Abstract
The microbiome represents a vast resource for drug discovery as its members engage in constant conflict to outcompete one another by deploying diverse strategies for survival. Cutibacterium acnes (C. acnes) is one of the most common bacterial species on human skin and can promote the common disease acne vulgaris. By employing a combined strategy of functional screening, genetics and proteomics we discovered a strain of Staphylococcus capitis (S. capitis E12) that selectively inhibited growth of C. acnes with potency greater than antibiotics commonly used in the treatment of acne. Antimicrobial peptides secreted from S. capitis E12 were identified as four distinct phenol soluble modulins acting synergistically. These peptides were not toxic to human keratinocytes and the S. capitis extract did not kill other commensal skin bacteria but was effective against C. acnes on pig skin and on mice. Overall, these data show how a member of the human skin microbiome can be useful as a biotherapy for acne vulgaris.
INTRODUCTION
Human skin is colonized by hundreds of diverse bacterial species. The complex and dynamic chemical interactions between bacteria are important for host health and protection from pathogen colonization and infection (Grice and Segre 2011). The skin microbial community is regulated in part through production of antimicrobials. In particular, the discovery of peptides from the human microbiome that selectively kill Staphylococcus aureus, including methicillin resistant strains (MRSA), represents a potential vanguard of new antimicrobials (Chu et al. 2016; Cogen et al. 2010; Nakatsuji et al. 2017; Saising et al. 2012; Zipperer et al. 2016). The major source for some of these new antimicrobials originate from the coagulase-negative staphylococci (CoNS), such as Staphylococcus epidermidis and Staphylococcus hominus. CoNS are a diverse bacterial species that frequently colonize human skin in great abundance and therefore have evolved the most effective strategies for survival in this environment. One mechanism responsible for their competitive success is that specific strains of CoNS harbor biosynthetic gene clusters that produce lantibiotics (Donia et al. 2014). Our group recently identified several undiscovered lantibiotics from a human skin S. hominus strain that targeted S. aureus (Nakatsuji et al. 2017). Another interesting recent discovery was a novel anti-S. aureus cyclic peptide produced by S. lugdunensis (Zipperer et al. 2016). Other examples include the production of phenol-soluble modulins gamma (PSMγ, also known as delta toxin) by S. epidermidis which inhibits growth of S. aureus and group A Streptococcus (GAS) (Cogen et al. 2010).
Human diseases are often associated with an imbalance in the microbial species that colonize the surface, a state known as dysbiosis. In the case of atopic dermatitis, patients are often colonized by S. aureus which then further drives inflammation and disease (Williams and Gallo 2015). Likewise, the common skin disease acne vulgaris is an inflammatory disorder of the pilosebaceous unit that is linked to colonization by Cutibacterium acnes (C. acnes). C. acnes is a lipophilic species which dominates sebaceous skin sites and produces a multitude of toxins, adhesions and immunomodulatory metabolites that activate innate immune responses (O’Neill and Gallo 2018). Like many other bacterial species in the skin microbiome, C. acnes has several defense mechanisms that help it to establish a dominant niche in the hair follicle. For example, the action of secreted C. acnes lipases on sebum can result in generation of free fatty acids and short chain fatty acids (SCFA) that can suppress the growth MRSA and GAS (Shu et al. 2013). In the disease acne, distinct strains of C. acnes are significantly enriched compared to healthy individuals and these organisms appear to exacerbate disease since both systemic and topical antibiotic therapy that inhibits C. acnes is therapeutically effective (Fitz-Gibbon et al. 2013). However, long-term use of pharmacologic antibiotics is undesirable due to the capacity to promote antibiotic resistance and their indiscriminate killing of beneficial members of the healthy microbiome (Chien et al. 2019; Ng et al. 2013; Shaw et al. 2019). It is therefore logical to explore use of the human skin microbiome as a source of antagonistic strains and/or undiscovered antimicrobials that could selectively target C. acnes. The selection of microbes that have evolved to adapt and be tolerated by humans optimizes the probability that they will promote establishment of normal homeostasis.
Here, we report discovery of a human skin commensal bacterial strain that produces antimicrobial molecules targeted against C. acnes. This strain of S. capitis exhibited potent activity against multiple isolates of C. acnes including acne-associated strains but did not kill other commensal skin bacteria. Identification of this unique member of the human skin microbial community represents an attractive alternative therapeutic approach to acne vulgaris.
RESULTS
Screening for antimicrobial activity from human skin bacteria identifies a S. capitis strain with potent and selective activity against C. acnes.
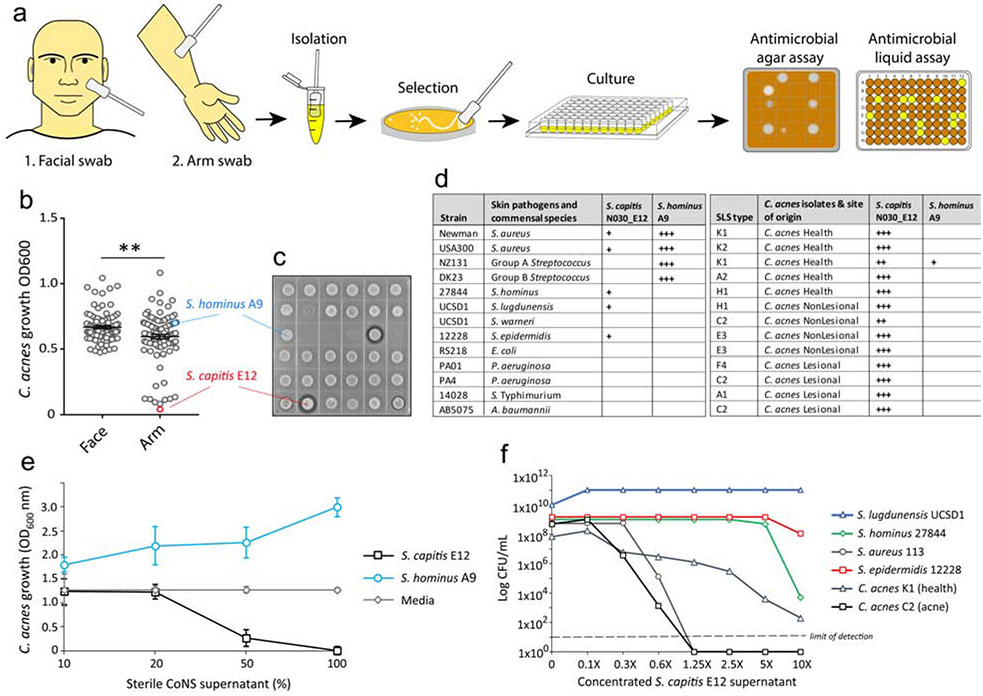
To discover antimicrobials from commensal bacteria on human skin that inhibit the growth of C. acnes a library of 288 CoNS isolates were collected from skin swabs of the forearm and face of healthy volunteers. An unbiased analysis of anti-C. acnes activity was performed by live coculture with the CoNS isolate growing on agar with C. acnes C2 and measuring the growth of C. acnes following the addition of 50% v/v conditioned CoNS supernatant that was sterile filtered (Fig. 1A). Results of the functional screen revealed a total of 13 isolates that were active against the growth of C. acnes C2 in both the liquid assays and agar coculture (Fig.1B,C). DNA extracted from the CoNS isolates with antimicrobial activity was subjected to 16S sequencing and several different species were identified, including S. epidermidis and S. hominus which are considered frequent colonizers of human skin. The most potent isolate was identified as Staphylococcus capitis (S. capitis strain E12) and was used for further characterization. Of the 13 antimicrobial strains identified, all were isolated from the forearm. None of the facial isolates were found to inhibit the growth of C. acnes C2. Interestingly, the lantibiotic-producing S. hominus A9 strain that is active against S. aureus (Nakatsuji et al. 2017) was ineffective against C. acnes C2, illustrating the selective nature of antimicrobials derived from the mixed bacterial community of the skin.
