Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infectivity is a major concern in coronavirus disease 2019 (COVID-19) prevention and economic reopening. However, rigorous determination of SARS-CoV-2 infectivity is very difficult owing to its continuous evolution with over 10,000 single nucleotide polymorphisms (SNP) variants in many subtypes. We employ an algebraic topology-based machine learning model to quantitatively evaluate the binding free energy changes of SARS-CoV-2 spike glycoprotein (S protein) and host angiotensin-converting enzyme 2 receptor following mutations. We reveal that the SARS-CoV-2 virus becomes more infectious. Three out of six SARS-CoV-2 subtypes have become slightly more infectious, while the other three subtypes have significantly strengthened their infectivity. We also find that SARS-CoV-2 is slightly more infectious than SARS-CoV according to computed S protein-angiotensin-converting enzyme 2 binding free energy changes. Based on a systematic evaluation of all possible 3686 future mutations on the S protein receptor-binding domain, we show that most likely future mutations will make SARS-CoV-2 more infectious. Combining sequence alignment, probability analysis, and binding free energy calculation, we predict that a few residues on the receptor-binding motif, i.e., 452, 489, 500, 501, and 505, have high chances to mutate into significantly more infectious COVID-19 strains.
Keywords: COVID-19, viral infectivity, spike protein, mutation, protein-protein interaction
Abbreviations: COVID-19, coronavirus disease 2019; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; ACE2, angiotensin-converting enzyme 2; RBD, receptor-binding domain; PPI, protein–protein interaction; BFE, binding free energy; RBM, receptor-binding motif
Graphical abstract
More than 8000 observed single mutations in the SARS-CoV-2 genomes have raised serious concerns about changes in infectivity. Qualitatively, such infectivity is proportional to the binding affinity between SARS-CoV-2 spike glycoprotein (S protein) and host ACE2 receptor. This work proposes a machine learning model to evaluate the relative infectivity following the mutations. We show that five out of six SARS-CoV-2 substrains have become more infectious, while the other one becomes less infectious. We found that a few potential future mutations on the S protein could lead to more dangerous new viruses.
Highlights
-
•
SARS-CoV-2 has had many mutations and evolved into six subtypes.
-
•
Three SARS-CoV-2 subtypes have significantly strengthened their infectivity.
-
•
A few future mutations have high chances to produce more contagious viruses.
Introduction
In December 2019, an outbreak of pneumonia due to coronavirus disease 2019 (COVID-19) was initially detected in Wuhan, China [1], due to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). It has now spread globally via travelers and breached the boundaries of 213 countries and territories, leading to more than 12 million infection cases and 553,000 deaths as of July 7, 2020. In the past two decades, there have been three major zoonotic disease outbreaks of betacoronaviruses: SARS-CoV in 2002, Middle East respiratory syndrome coronavirus in 2012, and SARS-CoV-2 in 2019. Similar to SARS-CoV and Middle East respiratory syndrome coronavirus, SARS-CoV-2 infections were observed in hospital personnel and family clusters in the early stages of the outbreak [2., 3., 4.]. SARS-CoV-2 is an enveloped non-segmented positive-sense RNA virus and belongs to the betacoronavirus genus. Although intensive investigation, the origin of SARS-CoV-2 remains elusive. Unfortunately, there are no specific antivirus drugs or effective vaccines developed to moderate this outbreak at present.
SARS-CoV-2 has undergone more than 10,000 recorded single mutations compared to the reference genome collected on January 5, 2020 [5,6]. In general, RNA viruses, except for Nidoviruses, are prone to random mutations because of the lack of the exonuclease proofreading activity of the virus-encoded RNA polymerases, RNA viruses. Nidoviruses, including coronaviruses, have an enzyme to excise erroneous mutagenic nucleotides inserted by RNA polymerases and thus maintain a relatively high accuracy in virus replication and transcription [7]. Human immune system intervention introduces viral mutations. Rapid global spread and transmission of COVID-19 provides the virus with substantial opportunities for the natural selection of rare-acted but favorable mutations. Although most viral mutations are benign, many mutations, such as D614G on the spike (S) protein, strengthen viral survival capability [8,9]. It is of paramount importance to understand SARS-CoV-2 infectivity changes following the existing mutations and predict the future infection tendency.
It is well known that like SARS-CoV, SARS-CoV-2 enters host cells through the interaction of spike glycoprotein (S protein) and host angiotensin-converting enzyme 2 (ACE2) receptor [10., 11., 12.]. In both SARS-CoV and SARS-CoV-2, the S protein receptor-binding domain (RBD) is recognized as on the S1 unit to bind directly to the ACE2. Compared to SARS-CoV, SARS-CoV-2 S protein harbors a furin cleavage site at the boundary between the S1/S2 subunits [6]. However, lessons learned from SARS-CoV are important in formulating hypotheses about SARS-CoV-2, as well as the receptor recognition when studying SARS-CoV-2 host range, cross-species transmission, and pathogenesis. In the studies of SARS-CoV, epidemiologic and biochemical studies show that the infectivity of different SARS-CoV strains in host cells is proportional to the binding free energy (BFE) between the RBD of each strain and the ACE2 expressed by the host cell [10,12., 13., 14., 15.]. Therefore, the assessment of BFE changes following mutations is vital for the understanding of SARS-CoV-2 infectivity evolution.
It is challenging to rigorously measure the relative viral infectivities of two dangerous viruses by experiments when one of them is evolving. There is a discrepancy in the literature about the relative S protein-ACE2 binding free energies of SARS-CoV and SARS-CoV-2. Wrapp et al. and Shang et al. reported that SARS-CoV-2 has a higher BFE than SARS-CoV does [16,17], whereas Walls et al. argued that SARS-CoV-2 and SARS-CoV bind with similar free energies to ACE2 [12].
