Abstract
Alzheimer's disease (AD) is a chronic neurodegenerative disorder. It is the most common type of dementia that has remained as an incurable disease in the world, which destroys the brain cells irreversibly. In this study, a systems biology approach was adopted to discover novel micro-RNA and gene-based biomarkers of the diagnosis of Alzheimer's disease. The gene expression data from three AD stages (Normal, Mild Cognitive Impairment, and Alzheimer) were used to reconstruct co-expression networks. After preprocessing and normalization, Weighted Gene Co-Expression Network Analysis (WGCNA) was used on a total of 329 samples, including 145 samples of Alzheimer stage, 80 samples of Mild Cognitive Impairment (MCI) stage, and 104 samples of the Normal stage. Next, three gene-miRNA bipartite networks were reconstructed by comparing the changes in module groups. Then, the functional enrichment analyses of extracted genes of three bipartite networks and miRNAs were done, respectively. Finally, a detailed analysis of the authentic studies was performed to discuss the obtained biomarkers. The outcomes addressed proposed novel genes, including MBOAT1, ARMC7, RABL2B, HNRNPUL1, LAMTOR1, PLAGL2, CREBRF, LCOR, and MRI1and novel miRNAs comprising miR-615-3p, miR-4722-5p, miR-4768-3p, miR-1827, miR-940 and miR-30b-3p which were related to AD. These biomarkers were proposed to be related to AD for the first time and should be examined in future clinical studies.
Subject terms: Biomarkers, Biomarkers, Genetics research, Systems biology
Introduction
Alzheimer is an incurable neurological disorder and is classified as an aging disease. It is one of the important neurological complications which can affect the whole society ranging from the patients themselves to the people who are around them. The aging population is growing in many countries, and the treatment costs of Alzheimer are dramatically high. These issues have drawn the attention of many researchers to the importance of the examination of this disease1. There are many organizations all over the world which work in the field of early diagnosis and prevention of Alzheimer2,3. National center for health statistics considers Alzheimer's disease as the sixth cause of death in the United States4. As a result, Alzheimer's disease is among the costliest diseases for various socioeconomic classes. As the population of the world grows, the number of inflicted people increases. Therefore, the control of the affected population becomes more difficult5. Significant advances in medical and neurological sciences have led to a longer life expectancy and have increased the number of Alzheimer's disease patients. Ultimately, the prevention of disease before its occurrence is regarded to be one of the most important pillars of treatment at different stages of this disease. Treatment or postponement of a disease depends on its discovery by identifying the biological pathways involved in the disease and adopting various drug-disease network approaches6 to control these pathways.
In recent decades, deep investigation of molecular mechanisms has become more prevalent as a research method for finding effective treatments for complex diseases such as cancer, diabetes, Alzheimer, and so on. Microarray and Next-Generation Sequencing (NGS) are the most common technologies utilized in the relevant research studies7. Related studies that use deep investigation methods focus on the preprocessing stage, which involves the different feature selection or dimension reduction algorithms7,8. Almost all of the previous studies in this field can be classified into two main categories. The studies in the first category have adopted image processing approaches based on brain images (e.g., MRI)9–17.
On the other hand, the studies in the second category have used gene expression data to predict the chance of developing Alzheimer’s disease1,6,18–28. An article that used Linear discriminant analysis as the best separation procedure is an example of the studies which fall into the first category. It used pathway analysis to distinguish between different stages of Alzheimer. More specifically, it classified different stages of Alzheimer’s disease using pathway analysis29. The investigation of meta-analysis studies and the examination of the studies which fall into these two categories highlight a significant gap in the relevant literature on Alzheimer’s disease and reveal that there is a need for further research on this disease.
A brief description of weighted gene co-expression network analysis (WGCNA) and some of the related studies which have adopted this approach is necessary to clarify our method. The WGCNA describes patterns that are constructed as a result of the correlation between the genes in microarray data. It is one of the system biology methods and is used in this study. It is a very useful R package that can be used to construct gene co-expression networks or to discover modules and correlations between genes. Moreover, it can also be utilized to identify Eigen genes or intra-modular hub genes or to calculate measurement values for the module memberships and topological properties30. A study adopted this method and used gene and miRNA expression data to discover some diagnostic biomarkers for the early detection of Colorectal Cancer. First, it utilized clustering to extract low preserved modules by constructing the co-expression networks for the different stages of colorectal cancer. Second, it reported two novel miRNAs that were related to colorectal cancer as biomarkers for this type of cancer by validating gene-miRNA interactions and constructing bipartite networks. These miRNAs were not reported in the previous studies31.