Figure.1.
Functional screen to identify antimicrobial CoNS species that target C. acnes. (A) Schematic for the high-throughput antimicrobial functional screen, outlining the collection and selection of CoNS strains from two distinct healthy skin sites and assays to detect antimicrobial activity against C. acnes via coculture on agar or growth in sterile conditioned supernatant of CoNS. (B,C) Growth of C. acnes C2 after 24 h incubation in 50% sterile filtered supernatant of the CoNS strain library (B) or growth of C. acnes C2 with CoNS coculture in agar (C). Each dot represent data from individual CoNS clone. The most potent antimicrobial CoNS isolate was selected and identified as S. capitis E12 (red), whilst S. hominus A9 (blue) did not exhibit activity against C. acnes C2. (D) Table showing the inhibitory activity of S. capitis E12 and S. hominus A9, against several skin commensal and pathogen strains, including several strains of C. acnes isolated from healthy and acne skin, as measured by size of zone inhibition by antimicrobial agar assay (+ small, ++ medium, +++ large). (E) Growth of C. acnes C2 after 24 h incubation in media alone or increasing concentrations of sterile supernatant of S. capitis E12 or S. hominus A9. (F) Survival of several species of staphylococci and C. acnes after 24 h post-treatment with increasing concentrations of sterile S. capitis E12 supernatant, as measured by the number of surviving CFU plated onto the appropriate selective agar.
Next, the selectivity of S. capitis E12 to inhibit the growth of several different CoNS commensal species and common skin pathogens was tested by agar assay. Whilst the S. hominus A9 potently inhibited the growth of S. aureus and GAS, S. capitis E12 exhibited only weak activity against CoNS and was ineffective against other skin pathogens (Fig.1D). Strikingly, S. capitis E12 exhibited potent activity against a wide range of C. acnes strains, including strains that were isolated from either lesional or non-lesional skin of acne patients. 50% dilution of unconcentrated media sterile supernatant from S. capitis E12 was sufficient to inhibit C. acnes growth, thus validating the findings from the functional screen and demonstrating that under normal growth conditions this strain of S. capitis can produce sufficient antimicrobial activity to outcompete C. acnes C2 (Fig.1E). To investigate if the antimicrobial supernatant is bactericidal, the number of surviving C. acnes colonies was enumerated after 24 h treatment with increasing concentrations of S. capitis E12. Consistent with results from the functional screen, S. capitis supernatant was generally not bactericidal against other CoNS species, with exception to the S. hominus strain 27844 only at the highest 10X concentration. However, the S. capitis E12 supernatant was bactericidal against C. acnes, showing a 6-log decrease of C. acnes C2 during exposure to 0.6X (60%) supernatant and complete sterilization of the culture at highest concentrations (Fig. 1F).
Purification and identification of PSMβ antimicrobial peptides.
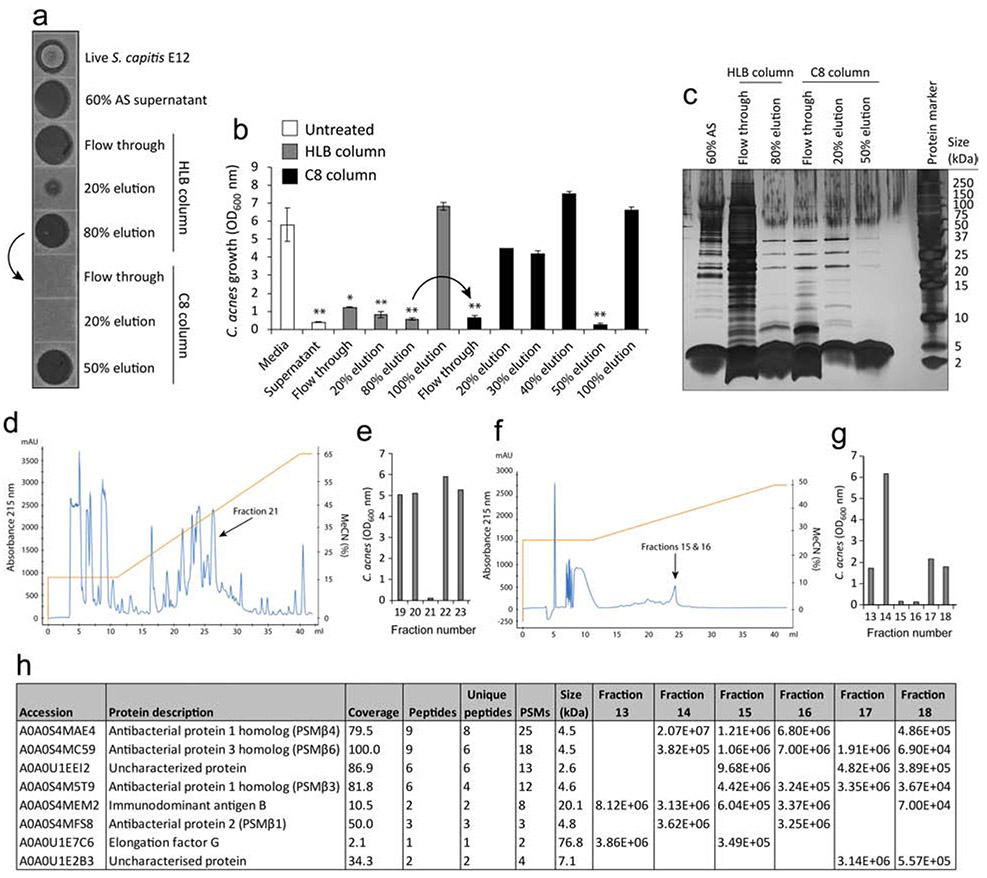
Initial examination into the nature of the antimicrobial factor revealed it to be resistant to heat treatment, but sensitive to proteolysis, suggestive of a proteinaceous molecule (Fig.S1A,B). Furthermore, the antimicrobial factor was precipitated from supernatant during incubation with 60-80% ammonium sulphate (AS) and concentrated by centrifugation (Fig.S1C). Next, sample preparation was carried out on the concentrated AS precipitate using solid phase extraction. Samples were first loaded onto a hydrophilic-lipophilic balanced (HLB) column and the anti-C. acnes molecule was found to be eluted at 80% acetonitrile, as determined 72 h after inoculation onto C. acnes C2 agar (Fig.2A) or with C. acnes C2 in liquid culture (Fig.2B). Next, the 80% active elution was subsequently loaded onto a hydrophobic C8 cartridge (indicated by arrow) and the anti-C. acnes molecule was eluted at 50% acetonitrile (Fig.2A,B). Total protein staining of the fractions revealed a highly enriched band(s) of roughly 4-5 kDa in size, with greater purity visible during subsequent elution steps (Fig.1C). Reverse phase high performance liquid chromatography (HPLC) revealed several peaks, of which a single fraction (fraction 21) was found to suppress the growth of C. acnes C2 in liquid culture (Fig.1D). To ensure the highest purity, the active fractions were then pooled together from four separate HPLC runs and a second step HPLC purification was performed. This time, several small peaks were visualized (Fig.2F), and two fractions (fractions 15 and 16) were found to suppress the growth of C. acnes C2 in liquid culture (Fig.2G). Mass spectrometry (MS) analysis of active fractions (15,16) and control non-active fractions (13,14,17,18) revealed four candidate peptides referred to as “antibacterial protein” each 5 kDa in size (Fig.2H). BLAST search of the peptide sequences identified these as belonging to the beta class family of phenol soluble modulins (PSMβ) present within multiple Staphylococcal species.
