Abstract
N6-methyladenosine (m6A) in messenger RNA (mRNA) regulates its stability, splicing, and translation efficiency. Here, we explored how the expression levels of small GTPase proteins are regulated by m6A modulators. We employed a high-throughput scheduled multiple-reaction monitoring (MRM)-based targeted proteomic approach to quantify systemically the changes in expression of small GTPase proteins in cells upon genetic ablation of METTL3 (the catalytic subunit of the major m6A methyltransferase complex), m6A demethylases (ALKBH5 and FTO), or m6A reader proteins (YTHDF1, YTHDF2, and YTHDF3). Depletions of METTL3 and ALKBH5 resulted in substantially diminished and augmented expression, respectively, of a subset of small GTPase proteins, including RHOB and RHOC. Our results also revealed that the stability of RHOB mRNA is significantly increased in cells depleted of METTL3, suggesting an m6A-elicited destabilization of this mRNA. Those small GTPases that are targeted by METTL3 and/or ALKBH5 also displayed higher discrepancies between protein and mRNA expression in paired primary/metastatic melanoma or colorectal cancer cells than those that are not. Together, this is the first comprehensive analysis of the alterations in small GTPase proteome regulated by epitranscriptomic modulators of m6A, and our study suggests the potential of an alternative therapeutic approach to target the currently “undruggable” small GTPases.
Graphical Abstract
INTRODUCTION
Small GTPases of the Ras superfamily are molecular switches that undergo conformational changes between the active guanosine triphosphate (GTP)-associated state and the inactive guanosine diphosphate (GDP)-occupied state.1 Members of this superfamily interact with effector proteins and modulate diverse essential cellular processes including proliferation, vesicular transport, and cytoskeleton organization.2–3 Mutations and aberrant expressions of small GTPases (e.g. KRAS) promote the progression of various types of cancer.4–5 Indeed, clinical studies have documented that numerous small GTPases, e.g. those in the RAS and RHO subfamilies, are differentially expressed in tumor vs. normal tissues, and metastatic vs. primary tumor cells.6 In this vein, targeting small GTPases has drawn substantial attention from academia and pharmaceutical industry for the development of cancer chemotherapeutic agents.7 Despite intensive efforts since the initial discovery of the RAS oncogenes over three decades ago, no effective inhibitors have been approved for cancer treatment.8–10 Therefore, alternative approaches to modulate the expression of dysregulated small GTPases may offer new venues for cancer therapy.
A growing number of studies in recent years have shown that RNA metabolism can be dynamically regulated by post-transcriptional modifications, where N6-methyladenosine (m6A) is the most prevalent internal modification in mRNA.11 Previous studies have established dynamic regulation of m6A in mRNA by the METTL3-METTL14 m6A methyltransferase complex and m6A demethylases (i.e. FTO and ALKBH5).12–14 When bound by its reader protein YTHDF2, the m6A in mRNAs could lead to their destabilization;15–16 the binding of m6A in mRNA by other reader proteins (e.g. YTHDF1 and YTHDF3), however, enhances the stability and/or translation efficiency of the mRNA.17–19 In addition, m6A within the 5′-untranslated region (UTR) of mRNA promotes cap-independent translation in mammalian cells.20 Interestingly, dysregulations of epitranscriptomic modulators were found to drive the abnormal expression of oncogenes at the protein level and thus promote the growth, survival, and invasion of cancer cells.21–23 We hypothesized that the expression of small GTPase proteins may be subjected to regulation by m6A modulators.
In the present study, we test the above hypothesis by analyzing comprehensively the alterations in small GTPase proteome in HEK293T cells upon CRISPR-Cas9-mediated ablation of METTL3, ALKBH5, FTO, YTHDF1, YTHDF2, and YTHDF3 genes.24 To achieve quantifications of small GTPase proteins in high sensitivity, reproducibility and throughput, we employed a previously developed scheduled multiple-reaction monitoring (MRM)-based quantitative proteomic approach, together with the use of synthetic stable isotope-labeled peptides as internal standards.25–27 We observed altered protein expression of several small GTPases, including the known tumor suppressor RHOB and metastasis driver RHOC,5 in cells depleted of METTL3 or ALKBH5.
EXPERIMENTAL SECTION
Cell culture and gene depletion
HEK293T human embryonic kidney epithelial cells (ATCC) were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Thermo Fisher) supplemented with 10% fetal bovine serum (FBS, Thermo Fisher) and 1% penicillin-streptomycin solution (PS, GE Healthcare). The cells were maintained at 37°C in a humidified chamber supplemented with 5% CO2. All knockout cells in the HEK293T background were generated by using the CRISPR-Cas9 genome editing method.28 In brief, plasmids (Addgene #62988) carrying required enzymes and selected guide sequence (Table S1) were transiently transfected into HEK293T cells and the single clones were selected. Successful knockouts of target genes were confirmed by Sanger sequencing and Western blot analyses (Figure S1).
Cell lysis and proteomic sample preparation
To extract total proteins, approximately 8×106 cells were incubated on ice with 100 μL of pre-chilled CelLytic M cell lysis reagent (Sigma) containing 1% protease inhibitor cocktail (Sigma) on-ice for 30 min. The cell lysate was centrifuged at 16000 g for 30 min at 4°C. Supernatants containing total proteins were transferred to a chilled vial. Protein concentration was measured by using Quick Start Bradford Protein Assay (Bio-Rad). Approximately 50 μg of total proteins was mixed with 4× Laemmli SDS loading buffer and the mixtures were boiled for 10 min. Total proteins were resolved by 10% SDS-PAGE gel and then stained with Coomassie Brilliant Blue R-250.