The first SARS-CoV-2 genome reported on January 5, 2020 [6] has about 80% sequence identity with that of SARS-CoV. However, compared with SARS-CoV, SARS-CoV-2 S protein has 725 mutations over its 1255 residues. Their sequence identity is only 76%. Among 725 mutations on SARS-CoV-2 S protein, 89 were on the RBD, which has a total of 194 residues, suggesting that the RBD is subject to more mutations. Our recent studies using over 15,000 genome samples show that SARS-CoV-2 S protein is among the most non-conservative ones in its genome [5]. Since early January 2020, hundreds of new mutations were found on different residue positions of SARS-CoV-2 S protein. Many of them are located on the RBD [5]. The existence of so many different S protein mutations indicates that there are many different SARS-CoV-2 subtypes that might have very different infectivities. Obviously, the relatively high mutation rate at the RBD poses a real threat to the occurrence of future SARS-CoV-2 strains that might be more infectious than the current SARS-CoV-2.
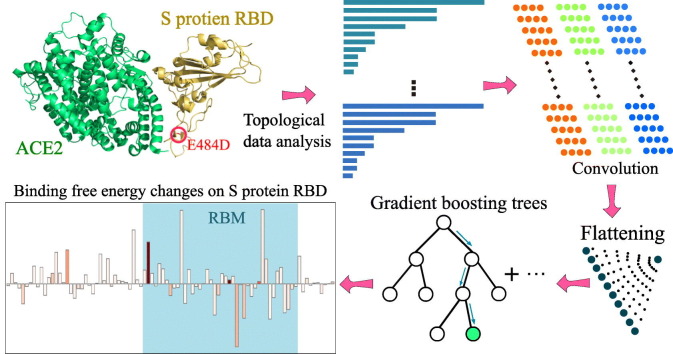
The computational estimation of mutation-induced protein–protein BFE changes is an important approach for understanding the impact of mutations on protein–protein interactions (PPIs). A variety of advanced methods have been developed [18., 19., 20., 21., 22., 23.]. There are many standard databases available, including the AB-Bind database of mutation-induced antibody–antigen complex BFE changes [24] and SKEMPI for protein–protein BFE changes upon mutation (∆∆G) [25,26]. These databases have been used as benchmarks for evaluating the predictive power of various computational methods [18., 19., 20., 21., 22., 23.]. To simplify the structural complexity of PPI complexes, we have recently introduced element-specific and site-specific persistent homology, a new branch of algebraic topology [27,28], to embed molecular mechanisms into topological invariants [29]. This approach was paired with a new deep learning algorithm called NetTree, to combined convolutional neural networks and gradient boosting trees. The resulting method, called TopNetTree, was about 22% better than the previous best result for the AB-Bind dataset and significantly outperformed the state-of-the-art in the literature on the SKEMPI database [29].
The objective of this work is 3-fold. First, we apply the TopNetTree to analyze the impacts of existing S protein RBD mutations on the BFE of the S protein and the ACE2. Since different SARS-CoV-2 subtypes have different mutation patterns, it is important to understand their mutation impacts accordingly. We carry out our analysis based on existing six mutation clusters [5], though more specific analysis can be easily done as well. Additionally, it is also extremely important to know whether future SARS-CoV-2 subtypes would pose an imminent danger to public health. To this end, we have conducted a systematic screening of all possible 3,686 future mutations on all 194 residues (residue IDs from 333 to 526 on S protein) on the RBD. We classify these mutations into three categories: the most likely ones that would happen by a single mutation at any one of three constitutive nucleotides; the likely mutations that would occur via two concurrent mutations at three constitutive nucleotides; and the unlikely mutations that would produce through three concurrent mutations at all of three constitutive nucleotides. Finally, we analyze how existing mutations on the RBD of the SARS-CoV-2 S protein with respect to SARS-CoV have changed its infectivity.
Results
Impacts of existing RBD mutations
Global analysis
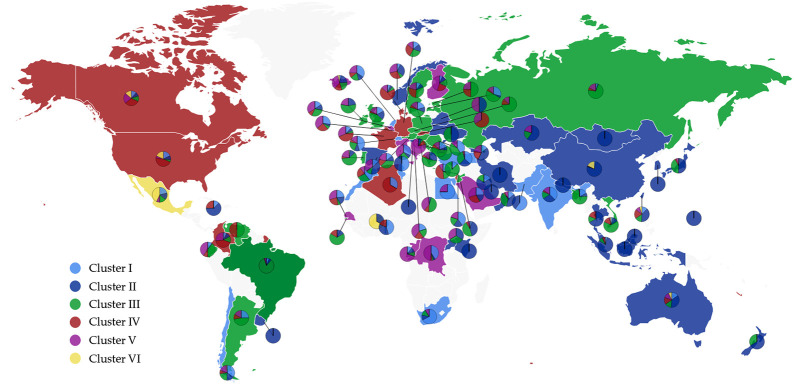
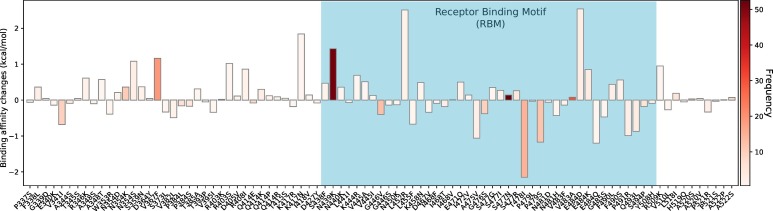
To investigate the influences of existing S protein RBD mutations on BFE of S protein and ACE2, the 24715 complete SARS-CoV-2 genome samples deposited at GISAID [30] are compared with the first genome sequence of SARS-CoV-2 collected on January 5, 2020 [6]. The resulting 11904 single mutations are found in six distinct clusters as shown in Table 1 [5] and Figure 1 . There are 725 existing non-degenerated mutations on SARS-CoV-2 S protein. Among them, 89 mutations occurred on the RBD, which are relevant to the binding of SARS-CoV-2 S protein and ACE2. Furthermore, 52 out of 89 mutations are on the receptor-binding motif (RBM), i.e., the region of RBD that is in direct contact with the ACE2.
Table 1.