Figure 2.
HPLC purification and identification of S. capitis E12 antimicrobial peptides. (A,B) 60% ammonium sulphate-precipitated supernatant of S. capitis E12 was loaded onto a HLB column and the antimicrobial factor eluted by 80% acetonitrile. The HLB 80% eluant was loaded onto a C8 cartridge (indicated by arrow) and the antimicrobial factor was eluted by 50% acetonitrile. Fractions were measured for activity against C. acnes C2 by agar assay (A) and liquid culture (B). Error bars depict mean ± SEM. *P < 0.05; **P < 0.01 t tests comparing values against media control. (C) Silver stain of the total protein content of S. capitis E12 treated or untreated supernatant and the SPE flow-through and eluted fractions. (D) HPLC purification of S. capitis supernatant identified several peptide peaks, of which a single fraction (21) was identified as having anti-C. acnes C2 activity (E) by liquid culture assay. (F) Fraction 21 was pooled from five separate HPLC runs and purified by second step HPLC resulting in two fractions (15 and 16) with anti-C. acnes C2 activity (G) by liquid culture assay. (H) Results of the top 8 peptide hits from MS detection of HPLC purified active fractions (15 and 16) and control non-active fractions (13,14, 17 and 18) revealing peptides corresponding to ‘antibacterial protein’ and later identified as PSMβ peptides by BLAST analyses.
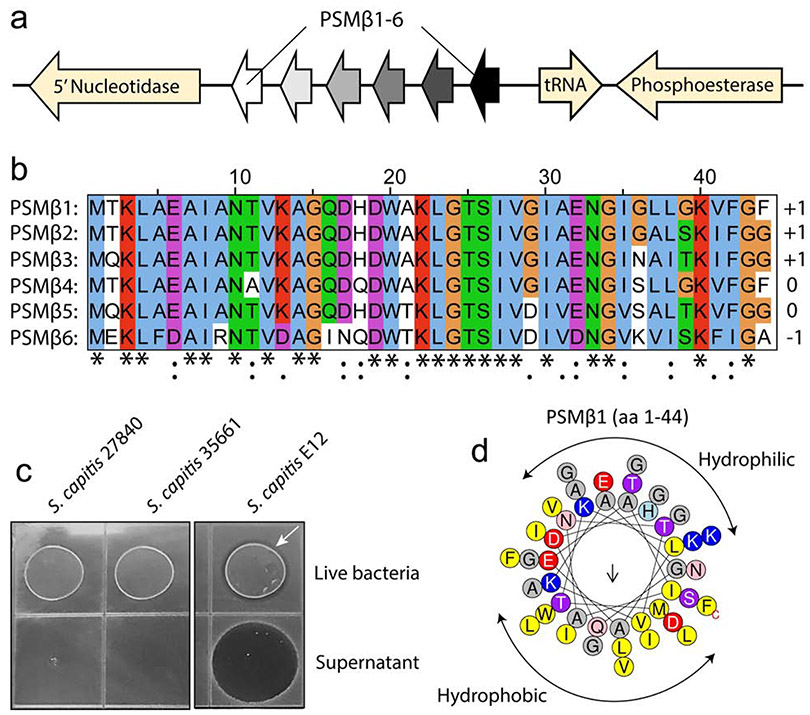
Whilst 4 distinct PSMβ peptides were detected by MS, analysis of the S. capitis E12 genome revealed up to 6 PSMβ-encoding genes, contained within an operon-like structure (Fig.3A). All six PSMβ-encoding genes are closely related, with amino acid sequence identity ranging from 56% to 91%, and predicted charge ranging from −1 to +1 (Fig.3B). Further analyses found these genes to be absent from genomes of ATCC strains of S. capitis 35661 and 27844 (data not shown) and indeed no antimicrobial activity was detected against C. acnes from these laboratory strains (Fig.3C). Alpha helical wheel plots (Heliquest) also predicted these peptides to form an α-helix, amphipathic-like structure for each PSMβ, with separate hydrophobic and hydrophilic faces on the opposite sides of the long axis of the peptide. This amphipathic structure is common amongst many well characterized antimicrobial peptides (Tossi et al. 2000) (Fig.3D).
Figure 3.
Whole genome sequencing of S. capitis E12 reveals sequence and predicted properties of the PSMβ peptides.
(A) Schematic highlighting the S. capitis E12 genetic cluster containing six gene-encoding PSMβ peptides (PSMβ1-6). (B) Multiple sequence alignment (ClustalW) of all six PSMβ peptides, including the predicted charge for each peptide. (C) Absence of antimicrobial activity against C. acnes C2 during live coculture or sterile supernatant exposure with ATCC S. capitis strains 35661 and 27840 that lack PSMβ genes. White arrow indicates zone of inhibition produced by live S. capitis E12 (D) Representative helical wheel plot (Heliquest) of PSMβ1 indicating a predicted amphipathic structure.
