the ILS and TOR pathways represent only a small fraction of those. In support of this argument, over 500 genes have been identified to influence longevity in a variety of models.
From our climbing ability GWAS, we identified a novel role for the gene dls in climbing ability and spontaneous activity. Our analyses of DGRP strain phenotypes showed that several polymorphisms in this gene were associated with climbing ability only under DR, and flies with a mutation in dls mirrored this diet-specific effect. No biological function has been suggested for dls, and the protein it encodes contains a conserved domain of unknown function (InterPro DUF1091). Interestingly, the longevity effect of this gene varied depending on the strain background. We found significantly increased lifespan on both diets in a w1118 background but significantly decreased lifespan on the AL diet in a Canton-S background. Despite these differing results, a significant increase in climbing ability and spontaneous activity under DR were observed in both backgrounds. Nutrient restriction leads to various responses including increase in physical activity, changes in fat metabolism, and lifespan extension. These data suggest the existence of genes that may differentially mediate these downstream effects, thus uncoupling the effect of DR on lifespan and age-related climbing decline.
We identified dcma through our lifespan analyses. The homology of dcma to human RGS7BP suggests it might regulate neuronal G-protein signaling [47]. We validated that expression of dcma alters lifespan in a diet-specific fashion. Although we observed that dcmaRNAi extended lifespan in AL, our GWAS effect was largest on DR. We attribute this difference to the difference between the effects of an intronic single-nucleotide polymorphism in the DGRP strains and RNAi knockdown [48]. A dietary link for RGS7BP has not previously been observed in humans.
In Drosophila, GPCR activity has been linked to insulin-like signaling [49]. We found that dcmaRNAi in the GABA receptor neurons reduced mRNA expression of dilp2, dilp3, and dilp5. Previous studies in flies have demonstrated how GABA receptor neurons regulate insulin-like peptide production [40], but our results show that dcma operates through a parallel pathway in the same cells in response to dietary protein. A previous study in mice has shown that Gβ5, which interacts with RGS7BP, is necessary for the insulin production [50]. Our data suggest that this additional component of the pathway is similarly necessary for this process. We also found that knocking down the longevity gene sNPF reduced both dcma and dilp2 expression. Previous studies have shown that pan-neuronal sNPFRNAi reduces dilp production and extends lifespan [39]. Our data suggests that sNPF’s induction of dilp production operates through dcma signaling. While pan-neuronal dcma knockdown was capable of extending lifespan under AL, it had no effect on climbing ability. These data further demonstrate the dichotomy that exists in the regulation of lifespan and functionality. This result was particularly intriguing, as the importance of activity levels in DR-mediated longevity has been previously reported [51]. Our data suggest that genotype plays an essential role in regulating these complex traits, and though some genes may impact both activity late in life and lifespan, other genes affect one but not the other.
Through this study, we identify genetic mechanisms that may operate in natural populations to modulate diet-dependent changes in longevity and functionality. By measuring both lifespan and age-related functionality in the same strains, we were able to simultaneously dissect the genetics of two age-related traits. Our experiments using diet manipulation have detailed how genotype and the diet-gene interaction impacts DR-mediated effects on lifespan and climbing ability. Most previous studies have examined how known longevity genes affect healthspan-related traits. These studies might be subject to confirmation bias if negative results where healthspan traits and lifespan were not correlated are under-reported. Our study utilized an unbiased approach to examine the relationship between climbing ability and lifespan in over 150 fly strains and validated the candidate genes that were identified from this analysis. Our findings argue for a genetic uncoupling of mechanisms that modulate longevity and functional ability and identify some genes that influence these traits. It is likely that depending on different contexts, other genes may be responsible for influencing these phenotypes. This uncoupling of functional ability from lifespan may have an important bearing in designing dietary interventions that delay the effects of aging in humans.
STAR METHODS
Resource Availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Pankaj Kapahi (pkapahi@buckinstitute.org).
Materials availability
All materials including fly strains will be made available upon request.
Data and Code Availability
The published article includes all datasets generated and analyzed during this study. Codes used will be available upon request to the Lead Contact.
Experimental model and subject details
Animals used in this study.
