Abstract
The rose is one of the most important ornamental woody plants because of its extensive use and high economic value. Herein, we sequenced a complete chloroplast genome of the miniature rose variety Rosa ‘Margo Koster’ and performed comparative analyses with sequences previously published for other species in the Rosaceae family. The chloroplast genome of Rosa ‘Margo Koster’, with a size of 157,395 bp, has a circular quadripartite structure typical of angiosperm chloroplast genomes and contains a total of 81 protein-coding genes, 30 tRNA genes and 4 rRNA genes. Conjunction regions in the chloroplast genome of Rosa ‘Margo Koster’ were verified and manually corrected by Sanger sequencing. Comparative genome analysis showed that the IR contraction and expansion events resulted in rps19 and ycf1 pseudogenes. The phylogenetic analysis within the Rosa genus showed that Rosa ‘Margo Koster’ is closer to Rosa odorata than to other Rosa species. Additionally, we identified and screened highly divergent sequences and cpSSRs and compared their power to discriminate rose varieties by Sanger sequencing and capillary electrophoresis. The results showed that 15 cpSSRs are polymorphic, but their discriminating power is only moderate among a set of rose varieties. However, more than 150 single nucleotide variations (SNVs) were discovered in the flanking region of cpSSRs, and the results indicated that these SNVs have a higher divergence and stronger power for profiling rose varieties. These findings suggest that nucleotide mutations in the chloroplast genome may be an effective and powerful tool for rose variety discrimination and DNA profiling. These molecular markers in the chloroplast genome sequence of Rosa spp. will facilitate population and phylogenetic studies and other related studies of this species.
Subject terms: Genetics, Plant sciences
Introduction
Chloroplasts are vital and unique components of photosynthetic cells in plants and algae. They are responsible for multiple functions, e.g., assimilation of carbon and nitrogen, synthesis of amino acids and fatty acids, etc., and play important roles in plant growth and development1. Chloroplasts may have originated from cyanobacteria through endosymbiosis2,3, and chloroplasts possess their own deoxyribonucleic acid (DNA). In the majority of plant species, the chloroplast genome (cp genome) is inherited maternally, with the exception of certain gymnosperms, in which the cp genome is paternally inherited4. In recent years, knowledge about the organization and evolution of the cp genome has rapidly improved because of the development of DNA sequencing technology and bioinformatics methods. The cp genome is relatively conserved in angiosperms, and its size ranges from 107 to 218 kb, including 100–200 encoding genes5. A typical angiosperm cp genome has circular DNA, including two small inverted repeats (IRa and IRb), a large single copy (LSC) region and a small single copy (SSC) region6. Compared to nuclear DNA, the cp genome is small, single-stranded, relatively conserved and has uniparental inheritance, and its nucleotide mutation rate is moderate. Variation in the cp genome arises from the expansion or contraction of IRs and the length of the intergenic spacers7,8. The cp genome is considered to be conserved in the genome organization, gene order and gene content9; however, large-scale genome rearrangement and gene loss have been reported10. In addition, genes in chloroplast genomes can transfer to nuclear genomes and this process is part of the evolutionary process11. Hence, diverse DNA sequences from the cp genome have been used to study the evolution of plants12. In addition, the chloroplast sequences have contributed to assessing population genetic diversity, identifying species and implementing plant conservation13–15.
The rose is one of the most important woody ornamental plants and belongs to the genus Rosa in the family Rosaceae. Rosa species are native to temperate and warm regions in the Northern Hemisphere, especially in Asia. These species from different regions hybridize easily, giving rise to types that overlap the parental forms, and it is difficult to determine basic species16. Roses are widely used as cut flowers and in gardening and medicine, and they contribute to maintaining livelihoods, improving the environment and meeting material and spiritual needs. Therefore, roses have attracted much attention from plant breeders and botanists, and a large number of rose varieties have been selected for and bred17,18. Previous reports indicate that over 20 species in the Rosa genus have been used to breed modern rose varieties19,20. These intraspecific and interspecific hybridizations and artificial selections have not only created abundant variations but also led to complex genetic components in roses21,22. The increased number of varieties and decreased morphologic and genetic differences among varieties make it difficult to manage and discriminate rose varieties according to phenotypic differences, which is considered as the technical basis of distinctness, uniformity, and stability (DUS) examination in the current UPOV system. For example, rose plants must be submitted in the form of young plants that meet specific standards for field trials; these plants must be observed and examined for at least one growing cycle, based on 50 morphological characteristics23. However, the emergence of molecular markers may provide new insight into understanding and solving these practical problems because molecular markers can reveal addtional information on genetic variation. Previous reports have indicated that DNA evidence may provide exciting insight into the evolutionary process of Rosa species and identify the root of the original Rosa species and modern varieties24–27. Previous studies have reported that analyses of molecular markers from the nuclear genome are powerful and effective approaches for discriminating rose varieties and establishing DNA profiles. For example, random-amplified polymorphic DNA (RAPD), amplified fragment length polymorphism (AFLP) and simple sequence repeat (SSR)28,29, and sequenced tagged microsatellite site (STMS) markers have been used to construct the rose DNA database30,31. However, to date, only a few cp genomes of Rosa spp. have been published in the GeneBank database32–35, which is a source of data on sequence diversity. Furthermore, few studies have focused on molecular markers from the cp genome of roses. To identify hybrids, chloroplast markers alone may be insufficient, as these markers are haploid and only provide maternal data; nevertheless, these markers may be useful tools for identifying a maternal parent of putative hybrid progeny. Using the associated cp genome sequence in combination with nuclear genomic markers to construct rose DNA profiles could be helpful in the management and discrimination of varieties.
In this paper, we hypothesized that the molecular markers and sequence diversity of the cp genome may assist in discriminating and profiling modern rose varieties. Therefore, we constructed a complete cp genome of Rosa ‘Margo Koster’ using next-generation sequencing with a de novo and reference-guided assembly strategy. The cp genome of Rosa ‘Margo Koster’ was compared and analyzed with the genomes of other species or varieties in the Rosa genus. Moreover, a complete cp genome of Rosa ‘Margo Koster’ and characterizations of SSRs and single nucleotide variations (SNVs) were screened and verified by capillary electrophoresis (CE) and Sanger sequencing, and their discriminating power was calculated and compared in a set of rose varieties.
Results
Genome sequence generation and PCR-based validation
More than 1,190.28 million reads (approximately 11 Gb of clean data) were generated by the Illumina HiSeq2500 platform for Rosa ‘Margo Koster’. These data were used to assemble the cp genome with a high mean coverage. After de novo and reference-guided assembly, the complete cp genome with a size of 157,395 bps was generated. Four regions and their conjunctions were validated using PCR-based Sanger sequencing, and we also corrected the errors via PCR-based validation. We designed 35 pairs of primers based on the variation in regions of alignments to validate these sequences (see Table S1); the gel photo of the PCR products are shown in Figure S1. The validated sequences amounted to 28,662 bp. We compared these sequences to the assembled genome and found two nucleotide mismatches that were corrected before submission to the GenBank database (accession No. MN435990).
General cp genome characterization and annotation
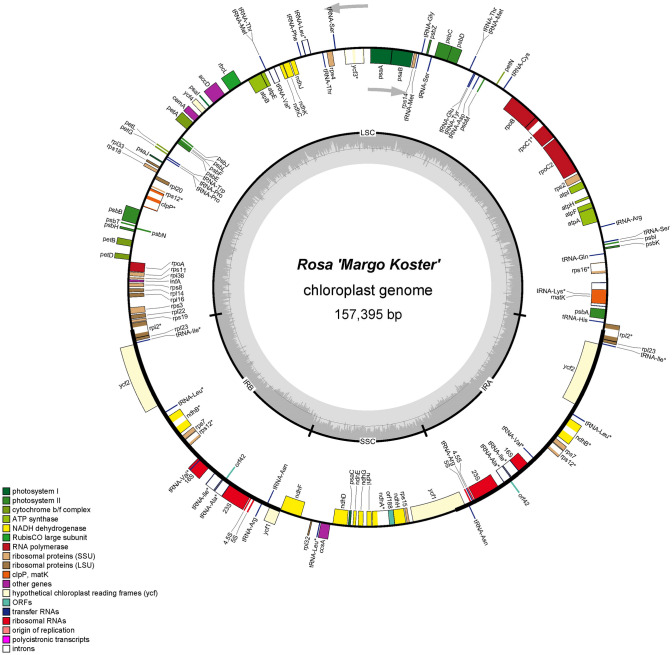
The cp genome displayed a typical quadripartite structure, consisting of one LSC region with a length of 87,710 bp, one SSC region with a length of 18,849 bp and a pair of IR regions (IRa and IRb) with a length of 25,418 bp each (Fig. 1). The overall GC content of the cp genome was 37.19%. Among the LSC, SSC and IR regions, the highest GC content was found in the IR regions (43.34%), and the GC content of the LSC and SSC regions was 35.19%, and 31.20%, respectively. More rRNA and tRNA genes, which have high GC content, could explain the higher GC content in the IR region than other regions. Thymine (T) and adenine (A) preferences in the third position of the codon were observed in this cp genome, and codon usage is shown in Table S2 and Figures S2 and S3. This event could be a result of an A + T rich genome, which has been also observed in other plants36–38.