Furthermore, another study used a similar method for discovering diagnostic biomarkers of the stratification of Breast Cancer molecular subtypes. It reported two or three miRNAs for each subtype and their target genes, which were significant and were highlighted in basic mechanisms of this cancer32. Moreover, there is a study that applied the WGCNA to find the key genes in Alzheimer’s disease and introduced them as potential targets in the therapy for this disease33. Another study in this field did not specifically deal with Alzheimer’s disease and concentrated on the aging of the brain. WGCNA was used in this study to identify the significant modules and effective biomarkers of the aging human brain34.
Many studies have been carried out in this field, and numerous proposed Alzheimer’s biomarkers have been introduced. However, the disorder remains incurable. This issue stems from the fact that its critical biological pathways, along with the involved functional genes, have not been fully discovered. For example, a recent research study applied WGCNA by focusing on gene targets and their pathways to investigate Alzheimer’s disease. However, it did not examine miRNAs33.
Therefore, our study broadens the scope of the previous studies and explores the relations between genes and their target miRNAs by constructing bipartite networks.
In this study, the exploration of the variations of genes expression between different steps by extracting related modules helps to find important interactions. Moreover, the examination of the related pathways helps us to develop a proper understanding of the development of Alzheimer’s disease. In the “Results” section, dataset details and the outcomes of the relevant experiments are illustrated and explained using the appropriate chart and tables. The “Discussion” section supports the applicability of the introduced method based on medical and clinical evidence. The proposed methods of our study are introduced in the “Methods” section, which is the last part of this article.
Results
First, the details of the used database are presented, and the adopted methodological approaches are discussed step by step.
Dataset and preprocessing
The dataset of this study was downloaded from the National Center for Biotechnology Information Gene expression Omnibus (GEO) using GSE63063 accession number. The platform of the chip analyzer was GPL6947.
First, before the preprocessing of the dataset, the non-gene transcripts were eliminated from the original file. Second, the remaining data were statically tested and preprocessed using “Limma” R package from the Bioconductor project, which was conducted in the RStudio ver.1.1.423 programming environment. Third, Benjamini & Hochberg’s false discovery rate method was applied to calculate the adjusted p-values. The genes were attributed to their related IDs, and the duplicated or ID-less genes were excluded. After sorting the genes according to their adjusted p-value, the significant genes with adjusted p-value < 0.01 were selected for further analyses, and the remaining ones were omitted. After preprocessing and removing outliers’ data, we determined 6,179 genes which were utilized as the gene list. This list enabled us to construct the network and perform further analysis.
Moreover, our database was narrowed down to 104 normal samples, including 80 samples of mild cognitive impairment, and 145 samples of Alzheimer’s disease cases. The samples which were related to three stages of the disease were summed up in 329 samples, including 200 female and 129 male patients. The boxplots of Fig. 1 show the range and dispersion of samples according to age and gender at every three stages. Moreover, Table 1 shows the number of samples at each stage based on gender.
Figure 1.
Dispersion of data samples according to their age and gender (Normal Healthy samples, MCI Mild Cognitive Impairment, AD Alzheimer’s Disease).
Table 1.
Number of data samples according to their gender.
| Gender | Alzheimer | Mild cognitive impairment | Normal |
|---|---|---|---|
| Male | 46 | 41 | 42 |
| Female | 99 | 39 | 62 |
| Total | 145 | 80 | 104 |
Weighted gene co-expression network analysis (WGCNA)
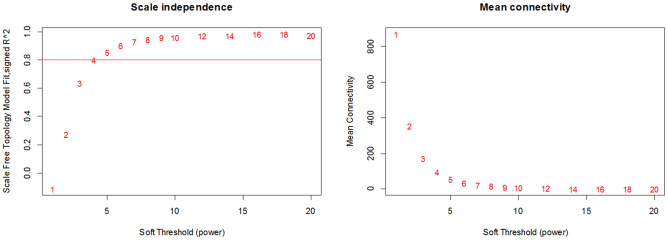
To construct the co-expression network, 6,179 genes from the 329 samples at three different stages were included. Figure 2 illustrates scale dependency by R2 and the mean connectivity, along with different values of the soft threshold. Among the powers which ranged from 1 to 20, the value of 4 was selected for β to gain the scale independency of the network at Normal stage, where the scale-free index R2 was 0.8. Consequently, the soft threshold values for the MCI stage and AD stage were set to 4 and 6 respectively (Supplementary Fig. S1 and S2).