DGRP and transgenic fly lines were obtained from Bloomington Drosophila Stock Center or Vienna Drosophila Resource Center, except where noted in the Key Resources Table. Mated females flies were used in this study except where noted, and were maintained in 8 oz. round bottom bottles when mating and progeny were transferred to 10 cm. long polystyrene fly vials when being placed onto DR or AL media for phenotyping. Flies were stored at 25°C and 65% relative humidity in a room that maintains a 12-hour light/dark cycle at all times.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Experimental Models: Drosophila melanogaster | ||
| Act5C-GS-Gal4 Driver P{w[+mC]=Act5C(FRT.y[+])GAL4.Switch.PR}X, y[1] w[*] |
Bloomington Drosophila Stock Center | #9431 [55, 56] |
| Elav-GS-Gal4 Driver w1118 ; P{w[+mC]=elav-Switch.O}GSG301 |
Bloomington Drosophila Stock Center | #43642 [56, 57] |
| Elav-Gal4 Driver P{w[+mW.hs]=GawB}elav[C155] |
Bloomington Drosophila Stock Center | #458 [58] |
| CG34351 RNAi w[1118]; P{GD15125}v30160 |
Vienna Drosophila Resource Center | #v30160 [59, 60] FBst0458367 |
| CG34351 RNAi w[1118]; P{GD15126}v30163/TM3 |
Vienna Drosophila Resource Center | #v30163 [59, 60] FBst0458368 |
| CG33690 Minos insertion mutation w[1118]; Mi{ET1}]CG33690[MB12096] |
Bloomington Drosophila Stock Center | #29277 [61] FBst0029277 |
| CR32111 RNAi w[1118]; P{GD2443}v40848 |
Vienna Drosophila Resource Center | #v40848 [59, 60] FBst0463796 |
| CG8312 RNAi P{KK107569}VIE-260B |
Vienna Drosophila Resource Center | #v109340 [59, 60] FBst0481057 |
| CG5888 RNAi w[1118]; P{GD5404}v12413 |
Vienna Drosophila Resource Center | #v12413 [59, 60] FBst0450490 |
| CG31221 RNAi y[1] sc[*] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.HMS02350}attP2 |
Bloomington Drosophila Stock Center | #41953 [62] FBst0041953 |
| Control Strain w[1118] |
Bloomington Drosophila Stock Center | #5905 [63] |
| KK Control Strain y,w[1118];P{attP,y[+],w[3`] |
Vienna Drosophila Resource Center | #v60100 [64] |
| TRiP Control Strain y1 sc* v1; P{VALIUM20-mCherry}attP2 |
Bloomington Drosophila Stock Center | #35785 [65] |
| RGS7 RNAi y1 v1; P{TRiP.HM05060}attP2/TM3, Sb1 |
Bloomington Drosophila Stock Center | #28574 [66] FBst0028574 |
| UAS-rpr w[1118]; P{w[+mC]=UAS-rpr.C}[14 |
Bloomington Drosophila Stock Center | #5824 [67] FBst0005824 |
| sNPF RNAi y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.JF01906}attP2 |
Bloomington Drosophila Stock Center | #25867 [66] FBst0025867 |
| sNPF-R RNAi y1 v1; P{TRiP.JF02657}attP2 |
Bloomington Drosophila Stock Center | #27507 [66] FBst0027507 |
| Crz RNAi y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.JF02023}attP2 |
Bloomington Drosophila Stock Center | #25999 [66] FBst0025999 |
| Tk RNAi y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.JF01818attP2 |
Bloomington Drosophila Stock Center | #25800 [66] FBst0025800 |
| Oamb RNAi y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.JF01732}attP2 |
Bloomington Drosophila Stock Center | #31233 [66] FBst0031233 |
| 5-HT1A RNAi y[1] sc[*] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.HMS00823}attP2 |
Bloomington Drosophila Stock Center | #33885 [66] FBst0033885 |
| GABA-B-R1 RNAi y1 v1; P{TRiP.HMC03388}attP2 |
Bloomington Drosophila Stock Center | #51817 [66] FBst0051817 |
| GABA-B-R2 RNAi y1 v1; P{TRiP.HMC02975}attP2 |
Bloomington Drosophila Stock Center | #50608 [66] FBst0050608 |
| GABA-B-R3 RNAi y1 v1; P{TRiP.HMC02989}attP40 |
Bloomington Drosophila Stock Center | #50622 [66] FBst0050622 |
| dilp2-Gal4 driver | Laboratory of Heinrich Jasper | [68] |
| dilp2-GS-Gal4 | Laboratory of Heinrich Jasper | [68] |
| UAS-dilp2 | Laboratory of Heinrich Jasper | [68] |
| GABA-B-R2-Gal4 driver | Laboratory of Norbert Perrimon | [69] |
| upd2 mutant yw; upd2del362/FM7, ubi::GFP |
Laboratory of Norbert Perrimon | [69] |
| UAS-upd2 UAS-upd2::eGFP-10D5 |
Laboratory of Norbert Perrimon | [69] |
| UAS-sNPF | Laboratory of David Anderson | [70] |
| UAS-sNPF-R | Laboratory of David Anderson | [70] |
Method Details
Fly lifespan phenotyping.