In-gel protein digestion was conducted as described previously.29 In brief, gel bands corresponding to a molecular weight range of 15–37 kDa were cut into 1 mm3 cubes, and sequentially destained with 25% and 50% CH3CN in 50 mM ammonium bicarbonate (pH 7.8). Reduction and alkylation were performed by incubating the gel pieces in 10 mM dithiothreitol (DTT) at 37°C for 1 h and 55 mM iodoacetamide at room temperature in dark for 20 min, respectively. The proteins were digested with trypsin at 37°C for 18 h with an enzyme/protein ratio of 1:100. Peptides were extracted from the gel pieces by sonicating in a solution containing CH3CN/H2O/acetic acid (45/45/5, v/v) and concentrated by Speed-Vac. Prior to LC-MRM analysis, the tryptic peptides were desalted using C18 ZipTip (Agilent), concentrated by Speed-Vac, and reconstituted in 30 μL of 0.1% formic acid. Approximately 8% of the digestion mixture was spiked-in with a crude pool of synthetic small GTPase peptides (New England Peptide, Inc.) (~4 fmol each) with a C-terminal [15N2, 13C6]-labeled lysine (+8.0 Da) or [15N4, 13C6]-labeled arginine (+10 Da).25
Liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis and data processing
The MRM-based LC-MS/MS experiments were performed on a TSQ Altis triple-quadrupole mass spectrometer (Thermo Fisher) coupled with an UltiMate 3000 UPLC (Thermo Fisher) and a Flex nanoelectrospray ion source (Thermo Fisher). The spiked-in sample was loaded onto a 4-cm trapping column (150 μm i.d.) packed in-house with C18 resin (5 μm in particle size and 120 Å in pore size, Dr. Maisch GmbH HPLC) with buffer A (0.1% formic acid in water) at a flow rate of 3 μL/min. The peptides were eluted from the trapping column and separated on an analytical column (75 μm i.d., ~25 cm in length) packed in-house with C18 resin (3 μm in particle size and 120 Å in pore size, Dr. Maisch GmbH HPLC), using a gradient of 12–40% buffer B (0.1% formic acid in 80% CH3CN) and a flow rate of 300 nL/min. The peptides were ionized with a spray voltage of 2200 V and an ion transport tube temperature of 325°C. A resolution of 0.7 F.W.H.M. was set for both Q1 and Q3. Fragmentation of precursor ions in Q2 was conducted with 1.5 mTorr argon and the collision energy was derived from default settings in Skyline (version 19.1.0.193).30
The mass spectrometer was scheduled to monitor the precursor to product ion transitions of 144 unique peptides of human small GTPases with a cycle time of 3 sec in a 4-min retention time (RT) window. The retention time of the monitored peptides was predicted from its normalized retention time (iRT) and the iRT-RT calibration curve determined from 10 tryptic peptides of BSA.25, 31 The three most abundant y ions found in MS/MS acquired from shotgun proteomic analyses were selected to quantify each peptide.
To enable robust peak detection, all target peptides were manually inspected and filtered with dot-product (dotp) > 0.7.32 To calculate the relative quantity of small GTPases, the sum of peak areas in the extracted-ion chromatograms (XICs) for the three monitored transitions were normalized against those of corresponding spiked-in heavy isotope-labeled peptide. The derived ratio of the knockout cells was further normalized against that of parental HEK293T cells. A total of three and four biological replicates were conducted for METTL3−/− and other knockout cells, respectively.
Real-time quantitative PCR (RT-qPCR) and mRNA stability assay
RNA was extracted from approximately 1×106 cells with Total RNA Kit I (Omega) and purified with HiBind RNA mini columns (VWR). Total RNA (1 μg) was subjected to reverse transcription and 66 ng of the resulting cDNA was mixed with Luna qPCR Master Mix (New England Biolabs) in a 96-well optical reaction plate (Bio-Rad). RT-qPCR was conducted on a CFX real-time PCR detection system (Bio-Rad). For mRNA stability assay, cells were treated with 5 μg/mL of actinomycin D in a reverse chronological order prior to harvesting cells for RNA extraction. The primer sequences used for RT-qPCR were: GAPDH forward, GGTCGGAGTCAACGGATT; GAPDH reverse, ATCGCCCCACTTGATTTTG; RHOB forward, CGGACTCGCTGGAGAACA; RHOB revere, GAGGTAGTCGTAGGCTTGGAT; RHOC forward, CAGTCTGAGCCTCCGGCAC; RHOC reverse, GAGGAGGCAGGTCTTCCCAC.
Hierarchical clustering
The hierarchical clustering was performed using the default setting of the ClustVis online tool with minor changes.33 Briefly, the Euclidean method was used for distance measurement, and the tightest is clustered first in the tree ordering.
RESULTS
MRM-based Targeted Proteomic Profiling of Differential Expression of Small GTPase Proteins in Cells Depleted of m6A Modulators
Recent studies revealed that dynamic and reversible formations of m6A in mRNA enable cells to adapt rapidly to environmental stresses.34–36 Since small GTPases serve as upstream molecular switches for numerous signal transduction cascades in response to extracellular stimuli, we reason that the expression of small GTPase proteins may be dynamically regulated by m6A modulators. To test this hypothesis, we systemically investigated how the protein expression levels of small GTPases are modulated by epitranscriptomic regulators. We utilized a previously established high-throughput MRM-based targeted proteomic workflow to quantify, at the proteome-wide scale, the changes in expression of small GTPase proteins in cells where the m6A writer (METTL3), eraser (ALKBH5 and FTO), or reader (YTHDF1, YTHDF2, and YTHDF3) proteins were individually knocked out by CRISPR-Cas9 (Figure S1).24–25
The MRM library contained a unique peptide for each of the 144 small GTPases, which represent approximately 85% of the human small GTPase proteome.27, 37 To fulfill sensitive and reproducible quantification, we selected three most abundant fragment ions for each peptide based on MS/MS obtained from shotgun proteomic analyses. We realized that the potential differences in efficiencies of tryptic digestion and peptide recovery from gels may affect the quantification accuracies for small GTPases. Thus, we always analyzed the lysates of the knockout cells in parallel with those of parental HEK293T cells in each batch of sample preparation (Figure S2). A mixture of synthetic stable isotope-labeled peptides with identical amino acid sequences to our target analytes was employed as internal standards for robust quantitation and was spiked into the samples prior to LC-MS/MS analysis (Figure 1A). Notably, the heavy peptides and the corresponding unlabeled sample peptides exhibit nearly identical retention time and ionization efficiency.38 MRM analyses facilitated quantification of > 80 small GTPases for each knocked-out background (Figure S3A–F, Table S2), and 78 small GTPases were commonly quantified in all genetic backgrounds (Figure 1B). It is of note that some peptides were only detected in the heavy form, suggesting that these small GTPases are not expressed or expressed at very low levels in HEK293T and the isogenic knockout cells. Additionally, the failure in detecting in some small GTPases may also arise from post-translational modifications that increase pronouncedly the molecular weight of the small GTPase protein (i.e. to above 37 kDa) and/or alter the mass of the small GTPase peptides selected for MRM analysis.
Figure 1. MRM-based targeted proteomic analysis for protein expression profiling of small GTPases in cells depleted of epitranscriptomic modulators.
