Abstract
Background –
To facilitate ablation of ventricular tachycardia (VT), an automated localization system to identify the site of origin of left ventricular (LV) activation in real time using the 12-lead ECG was developed. The objective of this study was to prospectively assess its accuracy.
Methods –
The automated site of origin localization (SOLO) system consists of three steps: (1) localization of ventricular segment based on population templates, (2) population-based localization within a segment, and (3) patient-specific site localization. Localization error was assessed by the distance between the known reference site and the estimated site.
Results –
In 19 patients undergoing 21 catheter ablation procedures of scar-related VT, SOLO accuracy was estimated using 552 LV endocardial pacing sites pooled together, and 25 VT-exit sites identified by contact mapping. For the 25 VT-exit sites, localization error of the population-based localization steps was within 10 mm. Patient-specific site localization achieved accuracy of within 3.5 mm after including up to 11 pacing (training) sites. Using three remotes (67.8 ± 17.0 mm from the reference VT-exit site), and then 5 close pacing sites, resulted in localization error of 7.2 ± 4.1 mm for the 25 identified VT-exit sites. In two emulated clinical procedure with 2 induced VTs, the SOLO system achieved accuracy within 4 mm.
Conclusions –
In this prospective validation study, the automated localization system achieved estimated accuracy within 10 mm and could thus provide clinical utility.
Keywords: ventricular tachycardia, mapping, ablation, ECG, pace-mapping
Journal Subject Terms: Catheter Ablation and Implantable Cardioverter-Defibrillator, Arrhythmias, Electrocardiology (ECG)
Graphical Abstract

Introduction
Catheter ablation has been demonstrated to be an effective therapeutic option to prevent and reduce recurrent scar-related VT1–2, but has imperfect success rates, limited, in part, by poorly tolerated, unmappable VT, and also by the use of point-by-point catheter mapping, even with newer multipolar catheters and substrate-based techniques. It remains a challenging procedure which may be restricted to specialized centers3.
The standard 12-lead ECG has demonstrated relatively accurate localization of sources of ventricular arrhythmias4, comparable to that achieved with electrocardiographic imaging (ECGI)5–8. Rapid ECG interpretation requires expertise and could be facilitated with a computer-assisted method to automatically localize the origin of early ventricular activation during an invasive EP study and ablation. Various algorithms based on the 12-lead ECG have been proposed for localizing the origin of early ventricular activation in human studies9–24. However, a simple intraprocedural automatic localization system is lacking. Therefore, we have previously developed an automated technique to provide site of origin localization (SOLO) of early left ventricular (LV) activation in real time using the 12-lead ECG4. The aim of this study was to prospectively evaluate the performance of the SOLO technique in a cohort of patients with scar-related VT undergoing catheter ablation.
Methods
Study population
Nineteen consecutive consenting patients with scar related VTs undergoing catheter ablation of VT were enrolled in a protocol approved by the Institutional Research Ethics Board; all patients provided written informed consent.
Clinical EP study and ablation
Scar-related VT ablation procedures were performed using standard ablation techniques2, or infusion-needle catheter ablation25. Use of the needle catheter was conducted through the Special Access Program, Health Canada. VT was induced by programmed ventricular stimulation from the right ventricular (RV) apex or outflow tract and the LV was mapped via the retrograde aortic or trans-septal atrial approach. Intracardiac electrograms were digitized and stored by CardioLab system (GE Healthcare, Barrington, Illinois), and three-dimensional (3D) electroanatomic (EA) maps were acquired by the Carto 3 system (Biosense Webster, Irvine, California). Substrate-based mapping and pace-mapping were used to identify scar and potential culprit sites within the scar, which were targeted for ablation. For each procedure, a complete LV endocardial EA map was created using a 3.5-mm tip irrigated catheter (Navistar SmartTouch, Biosense Webster, Diamond Bar, CA, USA) or a high-density multi-electrode catheter (PentaRay NAV Catheter, Biosense Webster). Pacing was performed with stable catheter position at multiple endocardial sites at minimum pacing output that ensured consistent focal myocardial capture.
Data acquisition and processing
Eight independent leads (I, II, V1—V6) of the 12-lead ECG were acquired via multichannel recording system (CardioLab, GE Healthcare, Barrington, Illinois) during the EP study, filtered (0.05 to 100 Hz), sampled at 1000 Hz with 16-bit resolution. The output of the amplifier and system which routinely processes the signal was cloned to an ancillary secured computer with which additional processing and analysis can be performed by the automated localization system without affecting the clinical signal. The 8-lead ECG data was recorded for 15 seconds during VT, sinus rhythm or pacing at LV endocardium; the other 4 leads (Lead III, aVF, aVL, aVR) were simultaneously computed by using Einthoven’s leads. Bipolar pacing stimuli were delivered with 2 ms duration at twice the diastolic threshold current. Within each recording, the integral of a capture pacing beat or VT beat with the best quality was calculated over the initial 120 ms of the QRS complex of the 12-lead ECG for real-time analysis4. For a recorded pacing site, one representative paced beat was selected in the corresponding 12-lead ECG, avoiding ectopic beats, motion artifacts, non-capture beats, and captured beats with stimulus-QRS delay > 40 ms (as these may represent capture of a channel within myocardial scar with exit remote from the site of pacing). In addition, the VTOL Export Tool (supplied by Carto 3, Biosense Webster, Irvine, California) as a real-time transfer protocol was used to transfer known 3D coordinates of recorded pacing sites from the Carto 3 mapping system (Biosense Webster, Irvine, California) to the automated localization system.
Intraprocedural automated localization system
The SOLO system4 consists of three consecutive steps to progressively approach a VT-exit site: 1. localization of a ventricular segment containing the VT-exit site based on population templates, 2. population-based exit site localization within a segment, and 3. patient-specific site localization. Briefly, after creating a LV endocardial EA map, VT induction was performed. The onset of one VT beat is automatically detected26, and can be manually adjusted. First, the initial 120-ms window of QRS is captured and the 12 QRS integrals are computed to form a 12-lead ECG integral pattern. The 12-lead ECG integral pattern is used to compare to template ECG integral patterns generated from each of 16 segments of a generic LV endocardial surface derived from a prior dataset4, producing a mean correlation coefficient value of match between the pattern and one of 16 template patterns; the best correlation segment provides an approximate localization. Second, to improve localization accuracy, population-based localization of the exit site within a segment uses a population-derived linear regression model that is fitted to a generic 238-element LV geometry. QRS integrals from the independent leads I, II and V1–V6, are used to provide a statistical estimate of the 3D location of the VT-exit site. Third, to achieve further precision, patient-specific site localization is performed using the patient’s own LV geometry, and substituting the population-derived regression coefficients with patient-specific coefficients calculated from the 3-lead ECG (III, V2, V6) while pacing at known sites (≥ 5) in the patient undergoing the ablation procedure.