DGRP lines were obtained from Bloomington Stock Center, Bloomington, IN [52]. Each line was mated and developed on a standard laboratory diet (1.5% yeast). Two to three days post-eclosion, mated female progeny were transferred to AL (5.0% yeast extract) or DR (0.5% yeast extract) diet via CO2, as previously described [4]. Eight vials of 25 flies were used per diet per strain. Living flies were transferred to fresh vials every other day, with dead flies being recorded, until all flies were dead. One biological replicate (200 animals) was recorded for 107 lines, two biological replicates for 52 other lines, and three biological replicates for two other lines. w1118 was also tested with each batch as an internal control. DGRP lines not tested were not viable long term in our lab.
Fly climbing phenotyping.
Throughout life, climbing ability was recorded weekly on days between vial transfers for all vials containing 20 or more living flies. The negative geotaxis climbing ability test was adapted from previous methods [17]. Flies were placed in an empty vial with a line 6 cm from the bottom. Flies were gently tapped to the bottom of the vial and the number able to cross the line within 10 seconds was recorded. This was repeated three times for each vial, and the percentage of live flies still climbing above the line was averaged for a given line at weekly timepoints throughout life. For normalized climbing values, weekly climbing values were normalized to the percentage of flies climbing one week following placement under AL or DR conditions. We used the day at which flies passed below 50% of their day seven climbing value for genome-wide analysis, as well as the day at which a 20% or less of a surviving population of flies were still able to climb.
Genome-wide association analysis.
We used DGRP release 2 genotypes, and FlyBase R5 coordinates for gene models. As in Nelson et al., 2016 [26], we used only homozygous positions and a minor allele frequency of ≥25% to ensure that the minor allele was represented by many observations at a given polymorphic locus. The collected phenotype and genotype data were used as input into an association test via ordinary least squares regression using the StatsModels module in Python [53]. The linear model was phenotype = β1 x genotype + β2 x diet + β3 x genotype x diet + intercept. Nominal p-values denoted as “genotype” in Table 1 report the probability that β1 ≠ 0, and those denoted as “interaction” report the probability that β3 ≠ 0. To avoid the potential for false positives at a given nominal cutoff owing to p-value inflation, we calculated false discovery rates via permutation as follows: for a given permutation i, we randomized phenotype values across DGRP lines, retaining the true diet assignment, and on this permuted data set we carried out association tests for each marker in turn as above. We counted the number of markers ni that scored above a given p-value threshold t. We tabulated the false discovery rate (FDR) at t as the ratio between the average ni across ten permutations and the number of markers called at t in the real data. We used an empirical FDR upper bound of 10% within a given analysis to call candidate loci of interest. Effect size was the coefficient for variables in each linear regression analysis.
Gene expression analysis.
To determine gene expression in a normal system, we sampled five whole flies, 50 heads, 50 thoraxes, or 50 abdomens from mated females of w1118 control strain after one week under AL or DR diet. We isolated RNA using Zymo Quick RNA MiniPrep kit (R1054) (Zymo Research, Irvine, CA). For qRT-PCR, we used Superscript III Platinum SYBR Green One-Step qRT-PCR kit from Invitrogen, Carlsbad, CA (11736-051) and followed manufacturer’s instructions with a Roche Lightcycler 480 II machine. To validate the effects of RNAi or mutation on gene expression, we collected five whole female bodies or 50 heads following one week under AL or DR. We then isolated RNA from these samples and performed qRT-PCR on the perturbed genes as described.
Gene alteration phenotyping.