PSMβ inhibits skin colonization by C. acnes
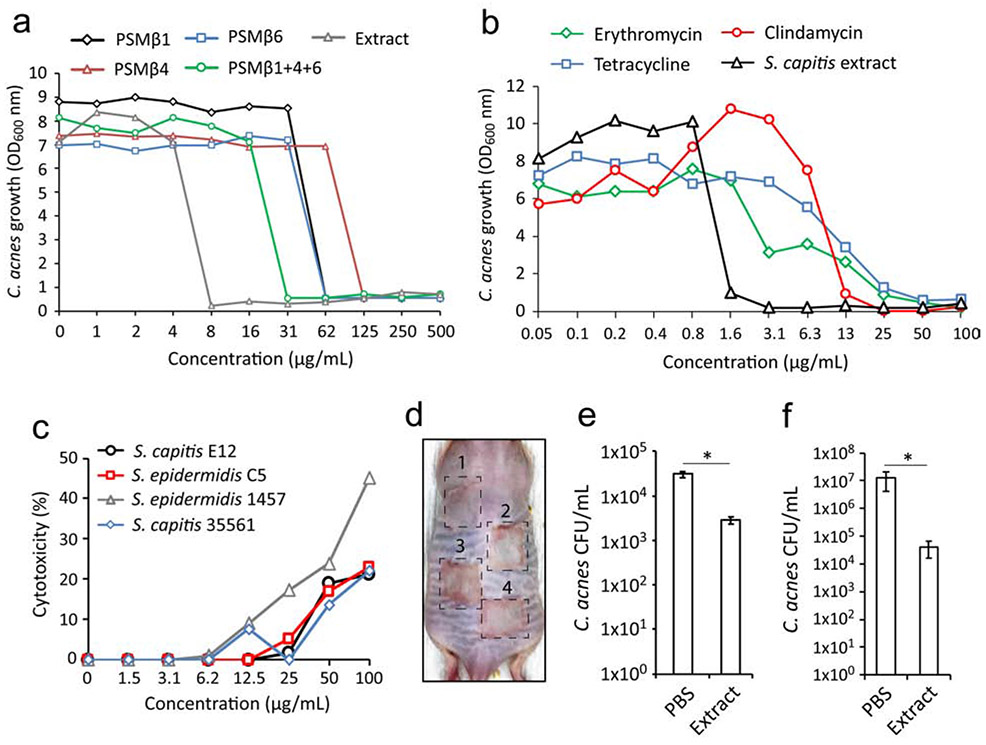
S. capitis E12 conditioned supernatant was extracted with n-Butanol, a procedure that enriches for PSM peptides (Joo et al. 2011). This resulted in a relatively pure extract enriched for all PSMβ peptides and retained activity against C. acnes C2 (Fig.S2). To confirm that PSMβ peptides were active as the anti-C. acnes molecule, and identify which peptide had optimal activity, we individually synthesized PSMβ1 and 6, the most abundant molecules from the purified active fractions (15 and 16, Fig2.H), as well as PSMβ4, which was less abundant. All three PSMs were shown to possess antimicrobial activity in vitro (Fig. 4A). PSMβ1 and 6 both effectively inhibited growth of C. acnesC2 at 62 μg/ml, whilst PSMβ4 was less potent, inhibiting at 125 μg/ml. Interestingly, the combination of all three peptides exhibited optimal activity against C. acnes, inhibiting growth in 31 μg/ml. However, this was not as inhibitory compared to the same concentration of the butanol extract, which inhibited at 8 μg/ml. This discrepancy could be due to the presence of the additional fourth PSMβ peptide (PSMβ3) found within the extract. Several bacterial antimicrobials have been shown to synergize with human antimicrobials such as defensins and LL37 (Nakatsuji et al. 2017). However, exposure of C. acnes C2 to S. capitis extract with or without LL-37 peptide did not result in enhanced killing activity (Fig.S3). Given the greater potency and relative purity of the PSMβ-containing extract compared to the synthetically produced PSMβ peptides, the extract was used in subsequent experiments to determine the efficacy and potency of S. capitis E12 in vitro and in vivo experiments.
Figure 4.
Efficacy of PSMβ peptides and extract as an alternative therapeutic for acne vulgaris. (A) Inhibition of C. acnes C2 after 72 h culture in the presence of S. capitis E12 extract or with synthetic peptides PSMβ1, PSMβ4, PSMβ6 individually or in combination of all three (B) Cytotoxicity of NHEKs treated with S. capitis E12, S. epidermidis C5, S. epidermidis 1457 and S. capitis 35561 supernatant for 24 h, presented as the percentage of maximal LDH release. (C) Visual representation of erythema on the back skin of SKH1 mouse after inoculation with 1x107 CFU of S. capitis E12 (1), S. epidermidis C5 (2), S. epidermidis 1457 (3) and S. capitis 35661 (4) for 24 h. (D) Total number of surviving CFU of C. acnes C2 on ex vivo pig skin explants 24 h post-treatment with S. capitis E12 extract (10 mg/mL) or PBS control. (E) Total number of surviving CFU of C. acnes C2 on SKH1 mouse back skin 24 h post-treatment with S. capitis E12 extract (10 mg/mL) or PBS control. Data represents ± SEM from five pigskin sheets or five independent mice. *P < 0.05 by two tailed independent t test.
S. capitis E12 extract was next compared to several antibiotics commonly used for acne treatment. Amongst four distinct acne-associated strains tested, sensitivity to each antibiotic varied extensively, with three out of four strains insensitive to erythromycin at the maximum tested concentration of 100 μg/ml (Fig.S4). In contrast, all four strains were found to be equally sensitive to the extract (3-6 μg/ml). In three out of four C. acnes strains tested, the extract exhibited greater potency than erythromycin, tetracycline and clindamycin (Fig.S4A,B,C), with one strain (C. acnes K2) found have greater sensitivity to antibiotics than the extract (Fig.S4D). Moreover, no increase in the MIC above 16 μg/mL was recorded for C. acnes during treatment with the extract for up to 20 generations (data not shown), indicating relative inability to promote resistance. Furthermore, exposure of primary human keratinocytes (NHEKs) to extracts did not produce a significant cytotoxic response from these epidermal cells as measured by lactate dehydrogenase (LDH) release (Fig. 4B). Likewise, mouse back skin exposed to different strains of CoNS for 24 h showed that S. capitis E12 did not induce observable erythema or injury (Fig. 4C). Finally, having confirmed the expected result that this commensal skin bacterium was well tolerated by the epidermis, C. acnes was epicutaneously applied to colonize pig skin ex vivo or back skin of live SKH1 mouse, and then extract from S. capitis E12 was applied. A single treatment resulted in a significant reduction in C. acnes CFU in both models (Fig. 4D,E). Overall, these results demonstrate PSMβ from S. capitis E12 has capacity to kill C. acnes and illustrates the potential of exploiting naturally occurring microbial events in the human skin for development of innovative therapeutics.