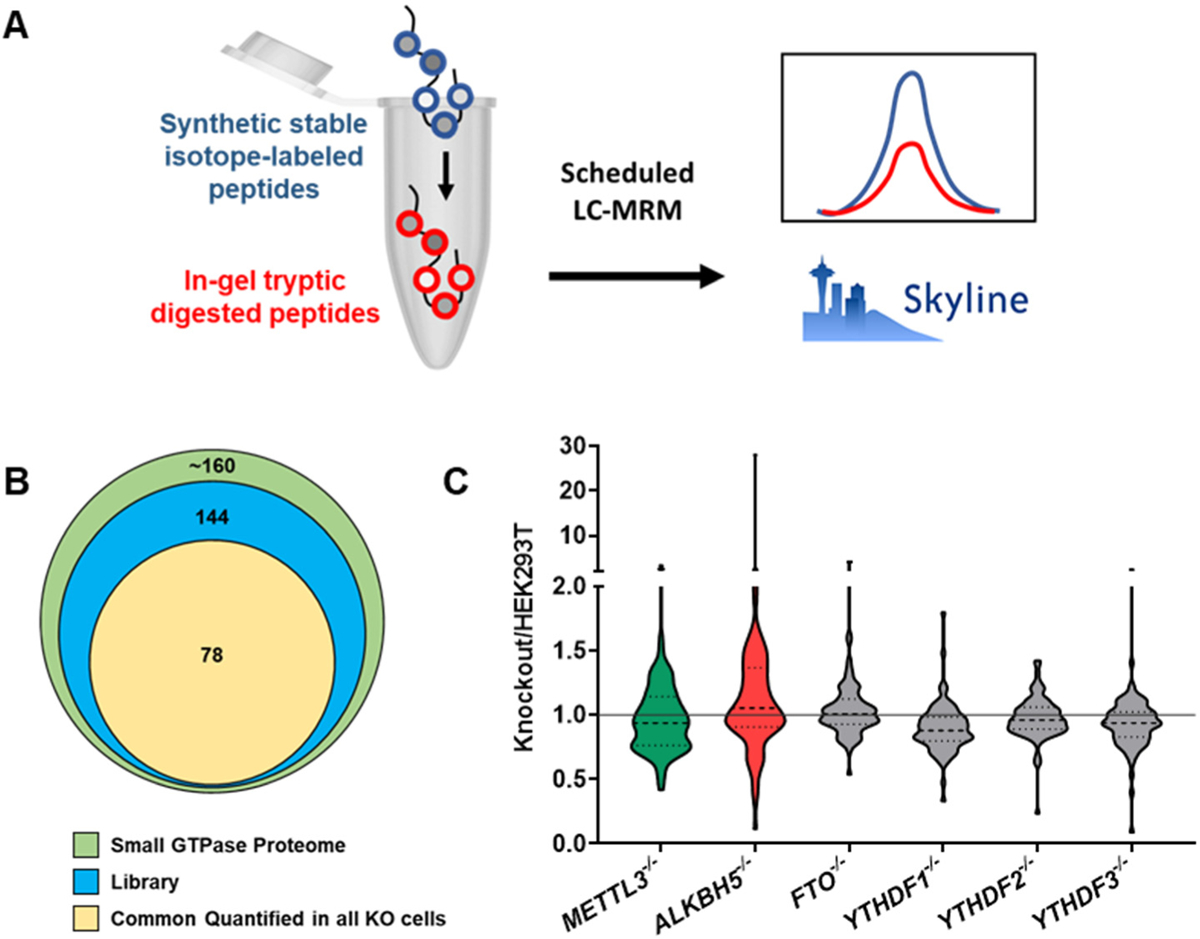
(A) A schematic diagram illustrating the workflow of MRM-based targeted proteomic analysis of the small GTPase proteome. In-gel tryptic digested peptides were spiked-in with a mixture of synthetic stable isotope-labeled peptides of identical amino acid sequences prior to LC-MRM analysis. The LC-MS/MS data were processed using Skyline. (B) A Venn diagram showing the number of small GTPases in human proteome, the current MRM library, and the commonly quantified in all knockout cells studied herein. (C) Violin plots displaying the ratio distributions of knockout/parental cells of the quantified small GTPases in cells depleted of m6A writer, eraser, or reader proteins.
Our results showed that depletion of METTL3 and ALKBH5 altered the expression of small GTPase proteins in HEK293T cells (Figure 1C). By imposing a cutoff ratio of 1.5-fold, we found that the expression levels of 13 and 20 small GTPase proteins were substantially altered in HEK293T cells upon genetic ablations of METTL3 and ALKBH5, respectively (Figure 2A). By using the same threshold, only a few small GTPases were, however, up- or down-regulated at the protein level in cells depleted of FTO or any of the three m6A reader proteins (Figure S3G–J). Our observation about the three m6A reader proteins is in keeping with a recent study showing that YTHDF1, YTHDF2, and YTHDF3 assume redundant functions, and their abilities in regulating mRNA stability becomes evident only when all three paralogs are simultaneously depleted.39
Figure 2. Modulations of the differential expression of the small GTPase proteome by m6A writer and eraser proteins.
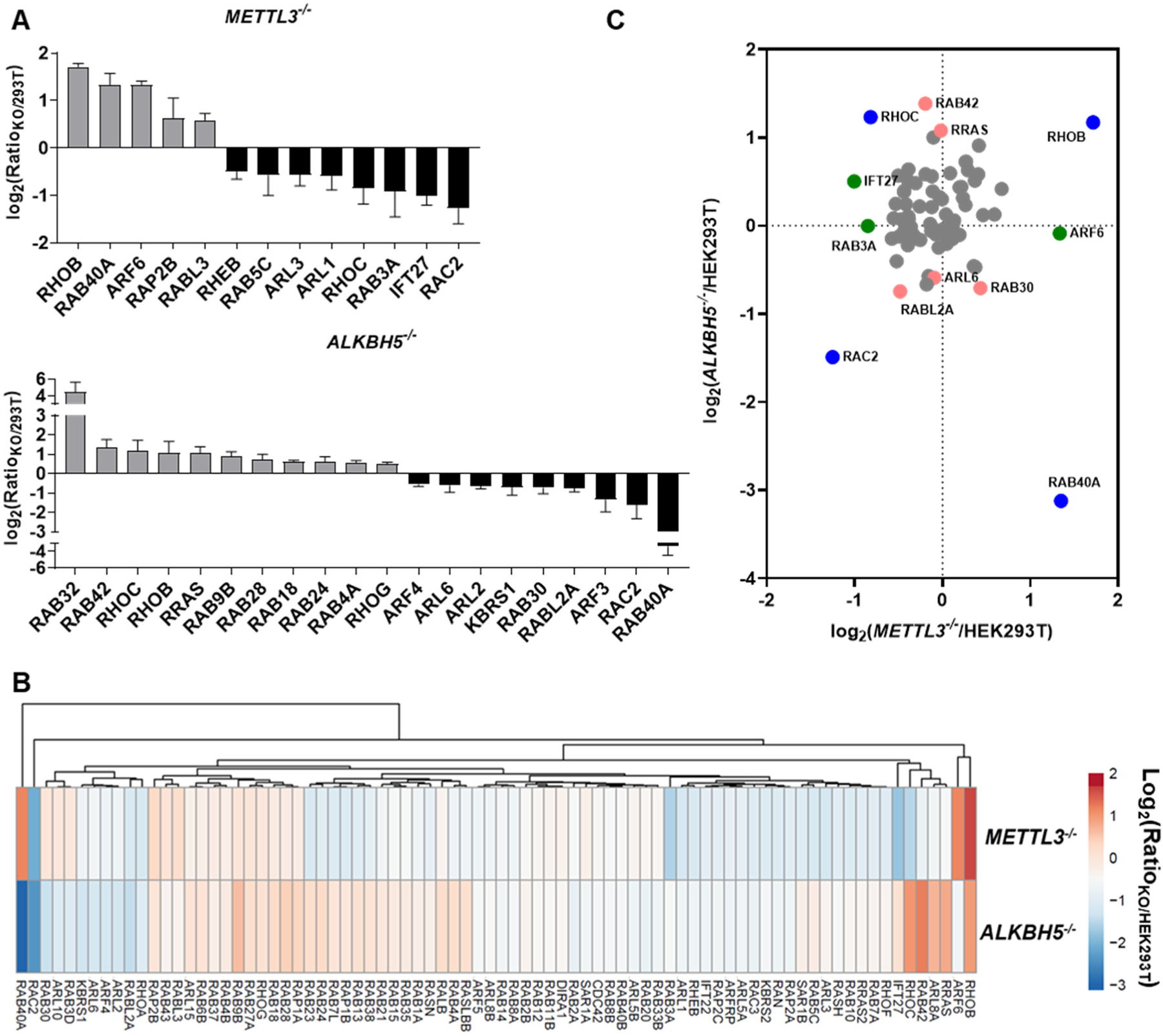
(A) A bar chart showing the substantially (>1.5-fold) up- (grey) or down-regulated (black) small GTPases quantified from three (for METTL3−/−) or four (for ALKBH5−/−) independent LC-MRM experiments. (B) Hierarchical clustering of the differential expression of small GTPase proteins between METTL3−/− vs. HEK293T and ALKBH5−/− vs. HEK293T cells. (C) A dot plot showing the significantly up- and down-regulated small GTPases in the METTL3−/− (green), ALKBH5−/− (red), or both (blue) cell lines relative to the isogenic parental HEK293T cells.