For candidate gene validation, all lines were obtained from Bloomington Stock Center [52] or Vienna Drosophila RNAi Center, Vienna, Austria [54] (see Key Resources Table for list of lines used). To validate the GWAS-predicted effects of candidate genes, we used the whole-body GeneSwitch driver Act5C-GS-Gal4 and the neuron-specific driver Elav-GS-Gal4 for directed RNAi. 15 virgin driver females were mated with three transgene line males in four bottles containing a standard diet. Two-three days following progeny eclosion, mated females were sorted onto AL or DR media with or without 200 μM RU486 (final concentration) for RNAi activation [3], and flies were maintained on these media for life. For dls analysis, a Minos element mutant line was used for gene disruption. This line was outcrossed to w1118 or Canton-S control strains for six generations using a GFP tag associated with the inserted element. Spontaneous activity was measured on day 5 after flies were placed on either the AL or DR diet. Three vials of 25 female flies for each condition were placed for 48 hours in a 12-hour light-dark cycle at 25°C and 65% relative humidity in a TriKinetics Drosophila Activity Monitor system (TriKinetics, Waltham, MA), and beam crosses were recorded for 24 hours. Activity was recorded for three separate biological replicates. Body mass was measured in 5 replicates of 3 flies using a Radwag 82/220.X2 analytical balance. Flies were then frozen and triglycerides were measured using Stanbio Triglycerides LiquiColor Test. Glucose was measured using Stanbio Liqui-UV Hexokinase kit. For both assays, absorbance was measured using SpectroMax M2 spectrophotometer. Starvation was performed as previously described [3] by rearing flies for 10 days under AL diet and then transferring to medium containing water and 1% agarose. Flies were checked for deaths every 4 hours until all flies were dead. Pupariation rate was determined by mating flies and placing 60 resulting embryos under AL diet. Number of pupae formed was recorded daily.
Quantification and statistical analysis
Significance was determined between different gene expression (qRT-PCR) data and phenotypic data at specific time points using two-tailed unpaired t-test. Significance of differences between survival curves was assessed by log rank test. p<0.05 was considered statistically significant. Significant differences between experimental groups and controls are indicated by *. * = p < 0.05, ** = p < 0.005, *** = p < 0.0005, determined by unpaired t test. nc = no change, ns = not significant. Statistical analyses were calculated with GraphPad Prism 4. Statistics for genome-wide analyses were performed as previously stated. Correlation between traits was determined by identifying best-fit trendline across all data points and calculating R2 value to determine how well the data adheres to the trendline.
Supplementary Material
Data S1. Median lifespan data for DGRP fly strains. Related to STAR Methods and Figure 1. DGRP flies are arranged in ascending order by Bloomington Stock ID number. Median lifespan values for a given strain and starting n are detailed in each row, with replicate 1 shown in Columns B-E, replicate 2 in Columns F-I, and replicate 3 in Columns J-M.
Data S2. Climbing data for DGRP fly strains. Related to STAR Methods and Figure 1. DGRP flies are arranged in ascending order by Bloomington Stock ID number. Day at which each strain population reach 50% of its initial climbing ability is shown in Sheet1. Day at which each strain population fell below 20% absolute climbing ability is shown in Sheet2. Replicate 1 is shown in Columns B-C and Replicate 2 is shown in Columns D-E. Starting n for each strain is shown in Data S1.
Data S3. Statistical analysis of data. Related to STAR Methods and Figures 3-6. p values are shown for each statistical comparison between data groups in Column C. Column D shows statistical denotation: ns = not significant, * = p<0.05, ** = p<0.005, *** = p<0.0005.
Data S4. Effect size for candidate gene loci. Related to STAR Methods and Table 1. Effect size for each of the tested lifespan and climbing traits are shown in Column A. Gene and Loci are shown in Columns B and C respectively. Genotype effect size refers to value associated with effect independent of diet. Interaction effect size refers to effect taking diet into consideration.