Data analysis
All VT ablation procedures were performed by clinicians (AA and JS), blinded to the prediction results of the SOLO system. The VT-exit site for each mapped VT was identified and confirmed by a combination of activation mapping27 and entrainment mapping during VT28, supplemented by pace-mapping at the scar margin29–30. Locations were defined as exit sites if: 1. entrainment resulted in concealed fusion with a difference of <30 msec between post-pacing interval and tachycardia cycle length, and if stimulus-QRS interval was <40 msec; OR 2. activation time was 0–40 msec pre-QRS; OR if a conducting corridor was identified within scar at which pacing resulted in >40 msec latency and >95% quantitative morphologic match and further mapping identified a site at or near the scar margin with <40 msec latency but preserved morphology match. Induced VTs for which an accurate scar exit site was not verifiable by contact mapping methods were excluded. Intraprocedural localization of an induced VT was performed on an ancillary secured computer with the SOLO system during the ablation procedure, showing the estimated site on the screen of the ancillary secured computer. Localization errors of each method (population-based localization within a segment and patient-specific site localization) were quantitatively estimated by one investigator who was blinded to the results of the VT ablation procedure.
Estimation of localization accuracy of the VT exit using population-based localization within a segment
Two observers, blinded to the results of the automated localization system, reviewed all identified VT-exit sites considered as the “ground truth” for comparisons and manually registered them from the patient-specific EA map onto the generic LV endocardial surface for each patient4. Each identified VT-exit site was associated with one of the generic LV endocardium’s 238 triangular area elements as shown in Figure 1; two-observer variability of this registration was assessed. Population-based localization within a segment was used to predict each identified VT on the generic LV endocardial surface. The localization accuracy was then estimated from the Euclidean distance, in mm, between the known and the estimated site, both located on the generic LV endocardial surface.
Figure 1.
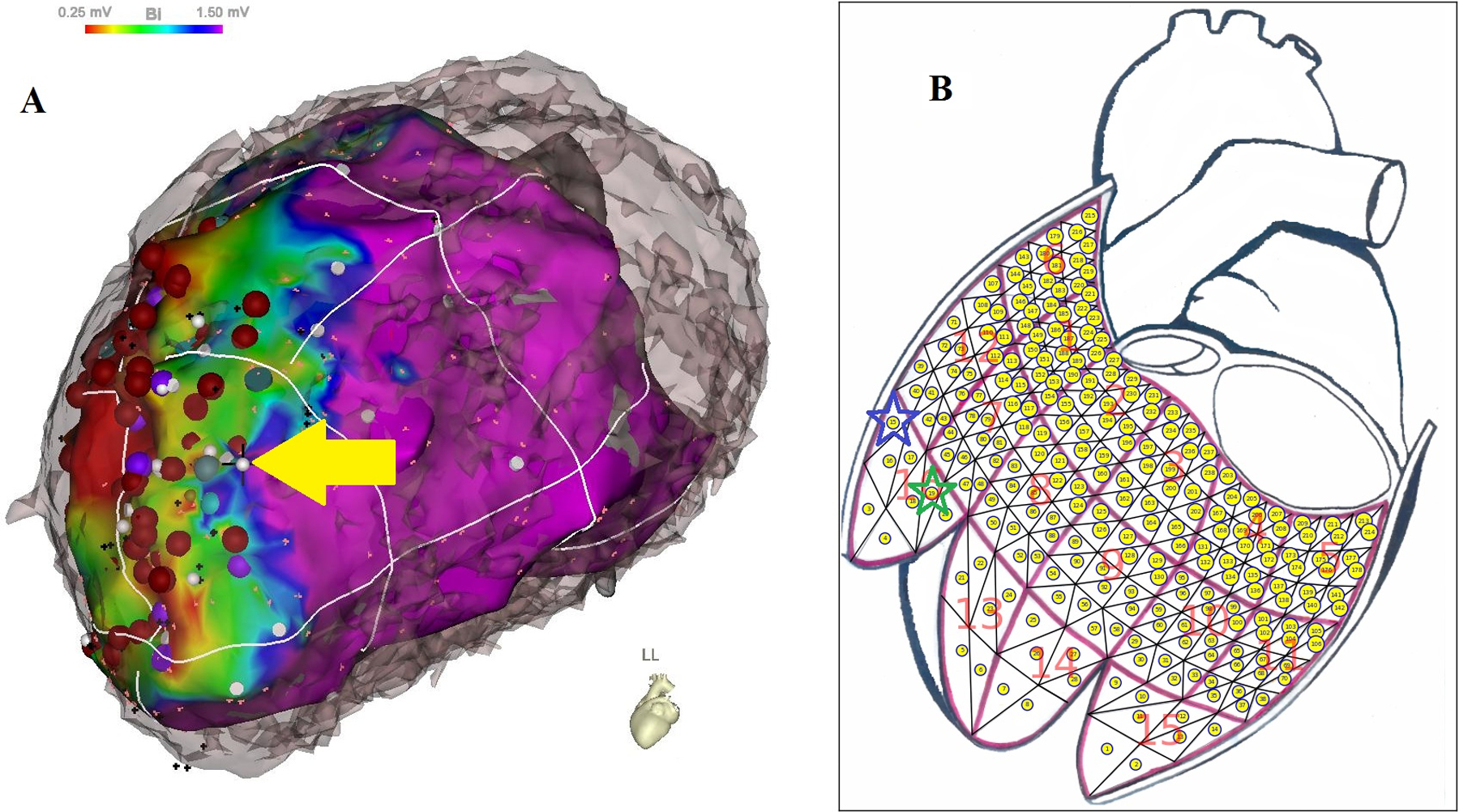
Registration of an identified VT-exit site. (A) A VT-exit site was identified by a combination of activation mapping (27) and entrainment mapping during VT (28), supplemented by pace-mapping at the scar margin (29–30); the identified VT-exit site marked by a white dot and yellow arrow) was located on the bipolar potential map aligned with patient’s own CT geometry. (B) The identified VT-exit site was registered from the bipolar potential map to one of 238 triangular area elements within a 16-segment generic LV endocardial surface by observer #1 with green star and observer #2 with blue star, respectively.
Estimation of localization performance of patient-specific site localization
To estimate the performance of patient-specific site localization using the patient’s own EA coordinates, localization of pacing sites was performed. Pacing at multiple endocardial sites had been performed in each patient, and ‘target’ sites were identified as those that had between 5 and 15 pacing site neighbors within radius of 10, 15, 20, 25, 30, 35, 40, 45, 50 mm. The neighboring pacing sites were used to calculate patient-specific regression coefficients by combining their known sites and ECG integrals from leads III, V2 and V6. These were then used to estimate the location of the “target” site, and accuracy was calculated as the distance between the estimated and actual sites.