DISCUSSION
In acne vulgaris, C. acnes is the dominant bacteria that plays an important role in disease (Barnard et al. 2016; Hall et al. 2018). Treatment approaches for moderate to severe acne typically consist of an extended regiment of topical and/or oral application of antibiotics (Zaenglein et al. 2016). However, such approaches can be ineffective due to several reasons, including poor activity and penetration on skin. Furthermore, there are negative consequences of long term antibiotic treatment such as the ability to negatively disrupt the microbiome thereby eliminating commensal bacteria important for skin health, and the increased risk for development of antibiotic resistance (Chien et al. 2019; Kelhälä et al. 2018). Here we report the discovery of several bacterial species from healthy human skin that exhibit potent and selective antimicrobial activity against C. acnes. These results demonstrate an alternative approach for discovery of effective antimicrobials that avoid the negative consequences of current non-specific antimicrobial therapies.
There have been several recent discoveries of novel antimicrobials from skin bacteria, one example is Lugdunin, a cyclic peptide antibiotic produced by S. lugdunensis that is active against S. aureus (Zipperer et al. 2016). Interestingly, it was reported that lugdunin was active only during agar coculture and not in liquid assay. Therefore, our functional screen and several antimicrobial assays were conducted using both independent approaches to ensure validity and reproducibility. Using such approaches, our group have also identified several unique antimicrobial peptides from CoNS including Sh-lantibiotics α/β from S. hominus (Nakatsuji et al. 2017). This strain was found to kill S. aureus in vitro and in vivo. The selectivity of bacteriocins made by the skin microbiome was evident with our observations that these strains of CoNS with anti-S. aureus activity were not inhibitory against C. acnes. Discovery of therapeutically useful organisms must be designed with the specific target in mind.
To discover potentially undiscovered antimicrobials effective against C. acnes, we conducted an expanded screen of CoNS from different healthy skin sites. Swabs were collected from the face and forearm; the objective to capture not only a diverse collection of CoNS species but to also sample an area of skin that C. acnes is less dominant, potentially showing the success of CoNS against C. acnes. Interestingly, results from our screen showed that a significantly greater number of anti-C. acnes strains were identified from the forearm compared to the face. One explanation for this could be a greater diversity CoNS species on the forearm. Furthermore, C. acnes can produce antimicrobial molecules, such as short chain fatty acids, from the breakdown of sebum, which have been shown to inhibit staphylococci (Shu et al. 2013). Such that, the increase in CoNS on the forearm may be aided by the lesser abundance of C. acnes. Together, these observations suggest that the regional differences described in microbiomes at different body sites (Bouslimani et al. 2015; Oh et al. 2014) are not only due to the host moisture or lipid levels but also reflect the outcome of microbial strategies to outcompete their community members.
Interestingly, in several acne microbiome studies S. epidermidis is reported to be most frequent CoNS species in health and disease (Barnard et al. 2016; Fitz-Gibbon et al. 2013), but only S. epidermidis strains located from forearm were antimicrobial in our study. Nevertheless, an important finding originating from the acne microbiome studies showed that distinct strain-types of C. acnes are associated with disease. A valid hypothesis is that these strains are more pathogenic and may drive inflammation and disease. Crucially, we’ve shown that S. capitis E12 was capable of inhibiting all C. acnes strains tested, including lesional strains isolated from acne patients, but not other commensal bacteria. This selective antimicrobial activity would be an attractive therapeutic approach for acne, targeting disease-causing bacteria whilst leaving other essential commensal bacteria unperturbed.
S. capitis E12 constitutively produces antimicrobial peptides in sufficient quantities to kill C. acnes. Strikingly, incubation of a 0.6X concentration of S. capitis E12 supernatant with a strain of C. acnes that is associated with acne resulted in greater than 4 log reduction in survival of the disease-associated bacterium, and complete sterilization of the culture after 24 h. Moreover, for many acne-associated C. acnes strains, the S. capitis E12 extract showed greater potency compared to other antibiotics commonly prescribed for acne. This could be particularly important for patients who are colonized with strains that are insensitive and/or have developed genetic resistance. Another desirable feature of the extract was that C. acnes C2 did not show acquisition of resistance up to 20 generations. The potency and poor capacity to induce resistance from the PSMs produced by S. capitis E12 is consistent with the mode of action of the α-helix, amphipathic structure of these peptides. Such a structure is common amongst many antimicrobial peptides, and enables insertion into and destabilization of bacterial membranes (Tossi et al. 2000). Such a mechanism of direct membrane disruption is ideal for an antimicrobial that works on an epithelial surface.
Whilst S. capitis genome contains a cluster of up to six PSM-encoding genes, only PSMβ1, 3, 4 and 6 were detected by MS analysis of the purified fractions. Antimicrobial assays with synthetic peptides PSMβ1, 4 and 6 revealed greater activity against C. acnes when the bacteria were treated with a combination of all three. Likewise, the expected overall charge of each PSMβ is subtly different, thus likely impacting binding capacity to a hydrophobic column as well as interaction with the bacterial membrane. Individually, the synthetic PSM peptides were far less potent than the extract. It remains to be determined if PSMβ2 and PSMβ5 were also secreted by S. capitis E12 but eluted in different fractions that were not biologically active by themselves but would increase the activity of PSMβ1, 3, 4 and 6. The β class PSMs have been found in other staphylococci and typically number between two and four (Wang et al. 2011). However, their exact role has remained elusive and unlike the smaller PSMα, are not thought to be involved in virulence (Cheung et al. 2014). S. aureus contains two βPSMs (PSMβ1/2), but neither are reported to be antimicrobial. Other studies have shown other synthetic PSMβ peptides to be inhibitory against S. aureus in vitro but not known to be active against C. acnes (Kumar et al. 2017). Therefore, adopting the present screening approach identified activity that would not have been detectable by using a genetic screening approach alone. The optimum killing efficacy and specificity of the combination, rather than individual peptides, suggests that a therapeutic approach combining multiple PSMs or the live organism itself may be most effective. In conclusion, we’ve shown that the skin microbiome is a valuable resource for discovering antimicrobials that can be remarkably selective against specific pathogens and valuable in combating skin disease such as acne vulgaris.
MATERIALS AND METHODS
Human subjects and sample swab collection
All experiments involving human subjects were carried out according to protocols approved by the University of California, San Diego (UCSD) IRB (Project#140144). Written informed consent was obtained from all subjects. Swab sample collection was performed as previously described (Nakatsuji et al. 2017) on 12 healthy adults with swabs taken from the upper arm and the face. Healthy adults included both males and females aged 22-34 years with normal healthy skin and were instructed to avoid washing and cosmetic application to the skin sites 24 hours prior to sampling. Swabs were incubated in 3% Tryptic Soy Broth (TSB), vortexed briefly and plated onto mannitol salt agar containing egg yolk (MAY) plate for selective growth of staphylococcal species (Parisi and Hamory 1986). S. aureus were distinguished from CoNS according to mannitol metabolism and the egg yolk reaction (Carter 1960). C. acnes clinical isolates were obtained by swabbing lesional and non-lesional facial skin sites of 5 acne patients as well as facial skin from 4 healthy volunteers. Subjects included both males and females, aged 18-24 years with acne patients diagnosed with mild-moderate facial acne. Acne patients at the time of sample collection were not using any known topical/systemic antibiotic or retinoid and were instructed to avoid any washing and cosmetic application to the skin sites in the preceding 24 hours. For C. acnes strain collection, facial swabs were incubated in reinforced clostridial media (RCM), vortexed and plated onto brucella blood agar plates supplemented with Vitamin K, hemin and 5% sheep’s blood and incubated for 5 days at 37°C in an anaeroPack (ThermoFisher).