Highlights:
Genetic variation alters lifespan and functional decline with age in response to DR
There is no correlation between lifespan and length of functionality
daedalus modulates climbing ability in diet-restricted conditions
decima modulates lifespan by regulating insulin-like peptide production
Acknowledgements
K.A.W. was supported by NIH/NIA award F31AG052299 and is currently supported by NIH/NIA training grant T32AG000266-21. C.S.N. was supported by NIH/NIA F32 award AG047024. This work was funded by grants from the American Federation of Aging Research (R.B.B. and P.K.), NIH grants R01AG038688 and AG045835 (to P.K.) and R01AG049494 (to Dr. Daniel Promislow) and the Larry L. Hillblom Foundation. We thank the Bloomington Drosophila Stock Center and the Vienna Drosophila Stock Center and the laboratories of Dr. Pejmun Haghighi, Dr. Henri Jasper, Dr. Norbert Perrimon, and Dr. Akhila Rajan for providing the flies used. We thank Geoffrey Meyerhof and the other members of the Kapahi lab for helpful discussions, as well as Dr. Geetanjali Chawla, Dr. John Newman, and Kelly Jin for their feedback.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
References
- 1.Mattison JA, Colman RJ, Beasley TM, Allison DB, Kemnitz JW, Roth GS, Ingram DK, Weindruch R, de Cabo R, and Anderson RM (2017). Caloric restriction improves health and survival of rhesus monkeys. Nature communications 8, 14063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Solon-Biet SM, McMahon AC, Ballard JW, Ruohonen K, Wu LE, Cogger VC, Warren A, Huang X, Pichaud N, Melvin RG, et al. (2014). The ratio of macronutrients, not caloric intake, dictates cardiometabolic health, aging, and longevity in ad libitum-fed mice. Cell metabolism 19, 418–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Katewa SD, Akagi K, Bose N, Rakshit K, Camarella T, Zheng X, Hall D, Davis S, Nelson CS, Brem RB, et al. (2016). Peripheral Circadian Clocks Mediate Dietary Restriction-Dependent Changes in Lifespan and Fat Metabolism in Drosophila. Cell Metab 23, 143–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zid BM, Rogers AN, Katewa SD, Vargas MA, Kolipinski MC, Lu TA, Benzer S, and Kapahi P (2009). 4E-BP extends lifespan upon dietary restriction by enhancing mitochondrial activity in Drosophila. Cell 139, 149–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kapahi P, Zid BM, Harper T, Koslover D, Sapin V, and Benzer S (2004). Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr Biol 14, 885–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Igarashi M, and Guarente L (2016). mTORC1 and SIRT1 Cooperate to Foster Expansion of Gut Adult Stem Cells during Calorie Restriction. Cell 166, 436–450. [DOI] [PubMed] [Google Scholar]
- 7.Broughton SJ, Slack C, Alic N, Metaxakis A, Bass TM, Driege Y, and Partridge L (2010). DILP-producing median neurosecretory cells in the Drosophila brain mediate the response of lifespan to nutrition. Aging cell 9, 336–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harper JM, Leathers CW, and Austad SN (2006). Does caloric restriction extend life in wild mice? Aging cell 5, 441–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olson MV (2012). Human genetic individuality. Annual review of genomics and human genetics 13, 1–27. [DOI] [PubMed] [Google Scholar]
- 10.Liao CY, Rikke BA, Johnson TE, Diaz V, and Nelson JF (2010). Genetic variation in the murine lifespan response to dietary restriction: from life extension to life shortening. Aging Cell 9, 92–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liao CY, Rikke BA, Johnson TE, Gelfond JA, Diaz V, and Nelson JF (2011). Fat maintenance is a predictor of the murine lifespan response to dietary restriction. Aging Cell 10, 629–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang WB, Sinha DB, Pittman WE, Hvatum E, Stroustrup N, and Pincus Z (2016). Extended Twilight among Isogenic C. elegans Causes a Disproportionate Scaling between Lifespan and Health. Cell Syst 3, 333–345 e334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bansal A, Zhu LJ, Yen K, and Tissenbaum HA (2015). Uncoupling lifespan and healthspan in Caenorhabditis elegans longevity mutants. Proceedings of the National Academy of Sciences of the United States of America 112, E277–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rollins JA, Howard AC, Dobbins SK, Washburn EH, and Rogers AN (2017). Assessing Health Span in Caenorhabditis elegans: Lessons From Short-Lived Mutants. The journals of gerontology. Series A, Biological sciences and medical sciences 72, 473–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischer KE, Hoffman JM, Sloane LB, Gelfond JA, Soto VY, Richardson AG, and Austad SN (2016). A cross-sectional study of male and female C57BL/6Nia mice suggests lifespan and healthspan are not necessarily correlated. Aging (Albany NY) 8, 