Figure 2.
Scale independence measure is shown by R2 and the mean connectivity value to determine the optimal value of the soft threshold, which ensures a scale-free network and retains important correlations.
Next, the adjacency matrix of the expression data was generated. Based on this matrix, we calculated our topological overlap matrix (TOM). The modules were selected using a tree-cut algorithm. Moreover, 4 and 20 values were used as deepSplit and minimal module size parameters respectively by examining different parameters. The extracted modules were merged in the following step and were labeled with colors. The threshold of 0.14 was chosen to merge the modules. The merged modules of all stages were presented in the Supplementary file (Supplementary Figs. S3–S5). The preservation measure, which is indicated in the Z_summary index, was used to select the effective modules. The modules were strongly preserved when the Z_summary value was equal to or larger than 10. The values which were between 2 and 10, were regarded to be moderately preserved. Finally, the values, which were equal to or smaller than 2, indicated the lack of preservation. The modules, whose Z_summary values were larger than 10, were strongly preserved and did not give us any information. Therefore, we did not use them.
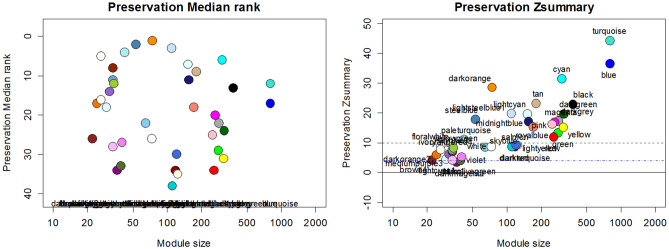
However, according to the obtained values, there were not any modules with z_summary values smaller than 2. Consequently, we selected 3, 5, and 3 as thresholds for Normal-MCI, MCI-AD, and Normal-AD module groups respectively. After choosing the thresholds, the Z_summary values of the selected modules were the values that ranged from 3.45 to 4.56 for Normal-MCI modules. Moreover, they ranged from 5.25 to 6.91 for MCI-AD modules and ranged from 3.41 to 5.33 for Normal-AD modules. To gain proper Zsummary values, we examined the deep-split parameter and the types of networks using separate execution procedures. Finally, the signed-hybrid network was set as the type of networks, and the value of deep-split was set to 4. Figure 3 illustrates the preservation of median rank and preservation of Z_summary along with the module size.
Figure 3.
Modules of the Normal stage against the MCI expression data according to their Zsummary (The charts including Supplementary Figs. S6 and S7, which are related to the other two stages are available in the Supplementary file).
Five modules of the normal stage were selected in comparison with the MCI expression data (Normal-MCI modules). Zsummary values of these five modules were smaller than 4.5. Six modules of the MCI stage were chosen with Zsummary values smaller than 6.9 compared with the AD expression data (MCI-AD modules). Furthermore, six modules of the normal stage were selected compared with the AD expression data (Normal-AD modules) that had Zsummary values smaller than 5.3. The selected modules are illustrated by their attributes in Table 2.
Table 2.
Extracted modules and their properties at three stages.