Figure 1.
Circular map of the cp genome for Rosa ‘Margo Koster’. The gray arrows show that genes inside the circle are transcribed clockwise, and genes outside the circle are transcribed counterclockwise. The innermost shaded areas inside the inner circle correspond to the GC content in the cp genome. Genes in different functional groups are color coded. The boundaries of four regions (IRa, IRb, LSC, SSC) are noted in the inner circle.
The gene content and order are illustrated in Fig. 1 and Table 1. There are a total of 115 genes, including 81 protein-coding genes, 30 tRNA genes and 4 rRNA genes. Seven protein-coding genes, 7 tRNA genes, and 4 rRNAs were duplicated in the IR regions. The LSC region contained 22 tRNA genes and 63 protein-coding genes, and the SSC region included 11 protein-coding genes and 1 tRNA gene. Most of these protein-coding genes did not contain introns, except that nine genes (rps16, rpoc1, trnI-GAU, trnA-UGC, rps12, rpl16, ndhA, rpl2, ndhB) contained a single intron and two genes (ycf3, clpP) contained two introns. The longest intron was trnK-UUU, with a length of 2,499 bp, and was present in the matK gene.
Table 1.
The gene content and functional classification in the cp genome of Rosa ‘Margo Koster’.
| Function | Genes |
|---|---|
| RNA transfer | trnH-GUG, trnK-UUU, trnQ-UUG, tRNA-Gly, trnG-UCC, trnR-UCU,trnD-GUC, trnY-GUA, trnE-UUC, trnT-GGU, trnM-CAU, trnS-UGA, trnfM-CAU, trnS-GCU, trnS-GGA, trnT-UGU, trnL-UAA,trnF-GAA,trnV-UAC, trnW-CCA, trnP-UGG, trnP-GGG, trnI-CAU, trnL-CAA, trnV-GAC, trnI-GAU*, trnA-UGC*, trnN-GUU, trnR-ACG, trnL-UAG |
| RNA ribosomal | rrn23, rrn16, rrn5, rrn4.5 |
| RNA polymerase | rpoC1*, rpoC2, rpoA, rpoB |
| Clpp, Matk, ORFs | Clpp**, matk, orf42, orf188 |
| Ribosomal proteins (SSU) | rps2, rps3, rps4, rps7, rps8, rps11, rps12*, rps14, rps15, rps16*, rps18, rps19 |
| Ribosomal proteins (LSU) | rpl2*, rpl2Ψ, rpl14, rpl16, rpl20, rpl22, rpl23, rpl32, rpl33, rpl36 |
| Hypothetical chloroplast reading frames (ycf) | ycf1, ycf1Ψ, ycf2, ycf3**, ycf4 |
| ATP synthase | atpE, atpB, atpA, atpF, atpH, atpI |
| Photosystem I | psaI, psaB, psaA, psaC, psaJ |
| Photosystem II | psbD, psbC, psbZ, psbT, psbH, psbK, psbI, psbJ, psbF, psbE, psbM, psbN, psbL, psbA, psbB |
| RubisCO large subunit | rbcL |
| Cytochrome complex | petN, petA, petL, petG, petB, petD |
| NADH dehydrogenase | ndhB*, ndhI, ndhK, ndhC, ndhF, ndhD, ndhG, ndhE, ndhA*, ndhH, ndhJ |
| Others | infA, accD, cemA, ccsA |
*Genes containing one intron; **genes containing two introns; Ψpseudogene.
Phylogenetic analysis in Rosaceae
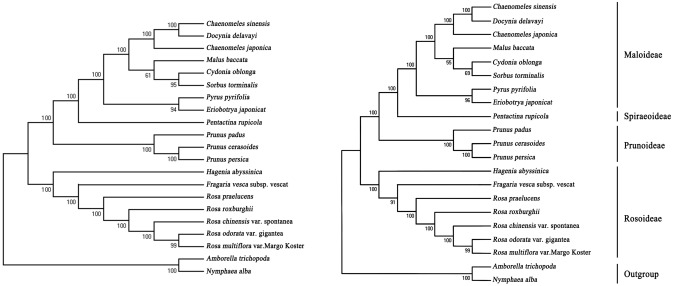
Sequencing alignment showed that the cp genomes of Rosa species are more homologous than those of other species of Rosaceae. Phylogenetic trees illustrate the partial relationship of some species in Rosaceae (Fig. 2) based on the maximum likelihood (ML) and neighbor-joining (NJ) methods. The results were consistent with previous studies of Rosaceae species39, and showed that Rosa ‘Margo Koster’ has a closer genetic similarity to Rosa odorata var. gigantea and Rosa chinensis var. spontanea than with the other Rosa species, which may be attributed to the selection of breeding material.
Figure 2.
Neighbor-joining (NJ) and maximum likelihood (ML) trees for the Rosaceae family inferred from the complete cp genomes of 19 species from 5 subfamilies. Bootstrap values are indicated at the nodes. The length scale behind the tree indicates substitutions per site.
Comparison with the cp genomes from Rosa species
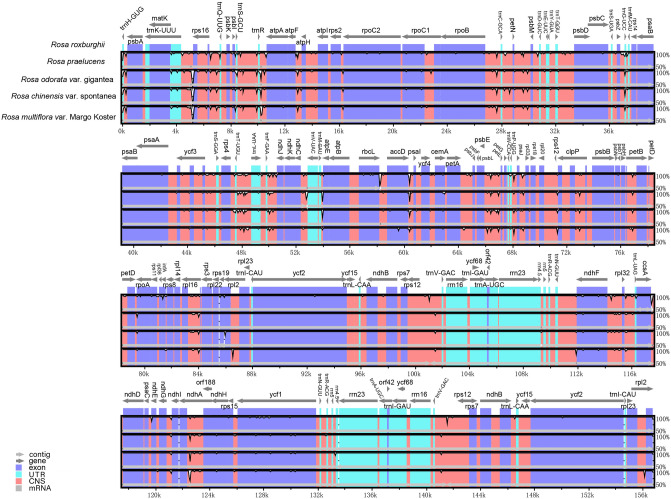
The whole cp genome of Rosa ‘Margo Koster’ was compared with those of four Rosa species, and the highly divergent regions are shown in Fig. 3. As a whole, the shared sequence identities were over 95% in pairwise comparisons for the five Rosa species. The sequences in two IR regions were more conserved than those in LSC and SSC regions, and protein-coding regions were less divergent than noncoding regions, such as introns and intergenic regions. The highly divergent regions were mainly found in intergenic regions, including trnD-GUC-trnY-GUA, rpoB-trnC-GCA, rpl12-clpp, aptF-aptH, rps2-rpoc2, trnS-GGA-rps4, psaJ-rpl33, rps16-trnQ-UUG, trnK-UUU-rps16, psbE-petL, trnT-UGU-trntL-UAA, trnP-UGG-psaJ, trnH-GUG-psbA, trnR-UCU-atpA, psbZ-trnG-UCC, trnG-UCC-trnfM-CAU, trnM-CAU-aptE, psbM-trnD-GUC, and psbL-trnS-GCU. Apart from those regions, some coding sequences, such as ndhA, clpp, ndhF, petB, and ycf1, had low similarity levels.
Figure 3.
Comparisons of sequence identity of cp genomes for five Rosa species. The vertical axis represents identity ranging from 50 to 100%. Each arrow indicates the annotated gene and its transcriptional direction. Genome regions are color coded as an exon, mRNA or tRNA, untranslated region (UTR) and conserved noncoding sequence (CNS).