The results from the MRM-based small GTPase quantification suggest that METTL3 and ALKBH5 may be important regulators for the expression of some small GTPases at the protein level (Figure 1C). In particular, 80 small GTPases were commonly quantified in METTL3−/− and ALKBH5−/− cells (Figure S4). A hierarchical clustering analysis for the commonly quantified small GTPases revealed the regulatory profiles of METTL3 and ALKBH5 in the expression of small GTPase proteins (Figure 2B).33 Notably, the protein expressions of several small GTPases were significantly altered in cells depleted of METTL3 and/or ALKBH5 (Figure 2C), including the well-established regulators for cancer metastasis, e.g., RHOB and RHOC, along with the less well-characterized RAB40A and RAC2.5, 40 Together, our results revealed the involvements of METTL3 and ALKBH5 in regulating the expression of small GTPase proteins.
Western Blot for the Validation of Differential Expression of Small GTPase Proteins in Cells Depleted of m6A Epitranscriptomic Modulators
To further validate the findings made from the scheduled MRM analysis, we employed Western blot analysis to quantify the small GTPase expression at the protein level. The Western blot results confirmed the relative expression levels of RHOB, RHOC and several other selected small GTPase proteins in METTL3−/− and ALKBH5−/− cells vs. the parental HEK293T cells (Figure 3A and B, Figure S5). Together, the quantification results obtained from Western blot analysis are highly consistent with the proteomic data with an R2 value of 0.991, and a slope of 0.934 in linear regression analysis (Figure 3C). This result demonstrates the excellent accuracy of the scheduled MRM approach in quantifying the protein expression of small GTPases in cells depleted of epitranscriptomic modulators.
Figure 3. Western blot validation of the quantification results obtained from scheduled LC-MRM analysis.
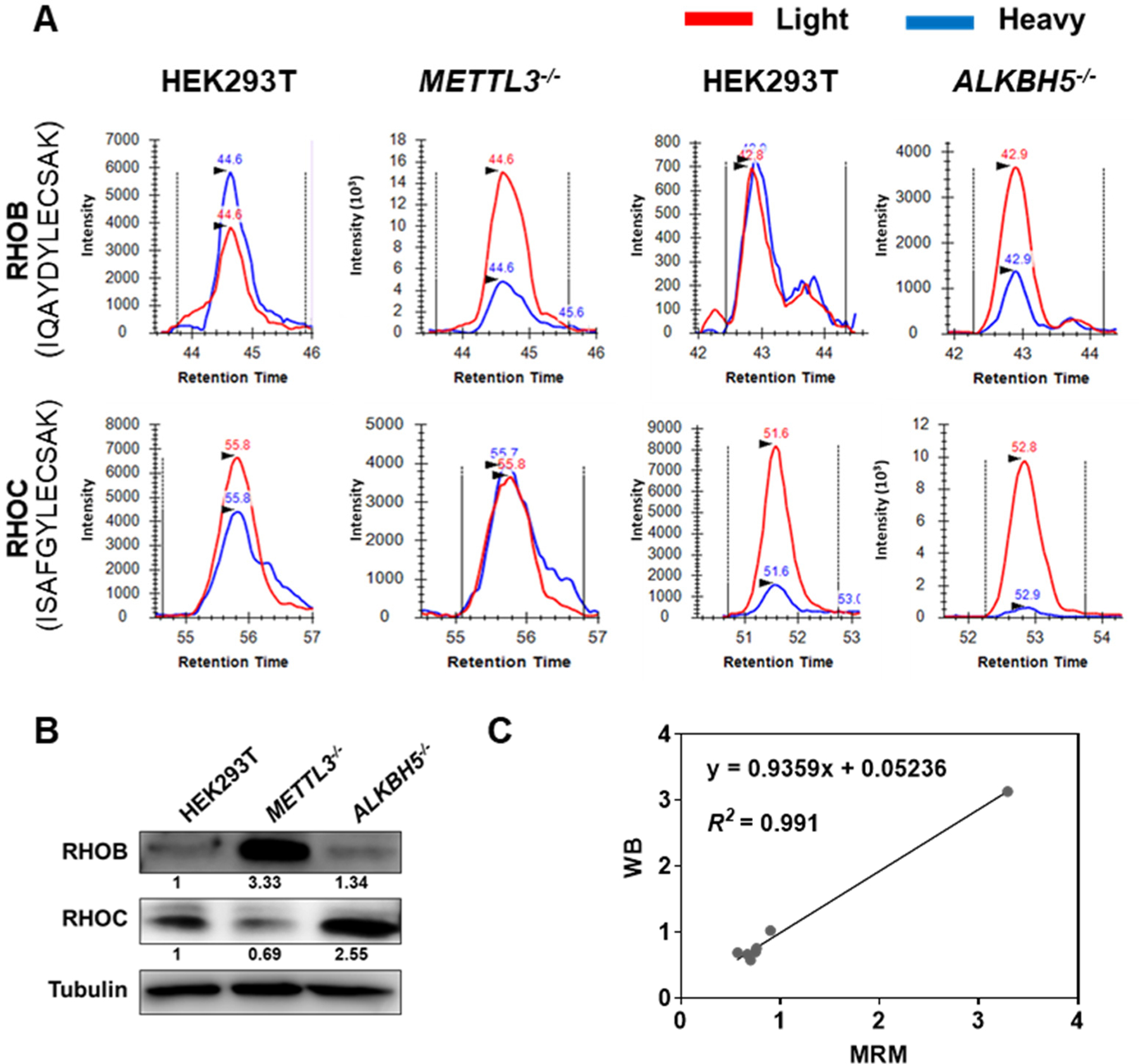
(A) Ion chromatograms showing the traces of the three monitored transitions for unique tryptic peptides of RHOB (y7, y8, and y9) (upper panel) and RHOC (y6, y8, and y9) (lower panel) in METTL3−/− and ALKBH5−/− in comparison with parental HEK293T cells. The traces for the unlabeled peptides in parental or knockout cells are shown in red and the corresponding traces for the spiked-in heavy isotope-labeled peptides are displayed in blue. (B) Western blot for validating the MRM results of RHOB and RHOC proteins in METTL3−/− and ALKBH5−/− cells vs. parental HEK293T cells. The band intensity of small GTPases was normalized against that of tubulin and further normalized against the corresponding ratio obtained for parental HEK293T cells. (C) Linear regression analysis of the quantification results of small GTPase expression in protein level obtained from the LC-MRM and Western blot analysis in METTL3−/− cells vs. parental HEK293T cells.