2370–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gill TM, Gahbauer EA, Han L, and Allore HG (2010). Trajectories of disability in the last year of life. The New England journal of medicine 362, 1173–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rhodenizer D, Martin I, Bhandari P, Pletcher SD, and Grotewiel M (2008). Genetic and environmental factors impact age-related impairment of negative geotaxis in Drosophila by altering age-dependent climbing speed. Exp Gerontol 43, 739–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newman JC, Covarrubias AJ, Zhao M, Yu X, Gut P, Ng CP, Huang Y, Haldar S, and Verdin E (2017). Ketogenic Diet Reduces Midlife Mortality and Improves Memory in Aging Mice. Cell metabolism 26, 547–557 e548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rera M, Clark RI, and Walker DW (2012). Intestinal barrier dysfunction links metabolic and inflammatory markers of aging to death in Drosophila. Proc Natl Acad Sci U S A 109, 21528–21533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fontana L, and Partridge L (2015). Promoting health and longevity through diet: from model organisms to humans. Cell 161, 106–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deelen J, Beekman M, Uh HW, Broer L, Ayers KL, Tan Q, Kamatani Y, Bennet AM, Tamm R, Trompet S, et al. (2014). Genome-wide association meta-analysis of human longevity identifies a novel locus conferring survival beyond 90 years of age. Human molecular genetics 23, 4420–4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Durham MF, Magwire MM, Stone EA, and Leips J (2014). Genome-wide analysis in Drosophila reveals age-specific effects of SNPs on fitness traits. Nature communications 5, 4338. [DOI] [PubMed] [Google Scholar]
- 23.Ivanov DK, Escott-Price V, Ziehm M, Magwire MM, Mackay TF, Partridge L, and Thornton JM (2015). Longevity GWAS Using the Drosophila Genetic Reference Panel. The journals of gerontology. Series A, Biological sciences and medical sciences 70, 1470–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carbone MA, Yamamoto A, Huang W, Lyman RA, Meadors TB, Yamamoto R, Anholt RR, and Mackay TF (2016). Genetic architecture of natural variation in visual senescence in Drosophila. Proc Natl Acad Sci U S A 113, E6620–E6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mackay TF, Richards S, Stone EA, Barbadilla A, Ayroles JF, Zhu D, Casillas S, Han Y, Magwire MM, Cridland JM, et al. (2012). The Drosophila melanogaster Genetic Reference Panel. Nature 482, 173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nelson CS, Beck JN, Wilson KA, Pilcher ER, Kapahi P, and Brem RB (2016). Cross-phenotype association tests uncover genes mediating nutrient response in Drosophila. BMC genomics 17, 867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jehrke L, Stewart FA, Droste A, and Beller M (2018). The impact of genome variation and diet on the metabolic phenotype and microbiome composition of Drosophila melanogaster. Scientific reports 8, 6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitchell SJ, Madrigal-Matute J, Scheibye-Knudsen M, Fang E, Aon M, Gonzalez-Reyes JA, Cortassa S, Kaushik S, Gonzalez-Freire M, Patel B, et al. (2016). Effects of Sex, Strain, and Energy Intake on Hallmarks of Aging in Mice. Cell Metab 23, 1093–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun J, Molitor J, and Tower J (2004). Effects of simultaneous over-expression of Cu/ZnSOD and MnSOD on Drosophila melanogaster life span. Mechanisms of ageing and development 125, 341–349. [DOI] [PubMed] [Google Scholar]
- 30.Wang J, Rousseau J, Kim E, Ehresmann S, Cheng YT, Duraine L, Zuo Z, Park YJ, Li-Kroeger D, Bi W, et al. (2019). Loss of Oxidation Resistance 1, OXR1, Is Associated with an Autosomal-Recessive Neurological Disease with Cerebellar Atrophy and Lysosomal Dysfunction. American journal of human genetics 105, 1237–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koudounas S, Green EW, and Clancy D (2012). Reliability and variability of sleep and activity as biomarkers of ageing in Drosophila. Biogerontology 13, 489–499. [DOI] [PubMed] [Google Scholar]
- 32.Vonesch SC, Lamparter D, Mackay TF, Bergmann S, and Hafen E (2016). Genome-Wide Analysis Reveals Novel Regulators of Growth in Drosophila melanogaster. PLoS genetics 12, e1005616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thurmond J, Goodman JL, Strelets VB, Attrill H, Gramates LS, Marygold SJ, Matthews B, Millburn G, Antonazzo G, Trovisco V, et al. (2019). FlyBase 2.0: the next generation. Nucleic acids research 47, D759–D765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shin SC, Kim SH, You H, Kim B, Kim AC, Lee KA, Yoon JH, Ryu JH, and Lee WJ (2011). Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science 334, 670–674. [DOI] [PubMed] [Google Scholar]