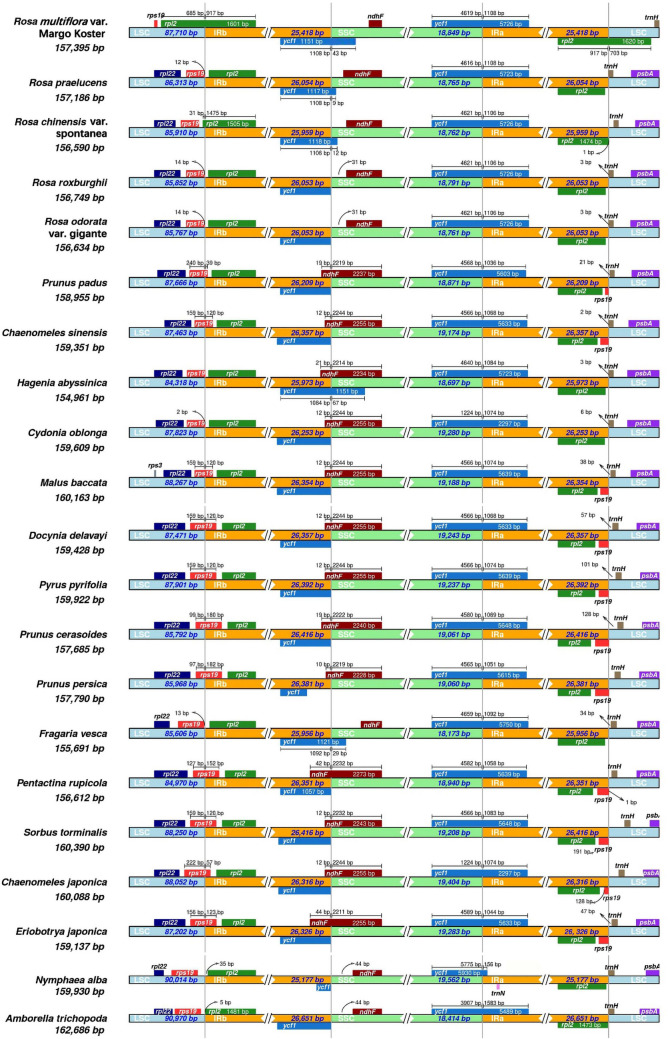
A comparison among boundary regions of cp genomes is illustrated in Fig. 4, and the list of 19 species in the family of Rosaceae and 2 basal angiosperm species is shown in Table S3. The results showed that the position of the SSC/IRb junction in all species was the ycf1 gene, and a pseudogene near the 5′ end of this gene (yfc1Ψ) was found in the IRa region. The size of yfc1Ψ was approximately 1,100 bp for the Rosaceae species. The rpl2 gene stretches across the boundary between the LSC and IRb regions and was only found in Rosa chinensis var. spontanea and Rosa ‘Margo Koster’, not in the other three species, which all contained a complete rpl2 gene in the IRb region. The rps19 gene was located in the LSC region in the five Rosa species and in one Fragaria species, but a partial sequence of this gene entered the IRb region in Malus, Pyrus, Prunus and other species. Therefore, a pseudogene (rps19Ψ) was designated in the IRa region. The ndhF gene was located completely within the SSC region for five Rosa species and the Fragaria species; however, the ndhF gene was located at the boundary of the LSC and IR regions in the other Rosaceae species, e.g., Pentactina rupicola, Chaenomeles japonica, and Sorbus torminalis.
Figure 4.
Comparison of the boundaries between LSC, SSC, and two IR regions among 19 cp genomes within the Rosaceae family and two species of basal angiosperms. The boundaries of five Rosa species are shown together at the top of the figure, and the differences in their boundaries are obvious. The number of base pairs (bp) represents the distance from the boundary to the end of the gene.
Repeat sequence analysis of Rosa species
A total of 84 repeat sequences were detected in the cp genome of Rosa ‘Margo Koster’, containing 27 tandem repeats, 26 forward repeats, one complementary repeat, 4 reverse repeats and 26 palindrome repeats. The detailed information is shown in Tables S3 and S4. SSRs, which are composed of a 1–6 bp low-repeat motif, were also widely found in the genome. A total of 49 perfect SSRs were detected, including 46 mononucleotides and 3 dinucleotides. Over 89% of the SSRs were mononucleotides, which contained a repeat motif of adenine (A) or thymine (T). For dinucleotide SSRs, the repeat motif was a repetition of only AT. The results showed that 85.71% of all the detected SSRs were located in the LSC regions, 6.12% in the IR regions, and 8.16% in the SSC regions. These findings indicated that the distribution of cpSSRs was not imbalanced in the cp genome of Rosa ‘Margo Koster’.
Highly divergent regions among Rosa species
The level of divergence among five Rosa species was variable in different regions of the cp genome. The results showed that the nucleotide diversity (π) ranged from 0.0012 (rps12-trnV-GAC, intergenic space) to 0.0524 (psbL-trnS-GCU, intergenic space) based on the comparative analysis of the regional sequence. The results showed that the average π value of the intergenic space was 0.0108, which was higher than that of the protein-coding region (0.0053), and genetic divergence did not exist in the rRNA sequences (π = 0). These findings indicated that the level of genetic variance depended on the region. Twenty regions with the highest π values are shown in Table 2, and their sequences included 19 IGSs and only one protein-coding gene—the ndhA gene—with a π value of 0.0094. Other regions with π values greater than zero are also illustrated in Figure S4.
Table 2.
Twenty most divergent regions of the cp genome based on a comparison of Rosa species.
| No. | Region | Nucleotide diversity (π) | Total number of mutations (η) | Region length (bp) |
|---|---|---|---|---|
| 1 | ndhA | 0.0094 | 41 | 2,381 |
| 2 | trnD-GUC-trnY-GUA | 0.0100 | 10 | 420 |
| 3 | rpoB-trnC-GCA | 0.0101 | 27 | 1,220 |
| 4 | rpl12-clpp | 0.0101 | 4 | 201 |
| 5 | aptF-aptH | 0.0103 | 12 | 510 |
| 6 | rps2-rpoc2 | 0.0109 | 5 | 247 |
| 7 | trnS-GGA-rps4 | 0.0110 | 7 | 292 |
| 8 | psaJ-rpl33 | 0.0113 | 13 | 469 |
| 9 | rps16-trnQ-UUG | 0.0120 | 21 | 883 |
| 10 | trnK-UUU-rps16 | 0.0121 | 20 | 827 |
| 11 | psbE-petL | 0.0124 | 35 | 1,314 |
| 12 | trnT-UGU-trntL-UAA | 0.0126 | 32 | 1,240 |
| 13 | trnP-UGG-psaJ | 0.0192 | 18 | 450 |
| 14 | trnH-GUG-psbA | 0.0244 | 11 | 312 |
| 15 | trnR-UCU-atpA | 0.0244 | 21 | 631 |
| 16 | psbZ-trnG-UCC | 0.0320 | 23 | 400 |
| 17 | trnG-UCC-trnfM-CAU | 0.0323 | 12 | 223 |
| 18 | trnM-CAU-aptE | 0.0368 | 9 | 214 |
| 19 | psbM-trnD-GUC | 0.0492 | 62 | 529 |
| 20 | psbL-trnS-GCU | 0.0524 | 18 | 183 |
Development and utilization of molecular markers in chloroplast genomes of Rosa species
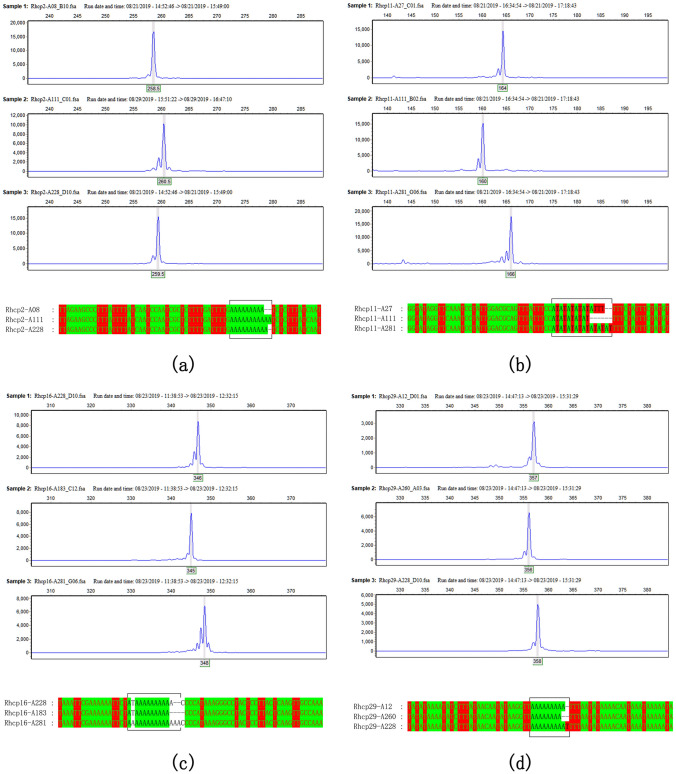
Based on repeat sequence analyses, a total of 49 candidate cpSSRs were subjected to genotyping using CE and Sanger sequencing in a set of different original rose varieties. The results showed that clear, stable and expected PCR products were obtained in all cpSSR loci; however, only 15 cpSSR loci were abundantly polymorphic, accounting for 31.64% (Table 3, Fig. 5). The results showed that the Dj, value, which is the discriminating power, ranged from 0.1043 to 0.9350, with an average value of 0.4018, based on the genotyping of cpSSRs (Table 3). These findings indicated that cpSSRs could be moderately polymorphic and have discriminating power in the set of test rose varieties. Over one hundred SNVs were also found in the flanking sequence of cpSSRs. The results showed that the π among the test rose varieties ranged from 0.0089 to 0.0744, with an average value of 0.0315, and the total number of mutations (η) ranged from 3 to 150. The Dj values ranged from 0.3446 to 0.9996, with an average value of 0.8303. These findings indicated that these SNVs in the cp genome have much more polymorphism and stronger discriminating power than traditional fragment-size molecular markers. Additionally, all the polymorphic cpSSRs and SNVs were used to examine and construct DNA profiles of 93 rose varieties.