To further explore the involvements of METTL3 and ALKBH5 in regulating small GTPase expression at the protein level, we compared the transcriptome datasets retrieved from the Broad Institute Cancer Cell Line Encyclopedia (CCLE) and the previously published proteomic results acquired from the paired primary/metastatic melanoma cells (WM115/WM266–4 and IGR39/IGR37) and colorectal cancer cells (SW480/SW620).26, 41–42 Interestingly, we observed a greater discrepancy between the mRNA and protein expression for the small GTPases found to be regulated by METTL3 and/or ALKBH5 in the present study than those that are not, for which a normal distribution of mRNA/protein ratio with the median value being 0.87, 0.97, and 1.17 were observed in the WM115/WM266–4, IGR39/IGR37, and SW480/SW620 paired cell lines, respectively (Figure S6). Notably, although the median ratio of the small GTPases regulated by METTL3 and ALKBH5 in the IGR39/IGR37 are close to 1, the ratio distribution is distinct from the non-targets (Figure S6B). Interestingly, the expression of the well-characterized driver genes for melanoma metastasis, e.g., ARF6 and RHOC, were down-regulated at the mRNA level but up-regulated at the protein level in the metastatic melanoma cells relative to the paired primary melanoma cells (Table S3).40, 43 Together, these results support that the differential expression of small GTPases during the metastatic progression may be regulated by epitranscriptomic modulators.
mRNA Metabolism Regulated by Epitranscriptomic Modulators
Aberrant protein expression of RHO small GTPases, e.g., RHOB and RHOC, drives the migration and invasion of cancer cells.40, 44–45 Our MRM analysis of small GTPase proteome revealed altered expressions of RHOB and RHOC proteins in the METTL3−/− and ALKBH5−/− cells. To understand how METTL3 and ALKBH5 regulate the protein expression of RHOB and RHOC, we carried out real-time quantitative PCT (RT-qPCR) analysis for RHOB and RHOC transcripts in METTL3−/− and ALKBH5−/− cells. The results revealed a similar trend in changes of mRNA and protein expression (Figure 4A). Thus, we hypothesized that METTL3 and ALKBH5 may alter the transcriptional efficiencies of RHOB and RHOC genes and/or the stabilities of the resulting transcripts in cells. To distinguish these two possibilities, we tested, by employing RT-qPCR analysis, the stabilities of RHOB and RHOC mRNAs by treating cells with a transcription inhibitor, actinomycin D, prior to harvesting cells for mRNA extraction. The results revealed an elevated stability of RHOB transcripts with half-life being increased by approximately 1.5-fold in cells depleted of METTL3 (Figure 4B). The mRNA of RHOC, on the other hand, was stable during the monitored time window with a marginal decrease in parental HEK293T cells (Figure S7). Together, our results demonstrated that the expression of RHOB and RHOC can be directly or indirectly regulated by epitranscriptomic modulators.
Figure 4. Expression and stability of selected small GTPase mRNA.
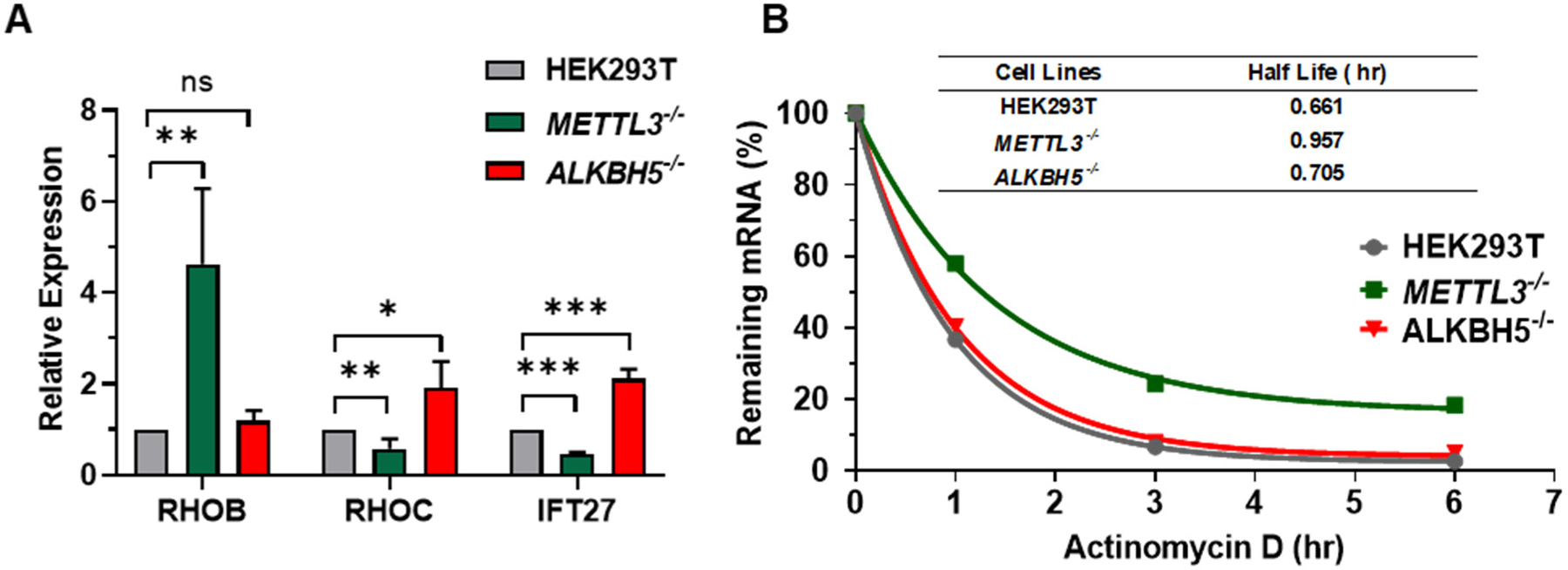
(A) RT-qPCR results for the steady-state mRNA expression levels of RHOB, RHOC, and IFT27 genes, which encode RHOB, RHOC, and IFT27 proteins, respectively, in HEK293T cells and the isogenic METTL3−/− and ALKBH5−/− cells. The levels of transcripts of small GTPase genes were normalized against that of the GAPDH gene. The data represent mean ± SD of results obtained from three biological replicates. The p values were calculated using two-tailed, unpaired t-test (*, 0.01 < p < 0.05; **, 0.001 < p < 0.01; ***, p < 0.001; ns, p > 0.05) (B) RT-qPCR results showing the stability of RHOB mRNA. Cells were treated with actinomycin D to block transcription in a reverse chronological order prior to RNA extraction. The level of RHOB mRNA at each time point was normalized against that of GAPDH and further normalized against that at 0 hr. The percentage of remaining mRNA was fit with single-phase exponential decay kinetics to calculate the half-life.