Screening for antimicrobial activity from skin-derived CoNS
24 individual isolated colonies of CoNS, that were selected from each skin site, were randomly picked from a MAY plate and transferred to TSB (1 mL) in a deep 96 well plate. Each plate contained a previously characterized CoNS strain, which included S. epidermidis ATCC1457 that failed to demonstrate antimicrobial activity against other staphylococci (negative control) as well as S. hominus A9 which was previously demonstrated to produce lantibiotics that kill S. aureus (positive control) (Nakatsuji et al. 2017). The CoNS plate was sealed with sterile Aeraseal film (sigma) and cultured at 37°C overnight, shaking at 250 rpm. Bacterial growth was evaluated by measuring OD600 and only CoNS grown to a high density (OD600>6.0) were used for subsequent analysis. To measure antimicrobial activity in the secreted supernatant, the CoNS supernatant from overnight culture was harvested and sterile filtered. To measure antimicrobial activity from live growing CoNS, bacteria from overnight culture were pelleted by centrifugation then the supernatant was discarded, and the bacteria were resuspended in fresh TSB.
in vitro antimicrobial assays
For the initial CoNS antimicrobial screen, the radial diffusion agar assays were performed using S. aureus 113 or either C. acnes HL110PA3 (health-associated, SLST K1) or C. acnes HL096PA1 (acne-associated, SLST C2). For the radial diffusion agar assays, melted TSB or RCM agar (12 mL) was mixed with S. aureus or C. acnes (1x106 CFU) and poured into a 10 cm square petri square. When the agar was solidified, a 10 μl aliquot of bacteria was inoculated onto a single grid. The plates were incubated at 37°C overnight shaking (for S. aureus) or incubated at 37°C for 3 days in an anaeroPack (for C. acnes) to allow visible growth of bacteria. Antibacterial activity was indicated by a clear zone of inhibition within the agar that surrounds the colony. The size of the inhibitory zone was recorded as a measure of antimicrobial activity (+ slight, ++ moderate, or +++ potent). For the liquid culture assay, conditioned supernatant from overnight cultures of CoNS were harvested and sterile filtered (0.22 μm). 50% of conditioned supernatant was mixed with 50% fresh TSB or RCM containing 1X105 CFU/mL of S. aureus or C. acnes, in a 96 well round bottom plate. S. aureus plates were incubated at 37°C shaking overnight and C. acnes plates incubated for 48 h standing at 37°C in an anaeroPack. Bacterial growth was measured by OD600 and positive antimicrobial strains were identified as those that suppressed bacterial growth to less than 50% (I50) of growth measured for negative control strains. Bacterial survival was measured by counting the number of CFU on TSB plates (S. aureus) or Brucella blood agar plates (C. acnes). At selected times post-treatment with bacterial supernatant or extracts, the number of CFU was determined by serial dilution in phosphate-buffered saline (PBS) and plating onto appropriate agar media. Bacterial survival was measured as the total number of CFU per milliliter.
Bacterial classification
Genomic DNA was isolated from CoNS and C. acnes isolates by MasterPure kit (Epicenter) according to manufacturer’s instructions. For species identification for CoNS, full-length 16S rRNA genes were amplified from representative isolates with universal 16S primers, 27-F and 1525-R. Amplicons were sequenced from both ends by Sanger method. Obtained 16S rRNA gene sequence was searched against Ribosomal Database Project (http://rdp.cme.msu.edu/) (Cole et al. 2014). For C. acnes strain typing, single locus sequence typing (SLST) was used with primers SLSTFor and SLSTRev (Scholz et al. 2014). Amplicons were sequenced from both ends by Sanger method. C. acnes strain types were assigned to amplicons using the SLST database (http://medbac.dk/slst/pacnes).
Solid Phase Extraction (SPE) and HPLC purification of supernatant
S. capitis E12 sterile supernatant that had been retained on a 10 kDa MWCO column and precipitated in 60% AS was applied on an Oasis HLB cartridge (Waters). The cartridge was washed in 20% acetonitrile in H2O and eluted in 80% acetonitrile in H2O. The eluted fractions were lyophilized and reconstituted in H2O. The 80% fraction was then loaded onto a C8 Sep-Pak cartridge (Waters), washed in 20% acetonitrile and eluted in 50% acetonitrile. The fractions were lyophilized, reconstituted in H2O and then assessed for antimicrobial activity and visualized by silver stain (Pierce) according to manufacturer’s instructions. First step HPLC purification was carried out with 1 mg of S. capitis E12 butanol extract loaded into a CapCel Pak C8 (5 μm, 300 Å, 4.6 × 250mm) (Shiseido Co.) with a linear gradient of acetonitrile from 10% to 60% in 0·1% (v/v) TFA at 0.8 mL/min. Fractions were lyophilized then resuspended in H2O and antimicrobial activity assessed by liquid culture assay. Up to five sequential purifications were carried out with each single antimicrobial fraction pooled together for second step HPLC. For the second purification, a linear gradient of acetonitrile from 25% to 50% was adopted.
Purification of antimicrobials produced by CoNS strains
Conditioned media from overnight cultures of the selected antibacterial S. capitis E12 strain, including positive control antimicrobial S. hominus A9 strain, were first sterilized by filtration through a 0,22um Millipore filter. Initial characterization involved treating the sterile bacterial supernatant to boiling at 100°C for 15 mins or incubation with proteolytic enzymes papain and proteinase K, at 200 μg/mL at 37°C for 60 mins. Antimicrobial activity was measured by radial diffusion assay. For size exclusion of the antimicrobial molecule, 20 mL of conditioned media was loaded onto a 3, 10 and 30 kDa molecular weight cutoff (MWCO) columns (Pierce) and centrifuged at 4000 x g for 15 mins. Whilst the flow-through fraction was set aside, and the retained fraction was washed 2X in PBS and resuspended with 20 mL TSB. Antimicrobial activity was assessed for both fractions. To precipitate the active molecule, ammonium sulphate was added to the sterile supernatant (30-80% saturation) for 1 hour under constant rotation at room temperature, followed by centrifugation at 4000 x g for 45 mins. The resulting pellet was washed 3X with H2O and reconstituted in H2O. Antimicrobial activity was determined by S. aureus and C. acnes growth in agar and liquid culture.