| Number of modules | Value of zsummary | Module color | Number of genes in module | |
|---|---|---|---|---|
| Normal | ||||
| 1 | 3 | 4.121447 | Brown4 | 302 |
| 2 | 7 | 3.456988 | Darkmagneta | 36 |
| 3 | 8 | 4.066048 | Darkolivegreen | 39 |
| 4 | 19 | 4.568499 | Lightcyan1 | 29 |
| 5 | 28 | 4,190,195 | Plum1 | 33 |
| MCI | ||||
| 1 | 7 | 5.254516 | Darkmagneta | 26 |
| 2 | 8 | 6.120115 | Darkolivegreen | 27 |
| 3 | 20 | 6.261032 | Paleturquoise | 31 |
| 4 | 26 | 6.434162 | Sienna3 | 26 |
| 5 | 28 | 6.918607 | Steelblue | 36 |
| 6 | 29 | 5.797834 | Violet | 28 |
| Alzheimer | ||||
| 1 | 3 | 4.455773 | Brown4 | 22 |
| 2 | 7 | 3.410899 | Darkmagneta | 36 |
| 3 | 8 | 4.958944 | Darkolivegreen | 39 |
| 4 | 19 | 4.924977 | Lightcyan1 | 29 |
| 5 | 28 | 4.263592 | Plum1 | 33 |
| 6 | 37 | 5.335066 | Violet | 40 |
Gene-miRNA bipartite network
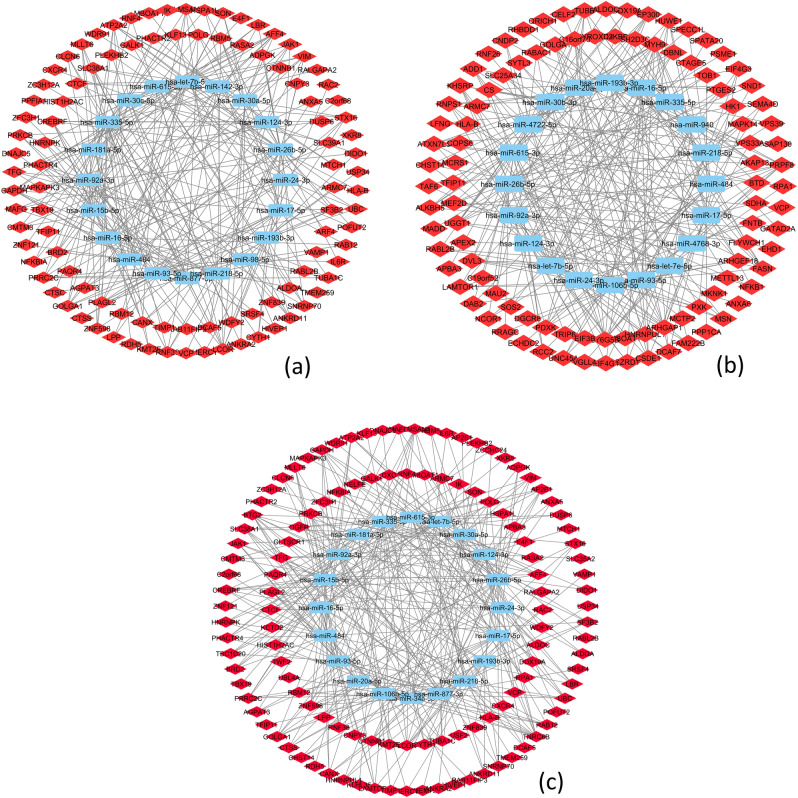
This part aims to analyze the relations among the obtained genes and their related miRNAs by which they are regulated. However, after constructing three bipartite networks, hub miRNAs with the highest connectivity degree were selected to reduce the complexity and to focus on the important connections. In this section, 20 miRNAs and their connections were selected. Therefore, the genes were also filtered by the ones that were at the end of this connection. In the obtained subnetworks, which are shown in Fig. 4, there were 116 genes of the Normal-MCI subnetwork, 131 genes of the MCI-AD subnetwork, and 145 genes of Normal-AD subnetwork.
Figure 4.
Bipartite gene-miRNA subnetworks for all of the three stages. Each subnetwork includes 20 miRNAs. (a) Normal-MCI subnetwork. (b) MCI-AD subnetwork and (c) Normal-AD subnetwork.
Enrichment analysis
Enrichment analysis of genes
To extract the important pathways using the DAVID database, the pathways with normal p-value < 0.05 were selected as significant on the studied gene list. The p-value of the most significant pathway in our experiment was equal to 0.0028 and involved six genes. This pathway is called Spliceosome and is found in Normal-MCI subnetwork. In MCI-AD subnetwork, the Herpes simplex infection was our substantial pathway. Its p-value was equal to 0.0063, and it included seven significant genes. Similar to the first group, in the third subnetwork, called Normal-AD, the important pathway was Spliceosome. Its p-value was equal to 0.005 and involved six genes. The tables which were extracted for biological pathways are available in the Supplementary File (Supplementary Tables S1–S3).