- 35.Brogiolo W, Stocker H, Ikeya T, Rintelen F, Fernandez R, and Hafen E (2001). An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Current biology : CB 11, 213–221. [DOI] [PubMed] [Google Scholar]
- 36.Broughton SJ, Piper MD, Ikeya T, Bass TM, Jacobson J, Driege Y, Martinez P, Hafen E, Withers DJ, Leevers SJ, et al. (2005). Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proc Natl Acad Sci U S A 102, 3105–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ostrovskaya O, Xie K, Masuho I, Fajardo-Serrano A, Lujan R, Wickman K, and Martemyanov KA (2014). RGS7/Gbeta5/R7BP complex regulates synaptic plasticity and memory by modulating hippocampal GABABR-GIRK signaling. eLife 3, e02053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nassel DR, Kubrak OI, Liu Y, Luo J, and Lushchak OV (2013). Factors that regulate insulin producing cells and their output in Drosophila. Frontiers in physiology 4, 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee KS, Kwon OY, Lee JH, Kwon K, Min KJ, Jung SA, Kim AK, You KH, Tatar M, and Yu K (2008). Drosophila short neuropeptide F signalling regulates growth by ERK-mediated insulin signalling. Nature cell biology 10, 468–475. [DOI] [PubMed] [Google Scholar]
- 40.Rajan A, and Perrimon N (2012). Drosophila cytokine unpaired 2 regulates physiological homeostasis by remotely controlling insulin secretion. Cell 151, 123–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rikke BA, Liao CY, McQueen MB, Nelson JF, and Johnson TE (2010). Genetic dissection of dietary restriction in mice supports the metabolic efficiency model of life extension. Exp Gerontol 45, 691–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simpson SJ, Le Couteur DG, Raubenheimer D, Solon-Biet SM, Cooney GJ, Cogger VC, and Fontana L (2017). Dietary protein, aging and nutritional geometry. Ageing research reviews 39, 78–86. [DOI] [PubMed] [Google Scholar]
- 43.Vargas MA, Luo N, Yamaguchi A, and Kapahi P (2010). A role for S6 kinase and serotonin in postmating dietary switch and balance of nutrients in D. melanogaster. Current biology : CB 20, 1006–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hahm JH, Kim S, DiLoreto R, Shi C, Lee SJ, Murphy CT, and Nam HG (2015). C. elegans maximum velocity correlates with healthspan and is maintained in worms with an insulin receptor mutation. Nature communications 6, 8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jacob ME, Yee LM, Diehr PH, Arnold AM, Thielke SM, Chaves PH, Gobbo LD, Hirsch C., Siscovick D, and Newman AB (2016). Can a Healthy Lifestyle Compress the Disabled Period in Older Adults? Journal of the American Geriatrics Society 64, 1952–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mattison JA, Roth GS, Beasley TM, Tilmont EM, Handy AM, Herbert RL, Longo DL, Allison DB, Young JE, Bryant M, et al. (2012). Impact of caloric restriction on health and survival in rhesus monkeys from the NIA study. Nature 489, 318–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jayaraman M, Zhou H, Jia L, Cain MD, and Blumer KJ (2009). R9AP and R7BP: traffic cops for the RGS7 family in phototransduction and neuronal GPCR signaling. Trends in pharmacological sciences 30, 17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Talbert ME, Barnett B, Hoff R, Amella M, Kuczynski K, Lavington E, Koury S, Brud E, and Eanes WF (2015). Genetic perturbation of key central metabolic genes extends lifespan in Drosophila and affects response to dietary restriction. Proceedings. Biological sciences / The Royal Society 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jaszczak JS, Wolpe JB, Bhandari R, Jaszczak RG, and Halme A (2016). Growth Coordination During Drosophila melanogaster Imaginal Disc Regeneration Is Mediated by Signaling Through the Relaxin Receptor Lgr3 in the Prothoracic Gland. Genetics 204, 703–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Q, Levay K, Chanturiya T, Dvoriantchikova G, Anderson KL, Bianco SD, Ueta CB, Molano RD, Pileggi A, Gurevich EV, et al. (2011). Targeted deletion of one or two copies of the G protein beta subunit Gbeta5 gene has distinct effects on body weight and behavior in mice. FASEB journal : official publication of the Federation of American Societies for Experimental Biology 25, 3949–3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Katewa SD, Demontis F, Kolipinski M, Hubbard A, Gill MS, Perrimon N, Melov S, and Kapahi P (2012). Intramyocellular fatty-acid metabolism plays a critical role in mediating responses to dietary restriction in Drosophila melanogaster. Cell Metab 16, 97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cook KR, Parks AL, Jacobus LM, Kaufman TC, and Matthews KA (2010). New research resources at the Bloomington Drosophila Stock Center. Fly (Austin) 4, 88–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perktold J, et al. StatsModels: Statistical Modeling and Econometrics in Python. [Google Scholar]