The cluster distributions of samples (NNS) and total mutation counts (NTF) for 17 countries [5]
| Cluster I |
Cluster II |
Cluster III |
Cluster IV |
Cluster V |
Cluster VI |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Country | NNS | NTF | NNS | NTF | NNS | NTF | NNS | NTF | NNS | NTF | NNS | NTF |
| US | 1171 | 13,725 | 603 | 4154 | 309 | 2937 | 2996 | 23,445 | 310 | 2014 | 1191 | 8744 |
| CA | 38 | 373 | 27 | 235 | 38 | 359 | 105 | 789 | 73 | 454 | 44 | 265 |
| AU | 188 | 2247 | 452 | 4072 | 173 | 1562 | 189 | 1353 | 152 | 795 | 89 | 604 |
| UK | 1353 | 14,667 | 1485 | 10,140 | 3362 | 33,196 | 257 | 2160 | 1331 | 8074 | 4 | 26 |
| IS | 14 | 119 | 89 | 474 | 89 | 870 | 71 | 482 | 147 | 884 | 15 | 127 |
| ES | 55 | 493 | 161 | 1192 | 45 | 392 | 5 | 43 | 138 | 850 | 2 | 6 |
| CN | 8 | 69 | 210 | 944 | 3 | 31 | 1 | 7 | 3 | 11 | 50 | 129 |
| DE | 22 | 171 | 25 | 121 | 43 | 369 | 44 | 298 | 22 | 110 | 0 | 0 |
| FR | 28 | 284 | 14 | 55 | 12 | 105 | 109 | 808 | 74 | 465 | 0 | 0 |
| IN | 313 | 3834 | 239 | 2698 | 131 | 1477 | 28 | 210 | 126 | 752 | 0 | 0 |
| RU | 8 | 68 | 2 | 32 | 122 | 1041 | 9 | 72 | 30 | 171 | 0 | 0 |
| BE | 108 | 978 | 79 | 356 | 201 | 1939 | 64 | 491 | 229 | 1376 | 1 | 3 |
| SA | 14 | 126 | 9 | 61 | 1 | 7 | 16 | 110 | 17 | 133 | 0 | 0 |
| IT | 41 | 687 | 8 | 104 | 32 | 304 | 0 | 0 | 47 | 232 | 0 | 0 |
| JP | 9 | 79 | 243 | 998 | 206 | 1858 | 18 | 134 | 23 | 139 | 0 | 0 |
| TR | 41 | 385 | 28 | 339 | 37 | 335 | 2 | 17 | 3 | 20 | 0 | 0 |
| KR | 0 | 0 | 48 | 272 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
The listed countries are United States (US), Canada (CA), Australia (AU), United Kingdom (UK), Iceland (IS), Spain (ES), China (CN), Germany (DE), France (FR), India (IN), Russia (RU), Belgium (BE), Saudi Arabia (SA), Italy (IT), Japan (JP), and Turkey (TR), and Korean (KR).
Figure 1.
The scatter plot of six distinct clusters in the world. The light blue, dark blue, green, red, pink, and yellow represent Cluster I, Cluster II, Cluster III, Cluster IV, Cluster V, and Cluster VI, respectively. The color of the dominated cluster decides the base color of each country. The world map is generated by the Highcharts.
We examine the BFE changes following the existing site-specific mutations. Our studies are based on the X-ray crystal structure of SARS-CoV-2 S protein and ACE2 (PDB 6M0J) [31] (see Figure S1), whose S protein gene sequence is consistent with that of the reference SARS-CoV-2 [6]. The BFE change following mutation (∆∆G) is defined as the subtraction of the BFE of the mutant from the BFE of wild-type, ∆∆G = ∆G W − ∆ G M, where ∆G W is BFE of the wild-type and ∆G M is BFE of mutant type. Therefore, a positive BFE change means that the mutation increases free energy of the binding complex, making the virus more infective.
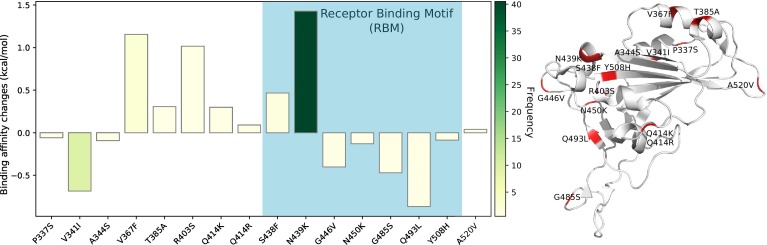
We present the overall BFE changes ∆∆G of SARS-CoV-2 S protein RBD in Figure 2 . Most mutations have small changes in their binding free energies, while some of them have large changes. There are 54% mutations on the RBD having positive BFE changes (i.e., 48 over 89), including N439L and S477N that have the highest frequencies. This statistic implies that the evolution of SARS-CoV-2 is mostly driven by selection and COVID-19 evolves toward more infectious. It is noted that many mutations on the RBM, such as N439K, L452R, T478I, and E484D, have significant free energy changes. The mutations on the RBM take 58% (52 over 89) of all mutations on the RBD, which potentially increases the complexity of antiviral drug and vaccine development. This global analysis indicates that mutations on the RBD strengthen the binding of S protein and ACE2, leading to more infectious SARS-CoV-2.
Figure 2.
Overall BFE changes ∆∆G on the RBD. The blue color region marks the BFE changes on the RBM. The height of each bar indicates the predicted ∆∆ G. The color indicates the occurrence frequency in the GISAID genome dataset.
We hypothesize that natural selection favors those mutations that enhance the viral transmission and if our predictions are correct, the predicted infectivity strengthening mutations will outpace predicted infectivity weakening mutations over time. Figure 3 illustrates the increase in the frequency of each mutation on the SARS-CoV-2 S protein RBD. The red and blue lines represent the mutations that strengthen and weaken the infectivity of SARS-CoV-2, respectively. In the first 2 months, only a few infectivity-strengthening mutations were detected on the S protein RBD. Later on, a few infectivity-weakening mutations gradually appeared, while more infectivity-strengthening mutations occurred. It is interesting to note that overall, infectivity-strengthening mutations grow faster than infectivity-weakening mutations, which also reveals that SARS-CoV-2 subtypes having infectivity-strengthening mutations are able to infect more people. Specifically, frequencies of S477N, N439K, V483A, and V367F are higher than those of other mutations, indicating these mutations have a stronger transmission capacity.
Figure 3.