Optimal locations of training-set pacing sites in the patient-specific site localization.
When a VT exit location is estimated by population-based localization within a segment, a clinician can then pace in the neighbourhood of that region to improve the localization accuracy by generating and using patient-specific regression coefficients, in order to perform patient-specific site localization. Localization accuracy was only moderate in the absence of training pacing sites which were remote from the target site. Therefore, we estimated how many pacing sites distributed around an induced VT exit site were needed, including both remote and neighboring sites, while minimizing the total number of pacing sites.
In this validation cohort, each procedure included recordings from a mean of 26 pacing sites for mapping, and at least one VT. For each identified VT-exit site, we first calculated distances between the identified VT-exit site and all pacing sites of the procedure. The farthest 5 pacing sites defined an initial larger remote sampling area, and the closest 6 pacing sites were defined as neighbouring sites. To identify an optimal pacing strategy, five groups of training-set pacing-site distributions were generated for comparison: the first group had 7 pacing sites that contained the most remote pacing site and the neighboring 6 pacing sites; the second group had 8 pacing sites that contained the 2 most remote pacing sites and the neighboring 6 pacing sites; this was repeated for groupings utilizing 3, 4 and 5 remote pacing sites. The localization accuracy using patient-specific site localization was assessed for each group.
Statistical Analysis
The mean and standard deviation (SD) were used to report statistical analysis results. Five pacing-site groups based on all possible pairings were compared by using paired t-test with the Bonferoni correction for multiple testing. Accuracy between two observers was compared using a paired t-test. A double-sided P < 0.05 was considered statistically significant. Statistical analysis was performed with the Minitab 18 Statistical Software (Minitab Inc, State College, PA). The data supporting the results of this study will be made available from the corresponding author upon reasonable request.
Results
Patient population
Among 19 consecutive patients enrolled (89.5% male, 66.6 ± 9.1 years), 21 separate VT procedures were performed, including 2 needle ablations and 19 standard ablation procedures. Table 1 summarized the baseline characteristics. For the 21 procedures, 552 LV endocardial pacing sites and 48 induced VTs were recorded by the automated localization system in real time (Table 2). Twenty-five VT-exit sites of the mapped 48 VTs were identified and were used as the “ground truth” for comparisons as shown in Table 3.
Table 1.
Baseline characteristics of VT patients
| Clinical Characteristics (N=19) | |
|---|---|
| Male (%) | 17 (89.5) |
| Age, years | 66.6 ± 9.1 |
| Non-ischemic | 3(15.8) |
| Ischemic cardiomyopathy | 15(79.0) |
| Cardiac sarcoid | 1(5.3) |
| LVEF(%) | 37.8 |
| Heart failure (%) | 14(73.7) |
| ICD present | 17(89.5) |
| Medication (%) | |
| Beta blocker | 13(68.4) |
| Amiodarone | 10(52.6) |
| Sotalol | 3(15.8) |
| Mexiletine | 4(21.1) |
| ACEi | 11(57.9) |
| Statin | 15(79.0) |
VT =ventricular tachycardia; LVEF = left ventricular ejection fraction; ICD = implantable cardioverter-defibrillator; ACEi = Angiotensin-converting enzyme inhibitors
Table 2.
Description of all recorded VTs from 21 ablation procedures
| Case # | Sex | Age | Etiology | # of pacing sites | Mapped VT | Characteristics of VT morphology |
|---|---|---|---|---|---|---|
| 1*† | M | 51 | NICM | 17 | VT1 | RB/right axis/CL 520 ms |
| 2 | M | 67 | ICM | 26 | ||
| VT3 | LB/left axis/CL 700 ms | |||||
| 3 | M | 72 | NICM | 20 | — | No-inducible VT |
| 4 | M | 70 | ICM | 30 | VT1 | RB/normal axis/CL 480 ms |
| 5 | F | 65 | ICM | 33 | VT1 | LB/left axis/CL 360 ms |
| 6 | M | 68 | ICM | 25 | VT1 | RB/left axis/CL 260 ms |
| 7 | M | 80 | ICM | 32 | ||
| VT2 | RB/left axis/CL 310 ms | |||||
| 8 | M | 69 | ICM | 23 | ||
| VT2 | LB/left axis/CL 440 ms | |||||
| 9* | M | 71 | ICM | 17 | ||
| VT2 | LB/normal axis/CL 260 ms | |||||
| 10 | M | 61 | ICM | 21 | — | No-inducible VT |
| 11* | M | 61 | ICM | 18 | — | No-inducible VT |
| 12 | M | 58 | ICM | 42 | ||
| VT2 | RB/left axis/CL 300 ms | |||||
| 13 | M | 76 | ICM | 31 | ||
| VT3 | LB/left axis/CL 285 ms | |||||
| 14 | M | 71 | ICM | 26 | ||
| VT4 | LB/left axis/CL 440 ms | |||||
| 15 | M | 80 | ICM | 20 | ||
| VT3 | RB/left axis/CL 425 ms | |||||
| 16 | M | 63 | ICM | 29 | ||
| VT3 | LB/extreme axis/CL 430 ms | |||||
| 17† | M | 71 | ICM | 23 | ||
| VT7 | RB/left axis/CL 600 ms | |||||
| 18 | M | 44 | ICM | 27 | ||
| VT2 | RB/normal axis/CL irregular | |||||
| 19 | F | 73 | NICM | 36 | ||
| VT6 | very similar to VT5, CL 480 ms | |||||
| 20† | M | 59 | ICM | 22 | ||
| VT3 | RB/left axis/CL 490 ms | |||||
| 21 | M | 67 | CS | 34 | ||
| VT2 | RB/right axis/CL 315 ms |
The ‘*’ indicates that three cases had insufficient pacing sites to estimate localization performance using patient-specific site localization which were excluded. The ‘†’ indicates needle ablation procedures. M: male; ICM, Ischemic cardiomyopathy; NICM, Non-ischemic cardiomyopathy; CS, Cardiac sarcoidosis; RB and LB refer to right bundle and left bundle; ms: milliseconds. Description of VT morphology: VT morphology/axis based on limb leads (normal axis (−30 to 90), right axis (90 to180), extreme axis (−90 to −180) and left axis (−30 to −90))/ cycle length (CL).
Table 3.