Table 3.
The Primer information of 15 cpSSRs and nucleotide mutations of their flanking sequences.
| Locus Name | Forward primer | Reverse primer | Product size (bps) | Tm (°C) | Nucleotide diversity (π) | Total number of mutations (η) | Region length (bp) | Dja | Djb | Site |
|---|---|---|---|---|---|---|---|---|---|---|
| Rhcp2 | AACTCATCAACGGACTCTCCA | ATTAGTGCTTGATGCGGGAAA | 261 | 59.5 | 0.00193 | 3 | 205 | 0.4130 | 0.3446 | matK-trnK-UUU |
| Rhcp5 | GACTTGTGTTGGATTGGCACT | ACGGAACTTCGCCTTAACCAA | 289 | 60 | 0.02362 | 53 | 221 | 0.1231 | 0.9016 | trnK-UUU-rps16 |
| Rhcp6 | CATTCCTTCAGTTTGGAACCCA | TCTTGGTACTTGAAGAAGTGTGA | 337 | 58 | 0.06351 | 150 | 271 | 0.4544 | 0.9885 | rps16-trnQ-UUG |
| Rhcp11 | TGGAGTGAAAAGCGTCCATTG | AGCGCCTCTTATTCAAGTTATTCA | 167 | 59 | 0.04529 | 78 | 113 | 0.3111 | 0.5339 | trnR-UCU-aptA |
| Rhcp13 | CGGATGGCCAATAACCCAAG | GAGGTATTTCGCAACTGGCCT | 311 | 59 | 0.00906 | 58 | 266 | 0.3700 | 0.5196 | atpF-atpH |
| Rhcp14 | CGCACGTCGTAAACAAATCCA | AGCTTAGCCTGACGCAATGT | 331 | 60 | 0.05264 | 76 | 282 | 0.4642 | 0.8277 | atpH-aptL |
| Rhcp16 | CCGGCTCCAGTAGTTACACC | TAACCGTCGAGGCGAAGTAG | 350 | 60 | 0.00891 | 34 | 296 | 0.2170 | 0.9495 | rpoC1 |
| Rhcp19 | CACATATTGCGCACTTCCCG | AGGGCCTCTTCGATGGGTAT | 246 | 60 | 0.04801 | 48 | 200 | 0.3534 | 0.9996 | petN-psbM |
| Rhcp22 | CGCTATCCGCCCAGGATAAT | CCTTGAGGTCACGGGTTCAA | 243 | 60 | 0.01792 | 24 | 197 | 0.3020 | 0.9727 | trnG-UUC-trnfM-CAU |
| Rhcp24 | CGGGGATACACGACAGAAGG | CACCTATTACAGAGATGGTGCGA | 190 | 60 | 0.02778 | 23 | 114 | 0.1043 | 0.9084 | ycf3 |
| Rhcp29 | CGTGTAGAAACGTGTAGAAGGG | GAAACCATTGCAATTGCCGGA | 360 | 59 | 0.01671 | 48 | 322 | 0.4523 | 0.7457 | accD-psaL |
| Rhcp31 | AGCGAGTCAACCGCTAGAAC | GGAGAATGAACTCTGGGAAGGT | 296 | 60 | 0.03396 | 53 | 252 | 0.5046 | 0.9991 | rps18-rpl20 |
| Rhcp34 | CTCCGAGTAAAGATCCGCCC | TGAAGTATCCAGGCTCCGTT | 308 | 59 | 0.03085 | 92 | 272 | 0.3925 | 0.7929 | rpl 20 |
| Rhcp38 | TGTGTATCTAGGGAATCGTCGC | CTGCCCCCGAGGGTCTATAA | 319 | 59 | 0.07443 | 81 | 269 | 0.9350 | 0.9930 | petD-rpoA |
| Rhcp41 | AGCTCCTCGCGAATTAAACGA | TGGGAACGACAGAACCTGTG | 390 | 60 | 0.01791 | 39 | 352 | 0.6307 | 0.9790 | rpl 16-rps3 |
‘a’ indicates the discriminating power based on the genotype of cpSSR; ‘b’ indicates the discriminating power based on the genotype of nucleotide mutations.
Figure 5.
The cpSSR genotype of rose varieties based on capillary electrophoresis and Sanger sequencing. (a)–(d) Variation in fragment size of cpSSRs and their repeat motif in different rose varieties.
Discussion
In this paper, we constructed cp genomes for a rose variety (Rosa ‘Margo Koster’) using next-generation sequencing on the Illumina HiSeq2500 platform. The comparative analysis of this cp genome and those of other species in the Rosaceae family provided new insight into the characteristics of their cp genomes, and the identified polymorphic markers in these cp genomes could be suitable for use in future DNA profiling, variety discrimination and evolutionary studies in Rosa spp. and their varieties to manage and profile a large number of rose varieties, clarify genetic relationships, and assist in breeding selection.
Compared to previously published data in Rosaceae, the cp genome of Rosa ‘Margo Koster’ has a typical quadripartite structure, where the LSC and SSC regions are separated by two IRs and show a relative similarity in cp genome size, gene number and structure, as well as total GC content, which is an extremely significant indicator of species affinity40. Compared with genomes of other genera, no large fragment inversions or gene rearrangements were detected in the Rosa genus. Noncoding sequences in the IR regions were more conserved than others that have been found in the cp genomes of Asteraceae and Lamiales41,42. Therefore, more genetic variation has originated from the LSC and SSC regions than from IR regions.
The contraction and expansion of IR regions, as an important evolutionary event in the cp genome development, is one of the primary reasons for the variation in size of the cp genome43,44. Therefore, the length of the IR region is variable, and this region is even lost in some plant species, such as Pelargonium × hortrum, Pinus thunbergii, Cryptomeria japonica, Erodium and Metasequoia glyptostroboides45–49. This type of gene arrangement in the cp genome is different from that of most eudicots and monocots; for example, in eudicots, the trnH gene is located in the LSC region rather than the IR region, where this gene is located in most monocots50.
Another important aspect of the use of the cp genome in evolutionary studies is the variation found near the boundaries among the four regions. The IR contraction and expansion could provide evidence about the evolution of some plant taxa and lead to the size or sequence divergences of the cp genome. Our comparative analysis of 19 species in the Rosaceae family showed that the rpl2 gene in Rosa ‘Margo Koster’ has moved from an IR region to an LSC region, resulting in a shorter IR region than that of other Rosa species, and this phenomenon of IR contraction has also occurred in Rosa chinensis var. Spontanea but not in other three Rosa species. Our results also revealed that some pseudogenes contribute to the variation in length in the IR region, such as in rps19Ψ or ycf1Ψ, and their biological function is still unclear. For example, ycf1, which is one of the largest ORFs in the cp genome, has been considered a protein-coding gene in tobacco, and the protein encoded by the ycf1 gene has been found to be important for cell survival or related to the ABC transporter in other studies51,52; ycf1Ψ is present in all studied species. The rps19 gene is present in the cp genome of Rosaceae species, and its location varies from species to species; for example, the rps19 gene is located in the LSC region in Rosa and Fragaria species, whereas this gene stretches across the LSC and IR regions and may lead to incomplete duplication in Malus, Prunus and Pyrus species, resulting in rps19Ψ. Contraction and expansion of the IR region have taken place in Rosaceae, leading to the creation of pseudogenes.
Repetitive sequences play an important role in genome recombination and rearrangement; thus, they are major sources of genetic variation. Many repetitive sequences have been identified and applied in a wide range of studies in plant science53–56. However, little attention has been given to cpSSRs in the cp genome of Rosa species. Our results revealed a total of 49 candidate cpSSR loci in the cp genome, and most of these markers were located in the non-coding region, which is consistent with the cp genomes of other angiosperms57,58. A previous study reported that the variability of cpSSRs in non-coding regions is more abundant than in conserved coding regions59. A strong AT bias, which has been reported in other species60–62, also existed in the cp genome of Rosa ‘Margo Koster’. Additionally, distribution of cpSSRs was imbalanced in the cp genomes, with over 85% of cpSSRs in the LSC regions.
In addition to repetitive sequences, nucleotide mutation is one of the most important sources of genetic variation. In the comparison of nucleotide diversity among different regions of five cp genomes in Rosa species, a set of regions with high divergence was obtained, and most of these regions are intergenic. These highly divergent sequences may be used to develop potential molecular markers for geographical and population genetic studies in Rosa spp. Finding polymorphic nucleotide variation in the cp genome used to be time-consuming and labor-intensive work because the available sequences were insufficient. However, highly divergent sequences in Rosa spp. have been identified here and will contribute to genetic and evolutionary studies in Rosa spp. as well as to wider applications in DNA profiling of rose varieties.
Molecular markers from the cp genome have several advantages, such as a moderate mutant rate, clear genotype, etc. Hence, these nucleotide variations can be used to discriminate varieties including those with close relationships, and to unveil the genetic variations at the population and individual levels. Additionally, the chloroplast SNVs and SSRs could be used to explore the genetic structure and population gene flow63,64. Herein, approximately 15 cpSSRs were shown to be polymorphic, although their discriminating power in the test rose varieties is lower, this is likely due to the relatively conserved and haplotype genome of these varieties.