DISCUSSION
In this study, we employed a facile MRM-based targeted proteomic analysis to study the alterations in small GTPase proteome in cells depleted of m6A epitranscriptomic modulators. With this high-throughput method, we were able to monitor 144 small GTPases, which represent approximately 85% of the entire small GTPase proteome in human cells, and commonly quantify 78 small GTPases among the cells depleted of m6A writer, eraser or reader proteins. Moreover, we validated the MRM data by Western blot analysis (Figure 3C), and the quantification results obtained from the two methods are highly consistent, underscoring the accuracy of the MRM-based target proteomic method in quantifying small GTPase protein expression. To our knowledge, this is the first comprehensive analysis of the entire small GTPase proteome regulated by m6A writer, eraser, and reader proteins. Our results revealed the roles of epitranscriptomic modulators in regulating the expression of small GTPases at the protein level.
The expression of small GTPase genes is tightly regulated, and maintaining their expression levels within a proper range is pivotal for cell fitness.2 Mutations and abnormal expression of small GTPase proteins are among the key drivers for tumor development and progression.5, 40, 46 The RHO small GTPases such as RHOC and RAC2 are overexpressed in cancer cells and are associated with poor patient prognosis. In contrast, RHOB was found to suppress tumorigenesis.4–5 Despite the importance of targeting small GTPases in anti-cancer therapy and decades of intensive research efforts, no effective inhibitors for oncogenic small GTPases have reached the clinic.9
Here, we found that the expression of small GTPase proteins, including several known tumor drivers or suppressors, were significantly altered in the cells depleted of m6A epitranscriptomic modulators, especially the m6A methyltransferase METTL3 and demethylase ALKBH5 (Figure 1C, 2C). In addition, parallel RT-qPCR analyses further revealed that METTL3 and ALKBH5 directly or indirectly regulate the mRNA metabolism of small GTPases and thus modulate their expression at the protein level (Figure 4, Figure S7). These observations resemble recent findings that some epitranscriptomic regulators assume important roles in oncogenesis.47–48 Our findings that the tumor suppressor RHOB and metastatic driver RHOC are regulated by METTL3 and ALKBH5 provide potential targets for therapeutic interventions of human cancer. Along this line, several small molecules were discovered to modulate the activities of m6A methyltransferase (i.e., METTL3) or demethylases (i.e., ALKBH5 and FTO).23, 49–51
Interestingly, we found that those small GTPases regulated by METTL3 and ALKBH5 in HEK293T cells also possess higher discrepancy in the expression between mRNA and protein levels in paired primary/metastatic cancer cells derived from the same patients (Figure S6, Table S3). This finding, despite with limited size of the dataset, suggests that the m6A writer and eraser proteins may be involved in altering the protein expression of small GTPases during cancer progression. Moreover, this observation indicates that the m6A-mediated epitranscriptomic pathway may constitute a very important mechanism contributing to the discrepancy between the transcriptome and proteome. We realized that only a modest number of small GTPases were modulated by METTL3 and ALKBH5; hence, it will be important to examine, in the future, whether the m6A-mediated epitranscriptomic pathway constitutes a major mechanism contributing to the differences between the transcriptome and proteome. This can be achieved by assessing how these m6A writer and eraser proteins affect protein expression at the entire proteome level.
In summary, we demonstrated that depletion of METTL3 and ALKBH5 perturbed the expression of small GTPase proteins in cells. Interestingly, those small GTPases targeted by METTL3 and ALKBH5 also exhibited higher discrepancy between the transcriptome and proteome in paired primary/metastatic melanoma and colorectal cancer cells. To our knowledge, this is the first comprehensive analysis of the involvements of m6A epitranscriptomic modulators in the expression of small GTPases at the protein level, and our findings lay the foundation for alternative therapeutic approach to tackle the abnormal protein expression of the currently “undruggable” small GTPases.
Supplementary Material
ACKNOWLEDGMENT.
This work was supported by the National Institutes of Health (R35 ES031707), and M.H. was supported by an NRSA T32 Institutional Training Grant (T32 ES018827).
Footnotes
Supporting Information Available. Representative LC-MS/MS data, the quantification results are available free of charge via the Internet at http://pubs.acs.org.
Supporting figures (PDF).
Table S1-S3: Small GTPase quantification results (XLSX).