Accuracy of VT exit localization achieved by the automated localization system
| Case# | Mapped VT | VT Exit/Ablation Site | Localization Error (mm) | ||
|---|---|---|---|---|---|
| Population-based localization within a segment | Patient-specific site localization | ||||
| Observer 1 | Observer 2 | ||||
| 1 | VT1 | Basal anterolateral LV | 9.3 | 4.5 | 3.9 |
| 2 | |||||
| VT3 | Mid inferior septum | 11.2 | 10.7 | 7.3 | |
| 4 | VT1 | Inferolateral segment | * | * | * |
| 5 | VT1 | — | — | — | — |
| 6 | VT1 | Basal inferior LV | 12.2 | 6.1 | 14.4 |
| 7 | |||||
| VT2 | Basal inferior | 11.2 | 10.3 | 16.0 | |
| 8 | |||||
| VT2 | Basal inferior septum | * | * | * | |
| 9 | |||||
| VT2 | Septal margin of the scar | 6.4 | 0.0 | 9.3 | |
| 12 | |||||
| VT2 | Basal septal margin of the scar | 10.4 | 4.7 | 4.0 | |
| 13 | |||||
| VT3 | — | — | — | — | |
| 14 | |||||
| VT4 | Basal inferoseptal segment | 6.9 | 20.1 | 13.6 | |
| 15 | |||||
| VT3 | Apical scar margin | 9.8 | 0.0 | 7.8 | |
| 16 | |||||
| VT3 | Inferoseptal apical segment | * | * | * | |
| 17 | |||||
| VT7 | Superior basal septum | 7.2 | 11.9 | 3.6 | |
| 18 | |||||
| VT2 | Non-sustained | — | — | — | |
| 19 | |||||
| VT6 | His proximal region | * | * | * | |
| 20 | |||||
| VT3 | Mid inferoseptal region | * | * | * | |
| 21 | |||||
| VT2 | Mid-apical anterolateral wall inferior to the papillary muscle | 6.7 | 9.6 | 5.3 | |
The error of VT-exit localization was calculated as Euclidean distance between the estimated VT-exit site and the known VT-exit site. ‘—’ indicates that the VT was not mapped, and an exit site was therefore not identified. The ‘*’ indicates VTs that were terminated within the middle of the scar at sites proximal to the scar exit site (in which cases localization accuracy could not be precisely quantitated because VT was terminated at a mid-diastolic site).
Indicates that site of origin/ablation site was only approximately localized.
Accuracy using population-based localization within a segment
The 25 VT morphologies were used to estimate the accuracy of population-based localization within a segment. Registration of pacing sites to the generic LV endocardial surface triangles for the 25 identified VT-exit sites had interobserver variability in the vicinity of 3.7 mm. Table 3 (columns 4 and 5) shows errors of VT-exit localization by using the two-observer registration as different references. The localization accuracy was 9.5 ± 2.6 mm (mean ± SD) when using observer 1’s registration, which was not significantly different from observer 2’s registration (7.9 ± 4.7 mm, P > 0.05).
Accuracy using patient-specific site localization
Three cases (#1, #9 and #11) had insufficient pacing sites for analysis and were excluded; thus 18 procedures with 500 pacing sites were included to estimate the accuracy of pacing-site localization as shown in Figure 2. The accuracy of localization improved with the inclusion of additional known pacing sites to calculate patient-specific regression coefficients. Better accuracy was observed when the training-set pacing sites were within 25 mm of the target site.
Figure 2.

Estimation of the localization performance of patient-specific site localization. Out of 500 pacing sites (pooled from 18 ablation procedures), for which there were known electroanatomic (EA) coordinates together with corresponding 3-variable ECG data, sets of “target” pacing sites with 5, 6, 7, 8, 9, 10, 11, 12, 13, 14 and 15 neighbors (as different training sets) within radius of 10, 15, 20, 25, 30, 35, 40, 45, and 50 millimeters were identified. For each selected “target” site, EA coordinates of its each training set together with 3 ECG variables (lead III, V2, and V6) were used to calculate patient-specific regression coefficients and these were then used, in turn, to predict the location (in terms of EA coordinates) of the “target” site; the mean localization error of this prediction was calculated for each set of “target” sites (in millimeters).
Identification of optimal distribution of pacing sites to generate patient-specific coefficients
Twenty-five identified VT-exit sites were used to estimate the optimal pacing-site distribution of a training set when performing patient-specific site localization. Table 4 summarizes the number of pacing sites in each group needed to reach a convergent localization, and the remote and neighboring sampling space of each group based on the distance between the pacing site and the identified VT-exit site. The localization errors of the 5 groups of pacing site distributions are summarized in Figure 3, which graphically depicts localization accuracy. We took each group as a control group and computed paired t-tests between the control group and the other groups for a total of 10 tests. After Bonferonni correction for multiple comparisons (significant P value < 0.005), only one pair (Group 3 - Group 1) was significant (P=0.003). Group 3 (3 pacing sites with large sampling space (67.8 ± 17.0 mm) and 5 pacing sites closer to the estimated source (20.0 ± 8.8 mm) had the best localization accuracy (7.2 ± 4.1 mm) compared to the others as shown in Figure 3.
Table 4.
Five groups for studying optimal locations for training-set pacing sites
| Group # | # of remote + neighbouring pacing sites | Distance from the dispersed pacing sites to the reference VT-exit site (Mean ± SD mm) | Number of pacing sites needed to achieve convergence |
|---|---|---|---|
| #1 | 1 + 6 | 70.6 ± 17.2 | 6 |
| #2 | 2 + 6 | 69.3 ± 17.1 | 7 |
| #3 | 3 + 6 | 67.8 ± 17.0 | 8 |
| #4 | 4 + 6 | 66.2 ± 16.8 | 9 |
| #5 | 5 + 6 | 64.8 ± 16.6 | 10 |
Figure 3.