At present, under the framework of the UPOV convention, plant breeder’s rights mainly depend on the distinctness of plant’s phenotypes. Therefore, almost all DUS examinations that require intensive field trials, are based on the morphological characteristics of plants. Collecting a large number of rose varieties in a common database is necessary and prerequisite for DUS examinations; this work requires a great deal of labor, material and land, especially for rose, because of its abundant cultivars and worldwide distribution. Based on the available database, the DUS examiner will select the most similar varieties to start a field trial. Then, according to DUS test guidelines, potential morphological divergences between candidate variety and similar variety will be observed and recorded during at least one growth cycle23. For roses, no less than 9 healthy and uniform clones of the candidate variety are required to submit to the national DUS examination station, where these plants will be compared with similar varieties. Over 40 phenotypic traits involved in leaf, flower and fruit, need to be observed and measured during at least one growth cycle. Finally, the examiner will complete a technical questionnaire, which is the most import document to determinate a new variety. The most challenging aspect of this process is the selection of appropriate similar varieties from over ten thousand of existing varieties, and then providing an accurate judgment about the candidate variety in one growth cycle. Because of this challenge, at the end of last century, developing DNA markers for DUS testing, variety profiling or origin tracing was proposed, and case studies in crop, vegetables and fruit trees have been introduced in BMT sessions65 or reported in scientific journals66–68. For rose, codominant markers from the nuclear genome have been developed and used in genetic studies of diploid and tetraploid rose or DNA profiles69–73. Few studies have focused on the genetic variation rose chloroplast genome74,75. In this study, we found that there are a large number of highly divergent regions in the rose cp genome, which could provide potential haplotype’ DNA markers. Previous reports also have shown that chloroplast DNA diversity may be a new approach for distinguishing crops, fruits cultivars or tree varieties76–81. Our findings showed that the SSRs and SNVs from the cp genome could also distinguish the test rose varieties, although the discrimination power of cpSSR was relatively lower than that of genomic markers. Similar results have been found in Prunus mume cultivar and tetraploid alfalfa82,83. cpSSR has also been used in individual identification of Cupressaceae species and genetic diversity assessment of cultivated and wild Hevea rubber84,85. We believe that the cpSSRs markers of rose could also be applied to further genetic studies.
In addition to SSR markers, SNPs are a powerful tool for genetic studies in plants, and these markers have been used to evaluate the genetic differentiation among accessions as well as population structure and diversity in crops86. A large number of SNP markers have been mined from the nuclear genome of roses72,87,88, and an SNP array (WagRhSNP68k) was developed for genetic mapping in tetraploid cut roses73,89. Based on the comparative analysis of nucleotide diversity among regions in these Rosa species, a set of 20 regions with high divergence have been found and these regions could be used as a starting point for candidate molecular markers for phylogenetic and phylogeographic studies in the Rosa genus. Uncovering polymorphic sequences in the cp genome is difficult, particularly when no previous reference genome has been published. The lack of available and polymorphic sequences prevents us to utilize in a phylogenetic context; however, we still expect that the highly divergent sequences identified here by comparing Rosa cp genomes will offer new tools for genetic and evolutionary studies in the Rosa genus and other related taxa. Our findings indicate that nucleotide mutations in the cp genomes of Rosa species are abundant and unambiguous, and the discriminating power of SNVs in the cp genome is stronger than that of cpSSRs in the test rose varieties. Although nuclear SSRs and SNPs have been proven to be reliable and powerful markers for DNA profiling in roses, we also expect that these highly abundant and polymorphic nucleotide variations in the cp genome will become clearly identified, effective and reliable supplementary tools for managing and profiling rose varieties in the future.
Materials and methods
Sampling and DNA extraction
All rose varieties used in this study were planted and collected from the National Rose DUS examination station, which is responsible for the DUS testing for rose varieties (Kunming, Yunnan). Fresh leaves were collected from healthy and strong plants, wrapped and stored on dry ice (− 70 °C) until analysis; a list of tested rose varieties is shown in Table S5. Rosa ‘Margo Koster’ was used to perform next-generation sequencing to construct the whole cp genome, and other rose varieties were used to test and validate the polymorphism of molecular markers. Genomic DNA was isolated using a DNA extraction Kit (DP-305, Tiangen Biotech, Beijing CO. LTD); agarose gel electrophoresis and a one-drop spectrophotometer were used to detect DNA integrity and quality (Spectramax I3 Microplate Reader, Molecular Devices, USA).
DNA sequencing, genome assembly, annotation and validation
DNA from Rosa ‘Margo Koster’ was used to construct shotgun libraries (250 bp) and sequences on an Illumina HiSeq 2,500 platform. Raw data were trimmed from both ends, individual bases were removed, and entire reads with a median quality score lower than Q20 or less than 25 bp in length after trimming were also discarded. After quality filtering, reads were mapped to an available cp genome of a closely related species (Rosa roxburghii; Accession: KX768420.1 GI:1104307301) using Bowtie2 v2.2.6. Then, all putative chloroplast reads mapped to the Rosa roxburghii reference above were used for de novo assembly to reconstruct the Rosa ‘Margo Koster’ cp genome using SOAPdenovo v2.04 with different kmer sizes90. The local gap filling and base correction of contigs were performed by GapCloser v1.12. Finally, in accordance with the size of the contigs, the number of scaffolds and scaffold N50, the best kmer size was used for de novo assembly. Thirty-five primer pairs were used to validate junctions using PCR-based sequencing in Rosa ‘Margo Koster’. PCR was performed using a thermal cycler (Applied Biosystems, Foster, CA, USA) with a 20 µL reaction volume as follows: 10 μL of 2 × Taq MasterMix (CWbiotech, Beijing, China), approximately 50 ng of DNA, 5 pmol forward primer, 5 pmol reverse primer, and sterile double-distilled water were added to reach the 20 μL volume. The amplifications were performed using the following schedule: denaturation at 94°C for 5 min; 35 cycles of denaturation for 30 s at 94°C, annealing for 30 s at the optimal temperature, and then extension for 30 s at 72°C; and a final extension at 72°C for 5 min. After PCR amplification, fragments were sequenced and aligned with the assembly cp genome (Table S1). Finally, the corrected cp genome was deposited into GenBank. Predictions of gene, rRNA and tRNA sequences were performed using DOGMA91 and manual correction. Gene annotation of Rosa ‘Margo Koster’ was obtained from the Nr, KEGG, COG and GO databases using BLAST2.2.28+ based on the predicted protein sequences. A circular representation of the cp genome was drawn using Organellar GenomeDRAW92, and the results of annotations were visualized by CGV93.
Repeat sequence analyses in the chloroplast genome
Simple sequence repeats (SSRs) on the cp genome were mined using MISA software (MicroSAtellite, https://pgrc.ipk-gatersleben.de/misa/). The minimal repeat units were set as mono-12, dimer-6, trimer-5, tetramer-5, pentamer-4 and hexamer-4, and Primer 3 was used for designing primer pairs in the flanking region of each candidate locus94. Tandem repeat sequences were analyzed by Tandem Repeats Finder95, the alignment parameters were set at recommended values (Match-2, Mismatch-7, Delta-7), and the minimum alignment score and maximum period size were set as 80 and 500, respectively. Palindromic repeat sequences, dispersed repeat sequences (including the forward repeats and inverse repeats) and complement repeats were analyzed by REPuter (https://bibiserv.cebitec.uni-bielefeld.de/reputerl)96, and the minimum repeat size and maximum base mismatch were set as 30 and 3, respectively.
Comparative analyses of the chloroplast genome in Rosaceae species
To perform a comparative genomic analysis within the Rosaceae family and Rosa genus, 19 species in the Rosaceae family, which are available in the NCBI database, were chosen (Table S4). Then, we used mVISTA software97 in shuffle-LANGAN mode and with default parameters for other options to compare the cp genomes from five Rosa species, using the sequenced Rosa roxburghii annotated genome as a reference. To examine expansion or contraction of the IR regions, boundaries between the four main compositions of the annotated cp genome (LSC, IRa, SSC and IRb) were inspected among 21 species using IRSCOPE software98.
To identify regions of high genetic divergence among Rosa species that could potentially inform genetic studies of the genus, the genetic divergence among five Rosa species across the entire cp genome was calculated using nucleotide diversity (π), and the total number of mutations (η) for gene and intron sequences and intergenic spacers (IGS) was aligned with Verdant and using DnaSP 5.099.
Phylogenetic analysis
The phylogenetic relationship among the Rosaceae family members was reconstructed using the partial set of species sampled in our studies, including 20 species available in NCBI, one described in our studies, and two species of different orders as outgroups (Amborella trichopoda and Nymphaea alba). First, complete cp genome sequences were aligned using MAFFT software, and then maximum likelihood (ML) and neighbor-joining (NJ) were used to reconstruct the phylogenetic tree with 2000 bootstrap replicates using MEGA 7.0 software100.