The time evolution of 89 SARS-CoV-2 S protein RBD mutations. The red lines represent the mutations that strengthen the infectivity of SARS-CoV-2 (i.e., ∆∆ G is positive), and the blue lines represent the mutations that weaken the infectivity of SARS-CoV-2 (i.e., ∆∆ G is negative). Many mutations overlap their trajectories. Here, the collection date of each genome sequence that deposited in GISAID is applied.
The SARS-CoV-2 genotypes are clustered into six clusters or subtypes based on their single nucleotide polymorphism variants [5]. Accordingly, a more detailed analysis of mutation impacts on the BFE changes can be carried out on each cluster, which reveals the diversity of COVID-19 infection rates and provides evidence for transmission pathways and spread dynamics across the world.
It is worth noting that residue 414 has three mutations, namely, Q414P, Q414E, and Q414R, due to mutations at two adjacent nucleotides 22802 and 22803: 22803A > C, Q414P; 22802C > G, Q414E; and 22803A > G, Q414R. At the protein level, some or all of these mutations show up in different clusters. Similarly, each of residues 354 and 521 has two existing mutations.
Cluster I analysis
Figure 4 depicts the BFE changes ∆∆G of Cluster I. There are 47 mutations on Cluster I, where 20 mutations happen in RBM. Particularly, mutation S477N has a higher frequency of 18 and a positive free energy changes. Other two mutations, N439K and P479S, which have slightly higher frequencies, 11 and 12, and large free energy change amplitudes, attained positive and negative BFE changes, respectively. Cluster I has a slightly increase in its infectivity. Cluster I is associated with COVID-19 in most countries except for South Korea [5].
Figure 4.
Cluster I. Left: BFE changes ∆∆G induced by mutations in Cluster I. Right: mutation locations on the SARS-CoV-2 S protein RBD.
Cluster II analysis
Figure 5 illustrates the BFE changes following the mutations of Cluster II. As shown in the figure, there are many mutations on the RBD. However, most mutations are associated with small free energy changes. When only considering the absolute value of BFE changes greater than or equal to 0.5 kcal/mol, seven mutations have positive BFE changes, whereas four mutations have negative BFE changes. Mutation D364Y has the highest frequency and the highest positive free energy change, indicating the increase in infectivity. Overall, Cluster II has a minor increase in infectivity. Note that Cluster II COVID-19 is found in every country that has submitted SARS-CoV-2 genome samples.
Figure 5.
Cluster II. Left: BFE changes ∆∆G induced by mutations in Cluster II. Right: mutation locations on the SARS-CoV-2 S protein RBD.
Cluster III analysis
From Figure 6 , a slightly increasing trend of BFE is observed such that the largest change is positive while the second largest change is negative. Notice that mutation S477N is observed increasing of its frequency in Figure 3. Interestingly, mutation T478I changes from amino acid with polar uncharged side chains, Threonine, to an amino acid with hydrophobic side chain, Isoleucine, which significantly decreases free energy between the S protein and the ACE2 receptor. Another observation is that mutations that happened in the same residues have similar BFE changes such as P384L and P384S, S477R and S477N, or Q414P and Q414R. Cluster III has a minor increase in infectivity. This cluster involves genome samples from all countries except for South Korea. Notably, most Cluster III samples were submitted by the UK.
Figure 6.
Cluster III. Left: BFE changes ∆∆G induced by mutations in Cluster III.
Right: mutation locations on the SARS-CoV-2 S protein RBD.
Cluster IV analysis
Figure 7 shows the BFE changes of mutations in Cluster IV. Among all mutations in Cluster IV, mutation L452R has the largest free energy change and directly connects the ACE2 receptor. The most frequent mutation of negative change, Q414E, has the BFE change ∆∆G is −0.055 kcal/mol, which is negligible compared with others. The overall trend of this cluster is considered as significantly increasing the COVID-19 infectibility. Note that most genome samples in Cluster IV were submitted by the US.
Figure 7.
Cluster IV. Left: BFE changes ∆∆G induced by mutations in Cluster IV.
Right: mutation locations on the SARS-CoV-2 S protein RBD.
Cluster V analysis
Figure 8 presents the BFE changes in the fifth cluster. Eight of 16 mutations have positive free energy changes. Interestingly, the mutation, N439K, which has the second highest frequency among all clusters, also has the largest BFE change in Cluster V. In Figure 3, it indicates that this mutation happens rapidly from Apr 3 to Apr 13. Moreover, most samples in this cluster were submitted from the US. It can be considered that Cluster V and N439K play a vital role in virus spreading in the US. This cluster is considered as significantly increasing in its infectibility.
Figure 8.
Cluster V. Left: BFE changes ∆∆G induced by mutations in Cluster V. Right: mutation locations on the SARS-CoV-2 S protein RBD.
Cluster VI analysis
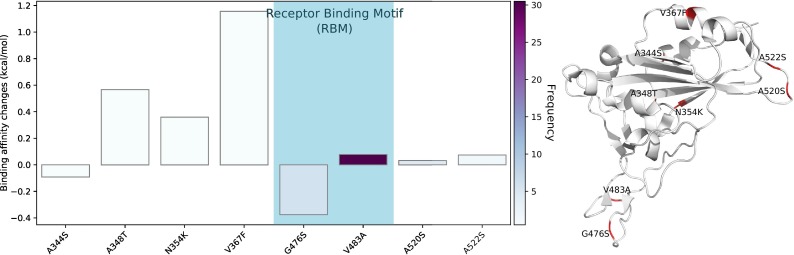
The BFE changes in the last cluster are shown in Figure 9 . Obviously, most mutations on Cluster VI have enhanced the BFE of the S protein and ACE2 receptor except A344S and G476S. The most significant positive free energy change is caused by mutation V367F. Overall, Cluster VI has strengthened infectivity. This cluster involves genome samples submitted from all countries except for Japan and South Korea. The US has submitted most samples. Cluster VI is a new cluster of SARS-CoV-2, and its mutations have relatively low frequencies.
Figure 9.
Cluster VI. Left: BFE changes ∆∆G induced by mutations in Cluster VI. Right: mutation locations on the SARS-CoV-2 S protein RBD.