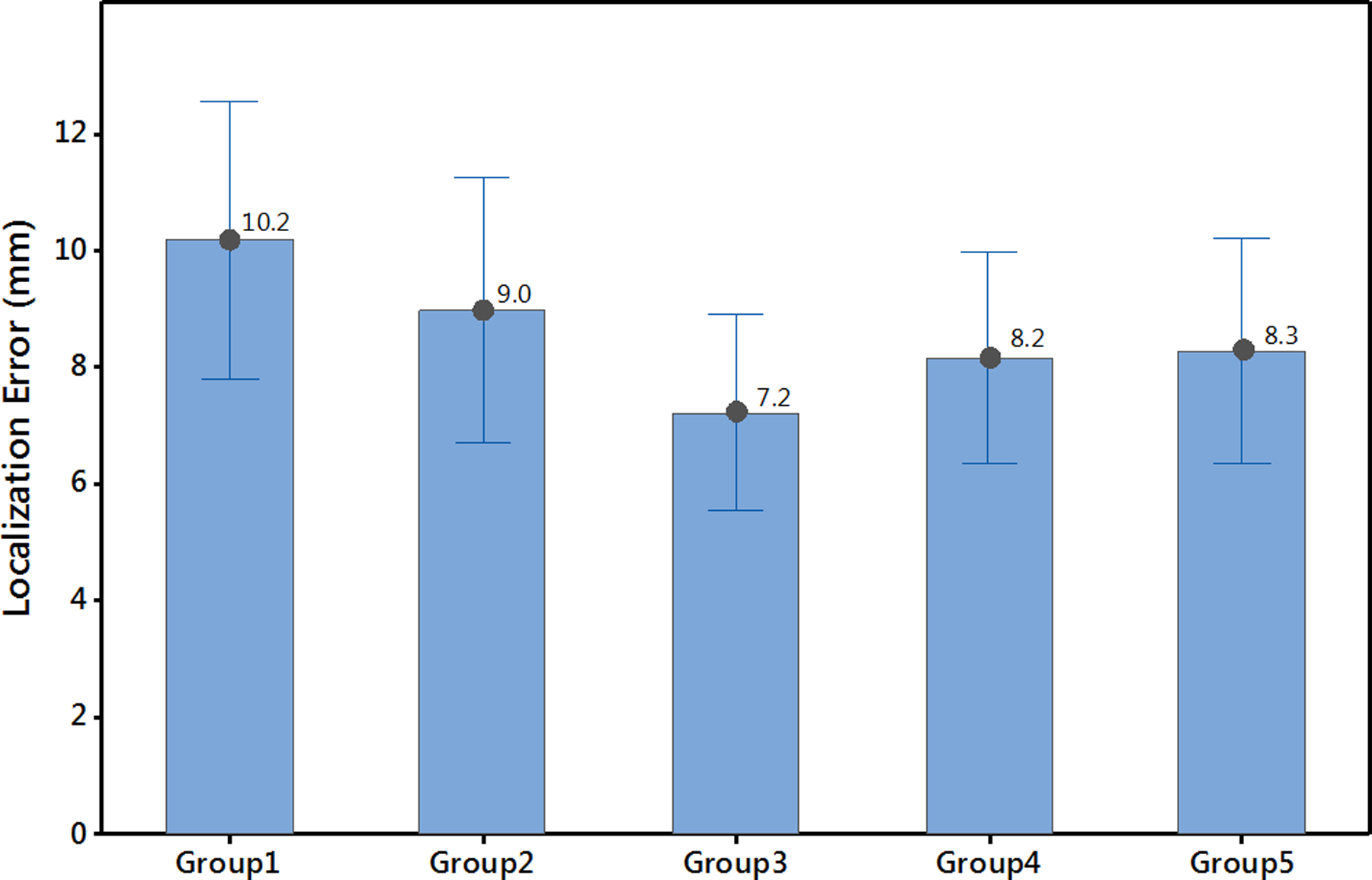
Identification of the optimal distribution of training-set pacing sites for performing patient-specific site localization. Localization accuracy of five groups of training-set pacing-site distributions in relation to remote and neighboring sampling space in the vicinity of a mapped VT. Details are discussed in the text.
Clinical emulation to localize the VT exit by the automated localization system
One patient was used to illustrate the ability of the proposed system to localize two induced VTs. For procedure #21, VT1 was mappable and exited scar at the mid-anterolateral wall superior to the papillary muscle (noted by the red star in Figure 4, panel C, and yellow arrow on panel D). Figure 4 (panel A) shows the recorded VT1 morphology. Using population-based localization within a segment, this VT1 was assigned to the nearest of 238 triangle elements and marked by the bullseye icon on the generic endocardial LV surface (Figure 4, panel B, mid-anterolateral wall). Pacing at 3 sites remote from the region of interest, followed by pacing at 5 more sites close to the estimated location, patient-specific regression coefficients are generated, which was used to localize the VT1-exit site, marked in blue on the EA geometry (Figure 4, panel C). This was 0.0 mm from the site identified by contact mapping. (Table 3).
Figure 4.
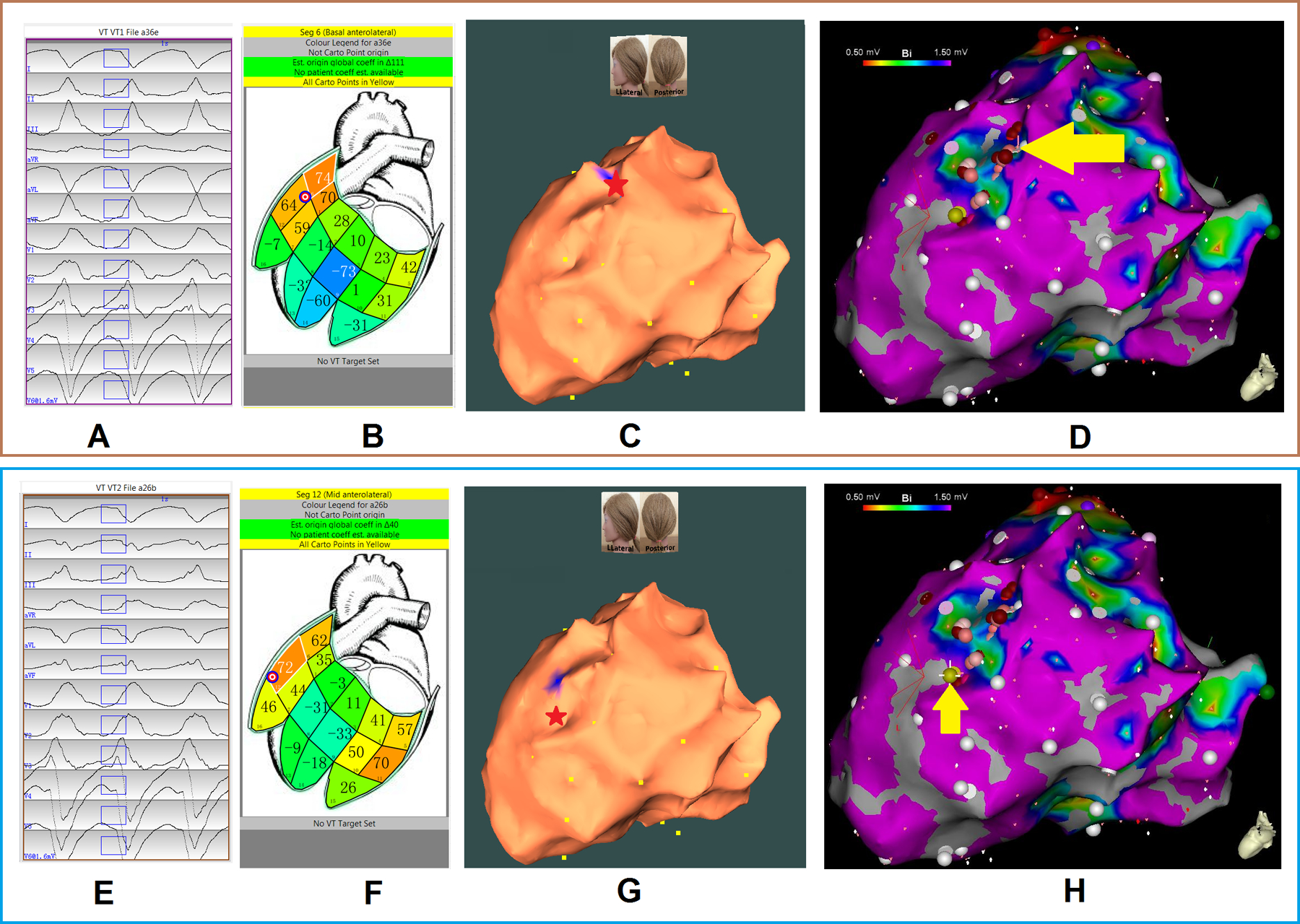
Localization of two VT exits by the automated localization system: (A, E panels) show the recorded 12-lead ECG of two induced monomorphic VTs during the procedure for patient #19. The onset of one VT beat was automatically detected (26); the user can edit the onset of the 120-ms window (rectangle box) if correction is necessary. (B, F panels) show bull’s eye icons that indicate the estimated VT-exit locations using population-based localization within a segment. The large number within each segment is the correlation coefficient (%) for match by the 12-lead ECG VT pattern with population based 12-lead ECG templates; the small number identifies the segment. (C, G panels) show that using patient-specific site localization to predict a VT-exit site colored blue on the electroanatomic (EA) geometry with the actual site of VT exit marked by the red star. Localization errors of VT 1 and 2 exits are 0.0 mm and 5.3 mm, respectively. Yellow dots indicate recorded pacing sites on the EA geometry. (D, H panels) illustrate the endocardial EA substrate maps for this patient, with areas featuring bipolar signal amplitude ≥ 1.50 mV in purple, and the site of VT exit (identified by contact mapping) depicted by the yellow arrow and gold ball.