Mass spectrometry
A portion of the fractions of interest (< 1 μg) were dried under vacuum and resuspended in 5 μL of 5% acetonitrile with 5% formic acid. Next, individual LC-MS (Elias and Gygi 2007) experiments were conducted on 1/5 of each sample through 85 minutes of data acquisition on an Orbitrap Fusion (Thermo Fisher Scientific) mass spectrometer with an in-line Easy-nLC 1000 (Thermo Fisher Scientific). A home-pulled and packed 30 cm column was triple-packed with 0.5 cm, 0.5 cm and 30 cm of 5 μm C4, 3 μm C18, and 1.8 μm C18 respectively and heated to 60 °C for use as the analytical column. Peptides were first loaded at 500 bar which was followed by a chromatography gradient ranging from 6 to 25% acetonitrile over 70 minutes followed by a 5-minute gradient to 100% acetonitrile, which was held for 10 minutes. Electrospray ionization was performed by applying 2000V through a stainless-steel T-junction connecting the analytical column and Easy-nLC system. Each sample was followed by two washes starting with a gradient from 3 to 100% acetonitrile over 15 minutes with an additional 10 minutes at 100% acetonitrile. An m/z range of 375-1500 was scanned for peptides with charge states between 2-6. Centroided data was used for quantitation of peaks. Acquisition was run in a data-dependent positive ion mode. Raw spectra was searched in Proteome Discoverer Version 2.1 against a uniprot reference database for Staphylococcus capitis (Uniprot proteome UP000042965, accessed 10/01/2018) using the Sequest algorithm (Eng et al. 1994) alongside a reverse database approach used to control peptide and protein false discoveries to 1% (Elias and Gygi 2007). No enzyme was specified in the search and a minimum peptide length was set to 6 amino acids. Search parameters included a precursor mass tolerance of 50 ppm and fragment mass tolerance of 0.6 Da and variable oxidation for modifications.
Transplantation of antimicrobial CoNS on pigskin and mice
All experiments involving live animal work were in accordance with the approval of the Institutional Animal Care and Use Guidelines of the University of California, San Diego (protocol no. S09074). Fresh-frozen pig skin sheets were obtained from Loretta Tomlin Animal Technologies (Livermore, CA) and sanitized by surgical brush with 3% chloroxylenol. The skin sheet was cut into 2.5cm × 2.5cm and rinsed 20X times with sterile PBS more. 1x107 CFU C. acnes (ATCC6919) was epicutaneously challenged on pig skin for 1 hour and 100 μl of S. capitis E12 extract (10 mg/mL) or PBS control was applied for 24 h. Live bacteria were harvested by swabbing to measure C. acnes survival. For mouse experiments, the backs of hairless SKH1 mice were scrubbed with alcohol wipes and 1x107 CFU C. acnes was inoculated onto sterile gauze pads which were placed onto the dorsal skin and secured with tegaderm film for 24 h. C. acnes-containing gauze pad was removed and 100 μl of S. capitis E12 extract (10 mg/mL) or PBS control was inoculated onto fresh gauze pad and placed back onto the same dorsal skin site and secured with tegaderm film for an additional 24 h. 48 h post-inoculation, skin was swabbed to measure surviving CFU of C. acnes.
Statistical analysis
Statistical analysis was performed by using Graphpad Prism (GraphPad Software, Inc., La Jolla, USA; version 7.03). Statistically significant differences were calculated by appropriate statistical methods as outlined. P values of ≤0.05 were considered significant.
Supplementary Material
Supplementary figure.1. Characterizing the biological nature of the S. capitis active molecule(s). (A) Growth of C. acnes C2 was measured in liquid culture with or without S. capitis E12 supernatant untreated, boiled at 90°C for 15 mins or separated through 10 kDa or 3 kDa MWCO columns into retained or flow-through fractions for 24 h. (B) S. capitis E12 and control S. hominus A9 supernatant was subjected to papain or proteinase K proteolytic digestion (200 μg/ml) for 45 min at 37°C and inoculated onto C. acnes-containing agar to measure antimicrobial activity. (C) CoNS supernatant was subjected to precipitation in different saturation amounts of ammonium sulphate, and antimicrobial activity of each precipitate was measured by visualizing growth of C. acnes C2 on agar after 72 h.
Supplementary figure.2. PSMβ peptides are extracted by n-Butanol treatment of S. capitis E12 supernatant. (A) Antimicrobial activity against C. acnes C2 is retained and enhanced after butanol extraction of PSMβ from sterile supernatant of S. capitis E12 but not with the lantibiotic-producing S. hominus A9. (B) Silver stain of the total protein content of S. capitis E12 supernatant before and after butanol extraction showing enrichment of small PSMβ peptides.
Supplementary figure.3. PSMβ does not synergize with host LL-37 antimicrobial peptide. (A) Assessment of synergistic antimicrobial activity against C. acnes C2 from S. capitis E12 extract and/or human LL-37 antimicrobial peptide, at indicated concentrations for 72 h.
Supplementary figure.4. S. capitis E12 extract can exhibit greater potency against C. acnes compared to several antibiotics. (A-D) Several distinct strains of C. acnes, isolated from skin of acne patients (A1, C2, K1, K2), were cultured for 48 h in the presence of increasing concentrations of S. capitis extract, tetracycline, clindamycin or erythromycin.