In summary, three of six SARS-CoV-2 clusters, Clusters I, II, and III, have slightly increase infectivity, while other three clusters, IV, V, and VI, have become significantly more infectious. Please note that SARS-CoV-2 is evolving and its clustering is changing over time. As a result, the mutation pattern in each cluster changes too. However, the general tendency does not change: the evolution of SARS-CoV-2 makes it more infectious by adapting mutations that increase the BFE with the ACE2 receptor.
Impacts of most likely future RBD mutations
In this section and the next section, we analyze the impacts of all of 3686 possible mutations on 194 residues of the S protein RBD. On each amino acid, we classify all 19 possible mutations into most likely future mutations, likely future mutations, and unlikely future mutations. Here, most likely, likely, and unlikely future mutations are defined by the protein mutations induced by only one, simultaneous two, and simultaneous three of genetic changes on three underlying nucleotides on a codon. Based on the codon analysis of all 194 amino acid residues on the RBD, we have 1149 most likely, 1912 likely, and 625 unlikely mutations. We note that the above definitions include existing mutations.
We compute the ∆∆Gs following most likely future mutations on the RBD. Figure 10 depicts 20 most likely future mutations that can have high adversarial impacts on COVID-19 infectivity. First, it is noted that mutation Y495N on the RBM has the highest free energy change and if it occurs, it will make the virus significantly more infectious. Additionally, mutation Y489H on the RBM would incur another large infectivity strengthening. It is worthy to note that residue 489 is a potentially hot spot, where five possible mutations, Y489H, Y489D, Y489F, Y489C, and Y489N, will lead to the strengthened S protein-ACE2 binding. The other potentially hot spot is residue 423 with Y423C, Y423F, and Y423S being infectivity-strengthening mutations. Residue 452 on the RBM has been proven to be a hot spot as it already has had an existing mutation L452R (see Figure 7) and another infectivity strengthening mutation, L452P. In general, the highest free energy changes are due to mutations on the RBM. However, mutations away from the RBM can have a considerable impact on the infectivity as well.
Figure 10.
Top 20 most likely future mutations that will strengthen the SARS-CoV-2 infectivity. Left: BFE changes ∆∆G. Right: mutations on the RBD. Red color indicates mutations on the RBM and blue color indicates mutations away from the RBM.
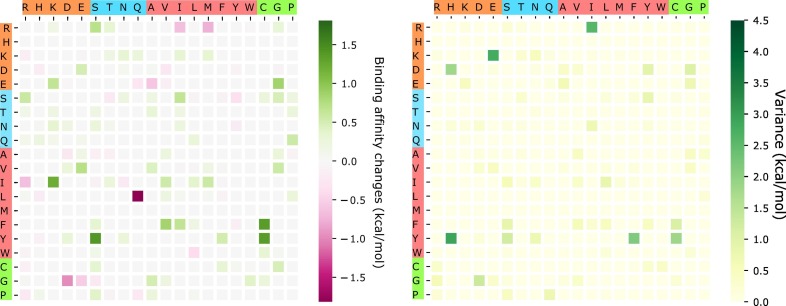
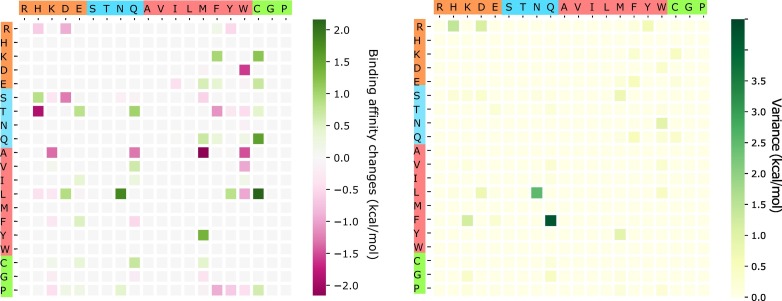
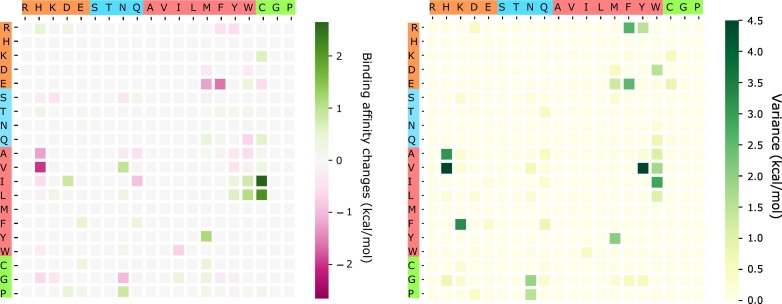
The above analysis considers only BFE strengthening mutations. To have a global view of how future mutations would change the COVID-19 infectivity, we analyze the general trend of the free energy changes of most likely mutations according to 400 possible mutation types. The ∆∆G values following mutations on each amino acid are predicted and averaged by their mutation types. Figure 11 shows the average and variance of ∆∆G (kcal/mol) of each mutation type for most likely mutations on the RBD. Here, y-axes stand for wild-type residues, and x-axes are mutant type residues. The colors on the axes are the residue types, namely, charged, polar uncharged, hydrophobic, and special cases. The colors in the heat maps indicate the BFE changes strengths and directions. It is worthy to note that there are more positive BFE changes (green cubes) than negative changes (pink cubes) on the heat map, showing a trend of more infectious COVID-19 strains due to most likely future mutations. For example, if a wild-type mutation takes place from wide type K, T, N, Q, L, F, or Y to any other residue type except for W, it will end up with a more infectious COVID-19 strain. However, mutations from R to T, or from A to many other residue types might lead to a less infectious COVID-19. The large values on the variance map indicate where the above average values might not be reliable. It is seen that the variances are general small. Figure 12 shows a similar trend for the most likely mutations away from the RDM.
Figure 11.
An illustration of the average and variance of ∆∆G (kcal/mol) for most likely mutation types on the RBM. y-axes: wild-type residues; x-axes: mutant type residues. Colors on the axes indicate residue types.
Figure 12.