Representative VT2 (Procedure #21): This VT2 had a cycle length of 315 ms, with right bundle branch block-type morphology in lead V1, and a rightward axis (Figure 4, panel E). The site of exit was identified at the mid-apical anterolateral wall inferior to the anterolateral papillary muscle. The VT2 exit site was localized to the more apical portion of the mid-anterolateral segment identified by the bull’s eye in the population-based localization steps (Figure 4, panel F). To refine the localization accuracy, training-set pacing sites were acquired; Figure 4 (panel G) shows that the patient-specific site localization becomes increasingly accurate with the addition of pacing sites, achieving reproducibility within 6 mm when pacing at the 8th site. The EA substrate map is shown in Figure 4 (panel H), with the site of VT exit identified (yellow arrow and gold ball).
Discussion
The purpose of this study was to prospectively assess the performance of the SOLO technique4 for localizing the origin of early LV activation during catheter ablation of VT. The results show (i) a mean error in VT-exit site localization of approximating 9 mm in identifying scar-related VT-exit sites using the population-based localization within a segment method; (ii) that the patient-specific site localization achieved mean accuracy of within 7 mm after including up to 7 pacing sites, and that accuracy increased by adding more pacing sites when they were focused on a suspected area of activation origin (radius < 10 mm); (iii) that the optimal training pacing-site distribution includes at least 3 sites from a large sampling area (mean 67.8 ± 17.0 mm radius from VT estimated VT exit site) followed by at least 5 pacing sites which are closer (within 20.0 ± 8.8 mm) on the LV endocardial surface to achieve localization accuracy of 7.2 mm, as illustrated in Figure 2.
The SOLO system might be useful as an advancement of conventional pace-mapping to predict the origin of an unknown VT site of origin. A clinical procedure might include programmed stimulation to induce VT(s), electroanatomic substrate mapping, and further pacing at sites anticipated to be remote and neighboring to the VT exit sites to rapidly identify scar exit sites graphically, and permit the design of an appropriate ablation strategy. The major difference between commercially available QRS matching and the proposed localization system is that this system predicts a site of origin and combines information from ECG recordings and anatomic geometry which can be personalized with the incorporation of pace mapping to accurately identify VT-exit sites31. Furthermore, QRS correlation coefficient matching may be nonlinear, particularly with increasing distance from the VT-exit site32.
To determine how many neighbouring vs. remote pacing sites are needed for derivation of patient-specific coefficients (to enable patient-specific site localization), while minimizing the total number of pacing sites, we studied the effect of the distribution of the pacing ‘training’ sites. Figure 2 illustrates the localization ability of patient-specific site localization, and shows that using 7 training-set pacing sites in the neighboring areas (radius of ≤ 20 mm) achieved localization accuracy within 10 mm.
To estimate the optimal locations of the training-set pacing sites for patient-specific site localization, we tested varying combinations of remote and neighboring sites. Optimal results were obtained with the combination of 3 widely distributed sites, a mean of 68 mm from the estimated VT exit site, combined with data derived by pacing from at least 5 sites clustered more closely to the estimated site, within mean radius 20 mm.
Accurate localization of the VT-exit site using the SOLO system could facilitate VT ablation procedures and assist clinicians in the specific targeting of any particular induced VT circuits during the ablation procedure. Once the LV EA map is created, only a few beats for each VT morphology are needed to identify their exit sites. This allows for targeting unstable or non-sustained VTs without significantly prolonging the procedure. It is possible that limiting RF energy delivery to predicted VT target sites may reduce the risk of heart failure worsening related to unnecessary myocardial damage during the procedure, and may permit greater attention to achieving effective ablation at the most important sites, although consideration must be given to targeting non-inducible VTs by ablating identifiable potential culprit substrate. The first two population-based localization steps outlined here do not require catheterization of the LV, and yet showed moderate accuracy in localizing VT. It is possible that this may be useful for non-invasive cardiac radiation for ablation of VT33. Also, recent progress in virtual-heart simulation34, e.g. incorporating magnetic resonance imaging of scar tissue with the population-based localization within a segment method, may improve understanding and interpretation of VT circuits and could aid in pre-procedural planning.
Study Limitations
The current study focused only on patients who underwent LV catheter ablation, using the SOLO system to estimate its localization accuracy. Further work will be required for right ventricular (RV) and epicardial sites of early activation. Notably, the accuracy of patient-specific site localization step relies on acquisition of a complete EA map which has to be imported into the system for localization. This hinders the use of the step particularly at the beginning of procedure, when a full 3D geometry has not yet been acquired. Furthermore, analysis suggests that local concave/convex geometry can lead to larger localization errors. This method is based on projection from the center of the ventricular geometry to the LV endocardial surface. The use of integrated, registered 3D imaging may improve this methodology35. It is worth noting that this technique can identify the site of activation of left ventricular myocardium; this may not be an ideal site for ablation but rather a guide for further substrate analysis. Finally, the methodology has only been validated in the Carto EA system, and may require further validation using different EA mapping systems.
Conclusions
The study prospectively validated the accuracy of an intraprocedural automated VT localization during catheter ablation procedures for VT. The localization performance was promising and could facilitate ablation procedures.