Polymorphism validation of candidate molecular markers
To verify the polymorphism of candidate cpSSRs and SNVs, genotyping PCR was performed using a thermal cycler (Applied Biosystems, Foster, CA, USA) with a 20 µL reaction volume as follows: 10 μL of 2 × Taq MasterMix (CWbiotech, Beijing, China), approximately 50 ng of DNA, 5 pmol forward primer, 5 pmol reverse primer, and sterile double-distilled water were added to reach the 20 μL volume. The amplifications were performed using the following schedule: denaturation at 94°C for 5 min; 35 cycles of denaturation for 30 s at 94°C, annealing for 30 s at the optimal temperature, and then extension for 30 s at 72°C; and a final extension at 72°C for 5 min. The PCR products were purified, analyzed by capillary electrophoresis and sequenced by Sanger’s method. Finally, polymorphisms of candidate cpSSRs and SNVs from flanking regions were aligned and analyzed using MEGA100 and DNASP5.099. Dj is the value of assessing the discriminating power of molecular marker101, and is calculated as .
Supplementary information
Acknowledgements
We thank the financial support from National Natural Science Foundation of China (Grant Number 31500538), Fundamental Research Funds for the Central Nonprofit Research Institution of CAF (Grant Number CAFYBB2016QB007), Special Funds from Laboratory of Forest Silviculture and Tree Cultivation [Grand Number. ZDRIF201713] and grant from Scientific & Development Center of National Forestry and Grassland Administration; we thank plant material supports from National Forestry and Grassland Genetic Resources Center and National Rose DUS examination station in Kunming, China. We also thank the editors and reviewers for critically evaluating the manuscript and providing constructive comments for its improvement.
Author contributions
C.L. contributed to the data analysis, experimental work and original draft; Y.Z. contributed to the conceptualization of the work, and review of the manuscript; P.H. contributed to conceptualizing the work, data analysis, editing and review of the original draft.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yongqi Zheng, Email: zhengyq@caf.ac.cn.
Ping Huang, Email: huangping@caf.ac.cn.
Supplementary information
is available for this paper at 10.1038/s41598-020-68092-1.
References
- 1.Hoober JK. Chloroplast metabolism: the structure and function of chloroplasts in green leaf cells. Bioscience. 1984 doi: 10.2307/1309996. [DOI] [Google Scholar]
- 2.Gray MW. The evolutionary origins of organelles. Trends Genet. 1989;5:294–299. doi: 10.1016/0168-9525(89)90111-X. [DOI] [PubMed] [Google Scholar]
- 3.Howe CJ, et al. Evolution of the chloroplast genome. Philos. Trans. R. Soc. B Biol. Sci. 2003;358:99–107. doi: 10.1098/rstb.2002.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sears BB. Elimination of plastids during spermatogenesis and fertilization in the plant kingdom. Plasmid. 1980;4:233–255. doi: 10.1016/0147-619X(80)90063-3. [DOI] [PubMed] [Google Scholar]
- 5.Daniell H, Lin C-S, Yu M, Chang W-J. Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 2016 doi: 10.1186/s13059-016-1004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palmer JD. Plastid chromosomes: structure and evolution. Cell Cult. Somat. Cell Genet. Plants. 1991;7:5–53. [Google Scholar]
- 7.Palmer JD, Jansen RK, Michaels HJ, Chase MW, Manhart JR. Chloroplast DNA variation and plant phylogeny. Ann. Mo. Bot. Gard. 1988;75:1180–1206. doi: 10.2307/2399279. [DOI] [Google Scholar]
- 8.Raubeson L, Jansen R, Henry RJ. Chloroplast genomes of plants. In: Henry RJ, editor. Diversity and Evolution of Plants-Genotypic and Phenotypic Variation in Higher Plants. Wallingford: CABI Publishing; 2005. [Google Scholar]
- 9.Wicke S, Schneeweiss GM, de Pamphilis CW, Muller KF, Quandt D. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol. Biol. 2011;76:273–297. doi: 10.1007/s11103-011-9762-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hae-Lim L, Jansen RK, Chumley TW, Ki-Joong K. Gene relocations within chloroplast genomes of Jasminum and Menodora (Oleaceae) are due to multiple, overlapping inversions. Mol. Biol. Evol. 2007;24:1161–1180. doi: 10.1093/molbev/msm036. [DOI] [PubMed] [Google Scholar]
- 11.Stegemann S, Hartmann S, Ruf S, Bock R. High-frequency gene transfer from the chloroplast genome to the nucleus. Proc. Natl. Acad. Sci. U. S. A. 2003;100:8828–8833. doi: 10.1073/pnas.1430924100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jose C-C, et al. A Phylogenetic analysis of 34 chloroplast genomes elucidates the relationships between wild and domestic species within the genus Citrus. Mol. Biol. Evol. 2015;32:2015–2035. doi: 10.1093/molbev/msv082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramamonjisoa L, Favreau B, Bouvet J-M, Andrianoelina O. Small effect of fragmentation on the genetic diversity of Dalbergia monticola, an endangered tree species of the eastern forest of Madagascar, detected by chloroplast and nuclear microsatellites. Ann. Bot. 2009;104:1231–1242. doi: 10.1093/aob/mcp231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao Y, et al. The complete chloroplast genome provides insight into the evolution and polymorphism of Panax ginseng. Front. Plant Sci. 2015;5:696. doi: 10.3389/fpls.2014.00696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiao-Ming Z, et al. Inferring the evolutionary mechanism of the chloroplast genome size by comparing whole-chloroplast genome sequences in seed plants. Sci. Rep. 2017;7:1555. doi: 10.1038/s41598-017-01518-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Vries DP, Dubois LAM. Rose breeding: past, present, prospects. Acta Hortic. 1996;424:241–248. doi: 10.17660/ActaHortic.1996.424.43. [DOI] [Google Scholar]
- 17.Gudin S. Rose breeding technologies. Acta Hortic. 2001;547:23–26. doi: 10.17660/ActaHortic.2001.547.2. [DOI] [Google Scholar]
- 18.Scalliet G, et al. Scent evolution in Chinese roses. Proc. Natl. Acad. Sci. U. S. A. 2008;105:5927–5932. doi: 10.1073/pnas.0711551105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin M, Piola F, Chessel D, Jay M, Heizmann P. The domestication process of the modern rose: genetic structure and allelic composition of the rose complex. Theor. Appl. Genet. 2001;102:398–404. doi: 10.1007/s001220051660. [DOI] [Google Scholar]
- 20.Guoliang, W. History of roses in cultivation. Ancient Chinese Roses. Encyclopedia of Rose Science, 387–395. 10.1016/B0-12-227620-5/00045-8 (2003).