References
- 1.van Dam TJ; Bos JL; Snel B, Evolution of the Ras-like small GTPases and their regulators. Small GTPases 2011, 2, 4–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ridley AJ, Rho GTPase signalling in cell migration. Curr. Opin. Cell Biol 2015, 36, 103–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saxena S; Bucci C; Weis J; Kruttgen A, The small GTPase Rab7 controls the endosomal trafficking and neuritogenic signaling of the nerve growth factor receptor TrkA. J. Neurosci 2005, 25, 10930–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sahai E; Marshall CJ, RHO-GTPases and cancer. Nat. Rev. Cancer 2002, 2, 133–42. [DOI] [PubMed] [Google Scholar]
- 5.Svensmark JH; Brakebusch C, Rho GTPases in cancer: friend or foe? Oncogene 2019, 38, 7447–7456. [DOI] [PubMed] [Google Scholar]
- 6.Fernandez-Medarde A; Santos E, Ras in cancer and developmental diseases. Genes Cancer 2011, 2, 344–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCormick F, KRAS as a Therapeutic Target. Clin. Cancer Res 2015, 21, 1797–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duan X; Zhang Y; Chen KL; Zhang HL; Wu LL; Liu HL; Wang ZB; Sun SC, The small GTPase RhoA regulates the LIMK1/2-cofilin pathway to modulate cytoskeletal dynamics in oocyte meiosis. J. Cell Physiol 2018, 233, 6088–6097. [DOI] [PubMed] [Google Scholar]
- 9.Cox AD; Fesik SW; Kimmelman AC; Luo J; Der CJ, Drugging the undruggable RAS: Mission possible? Nat. Rev. Drug Discov 2014, 13, 828–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kessler D; Gmachl M; Mantoulidis A; Martin LJ; Zoephel A; Mayer M; Gollner A; Covini D; Fischer S; Gerstberger T; Gmaschitz T; Goodwin C; Greb P; Haring D; Hela W; Hoffmann J; Karolyi-Oezguer J; Knesl P; Kornigg S; Koegl M; Kousek R; Lamarre L; Moser F; Munico-Martinez S; Peinsipp C; Phan J; Rinnenthal J; Sai J; Salamon C; Scherbantin Y; Schipany K; Schnitzer R; Schrenk A; Sharps B; Siszler G; Sun Q; Waterson A; Wolkerstorfer B; Zeeb M; Pearson M; Fesik SW; McConnell DB, Drugging an undruggable pocket on KRAS. Proc. Natl. Acad. Sci. U. S. A 2019, 116, 15823–15829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perry RP; Kelley DE, Existence of Methylated Messenger-Rna in Mouse L Cells. Cell 1974, 1, 37–42. [Google Scholar]
- 12.Liu J; Yue Y; Han D; Wang X; Fu Y; Zhang L; Jia G; Yu M; Lu Z; Deng X; Dai Q; Chen W; He C, A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol 2014, 10, 93–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao X; Yang Y; Sun BF; Shi Y; Yang X; Xiao W; Hao YJ; Ping XL; Chen YS; Wang WJ; Jin KX; Wang X; Huang CM; Fu Y; Ge XM; Song SH; Jeong HS; Yanagisawa H; Niu Y; Jia GF; Wu W; Tong WM; Okamoto A; He C; Rendtlew Danielsen JM; Wang XJ; Yang YG, FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014, 24, 1403–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng G; Dahl JA; Niu Y; Fedorcsak P; Huang CM; Li CJ; Vagbo CB; Shi Y; Wang WL; Song SH; Lu Z; Bosmans RP; Dai Q; Hao YJ; Yang X; Zhao WM; Tong WM; Wang XJ; Bogdan F; Furu K; Fu Y; Jia G; Zhao X; Liu J; Krokan HE; Klungland A; Yang YG; He C, ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 2013, 49, 18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Du H; Zhao Y; He J; Zhang Y; Xi H; Liu M; Ma J; Wu L, YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat. Commun 2016, 7, 12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X; Lu Z; Gomez A; Hon GC; Yue Y; Han D; Fu Y; Parisien M; Dai Q; Jia G; Ren B; Pan T; He C, N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014, 505, 117–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi H; Wang X; Lu Z; Zhao BS; Ma H; Hsu PJ; Liu C; He C, YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 2017, 27, 315–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X; Zhao BS; Roundtree IA; Lu Z; Han D; Ma H; Weng X; Chen K; Shi H; He C, N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 2015, 161, 1388–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li A; Chen YS; Ping XL; Yang X; Xiao W; Yang Y; Sun HY; Zhu Q; Baidya P; Wang X; Bhattarai DP; Zhao YL; Sun BF; Yang YG, Cytoplasmic m6A reader YTHDF3 promotes mRNA translation. Cell Res. 2017, 27, 444–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meyer Kate D.; Patil Deepak P.; Zhou J; Zinoviev A; Skabkin Maxim A.; Elemento O; Pestova Tatyana V.; Qian S-B; Jaffrey Samie R., 5′ UTR m6A promotes cap-independent translation. Cell 2015, 163, 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin S; Choe J; Du P; Triboulet R; Gregory RI, The m6A methyltransferase METTL3 promotes translation in human cancer cells. Mol. Cell 2016, 62, 335–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang S; Zhao BS; Zhou A; Lin K; Zheng S; Lu Z; Chen Y; Sulman EP; Xie K; Bogler O; Majumder S; He C; Huang S, m6A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell 2017, 31, 591–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang Y; Su R; Sheng Y; Dong L; Dong Z; Xu H; Ni T; Zhang ZS; Zhang T; Li C; Han L; Zhu Z; Lian F; Wei J; Deng Q; Wang Y; Wunderlich M; Gao Z; Pan G; Zhong D; Zhou H; Zhang N; Gan J; Jiang H; Mulloy JC; Qian Z; Chen J; Yang CG, Small-molecule targeting of oncogenic FTO demethylase in acute myeloid leukemia. Cancer Cell 2019, 35, 677–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miao W; Li L; Zhao Y; Dai X; Chen X; Wang Y, HSP90 inhibitors stimulate DNAJB4 protein expression through a mechanism involving N(6)-methyladenosine. Nat. Commun 2019, 10, 3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang M; Darvas M; Keene CD; Wang Y, Targeted quantitative proteomic approach for high-throughput quantitative profiling of small GTPases in brain tissues of Alzheimer’s disease patients. Anal. Chem 2019, 91, 12307–12314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang M; Qi TF; Li L; Zhang G; Wang Y, A targeted quantitative proteomic approach assesses the reprogramming of small GTPases during melanoma metastasis. Cancer Res. 2018, 78, 5431–5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang YY; Huang M; Wang Y, Targeted Proteomic Analysis of Small GTPases in Murine Adipogenesis. Anal. Chem 2020, 92, 6756–6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ran FA; Hsu PD; Wright J; Agarwala V; Scott DA; Zhang F, Genome engineering using the CRISPR-Cas9 system. Nat. Protoc 2013, 8, 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shevchenko A; Tomas H; Havlis J; Olsen JV; Mann M, In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc 2006, 1, 2856–60. [DOI] [PubMed] [Google Scholar]