Gene ontology analysis of these three subnetworks indicated that in the Normal-MCI subnetwork, the regulation of the gene metabolic process (p-value = 2.18e−04), which involved seven genes, was the most important process. Vesicle-mediated transport (p-value = 4.87e−04) with 22 genes, apoptotic process (p-value = 6.33e−04) with 24 genes, and regulation of RNA splicing (p-value = 6.89e−04) with 6 genes were the following important processes respectively. In the second subnetwork, called MCI-AD, posttranscriptional regulation of gene expression (p-value = 3.01e−05) with 14 genes, regulation of translation (p-value = 1.65e−04) with 11 genes, regulation of cellular amide metabolic process (p-value = 3.29e−04) with 11 genes, and regulation of cellular protein metabolic process (p-value = 7.70e−04) with 31 genes were the most important processes respectively. In the third subnetwork, called Normal-AD, regulation of gene metabolic process (p-value = 1.04e−06) with 10 genes, gene metabolic process (p-value = 7.60e−05) with 17 genes, viral process (p-value = 1.96e−04) with 20 genes, multi-organism cellular process (p-value = 2.15e−04) with 20 genes, vesicle-mediated transport (p-value = 2.28e−04) with 26 genes, interspecies interaction between organisms (p-value = 2.96e−04) with 20 genes, symbiosis, encompassing mutualism through parasitism (p-value = 2.96e−04) with 20 genes, and positive regulation of gene metabolic process (p-value = 6.85e−04) with 5 genes were the most significant processes respectively. All of the biological processes by related genes are available in Supplementary File (Supplementary Tables S4–S6).
Enrichment analysis of miRNAs
At this stage, the hub miRNAs were evaluated using TAM tool35, and the type of p-value was normal. miRNAs found in Normal-MCI subnetwork were almost from the miR-15 and miR-17 family, and their p-values were 0.0097 and 0.0032, respectively. The most important functions in this subnetwork were Apoptosis, Cell cycle-related, HIV latency, Hormones regulation, Human embryonic stem cell (hESC) regulation, cell proliferation, and immune system. The second subnetwork was almost from the miR-17 family. Its p-value was equal to 1.54e−06. The important functions related to this subnetwork were Angiogenesis, HIV latency, Hormones regulation, Human embryonic stem cell (hESC) regulation, cell proliferation, and immune system. Finally, the third subnetwork was almost from the miR-15 family and miR-17 family. Its p-values were equal to 0.0097 and 0.0002, respectively. The important functions of this subnetwork were Apoptosis, HIV latency, Hormones regulation, Human embryonic stem cell (hESC) regulation, cell proliferation, and immune system. The related tables are available in Supplementary file (Supplementary Tables S7–S12).
Detailed investigation over obtained biomarkers
First, the genes, which were regulated by the obtained miRNAs, were investigated. The first group involved nine genes that were obtained as a result of the intersection of three module groups. These genes were GOLGA1, HLA-B, MBOAT1, RABL2B, ARMC7, IL10RB, STX5, TFIP11, and VCP. The first one, GoLGA1, was introduced as one of the age-regulated genes36. The other gene, HLA-B, had high sensitivity and high specificity measurement values and was regarded to be a signal that showed the patients who suffered from hypersensitivity syndrome (HSS)37. One study compared AD and normal samples and revealed a significant difference in their HLA-B frequency38. The next gene, RABL2B, was introduced as a gene that is involved in a kind of neurological deficit called Phelan-McDermid Syndrome (PMS) 39,40.
IL10RB was obtained as one of the cell signaling molecules in the aging disease of the young population in 201441. Another study used blood samples to determine early Alzheimer markers. It mentioned IL10RB as one of the best discriminators which distinguished between AD and normal samples42. A study in 2015 listed some previous studies that presented STX5 as the protein which plays a role in Alzheimer and Parkinson diseases43. Another study examined protein–protein interaction networks and their impact on gene-network analysis using AD gene expression data. It introduced TFIP11 as a significant hub gene44. The VCP gene is known to have a positive relationship with AD development risk based on the investigation of different types of dementia45. VCP mutations were investigated in another study that illustrated the vital role of these mutations in frontotemporal dementia46. Another study showed different genetic variants in genes like VCP which were associated with frontotemporal dementia and its related diseases47. According to another study, a mutation of VCP is related to Parkinson and Alzheimer diseases48.
The next group involved the genes which belonged to the overlap between MCI-AD module groups and CTL-AD module groups. There were eight genes in this group, including ALDOC, APBA3, CHST14, DDX19A, HNRNPUL1, KCTD2, LAMTOR1, and RPA1. The first one, ALDOC, was found to be related to Alzheimer’s disease in a study that investigated the proteomics in Alzheimer’s brain49. Another study, which discussed this gene, examined AD pathogenesis and its related key regulators50. The second gene, APBA3, and its interaction with beta-amyloid highlight the importance of the examination of its genomic structure51.