- 21.Debener T, Linde M. Exploring complex ornamental genomes: the rose as a model plant. Crit. Rev. Plant Sci. 2009;28:267–880. doi: 10.1080/07352680903035481. [DOI] [Google Scholar]
- 22.Olivier R, et al. The Rosa genome provides new insights into the domestication of modern roses. Nat. Genet. 2018;50:772–777. doi: 10.1038/s41588-018-0110-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.International Union for the Protection of New Varieties of Plants. ROSE UPOV Code: ROSAA Rosa L. Guidelines for the conduct of tests for distinctness, uniformity and stability. https://www.upov.int/edocs/tgdocs/en/tg011.pdf (2010)
- 24.Rusanov K, et al. Microsatellite analysis of Rosa damascena Mill. accessions reveals genetic similarity between genotypes used for rose oil production and old Damask rose varieties. Theor. Appl. Genet. 2005;111:804–809. doi: 10.1007/s00122-005-2066-9. [DOI] [PubMed] [Google Scholar]
- 25.Bruneau A, Starr JR, Joly S. Phylogenetic relationships in the genus Rosa: New evidence from chloroplast DNA sequences and an appraisal of current knowledge. Syst. Bot. 2007;32:366–378. doi: 10.1600/036364407781179653. [DOI] [Google Scholar]
- 26.Takeuchi S, Nomura K, Uchiyama H, Yoneda K. Phylogenetic relationship in the genus Rosa based on the restriction enzyme analysis of the chloroplast DNA. Engei Gakkai zasshi. 2000;69:598–604. doi: 10.2503/jjshs.69.598. [DOI] [Google Scholar]
- 27.Zhu Z-M, Gao X-F, Fougère-Danezan M. Phylogeny of Rosa sections Chinenses and Synstylae (Rosaceae) based on chloroplast and nuclear markers. Mol. Phylogenet. Evol. 2015;87:50–64. doi: 10.1016/j.ympev.2015.03.014. [DOI] [PubMed] [Google Scholar]
- 28.Debener T, Bartels C, Mattiesch L. RAPD analysis of genetic variation between a group of rose cultivars and selected wild rose species. Mol. Breed. 1996;2:321–327. doi: 10.1007/BF00437910. [DOI] [Google Scholar]
- 29.Debener T, Janakiram T, Mattiesch L. Sport and seedlings of rose varieties analysed with molecular markers. Plant Breed. 2008;119:71–74. doi: 10.1046/j.1439-0523.2000.00459.x. [DOI] [Google Scholar]
- 30.Esselink G, Smulders M, Vosman B. Identification of cut rose (Rosa hybrida) and rootstock varieties using robust sequence tagged microsatellite site markers. Theor. Appl. Genet. 2003;106:277–286. doi: 10.1007/s00122-002-1122-y. [DOI] [PubMed] [Google Scholar]
- 31.Esselink D, Vosman B, Smulders MJM, Voorrips RE. Analysis of a database of DNA profiles of 734 hybrid tea rose varieties. Acta Hort. 2009;2009:169–174. doi: 10.17660/ActaHortic.2009.836.24. [DOI] [Google Scholar]
- 32.Jian HY, et al. Characterization of the complete chloroplast genome of a critically endangered decaploid rose species, Rosa praelucens (Rosaceae) Conserv. Genet. Resour. 2018;10:851–854. doi: 10.1007/s12686-017-0946-3. [DOI] [Google Scholar]
- 33.Hong-Ying J, et al. The complete chloroplast genome of a key ancestor of modern roses, Rosa chinensis var. spontanea, and a comparison with congeneric species. Molecules. 2018;23:389. doi: 10.3390/molecules23020389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qian W, Hu H, An J, Bai G, Liu J. Complete chloroplast genome sequence of Rosa roxburghii and its phylogenetic analysis. Mitochondrial DNA Part B. 2018;3:149–150. doi: 10.1080/23802359.2018.1431074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang J-B, Li D-Z, Li H-T. Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs. Mol. Ecol. Resour. 2014;14:1024–1031. doi: 10.1111/1755-0998.12251. [DOI] [PubMed] [Google Scholar]
- 36.Yang H, et al. The complete chloroplast genome sequences of the medicinal plant Pogostemon cablin. Int. J. Mol. Sci. 2016;17:820. doi: 10.3390/ijms17060820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jansen RK, Christopher S, Seung-Bum L, Hansen AK, Henry D. Complete plastid genome sequences of three Rosids (Castanea, Prunus, Theobroma): evidence for at least two independent transfers of rpl22 to the nucleus. Mol. Biol. Evol. 2010;28:835–847. doi: 10.1093/molbev/msq261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gonçalves NA, Monica C, Garcez LL. Complete chloroplast genome of Tanaecium tetragonolobum: the first Bignoniaceae Plastome. PLoS ONE. 2015;10:e0129930. doi: 10.1371/journal.pone.0129930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shu-Dong Z, et al. Diversification of Rosaceae since the Late Cretaceous based on plastid phylogenomics. New Phytol. 2017;214:1355–1367. doi: 10.1111/nph.14461. [DOI] [PubMed] [Google Scholar]
- 40.Shuai G, Jiang X, Xiaogai H. Complete chloroplast genome sequence and phylogenetic analysis of Paeonia ostii. Molecules. 2018;23:246. doi: 10.3390/molecules23020246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiaojun N, et al. Complete chloroplast genome sequence of a major invasive species, crofton weed (Ageratina adenophora) PLoS ONE. 2012;7:e36869. doi: 10.1371/journal.pone.0036869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jun Q, et al. The complete chloroplast genome sequence of the medicinal plant Salvia miltiorrhiza. PLoS ONE. 2013;8:e57607. doi: 10.1371/journal.pone.0057607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goulding SE, Wolfe KH, Olmstead RG, Morden CW. Ebb and flow of the chloroplast inverted repeat. MGG Mol. Gen. Genet. 1996;252:195–206. doi: 10.1007/bf02173220. [DOI] [PubMed] [Google Scholar]
- 44.Wang R-J, et al. Dynamics and evolution of the inverted repeat-large single copy junctions in the chloroplast genomes of monocots. BMC Evol. Biol. 2008;8:36. doi: 10.1186/1471-2148-8-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chumley TW, et al. The complete chloroplast genome sequence of Pelargonium × hortorum: organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol. Biol. Evol. 2006;23:2175–2190. doi: 10.1093/molbev/msl089. [DOI] [PubMed] [Google Scholar]
- 46.Wakasugi T, et al. Loss of all ndh genes as determined by sequencing the entire chloroplast genome of the black pine Pinus thunbergii. Proc. Natl. Acad. Sci. U. S. A. 1994;91:9794–9798. doi: 10.2307/2365708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tomonori H, Atsushi W, Manabu K, Teiji K, Katsuhiko T. Complete nucleotide sequence of the Cryptomeria japonica D. Don. chloroplast genome and comparative chloroplast genomics: diversified genomic structure of coniferous species. BMC Plant Biol. 2008;8:70. doi: 10.1186/1471-2229-8-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guisinger MM, Kuehl JV, Boore JL, Jansen RK. Extreme reconfiguration of plastid genomes in the angiosperm family Geraniaceae: rearrangements, repeats, and codon usage. Mol. Biol. Evol. 2011;28:1543. doi: 10.1093/molbev/msq229. [DOI] [PubMed] [Google Scholar]
- 49.Chen J, et al. The complete chloroplast genome sequence of the relict woody plant Metasequoia glyptostroboides Hu et Cheng. Front. Plant Sci. 2015;6:447. doi: 10.3389/fpls.2015.00447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huotari T, Korpelainen H. Complete chloroplast genome sequence of Elodea canadensis and comparative analyses with other monocot plastid genomes. Gene. 2012;508:96–105. doi: 10.1016/j.gene.2012.07.020. [DOI] [PubMed] [Google Scholar]
- 51.Tobias D, Copley RR, Jörg S, Ponting CP, Peer B. Systematic identification of novel protein domain families associated with nuclear functions. Genome Res. 2002;12:47–56. doi: 10.1101/gr.203201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Drescher A, Ruf S, Calsa T, Jr, Carrer H, Bock R. The two largest chloroplast genome-encoded open reading frames of higher plants are essential genes. Plant J. 2000;22:104. doi: 10.1046/j.1365-313x.2000.00722.x. [DOI] [PubMed] [Google Scholar]
- 53.Edh K, Widén B, Ceplitis A. Nuclear and chloroplast microsatellites reveal extreme population differentiation and limited gene flow in the Aegean endemic Brassica cretica (Brassicaceae) Mol. Ecol. 2007;16:4772–4783. doi: 10.1111/j.1365-294X.2007.03585.x. [DOI] [PubMed] [Google Scholar]
- 54.Su CK, Gi CM, SeonJoo P. The complete chloroplast genome sequences of three Veroniceae species (Plantaginaceae): comparative analysis and highly divergent regions. Front. Plant Sci. 2016;7:355. doi: 10.3389/fpls.2016.00355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gong Y-Q, Gong X. Pollen-mediated gene flow promotes low nuclear genetic differentiation among populations of Cycas debaoensis (Cycadaceae) Tree Genet. Genomes. 2016;12:93. doi: 10.1007/s11295-016-1051-6. [DOI] [Google Scholar]
- 56.Sundar RP, et al. Nuclear and chloroplast DNA variation provides insights into population structure and multiple origin of native aromatic rices of Odisha India. PLoS ONE. 2016;11:e0162268. doi: 10.1371/journal.pone.0162268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jakobsson M, Säll T, Lind-Halldén C, Halldén C. Evolution of chloroplast mononucleotide microsatellites in Arabidopsis thaliana. Theor. Appl. Genet. 2007;114:223–235. doi: 10.1007/s00122-006-0425-9. [DOI] [PubMed] [Google Scholar]
- 58.Huang LS, et al. Development of high transferability cp SSR markers for individual identification and genetic investigation in Cupressaceae species. Ecol. Evol. 2018;8:4967–4977. doi: 10.1002/ece3.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bodin SS, Kim JS, Kim J-H. Complete chloroplast genome of Chionographis japonica (Willd.) Maxim. (Melanthiaceae): comparative genomics and evaluation of universal primers for Liliales. Plant Mol. Biol. Rep. 2013;31:1407–1421. doi: 10.1007/s11105-013-0616-x. [DOI] [Google Scholar]
- 60.Ji M, et al. The complete chloroplast genome sequence of Mahonia bealei (Berberidaceae) reveals a significant expansion of the inverted repeat and phylogenetic relationship with other angiosperms. Gene. 2013;528:120–131. doi: 10.1016/j.gene.2013.07.037. [DOI] [PubMed] [Google Scholar]
- 61.Dai-Yong K, et al. Complete chloroplast genome sequence of Magnolia kwangsiensis (Magnoliaceae): implication for DNA barcoding and population genetics. Genome. 2011;54:663–673. doi: 10.1139/g11-026. [DOI] [PubMed] [Google Scholar]
- 62.Do HDK, Kim JS, Kim J-H. Comparative genomics of four Liliales families inferred from the complete chloroplast genome sequence of Veratrum patulum O. Loes. (Melanthiaceae) Gene. 2013;530:229–235. doi: 10.1016/j.gene.2013.07.100. [DOI] [PubMed] [Google Scholar]
- 63.Perdereau AC, Kelleher CT, Douglas GC, Hodkinson TR. High levels of gene flow and genetic diversity in Irish populations of Salix caprea L. inferred from chloroplast and nuclear SSR markers. BMC Plant Biol. 2014;14:202. doi: 10.1186/s12870-014-0202-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Harada K, et al. Genetic variation and structure of Ubame oak, Quercus phillyraeoides, in Japan revealed by chloroplast DNA and nuclear microsatellite markers. Genes Genet. Syst. 2018;93:37–50. doi: 10.1266/ggs.17-00026. [DOI] [PubMed] [Google Scholar]
- 65.UPOV Working Group on Biochemical & Molecular Techniques & DNA-Profiling in Particular (BMT/2). https://www.upov.int/meetings/en/details.jsp?meeting_id=2398(1994).