- 54.Kaya-Copur A, and Schnorrer F (2016). A Guide to Genome-Wide In Vivo RNAi Applications in Drosophila. Methods in molecular biology 1478, 117–143. [DOI] [PubMed] [Google Scholar]
- 55.Rogulja D and Irvine KD, Regulation of cell proliferation by a morphogen gradient. Cell, 2005. 123(3): p. 449–61. [DOI] [PubMed] [Google Scholar]
- 56.Osterwalder T, et al. , A conditional tissue-specific transgene expression system using inducible GAL4. Proc Natl Acad Sci U S A, 2001. 98(22): p. 12596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ford D, et al. , Alteration of Drosophila life span using conditional, tissue-specific expression of transgenes triggered by doxycyline or RU486/Mifepristone. Exp Gerontol, 2007. 42(6): p. 483–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brand AH and Perrimon N, Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development, 1993. 118(2): p. 401–15. [DOI] [PubMed] [Google Scholar]
- 59.Dietzl G, et al. , A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature, 2007. 448(7150): p. 151–6. [DOI] [PubMed] [Google Scholar]
- 60.Mummery-Widmer JL, et al. , Genome-wide analysis of Notch signalling in Drosophila by transgenic RNAi. Nature, 2009. 458(7241): p. 987–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bellen HJ, et al. , The Drosophila gene disruption project: progress using transposons with distinctive site specificities. Genetics, 2011. 188(3): p. 731–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cook KR, et al. , New research resources at the Bloomington Drosophila Stock Center. Fly (Austin), 2010. 4(1): p. 88–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dorer DR and Henikoff S, Expansions of transgene repeats cause heterochromatin formation and gene silencing in Drosophila. Cell, 1994. 77(7): p. 993–1002. [DOI] [PubMed] [Google Scholar]
- 64.Green EW, et al. , A Drosophila RNAi collection is subject to dominant phenotypic effects. Nat Methods, 2014. 11(3): p. 222–3. [DOI] [PubMed] [Google Scholar]
- 65.Lee DM, et al. , PH Domain-Arf G Protein Interactions Localize the Arf-GEF Steppke for Cleavage Furrow Regulation in Drosophila. PLoS One, 2015. 10(11): p. e0142562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Perkins LA, et al. , The Transgenic RNAi Project at Harvard Medical School: Resources and Validation. Genetics, 2015. 201(3): p. 843–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim M, et al. , Drosophila Gyf/GRB10 interacting GYF protein is an autophagy regulator that controls neuron and muscle homeostasis. Autophagy, 2015. 11(8): p. 1358–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Karpac J, et al. , JNK signaling in insulin-producing cells is required for adaptive responses to stress in Drosophila. Aging Cell, 2009. 8(3): p. 288–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rajan A and Perrimon N, Drosophila cytokine unpaired 2 regulates physiological homeostasis by remotely controlling insulin secretion. Cell, 2012. 151(1): p. 123–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Inagaki HK, Panse KM, and Anderson DJ, Independent, reciprocal neuromodulatory control of sweet and bitter taste sensitivity during starvation in Drosophila. Neuron, 2014. 84(4): p. 806–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Median lifespan data for DGRP fly strains. Related to STAR Methods and Figure 1. DGRP flies are arranged in ascending order by Bloomington Stock ID number. Median lifespan values for a given strain and starting n are detailed in each row, with replicate 1 shown in Columns B-E, replicate 2 in Columns F-I, and replicate 3 in Columns J-M.
Data S2. Climbing data for DGRP fly strains. Related to STAR Methods and Figure 1. DGRP flies are arranged in ascending order by Bloomington Stock ID number. Day at which each strain population reach 50% of its initial climbing ability is shown in Sheet1. Day at which each strain population fell below 20% absolute climbing ability is shown in Sheet2. Replicate 1 is shown in Columns B-C and Replicate 2 is shown in Columns D-E. Starting n for each strain is shown in Data S1.
Data S3. Statistical analysis of data. Related to STAR Methods and Figures 3-6. p values are shown for each statistical comparison between data groups in Column C. Column D shows statistical denotation: ns = not significant, * = p<0.05, ** = p<0.005, *** = p<0.0005.
Data S4. Effect size for candidate gene loci. Related to STAR Methods and Table 1. Effect size for each of the tested lifespan and climbing traits are shown in Column A. Gene and Loci are shown in Columns B and C respectively. Genotype effect size refers to value associated with effect independent of diet. Interaction effect size refers to effect taking diet into consideration.
Data Availability Statement
The published article includes all datasets generated and analyzed during this study. Codes used will be available upon request to the Lead Contact.