Similarly, another study investigated APBA3 as the gene which had a regulatory role in Alzheimer’s disease52. It has been shown that the third gene in this group, CHST14, and its relationship with impaired cognitive function affect the learning and memory abilities53. The next one, DDX19A, is among the AD-associated genes. This issue has been proved by imaging-wide association study (IWAS) and transcriptome-wide association study (TWAS)54. The next one, KCTD2, was found to be related to AD based on the results of a relevant study55. Another study, which focused on the genetic similarity between AD and Ischaemic Stroke (IS), found that KCTD2 was associated with both of these diseases56. The last gene in this group, PRA1, was mentioned in a study that compared the expression of nucleonic excision repair (NER) in AD according to brain tissues and blood. In both of these cases, RPA1 showed lower expression in AD samples in comparison with the healthy ones57. Two genes of this group, including HNRNPUL1 and LAMTOR1, have not been found in clinical research studies.
In this step, ten genes with larger degrees were selected from among the 107 genes which belonged to the overlap between CTL-MCI module groups and CTL-AD module groups to extend the exploration of the obtained results. These genes were PLAGL2, CREBRF, LCOR, ALDOA, LPP, KLF13, CANX, MRI1, STX16, and SLC38A1. One study showed that the variations of ALDOA were associated with Alzheimer’s disease58. Another study, which indicated the biomarkers of AD pathology, revealed that ALDOA was one of the obtained ones59. A different study showed that LPP was suppressed considerably in MCI samples in comparison with the healthy ones60. Another study named some of the genes that had regulation changes between healthy and AD samples and argued that LPP was one of them61. KLF13 was introduced in a neurodegenerative disease study as one of the key regulators in Alzheimer’s disease62. A similar study, which examined this disease between male and female samples, found that KLF13 existed in four male clinical traits in male patients63. The next one, CANX, was mentioned in a study as a target that had an important role in protein folding in AD cases64.
Moreover, it was mentioned in a comparative study on brain samples as an AD-related gene65. The other one, STX16, was among the genes which had expression changes in AD Frontal Cortex66 and was indicated in another study as the gene that showed common expression changes in AD samples67. Pathological survey on the role of mammalian target of rapamycin complex showed that SLC38A1 is one of the significant genes in neurodegenerative disease68. Finally, a study examined the potential functions of Amyloid β peptide, which has an important role in Alzheimer’s disease. It found that SLC38A1 was an effective gen69. Four of the genes in this group, including PLAGL2, CREBRF, LCOR, and MRI1, have not been found in clinical research studies.
In the following section, target miRNAs are investigated based on clinical research studies of Alzheimer’s disease. The first group involves thirteen miRNAs that were obtained based on the intersection of three module groups. These miRNAs were miR-26b-5p, miR-335-5p, miR-92a-3p, miR-615-3p, miR-484, miR-16-5p, miR-17-5p, miR-218-5p, miR-24-3p, miR-124-3p, miR-93-5p, miR193-3p, and miR-20a-5p.
The first miRNA was mir-26b-5p. According to the studies published in 2013 and 2018, the identification of the key miRNAs, which are associated with AD and mir-26b-5p, is also reported as one of the down-regulated key miRNAs70, 71. In another study, the authors reported the existence of a relationship between mir-26b-5p and Alzheimer’s disease and argued that this miRNA was upregulated in Alzheimer’s disease72. The miR-26b-5p was introduced as one of the signals which helped to distinguish sporadic behavioral variant of frontotemporal dementia from Alzheimer and healthy cases73. In two other studies, the researchers used miR-26b-5p as a previously known miRNA in brain diseases, especially Alzheimer’s disease74,75. In another study, which was published in 2017, miR-26b-5p was one of the significant miRNAs because of the dysregulation that it caused between AD and normal samples76. In two other studies, AD samples and normal samples were compared. These studies found that miR-26b-5p was one of the significant regulators for the identified differentially expressed genes77,78.