Footnotes
CONFLICT OF INTEREST
A.O.N, T.N and R.L.G are co-inventors of UCSD technology related to the bacterial AMPs discussed herein, and R.L.G is a cofounder and consultant of MatriSys Bioscience. Funding for this project was provided by Almirall pharmaceutical company.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Barnard E, Shi B, Kang D, Craft N, Li H. The balance of metagenomic elements shapes the skin microbiome in acne and health. Sci. Rep 2016;6:srep39491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouslimani A, Porto C, Rath CM, Wang M, Guo Y, Gonzalez A, et al. Molecular cartography of the human skin surface in 3D. Proc. Natl. Acad. Sci. U.S.A 2015;112(17):E2120–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter CH. Egg yolk agar for isolation of coagulase-positive staphylococci. J. Bacteriol 1960;79:753–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung GYC, Joo H-S, Chatterjee SS, Otto M. Phenol-soluble modulins – critical determinants of staphylococcal virulence. FEMS Microbiol Rev. 2014;38(4):698–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien AL, Tsai J, Leung S, Mongodin EF, Nelson AM, Kang S, et al. Association of Systemic Antibiotic Treatment of Acne With Skin Microbiota Characteristics. JAMA Dermatol. 2019; 155(4):425–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu J, Vila-Farres X, Inoyama D, Ternei M, Cohen LJ, Gordon EA, et al. Discovery of MRSA active antibiotics using primary sequence from the human microbiome. Nat Chem Biol. 2016;12(12):1004–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogen AL, Yamasaki K, Muto J, Sanchez KM, Crotty Alexander L, Tanios J, et al. Staphylococcus epidermidis antimicrobial delta-toxin (phenol-soluble modulin-gamma) cooperates with host antimicrobial peptides to kill group A Streptococcus. PLoS One. 2010;5(1):e8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, et al. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014;42(Database issue):D633–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donia MS, Cimermancic P, Schulze CJ, Wieland Brown LC, Martin J, Mitreva M, et al. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell. 2014;158(6):1402–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 2007;4(3):207–14 [DOI] [PubMed] [Google Scholar]
- Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom 1994;5(11):976–89 [DOI] [PubMed] [Google Scholar]
- Fitz-Gibbon S, Tomida S, Chiu B-H, Nguyen L, Du C, Liu M, et al. Propionibacterium acnes strain populations in the human skin microbiome associated with acne. J. Invest. Dermatol 2013;133(9):2152–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice EA, Segre JA. The skin microbiome. Nat. Rev. Microbiol 2011;9(4):244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JB, Cong Z, Imamura-Kawasawa Y, Kidd BA, Dudley JT, Thiboutot DM, et al. Isolation and identification of the follicular microbiome: implications for acne research. J. Invest. Dermatol 2018; [DOI] [PubMed] [Google Scholar]
- Joo H-S, Cheung GYC, Otto M. Antimicrobial activity of community-associated methicillin-resistant Staphylococcus aureus is caused by phenol-soluble modulin derivatives. J Biol Chem. 2011;286(11):8933–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelhälä H-L, Aho VTE, Fyhrquist N, Pereira PAB, Kubin ME, Paulin L, et al. Isotretinoin and lymecycline treatments modify the skin microbiota in acne. Exp. Dermatol 2018;27(1):30–6 [DOI] [PubMed] [Google Scholar]
- Kumar R, Jangir PK, Das J, Taneja B, Sharma R. Genome analysis of Staphylococcus capitis TE8 reveals repertoire of antimicrobial peptides and adaptation strategies for growth on human Skin. Sci Rep. 2017;7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsuji T, Chen TH, Narala S, Chun KA, Two AM, Yun T, et al. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci Transl Med. 2017;9(378) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013;502(7469):96–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill AM, Gallo RL. Host-microbiome interactions and recent progress into understanding the biology of acne vulgaris. Microbiome. 2018;6(1):177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J, Byrd AL, Deming C, Conlan S, NISC Comparative Sequencing Program, Kong HH, et al. Biogeography and individuality shape function in the human skin metagenome. Nature. 2014;514(7520):59–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi JT, Hamory BH. Simplified method for the isolation, identification, and characterization of Staphylococcus epidermidis in epidemiologic studies. Diagn. Microbiol. Infect. Dis 1986;4(1):29–35 [DOI] [PubMed] [Google Scholar]
- Saising J, Dube L, Ziebandt A-K, Voravuthikunchai SP, Nega M, Götz F. Activity of Gallidermin on Staphylococcus aureus and Staphylococcus epidermidis Biofilms. Antimicrob. Agents Chemother 2012;56(11):5804–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz CFP, Jensen A, Lomholt HB, Brüggemann H, Kilian M. A novel high-resolution single locus sequence typing scheme for mixed populations of Propionibacterium acnes in vivo. PLoS One. 2014;9(8):e104199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw LP, Bassam H, Barnes CP, Walker AS, Klein N, Balloux F. Modelling microbiome recovery after antibiotics using a stability landscape framework. ISME J. 2019;13(7):1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu M, Wang Y, Yu J, Kuo S, Coda A, Jiang Y, et al. Fermentation of Propionibacterium acnes, a Commensal Bacterium in the Human Skin Microbiome, as Skin Probiotics against Methicillin-Resistant Staphylococcus aureus. PLoS One. 2013;8(2):e55380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tossi A, Sandri L, Giangaspero A. Amphipathic, alpha-helical antimicrobial peptides. Biopolymers. 2000;55(1):4–30 [DOI] [PubMed] [Google Scholar]
- Wang R, Khan BA, Cheung GYC, Bach T-HL, Jameson-Lee M, Kong K-F, et al. Staphylococcus epidermidis surfactant peptides promote biofilm maturation and dissemination of biofilm-associated infection in mice. J. Clin. Invest 2011;121(1):238–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams MR, Gallo RL. The Role of the Skin Microbiome in Atopic Dermatitis. Curr Allergy Asthma Rep. 2015;15(11):65. [DOI] [PubMed] [Google Scholar]
- Zaenglein AL, Pathy AL, Schlosser BJ, Alikhan A, Baldwin HE, Berson DS, et al. Guidelines of care for the management of acne vulgaris. J. Am. Acad. Dermatol 2016;74(5):945–973.e33 [DOI] [PubMed] [Google Scholar]
- Zipperer A, Konnerth MC, Laux C, Berscheid A, Janek D, Weidenmaier C, et al. Human commensals producing a novel antibiotic impair pathogen colonization. Nature. 2016;535(7613):511–6 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure.1. Characterizing the biological nature of the S. capitis active molecule(s). (A) Growth of C. acnes C2 was measured in liquid culture with or without S. capitis E12 supernatant untreated, boiled at 90°C for 15 mins or separated through 10 kDa or 3 kDa MWCO columns into retained or flow-through fractions for 24 h. (B) S. capitis E12 and control S. hominus A9 supernatant was subjected to papain or proteinase K proteolytic digestion (200 μg/ml) for 45 min at 37°C and inoculated onto C. acnes-containing agar to measure antimicrobial activity. (C) CoNS supernatant was subjected to precipitation in different saturation amounts of ammonium sulphate, and antimicrobial activity of each precipitate was measured by visualizing growth of C. acnes C2 on agar after 72 h.
Supplementary figure.2. PSMβ peptides are extracted by n-Butanol treatment of S. capitis E12 supernatant. (A) Antimicrobial activity against C. acnes C2 is retained and enhanced after butanol extraction of PSMβ from sterile supernatant of S. capitis E12 but not with the lantibiotic-producing S. hominus A9. (B) Silver stain of the total protein content of S. capitis E12 supernatant before and after butanol extraction showing enrichment of small PSMβ peptides.
Supplementary figure.3. PSMβ does not synergize with host LL-37 antimicrobial peptide. (A) Assessment of synergistic antimicrobial activity against C. acnes C2 from S. capitis E12 extract and/or human LL-37 antimicrobial peptide, at indicated concentrations for 72 h.
Supplementary figure.4. S. capitis E12 extract can exhibit greater potency against C. acnes compared to several antibiotics. (A-D) Several distinct strains of C. acnes, isolated from skin of acne patients (A1, C2, K1, K2), were cultured for 48 h in the presence of increasing concentrations of S. capitis extract, tetracycline, clindamycin or erythromycin.