- 30.Pino LK; Searle BC; Bollinger JG; Nunn B; MacLean B; MacCoss MJ, The Skyline ecosystem: Informatics for quantitative mass spectrometry proteomics. Mass Spectrom. Rev 2017, 39, 229–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Escher C; Reiter L; MacLean B; Ossola R; Herzog F; Chilton J; MacCoss MJ; Rinner O, Using iRT, a normalized retention time for more targeted measurement of peptides. Proteomics 2012, 12, 1111–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frewen B; MacCoss MJ, Using BiblioSpec for creating and searching tandem MS peptide libraries Curr. Protoc. Bioinformatics 2007, Chapter 13, Unit 13 7. [DOI] [PubMed] [Google Scholar]
- 33.Metsalu T; Vilo J, ClustVis: a web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ries RJ; Zaccara S; Klein P; Olarerin-George A; Namkoong S; Pickering BF; Patil DP; Kwak H; Lee JH; Jaffrey SR, m(6)A enhances the phase separation potential of mRNA. Nature 2019, 571, 424–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bai L; Tang Q; Zou Z; Meng P; Tu B; Xia Y; Cheng S; Zhang L; Yang K; Mu S; Wang X; Qin X; Lv B; Cao X; Qin Q; Jiang X; Chen C, m6A Demethylase FTO Regulates Dopaminergic Neurotransmission Deficits Caused by Arsenite. Toxicol. Sci 2018, 165, 431–446. [DOI] [PubMed] [Google Scholar]
- 36.Engel M; Eggert C; Kaplick PM; Eder M; Roh S; Tietze L; Namendorf C; Arloth J; Weber P; Rex-Haffner M; Geula S; Jakovcevski M; Hanna JH; Leshkowitz D; Uhr M; Wotjak CT; Schmidt MV; Deussing JM; Binder EB; Chen A, The Role of m(6)A/m-RNA Methylation in Stress Response Regulation. Neuron 2018, 99, 389–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wennerberg K; Rossman KL; Der CJ, The Ras superfamily at a glance. J. Cell Sci 2005, 118, 843–6. [DOI] [PubMed] [Google Scholar]
- 38.Zhang G; Wujcik CE, Overcoming ionization effects through chromatography: a case study for the ESI-LC-MS/MS quantitation of a hydrophobic therapeutic agent in human serum using a stable-label internal standard. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci 2009, 877, 2003–10. [DOI] [PubMed] [Google Scholar]
- 39.Zaccara S; Jaffrey SR, A unified model for the function of YTHDF proteins in regulating m6A-modified mRNA. Cell 2020, doi: 10.1016/j.cell.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thomas P; Pranatharthi A; Ross C; Srivastava S, RhoC: a fascinating journey from a cytoskeletal organizer to a Cancer stem cell therapeutic target. J. Exp. Clin. Cancer Res 2019, 38, 328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barretina J; Caponigro G; Stransky N; Venkatesan K; Margolin AA; Kim S; Wilson CJ; Lehar J; Kryukov GV; Sonkin D; Reddy A; Liu M; Murray L; Berger MF; Monahan JE; Morais P; Meltzer J; Korejwa A; Jane-Valbuena J; Mapa FA; Thibault J; Bric-Furlong E; Raman P; Shipway A; Engels IH; Cheng J; Yu GK; Yu J; Aspesi P Jr.; de Silva M; Jagtap K; Jones MD; Wang L; Hatton C; Palescandolo E; Gupta S; Mahan S; Sougnez C; Onofrio RC; Liefeld T; MacConaill L; Winckler W; Reich M; Li N; Mesirov JP; Gabriel SB; Getz G; Ardlie K; Chan V; Myer VE; Weber BL; Porter J; Warmuth M; Finan P; Harris JL; Meyerson M; Golub TR; Morrissey MP; Sellers WR; Schlegel R; Garraway LA, The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012, 483, 603–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang M; Wang Y, Targeted Quantitative Proteomic Approach for Probing Altered Protein Expression of Small GTPases Associated with Colorectal Cancer Metastasis. Anal. Chem 2019, 91, 6233–6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoo JH; Brady SW; Acosta-Alvarez L; Rogers A; Peng J; Sorensen LK; Wolff RK; Mleynek T; Shin D; Rich CP; Kircher DA; Bild A; Odelberg SJ; Li DY; Holmen SL; Grossmann AH, The Small GTPase ARF6 Activates PI3K in Melanoma to Induce a Prometastatic State. Cancer Res. 2019, 79, 2892–2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang K; Sun J; Cheng J; Djeu JY; Wei S; Sebti S, Akt mediates Ras downregulation of RhoB, a suppressor of transformation, invasion, and metastasis. Mol. Cell Biol 2004, 24, 5565–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haak AJ; Appleton KM; Lisabeth EM; Misek SA; Ji Y; Wade SM; Bell JL; Rockwell CE; Airik M; Krook MA; Larsen SD; Verhaegen M; Lawlor ER; Neubig RR, Pharmacological Inhibition of Myocardin-related Transcription Factor Pathway Blocks Lung Metastases of RhoC-Overexpressing Melanoma. Mol. Cancer Ther 2017, 16, 193–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Casalou C; Faustino A; Barral DC, Arf proteins in cancer cell migration. Small GTPases 2016, 7, 270–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Choe J; Lin S; Zhang W; Liu Q; Wang L; Ramirez-Moya J; Du P; Kim W; Tang S; Sliz P; Santisteban P; George RE; Richards WG; Wong KK; Locker N; Slack FJ; Gregory RI, mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis. Nature 2018, 561, 556–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barbieri I; Tzelepis K; Pandolfini L; Shi J; Millan-Zambrano G; Robson SC; Aspris D; Migliori V; Bannister AJ; Han N; De Braekeleer E; Ponstingl H; Hendrick A; Vakoc CR; Vassiliou GS; Kouzarides T, Promoter-bound METTL3 maintains myeloid leukaemia by m6A-dependent translation control. Nature 2017, 552, 126–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Selberg S; Blokhina D; Aatonen M; Koivisto P; Siltanen A; Mervaala E; Kankuri E; Karelson M, Discovery of Small Molecules that Activate RNA Methylation through Cooperative Binding to the METTL3–14-WTAP Complex Active Site. Cell Rep. 2019, 26, 3762–3771. [DOI] [PubMed] [Google Scholar]
- 50.Tzelepis K; De Braekeleer E; Yankova E; Rak J; Aspris D; Domingues AF; Fosbeary R; Hendrick A; Leggate D; Ofir-Rosenfeld Y; Sapetschnig A; Pina C; Albertella M; Blackaby W; Rausch O; Vassiliou GS; Kouzarides T, Pharmacological Inhibition of the RNA m6a Writer METTL3 As a Novel Therapeutic Strategy for Acute Myeloid Leukemia. Blood 2019, 134, 403–403. [Google Scholar]
- 51.Malacrida A; Rivara M; Di Domizio A; Cislaghi G; Miloso M; Zuliani V; Nicolini G, 3D proteome-wide scale screening and activity evaluation of a new ALKBH5 inhibitor in U87 glioblastoma cell line. Bioorg. Med. Chem 2020, 28, 115300. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