An illustration of the average and variance of ∆∆G (kcal/mol) for most likely mutation types away from the RBM. y-axes: wild-type residues; x-axes: mutant type residues. Colors on the axes indicate residue types.
Impacts of likely and unlikely future RBD mutations
As discussed above, likely and unlikely future mutations require two and three concurrent nucleotide mutations on each codon to happen, respectively. Figure 13 presents the top 20 likely future mutations that will strengthen the COVID-19 infectivity. The most energetic adversarial mutation is Y505G. Note that residue 505 has a most likely mutation Y505H shown in Figure 10. Therefore, residue 505 is a potentially hot spot on the RBM. The next few energetic adversarial mutations are away from the RBM. Among them, N423P and N422G are hot-spot mutations. Figure 10 shows that residue 423 has three most likely energetic mutations while in Figure 13, it has the other three likely energetic mutations. Similarly, residue 489 on the RBM has five most likely energetic mutations (see Figure 10) and four likely energetic mutations as shown in Figure 13. It is on our top surveillance list for the next generation of infectious COVID-19 strains. Another potentially hot spot is residue 495. Figure 14 shows the average and variance of ∆∆G (kcal/mol) of all likely mutations on the RBM where values of most likely mutations and unlikely mutations are excluded. About the same amount of mutations has positive and negative BFE changes. In Figure 15 , similar results are shown for the RBD, excluding the RBM. Interestingly, mutations on the RBM have larger magnitude changes rather than out of this region for second potential mutations. It again shows that RBM is the most important region to study.
Figure 13.
Top 20 likely future mutations that will strengthen the COVID-19 infectivity. Left: BFE changes.
∆∆G. Right: mutations on the RBD. Red color indicates mutations on the RBM and blue color indicates mutations away from the RBM.
Figure 14.
An illustration of the average and variance of ∆∆G (kcal/mol) for likely mutation types on the RBM. y-axes: wild-type residues; x-axes: mutant type residues. Colors on the axes indicate residue types.
Figure 15.
An illustration of the average and variance of ∆∆G (kcal/mol) for likely mutation types away from the RBM. y-axes: wild-type residues; x-axes: mutant type residues. Colors on the axes indicate residue types.
Figure 16, Figure 17 show the predictions of free energy changes due to unlikely mutations. These mutations have a balanced positive and negative BFE changes. We do not expect these mutations to occur in the near future.
Figure 16.
An illustration of the average and variance of ∆∆G (kcal/mol) for unlikely mutation types on the RBM. y-axes: wild-type residues; x-axes: mutant type residues. Colors on the axes indicate residue types.
Figure 17.
An illustration of the average and variance of ∆∆G (kcal/mol) for most likely mutation types away from the RBM. y-axes: wild-type residues; x-axes: mutant type residues. Colors on the axes indicate residue types.
Discussion
Conservation analysis via sequence alignment
To further understand the evolutionary trend and potential infectivity changes of the COVID-19, we carry out S protein sequence alignment analysis to analyze residue conservativeness. Figure 18 presents the alignment analysis of SARS-CoV-2 S protein sequence and those of the other four closely related species, namely, SARS-CoV [32], bat coronavirus RaTG13 [33], bat coronavirus BM48-31 [34], and bat coronavirus CoVZC45 [35]. We note that among the residues we discussed in the last section, 414, 422, 423, 492, and 495 are very conservative. They have not undergone any mutations among five related species, although 414 has three confirmed mutations with small BFE changes as shown in Figure 2. In contrast, RBM residues 452, 489, 500, 501, and 505 have a history of mutations and are non-conservative. Therefore, the predicted infectivity-strengthening mutations on these residues are more likely to happen.
Figure 18.
Sequence alignments of SARS-CoV-2 S protein with those of closely related species, including SARS-CoV [32], bat coronavirus RaTG13 [33], bat coronavirus BM48–31 [34], and bat coronavirus CoVZC45 [35]. Detailed numbering is given according to SARS-CoV-2. Residue 364 Ala (A) of bat coronavirus BM48–31 is omitted.
Relative infectivity change analysis for SARS-CoV and SARS-CoV-2
As mentioned earlier, there are inconsistent assessments about relative infectivities between SARS-CoV and SARS-CoV-2 in the literature [12,16,17]. Our validated computational method can be employed to resolve this discrepancy. Based on the sequence alignment shown in Figure 18, we conduct BFE change calculations for all relevant mutations on the SARS-CoV S protein RBD. Figure 19 illustrates the S protein-ACE2 BFE changes following the mutations from SARS-CoV to SARS-CoV-2. The SARS-CoV S protein and ACE2 complex 3D0G [36] is used as the wide type in our predictions. It is interesting to note that overall, there are more infectivity-strengthening mutations than infectivity-weakening mutations on the RBD. This is particularly true for mutations on the RBM. This result indicates that the SARS-CoV-2 sample collected on January 5, 2020 [6], is slightly more infectious than SARS-CoV found in 2003 [32]. This is consistent with experimental results that the binding free energies of ACE2 with the spike proteins of SARS-CoV-2 and SARS-CoV are 1.2 and 5.0 nM, respectively [12].
Figure 19.
Overall BFE changes ∆∆G on the S protein RBD from SARS-CoV to SARS-CoV-2. The blue color region marks the BFE changes on the RBM. The height of each bar indicates the predicted ∆∆G. Residues are labeled according to PDB ID 3D0G [36].
For a comparison between various SARS-CoV-2 subtypes and SARS-CoV of 2003, our results indicate that SARS-CoV-2 in all clusters is more infectious than SARS-CoV.
Compared with SARS-CoV, SARS-CoV-2 has four extra residues, i.e., PRRA from 681 to 684, as shown in Figure 18. It is believed that these extra residues might change SARS-CoV-2's behavior in ACE2 assisted entry of host cells [12]. However, this speculation has no qualitative nor quantitative validation at present. Experiments show that S protein mutation D614G also has made SARS-CoV-2 more infectious [9].