- 66.Singh RK, et al. Suitability of mapped sequence tagged microsatellite site markers for establishing distinctness, uniformity and stability in aromatic rice. Euphytica. 2004;135:135–143. doi: 10.1023/b:euph.0000014905.10397.08. [DOI] [Google Scholar]
- 67.Spooner DM, Nuñez J, Rodríguez F, Naik PS, Ghislain M. Nuclear and chloroplast DNA reassessment of the origin of Indian potato varieties and its implications for the origin of the early European potato. Theor. Appl. Genet. 2005;110:1020–1026. doi: 10.1007/s00122-004-1917-0. [DOI] [PubMed] [Google Scholar]
- 68.Meegahakumbura MK, et al. Domestication origin and breeding history of the tea plant (Camellia sinensis) in China and India based on nuclear microsatellites and cpDNA sequence data. Front. Plant Sci. 2018;8:2270. doi: 10.3389/fpls.2017.02270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Oyant HS, Crespel L, Rajapakse S, Zhang L, Foucher F. Genetic linkage maps of rose constructed with new microsatellite markers and locating QTL controlling flowering traits. Tree Genet. Genomes. 2008;4:11–23. doi: 10.1007/s11295-007-0084-2. [DOI] [Google Scholar]
- 70.Hong JH, Kwon Y-S, Suh J-N, Choi K-J. Identification of Rose (Rosa × hybrida) varieties using simple sequence repeat (SSR) markers. Korean J. Breed. Sci. 2013;45:96–103. doi: 10.9787/KJBS.2013.45.2.096. [DOI] [Google Scholar]
- 71.Heo MS, Han K, Kwon JK. Development of SNP markers using genotyping-by-sequencing for cultivar identification in rose (Rosa hybrida) Horticult. Environ. Biotechnol. 2017;58:292–302. doi: 10.1007/s13580-017-0268-0. [DOI] [Google Scholar]
- 72.Muqing Y, et al. Genotyping-by-sequencing application on diploid rose and a resulting high-density SNP-based consensus map. Horticult. Res. 2018;5:17. doi: 10.1038/s41438-018-0021-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vukosavljev M, et al. High-density SNP-based genetic maps for the parents of an outcrossed and a selfed tetraploid garden rose cross, inferred from admixed progeny using the 68k rose SNP array. Horticult. Res. 2016;3:16052. doi: 10.1038/hortres.2016.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jian HY, et al. The complete chloroplast genome of a key ancestor of modern roses, Rosa chinensis var. spontanea, and a comparison with congeneric species. Molecules. 2018;23:389. doi: 10.3390/molecules23020389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jeon J-H, Kim S-C. Comparative analysis of the complete chloroplast genome sequences of three closely related East-Asian Wild Roses (Rosa sect. Synstylae; Rosaceae) Genes. 2019 doi: 10.3390/genes10010023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mohanty A, Martín JP, Aguinagalde I. Chloroplast DNA diversity and distinguishing sweet cherry cultivars: a new approach. Acta Hortic. 2001;546:463–469. doi: 10.17660/ActaHortic.2001.546.63. [DOI] [Google Scholar]
- 77.Khan IA, Azim MK. Variations in intergenic spacer rpl20-rps12 of mango (Mangifera indica) chloroplast DNA: implications for cultivar identification and phylogenetic analysis. Plant Syst. Evol. 2011;292:249–255. doi: 10.1007/s00606-011-0424-4. [DOI] [Google Scholar]
- 78.Yue X, et al. Combined analyses of chloroplast DNA haplotypes and microsatellite markers reveal new insights into the origin and dissemination route of cultivated pears native to East Asia. Front. Plant Sci. 2018;9:591. doi: 10.3389/fpls.2018.00591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Intrieri MC, Muleo R, Buiatti M. Chloroplast DNA polymorphisms as molecular markers to identify cultivars of Olea europaea L. J. Pomol. Hortic. Sci. 2007;82:109–113. doi: 10.1080/14620316.2007.11512206. [DOI] [Google Scholar]
- 80.Cho MS, et al. Molecular and morphological data reveal hybrid origin of wild Prunus yedoensis (Rosaceae) from Jeju Island, Korea: implications for the origin of the flowering cherry. Am. J. Bot. 2014;101:1976–1986. doi: 10.3732/ajb.1400318. [DOI] [PubMed] [Google Scholar]
- 81.Yang J, et al. Indica-japonica differentiation of chloroplast DNA of weedy rice in the Changjiang and Huaihe River Valley of China. Chin. J. Rice Sci. 2009;23:391–397. [Google Scholar]
- 82.Lv, Y. M., Yin, J., Yang, G. & Zhang, Q. X. CpSSR analysis of Prunus mume cultivar resources. J. Beijing For. Univ. (2007).
- 83.Mengoni A, Ruggini C, Vendramin GG, Bazzicalupo M. Chloroplast microsatellite variations in tetraploid alfalfa. Plant Breed. 2000;119:509–512. doi: 10.1046/j.1439-0523.2000.00537.x. [DOI] [Google Scholar]
- 84.Huang L, et al. Development of high transferability cpSSR markers for individual identification and genetic investigation in Cupressaceae species. Ecol. Evol. 2018;8:4967–4977. doi: 10.1002/ece3.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Phumichai C, Phumichai T, Wongkaew A. Novel chloroplast microsatellite (cpSSR) markers for genetic diversity assessment of cultivated and wild Hevea Rubber. Plant Mol. Biol. Rep. 2015;33:1486–1498. doi: 10.1007/s11105-014-0850-x. [DOI] [Google Scholar]
- 86.Rafalski A. Applications of single nucleotide polymorphisms in crop genetics. Curr. Opin. Plant Biol. 2002 doi: 10.1016/S1369-5266(02)00240-6. [DOI] [PubMed] [Google Scholar]
- 87.Koning-Boucoiran CF, et al. Using RNA-Seq to assemble a rose transcriptome with more than 13,000 full-length expressed genes and to develop the WagRhSNP 68k Axiom SNP array for rose (Rosa L.) Front. Plant Sci. 2015;6:249. doi: 10.3389/fpls.2015.00249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Heo M-S, Han K, Kwon J-K, Kang B-C. Development of SNP markers using genotyping-by-sequencing for cultivar identification in rose (Rosa hybrida) Hortic. Environ. Biotechnol. 2017;58:292–302. doi: 10.1007/s13580-017-0268-0. [DOI] [Google Scholar]
- 89.Bourke PM, et al. Partial preferential chromosome pairing is genotype dependent in tetraploid rose. Plant J. 2017;90:330–343. doi: 10.1111/tpj.13496. [DOI] [PubMed] [Google Scholar]
- 90.Ru L. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wyman SK, Boore JL, Jansen RK. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20:3252–3255. doi: 10.1093/bioinformatics/bth352. [DOI] [PubMed] [Google Scholar]
- 92.Lohse M, Drechsel O, Bock R. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 2007;52:267–274. doi: 10.1007/s00294-007-0161-y. [DOI] [PubMed] [Google Scholar]
- 93.Tominski C, Abello J, Schumann H. CGV—an interactive graph visualization system. Comput. Gr. 2009;33:660–678. doi: 10.1016/j.cag.2009.06.002. [DOI] [Google Scholar]
- 94.Untergasser A, et al. Primer3—new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kurtz S, Schleiermacher C. REPuter: fast computation of maximal repeats in complete genomes. Bioinformatics (Oxford, England) 1999;15:426–427. doi: 10.1093/bioinformatics/15.5.426. [DOI] [PubMed] [Google Scholar]
- 97.Frazer KA, Lior P, Alexander P, Rubin EM, Inna D. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:273–279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ali A, Jaakko H, Peter P. IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics (Oxford, England) 2018;34:3030–3031. doi: 10.1093/bioinformatics/bty220. [DOI] [PubMed] [Google Scholar]
- 99.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 100.Sudhir K, Glen S, Koichiro T. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tessier C, David J, This P, Boursiquot JM, Charrier A. Optimization of the choice of molecular markers for varietal identification in Vitis vinifera L. Theor. Appl. Genet. 1999;98:171–177. doi: 10.1007/s001220051054. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.