What is known?
Localizing VT-exit sites can be helpful for ablation of scar-related VT.
An automated intraprocedural site of origin localization (SOLO) system has been developed for localizing the site of origin of early left ventricular activation in real time using the 12-lead ECG.
The SOLO system consists of three steps: (1) localization of ventricular segment based on population templates, (2) population-based localization within a segment, and (3) patient-specific site localization.
What the study adds?
The SOLO technique-maintained accuracy in a prospective cohort study.
The SOLO system achieved estimated accuracy on the order of 10 mm, and might be useful as an advancement of conventional pace-mapping to provide clinical utility.
Sources of Funding:
Department of Medicine (DoM) of Dalhousie University to AA, Maritime Heart Centre Research Grant to AA, Cardiac Arrhythmia Network of Canada Fellowship to SZ, Heart Rhythm Society Research Fellowship to SZ, NIH R01HL142496 to NT.
Nonstandard Abbreviations and Acronyms
- VT
ventricular tachycardia
- ECGI
electrocardiographic imaging
- SOLO
site of origin localization
- CT
computed tomography
- LV
left ventricle and/or left-ventricular
- RV
right ventricle and/or right-ventricular
- 3D
three-dimensional
- EA
electroanatomic
- EP
electrophysiology
- SD
standard deviation
- RF
radiofrequency
Footnotes
Disclosures: Dr. B. Milan Horáček is a co-holder of a patent for automated VT localization, but no licensing, royalties, or income currently or anticipated. Dr. John L. Sapp is a co-holder of a patent for automated VT localization, but no licensing, royalties, or income currently or anticipated. He receives research funding from Biosense-Webster, and Abbott (for clinical trial of catheter ablation of VT), and receives a modest speaker honorarium from Medtronic, Biosense Webster, and Abbott. Dr. Amir AbdelWahab receives speaker honoraria from Medtronic and Abbott. Dr. Ciorsti J. MacIntyre receives speaker honoraria Medtronic and Abbott.
References:
- 1.Sapp JL, Wells GA, Parkash R, Stevenson WG, Blier L, Sarrazin J-F, Thibault B, Rivard L, Gula L, Leong-Sit P, et al. Ventricular tachycardia ablation versus escalation of antiarrhythmic drugs. N Engl J Med. 2016;375:111–21. [DOI] [PubMed] [Google Scholar]
- 2.Cronin EM, Bogun EM, Maury P, Perichl P, Chen M, Namboodiri N, Aguinaga L, Leite LR, Al-khatib SM, Anter E, et al. 2019 HRS/EHRA/APHRS/LHRS expert consensus statement on catheter ablation of ventricular arrhythmias. Heart Rhythm. 2019;17:e2–e154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tilz RR, Lenarczyk R, Scherr D, Haugaa KH, lliodromitis K, Purerfellner H, Kiliszek M, Dagres N. Management of ventricular tachycardia in the ablation era: results of the European Heart Rhythm Association Survey. Europace. 2018;20:209–213. [DOI] [PubMed] [Google Scholar]
- 4.Sapp JL, Bar-Tal M, Howes AJ, Toma JE, El-Damaty A, Warren JW, MacInnis PJ, Zhou S, Horacek BM. Real-time localization of ventricular tachycardia origin from the 12-lead electrocardiogram. JACC Clin Electrophysiol. 2017;3:687–699. [DOI] [PubMed] [Google Scholar]
- 5.Sapp JL, Dawoud F, Clements JC, Horacek BM. Inverse Solution Mapping of Epicardial Potentials. Circ Arrhythm Electrophysiol. 2012;5:1001–1009. [DOI] [PubMed] [Google Scholar]
- 6.Zhou S, Horacek BM, Warren JW, Sapp JL. Rapid 12-lead Automated Localization Method: Comparison to Electrocardiographic Imaging (ECGI) in Patient-specific Geometry. J Electrocardiol. 2018;51:S92–S97. [DOI] [PubMed] [Google Scholar]
- 7.Zhou S, Sapp JL, AbdelWahab A, Stovicek P, Horacek BM. Localization of ventricular activation origin using patient-specific geometry: Preliminary results. J Cardiovasc Electrophysiol. 2018;29:979–986. [DOI] [PubMed] [Google Scholar]
- 8.Duchaterau J, Sacher F, Pambrun T, Derval N, Chamorro-Servent J, Denis A, Ploux S, Hocini M, Jais P, Bernus O, et al. Performance and limitations of noninvasive cardiac activation mapping. Heart Rhythm. 2019;16:435–422. [DOI] [PubMed] [Google Scholar]
- 9.Josephson ME, Horowitz LN, Waxman HL, Cain ME, Spielman SR, Greenspan AM, Marchlinski FE, Ezri MD. Sustained ventricular tachycardia: role of the 12-lead electrocardiogram in localizing site of origin. Circulation. 1981;64:257–72. [DOI] [PubMed] [Google Scholar]
- 10.Miller JM, Marchlinski FE, Buxton AE, Josephson ME. Relationship between the 12-lead electrocardiogram during ventricular tachycardia and endocardial site of origin in patients with coronary artery disease. Circulation. 1988;77:759–66. [DOI] [PubMed] [Google Scholar]
- 11.Kuchar DL, Ruskin JN, Garan H. Electrocardiographic localization of the site of origin of ventricular tachycardia in patients with prior myocardial infarction. J Am Coll Cardiol. 1989;13:893–903. [DOI] [PubMed] [Google Scholar]
- 12.Josephson ME, Miller JM. Endocardial and epicardial recordings. Correlation of twelve-lead electrocardiograms at the site of origin of ventricular tachycardia. Ann N Y Acad Sci. 1990;601:128–47. [DOI] [PubMed] [Google Scholar]
- 13.Sippensgroenewegen A, Spekhorst H, van Hemel NM, Kingma JH, Hauer RN, de Bakker JM, Grimbergen CA, Janse MJ, Dunning AJ. Localization of the site of origin of postinfarction ventricular tachycardia by endocardial pacing mapping: Body surface mapping compared with the 12-lead ECG. Circulation. 1993;88:2290–306. [DOI] [PubMed] [Google Scholar]
- 14.Sippensgroenewegen A, Spekhorst H, van Hemel NM, Kingma JH, Hauer RN, de Bakker JM, Grimbergen CA, Janse MJ, Dunning AJ. Value of body surface mapping in localizing the site of origin of ventricular tachycardia in patients with previous myocardial infarction. J Am Coll Cardiol. 1994;24:1708–24. [DOI] [PubMed] [Google Scholar]