Materials and Methods
Sequences and structures
Amino acid sequences and mutant data of the S protein used in the analysis were obtained from NCBI GenBank and GISAID [30]. After the first complete genome sequence of SARS-CoV-2 released on NCBI GenBank (accession number: NC 045512.2) [6], there has been a large number of genome sequences. Other sequences from GenBank are as follows: bat coronavirus RaTG13 (MN996532.1) [33], bat coronavirus BM48-31 (NC 014470.1) [34], and bat coronavirus CoVZC45 (MG772933.1) [37]. The mutant information of 15140 whole-genome sequences of S protein with high coverage of SARS-CoV-2 strains from the infected individuals around the world was obtained from the GISAID database [30] (https://www.gisaid.org/). Data without the exact submission date in GISAID were not considered. Sequence analysis and k-means clustering were described in detail elsewhere [5].
Beta-CoV S protein structures were obtained from the RCSB Protein Data Bank: SARS-CoV RBD with ACE2 (PDB 3D0G) [36] and SARS-CoV-2 RBD with ACE2 (PDB 6M0J) [31]. The structures were presented by using PyMOL [38]. Sequences alignments were performed on SARS-CoV-2 S protein sequences by using MAFFT v7.388 [37] and on SARS-CoV-2 genome by using the Clustal Omega multiple sequence alignment with default parameters [39].
TopNetTree model for PPI BFE changes upon mutation
The topology-based network tree (TopNetTree) was constructed by an innovative integration between the topological representation and NetTree for predicting PPI BFE changes following mutation ∆∆G [29]. In this work, TopNetTree is applied to predict the BFE changes of mutations that happened on the RBD with ACE2 of SARS-CoV-2 after January 5, 2020. As shown in Figure S1, topology-based feature generation is the first step followed by a convolutional neural network-assisted model. The topological representation uses element- and site-specific persistent homology to simplify the structural complexity of protein–protein complexes and encode vital biological information into topological invariants [40]. NetTree is a recently developed deep learning algorithm that integrates the advantages of convolutional neural networks and gradient-boosting trees [29]. Details of the method can be found in the literature [29]. New parametrization and validation are given in the Supporting Material.
Conclusion
The infectivity of SARS-CoV-2 is a vital factor for preventive measurements against COVID-19 and reopening the global economy [12,16,17]. However, it is very challenging to rigorously determine the viral infectivity of all SARS-CoV-2 substrains experimentally. These challenges are deteriorated by the continuous evolution of SARS-CoV-2 due to its over 10,000 single nucleotide polymorphisms variants in various distinct clusters [5]. In the present work, we employ an advanced TopNetTree method based on algebraic topology and deep learning to predict the spike glycoprotein (S protein) and the host ACE2 receptor BFE changes induced by mutations. Based on BFE changes, we reveal that mutations have made all clusters of SARS-CoV-2 more infectious than the original virus found in Wuhan [6]. Additionally, based on sequence alignment and mutation-induced BFE changes, we show that SARS-CoV-2 [6] is slightly more infectious than SARS-CoV found in 2003 [32]. This result is consistent with experiments [12]. Finally, we systematically compute the BFE changes of all possible 3686 future mutations to unveil that the most likely mutations will further strengthen SARS-CoV infectivity. Based on sequence alignment, probability estimation, and BFE analysis, we predict that residues 452, 489, 500, 501, and 505 on the RBM have high chances to mutate into significantly more infectious COVID-19 strains.
Acknowledgment
This work was supported in part by National Institutes of Health grant GM126189; NSF Grants DMS-1721024, DMS-1761320, and IIS1900473; Michigan Economic Development Corporation; Bristol-Myers Squibb; and Pfizer. The authors thank The IBM TJ Watson Research Center, The COVID-19 High Performance Computing Consortium, NVIDIA, and MSU HPPC for computational assistance. R.W. thanks Dr. Changchuan Yin for assistance.
Edited by Anna Panchenko
Footnotes
Supplementary Materials are available for S1: The new parametrization and validation of the TopNetTree model and S2: Supplementary tables. Supplementary data to this article can be found online at https://doi.org/10.1016/j.jmb.2020.07.009.
Appendix A. Supplementary data
Supplementary Materials are available for S1: The new parameterization and validation of the TopNetTree model and S2: Supplementary tables for the results in figures.
References
- 1.C. Huang, Y. Wang, X. Li, L. Ren, J. Zhao, Y. Hu, L. Zhang, G. Fan, J. Xu, X. Gu, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet, 395(10223):497–506, 2020. [DOI] [PMC free article] [PubMed]
- 2.J. F.-W. Chan, S. Yuan, K.-H. Kok, K. K.-W. To, H. Chu, J. Yang, F. Xing, J. Liu, C. C.-Y. Yip, R. W.-S. Poon, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet, 395(10223):514–523, 2020. [DOI] [PMC free article] [PubMed]
- 3.Gralinski L.E., Menachery V.D. Return of the coronavirus: 2019-nCoV. Viruses. 2020;12(2):135. doi: 10.3390/v12020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang R., Hozumi Y., Yin C., Wei G.-W. Decoding SARS-CoV-2 transmission and evolution and ramifications for COVID-19 diagnosis, vaccine, and medicine. J. Chem. Inf. Model. 2020 doi: 10.1021/acs.jcim.0c00501. 32530284 in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferron F., Subissi L., De Morais A.T.S., Le N.T.T., Sevajol M., Gluais L., Decroly E., Vonrhein C. Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc. Natl. Acad. Sci. 2018;115(2):E162–E171. doi: 10.1073/pnas.1718806115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Korber B., Fischer W., Gnanakaran S.G., Yoon H., Theiler J., Abfalterer W., Foley B., Giorgi E.E. Spike mutation pipeline reveals the emergence of a more transmissible form of SARS-CoV-2. bioRxiv. 2020 preprint article. [Google Scholar]
- 9.Zhang L., Jackson C.B., Mou H., Ojha A., Rangarajan E.S., Izard T., Farzan M., Choe H. The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity. bioRxiv. 2020 preprint article. [Google Scholar]