- 15.Potse M, Linnenbank AC, Peeters HA, SippensGroenewegen A, Grimbergen CA. Continuous localization of cardiac activation sites using a database of multichannel ECG recordings. IEEE Trans Biomed Eng. 2000;47:682–9. [DOI] [PubMed] [Google Scholar]
- 16.Segal OR, Chow AW, Wong T, Trvisi N, Lowe MD, Davies DW, Della Bella P, Packer DL, Peters NS. A novel algorithm for determining endocardial VT exit site from 12-lead surface ECG characteristics in human, infarct-related ventricular tachycardia. J Cardiovasc Electrophysiol. 2007;18:161–8. [DOI] [PubMed] [Google Scholar]
- 17.Yokokawa M, Liu TY, Yoshida K, Scott C, Hero A, Good E, Morady F, Bogun F. Automated analysis of the 12-lead electrocardiogram to identify the exit site of postinfarction ventricular tachycardia. Heart Rhythm. 2012;9:330–4. [DOI] [PubMed] [Google Scholar]
- 18.Betensky BP, Park RE, Marchlinski FE, Hutchinson MD, Garcia FC, Dixit S, Callans DJ, Cooper JM, Bala R, Lin D, Riley MP, et al. The V(2) transition ratio: a new ectrocardiographic criterion for distinguishing left from right ventricular outflow tract tachycardia origin. J Am Coll Cardiol. 2011;57:2255–62. [DOI] [PubMed] [Google Scholar]
- 19.Ito S, Tada H, Naito S, Kurosaki K, Ueda M, Hoshizaki H, Miyamori I, Oshima S, Taniguchi K, Nogami A. Development and validation of an ECG algorithm for identifying the optimal ablation site for idiopathic ventricular outflow tract tachycardia. J Cardiovasc Electrophysiol. 2003;14:1280–6. [DOI] [PubMed] [Google Scholar]
- 20.Efimova E, Dinov B, Acou WJ, Schirripa V, Kornej J, Kosiuk J, Rolf S, Sommer P, Richter S, Bollmann A, Hindricks G, Arya A. Differentiating the origin of outflow tract ventricular arrhythmia using a simple, novel approach. Heart Rhythm. 2015;12:1534–40. [DOI] [PubMed] [Google Scholar]
- 21.Li A, Davis JS, Grimster A, Wierwille J, Herold K, Morgan D, Behr ER, Shorofsky S, Saba M. Proof of concept study of a novel pacemapping algorithm as a basis to guide ablation of ventricular arrhythmias. Europace. 2018;20:1647–1656. [DOI] [PubMed] [Google Scholar]
- 22.Misra S, van Dam P, Chrispin J, Assis F, Keramati A, Kolandaivelu A, Berger R, Tandri H. Initial validation of a novel ECGI system for localization of premature ventricular contractions and ventricular tachycardia in structurally normal and abnormal hearts. J Electrocardiol. 2018;51:801–808. [DOI] [PubMed] [Google Scholar]
- 23.Odille F, Battaglia A, Hoyland P, Sellal JM, Voilliot D, de Chillou C, Felblinger J. Catheter treatment of ventricular tachycardia: a reference-less pace-mapping method to identify ablation targets. IEEE Trans Biomed Eng. 2019;66:3278–3287. [DOI] [PubMed] [Google Scholar]
- 24.Yang T, Yu L, Jin Q, Wu L, He B. Localization of Origins of Premature Ventricular Contraction by Means of Convolutional Neural Network from 12-lead ECG. IEEE Trans Biomed Eng. 2018;65:1662–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stevenson WG, Tedrow UB, Reddy V, AbdelWahab A, Dukkipati S, John RM, Fujii A, Schaeffer B, Tanigawa S, Elsokkari I, et al. Infusion needle radiofrequency ablation for treatment of refractory ventricular arrhythmias. J Am Coll Cardiol. 2019;73:1413–1425. [DOI] [PubMed] [Google Scholar]
- 26.Kemmelings JG, Linnenbank AC, Muilwijk SL, SippensGroenewegen A, Peper A, Grimbergen CA. Automatic QRS onset and offset detection for body surface QRS integral mapping of ventricular tachycardia. IEEE Trans Biomed Eng. 1994;41:830–836. [DOI] [PubMed] [Google Scholar]
- 27.Dixit S, Callans DJ. Mapping for ventricular tachycardia. Card Electrophysiol Rev. 2002;6:436–41. [DOI] [PubMed] [Google Scholar]
- 28.Stevenson WG, Friedman PL, Kocovic D, Sager PT, Saxon LA, Pavri B. Radiofrequency catheter ablation of ventricular tachycardia after myocardial infarction. Circulation. 1998;98:308–14. [DOI] [PubMed] [Google Scholar]
- 29.Bogun F, Good E, Reich S, Elmouchi D, Igic P, Lemola K, Tschopp D, Jongnarangsin K, Oral H, Chugh A, et al. Isolated potentials during sinus rhythm and pace-mapping within scars as guides for ablation of post-infarction ventricular tachycardia. J Am Coll Cardiol. 2006; 47:2013–2019. [DOI] [PubMed] [Google Scholar]
- 30.Soejima K, Stevenson WG, Maisel WH, Sapp JL, Epstein LM. Electrically unexcitable scar mapping based on pacing threshold for identification of the reentry circuit isthmus: feasibility for guiding ventricular tachycardia ablation. Circulation. 2002;106:1678–83. [DOI] [PubMed] [Google Scholar]
- 31.Széplaki G, Tahin T, Szilágyi Sz, Osztheimer I, Bettenbuch T, Srej M Merkely B, Geller L Ablation of premature ventricular complexes originating from the left ventricular outflow tract using a novel automated pace-mapping software. Interv Med Appl Sci. 2010;2:181–183. [Google Scholar]
- 32.Sapp JL, Gardner MJ, Parkash R, Basta M, Warren JW, Horack BM. Body-surface potential mapping to aid ablation of scar-related ventricular tachycardia. J Electrocardiol. 2006;39:S87–95. [DOI] [PubMed] [Google Scholar]
- 33.Cuculich PS, Schill MR, Kashani R, Mutic S, Lang A, Cooper D, Faddis M, Gleva M, Noheria A, Smith TW, et al. Noninvasive Cardiac Radiation for Ablation of Ventricular Tachycardia. N Engl J Med. 2017;377:2325–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prakosa A, Arevalo HJ, Deng D, Boyle PM, Nikolov PP, Ashikaga H, Blauer JJE, Ghafoori E, Park CJ, Blake RC 3rd, et al. Personalized virtual-heart technology for guiding the ablation of infarct-related ventricular tachycardia. Nat Biomed Eng. 2018;2:732–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou S, Sapp JL, Horacek BM, Warren JW, MacInnis PJ, Davis J, Elsokkari I, Choudhury R, Parkash R, Gray C, et al. Automated Intraprocedural Localization of Origin of Ventricular Activation Using Patient-Specific Computerized Tomography Imaging. Heart Rhythm. 2019;17:567–575. [DOI] [PubMed] [Google Scholar]