- 10.M. Hoffmann, H. Kleine-Weber, S. Schroeder, N. Kru¨ger, T. Herrler, S. Erichsen, T.S. Schiergens, G. Herrler, N.-H. Wu, A. Nitsche, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell, 2020. [DOI] [PMC free article] [PubMed]
- 11.Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309(5742):1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 12.Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310(5748):676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 14.Qu X.-X., Hao P., Song X.-J., Jiang S.-M., Liu Y.-X., Wang P.-G., Rao X., Song H.-D. Identification of two critical amino acid residues of the severe acute respiratory syndrome coronavirus spike protein for its variation in zoonotic tropism transition via a double substitution strategy. J. Biol. Chem. 2005;280(33):29588–29595. doi: 10.1074/jbc.M500662200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song H.-D., Tu C.-C., Zhang G.-W., Wang S.-Y., Zheng K., Lei L.-C., Chen Q.-X., Gao Y.-W. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc. Natl. Acad. Sci. 2005;102(7):2430–2435. doi: 10.1073/pnas.0409608102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;1–4 doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-em structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.M. Li, F. L. Simonetti, A. Goncearenco, and A. R. Panchenko. Mutabind estimates and interprets the effects of sequence variants on protein–protein interactions. Nucleic Acids Res, 44(W1):W494–W501, 2016. [DOI] [PMC free article] [PubMed]
- 19.M. Petukh, L. Dai, and E. Alexov. Saambe: webserver to predict the charge of binding free energy caused by amino acids mutations. Int. J. Mol. Sci, 17(4):547, 2016. [DOI] [PMC free article] [PubMed]
- 20.Pires D.E., Blundell T.L., Ascher D.B. mCSM-lig: quantifying the effects of mutations on protein-small molecule affinity in genetic disease and emergence of drug resistance. Sci. Rep. 2016;6:29575. doi: 10.1038/srep29575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodrigues C.H., Myung Y., Pires D.E., Ascher D.B. mCSM-PPI2: predicting the effects of mutations on protein–protein interactions. Nucleic Acids Res. 2019;47(W1):W338–W344. doi: 10.1093/nar/gkz383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schymkowitz J., Borg J., Stricher F., Nys R., Rousseau F., Serrano L. The FoldX web server: an online force field. Nucleic Acids Res. 2005;33(Suppl. 2):W382–W388. doi: 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang N., Chen Y., Lu H., Zhao F., Alvarez R.V., Goncearenco A., Panchenko A.R., Li M. Mutabind2: predicting the impacts of single and multiple mutations on protein–protein interactions. Iscience. 2020;100939 doi: 10.1016/j.isci.2020.100939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sirin S., Apgar J.R., Bennett E.M., Keating A.E. AB-Bind: antibody binding mutational database for computational affinity predictions. Protein Sci. 2016;25(2):393–409. doi: 10.1002/pro.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.J. Jankauskaite•, B. Jim´enez-Garc´ıa, J. Dapku¯nas, J. Ferna´ndez-Recio, and I.H. Moal. Skempi 2.0: an updated benchmark of changes in protein–protein binding energy, kinetics and thermodynamics upon mutation. Bioinformatics, 35(3):462–469, 2019. [DOI] [PMC free article] [PubMed]
- 26.I.H. Moal, J. Ferna´ndez-Recio. SKEMPI: a Structural Kinetic and Energetic database of mutant protein interactions and its use in empirical models. Bioinformatics, 28(20):2600–2607, 2012. [DOI] [PubMed]
- 27.Carlsson G. Topology and data. Bull. Am. Math. Soc. 2009;46(2):255–308. [Google Scholar]
- 28.Edelsbrunner H., Letscher D., Zomorodian A. Proceedings 41st Annual Symposium on Foundations of Computer Science. IEEE; 2000. Topological persistence and simplification; pp. 454–463. [Google Scholar]
- 29.Wang M., Cang Z., Wei G.-W. A topology-based network tree for the prediction of protein–protein binding affinity changes following mutation. Nat. Mach. Intell. 2020;2(2):116–123. doi: 10.1038/s42256-020-0149-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Y. Shu and J. McCauley. GISAID: Globalinitiative on sharing all influenza data–from vision to reality. Eurosurveillance, 22(13), 2017. [DOI] [PMC free article] [PubMed]
- 31.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;1–6 doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 32.Lee N., Hui D., Wu A., Chan P., Cameron P., Joynt G.M., Ahuja A., Yung M.Y. A major outbreak of severe acute respiratory syndrome in hong kong. N. Engl. J. Med. 2003;348(20):1986–1994. doi: 10.1056/NEJMoa030685. [DOI] [PubMed] [Google Scholar]
- 33.P. Zhou, X.-L. Yang, X.-G. Wang, B. Hu, L. Zhang, W. Zhang, H.-R. Si, Y. Zhu, B. Li, C.-L. Huang, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 579(7798):270–273, 2020. [DOI] [PMC free article] [PubMed]
- 34.Drexler J.F., Gloza-Rausch F., Glende J., Corman V.M., Muth D., Goettsche M., Seebens A., Niedrig M. Genomic characterization of SARS-related coronavirus in european bats and classification of coronaviruses based on partial RNA-dependent RNA polymerase gene sequences. J. Virol. 2010;84:11336–11349. doi: 10.1128/JVI.00650-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu D., Zhu C., Ai L., He T., Wang Y., Ye F., Yang L., Ding C. Genomic characterization and infectivity of a novel SARS-like coronavirus in Chinese bats. Emerg. Microbes Infect. 2018;7(1):1–10. doi: 10.1038/s41426-018-0155-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li F. Structural analysis of major species barriers between humans and palm civets for severe acute respiratory syndrome coronavirus infections. J. Virol. 2008;82(14):6984–6991. doi: 10.1128/JVI.00442-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schrödinger, LLC. The PyMOL molecular graphics system, version 1.8. November 2015.
- 39.Sievers F., Higgins D.G. Multiple Sequence Alignment Methods. Springer; 2014. Clustal omega, accurate alignment of very large numbers of sequences; pp. 105–116. [DOI] [PubMed] [Google Scholar]
- 40.Cang Z., Mu L., Wei G.-W. Representability of algebraic topology for biomolecules in machine learning based scoring and virtual screening. PLoS Comput. Biol. 2018;14(1):e1005929. doi: 10.1371/journal.pcbi.1005929. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials are available for S1: The new parameterization and validation of the TopNetTree model and S2: Supplementary tables for the results in figures.