The second miRNA was miR-335-5p. This miRNA was reported as one of the miRNAs which were related to Alzheimer’s disease and represented the classifiers of Parkinson’s disease using dementia and Alzheimer’ samples79. There is also a study that represented mir-335-5p as an upregulated biomarker of AD80. The same miRNA was introduced as an upregulated biomarker in a different study81. Another study, which utilized neuroimages and investigated the concordance of miRNA biomarkers related to AD, identified mir-335-5p as an upregulated miRNA82. There is another study that used Low-Frequency Pulsed Electromagnetic Field (LF-PEMF) and found that miR-335-5p was a target miRNA and had a role in biological pathways of the Alzheimer’s disease83. Huynh, R.A., et al. investigated biomarkers of Alzheimer’s diseases in the genome, blood and cerebrospinal fluid and compared the AD samples with the normal ones. They observed that the gene expression level of miR-335-5p increased in the normal samples84.
The third miRNA, miR-92a-3p, was indicated as one of the regulator molecules which influenced transcriptional changes in Alzheimer’s disease85. In another study, mir-92a-3p was identified as one of the miRNAs that showed significant upregulation86. Similarly, in another study, the researchers mentioned that mir-92a-3p was a downregulated miRNA in serum samples of Alzheimer’s disease patients in comparison with the MCI patients’ serum samples87. In a different study, which measured miRNAs in the cerebrospinal fluid (CSF) and the blood of AD and MCI patients, mir-92a-3p was detected as the most frequent miRNAs in dementia patients88.
The fourth miRNA was miR-615-3p. In a study by Hoss, A.G, et al., this miRNA was detected as a significant signal of Huntington disease, which is a progressive brain disorder89. Likewise, a study by Hoss, A.G, et al. confirmed that mir-615-3p was a differentially expressed miRNA in Huntington disease90. Finally, Karnati, H.H., et al., reviewed the key causes of neurodegenerative disorders and epilepsy and emphasized the role of this miRNA in Huntington disease91.
The fifth miRNA was miR-484. In a study, it was determined to be an important miRNA which differed considerably between healthy individuals and MS92. Similarly, one of the previously-mentioned studies indicated that miR-484 was an Alzheimer related miRNA86.
The sixth miRNA was miR-16-5p. It functioned as a deregulator in brain tissues based on a study that focused on Late-onset Alzheimer’s disease (LOAD)93. Another study investigated the effect of curcumin on Alzheimer’s disease and its neuroprotective role in this disease. The changes of the related miRNAs were assessed in this study. The study showed that miR-16-5p had a relationship with Alzheimer’s disease94. A different study investigated Frontotemporal Dementia (FTD). It used some circulating miRNAs and showed that miR-16-5p underwent significant changes from healthy individuals to FTD patients95. This miRNA was also recognized in Young-onset Alzheimer Disease (YOAD), which is recognized by clinical diagnosis before the age of 6596.
The next one, called miR-17-5p, has also been mentioned in many research studies that are related to Alzheimer’s disease. Mir-17-5p was considered as an important miRNA in recognition of FTD95. Study on the overlapping molecules of cancer and neurodegeneration showed that miR-17-5p and miR-18d are two gene regulators in neurotransmission97. The miR-17-5p was also determined as an AD-related miRNA98. Moreover, miR-17-5p was found to play an effective role in the production of the amyloid precursor protein (APP) and neuronal apoptosis which are two Alzheimer-related proteins99. A different study investigated miR-17-5p and its intersectional role in aging diseases and cancer100.
The next examined miRNA was miR-218-5p. As we examined clinical research studies, we found a study on the important miRNAs. The researchers of this study compared samples of MDD (Major Depressive Disorder), MCI, and AD patients. They argued that miR-218-5p was one of the ten top miRNAs whose expression differed conspicuously from MDD patients to MCI patients101.
One study used plasma exosomal miRNAs to find the effective miRNAs of Alzheimer. The researchers of this study argued that miR-24-3p was one of these miRNAs102. Another study that highlighted the diagnostic role of miRNAs in AD showed that miR-24-3p was an important signal in samples of cerebrospinal fluid (CSF) assays77. A different study expressed that miR-24-3p showed a considerable negative correlation between the expression levels in serum and CSF of the normal samples73. Another study examined the effects of the human microRNAome on modulating cellular prion protein (PrPC). The results of this study showed that miR-124-3p was an indirect regulator of PrPC72. An attractive study concluded that the regulation of miR-124-3p prevented the abnormal hyperphosphorylation of Tau protein103. In another study, the researchers found that mir-124-3p was among the other effective miRNAs and locus coeruleus (LC) was the most affected region that must be considered in future studies for further investigation104.
The mir-93-5p was the next miRNA which was found using serum data of AD and normal samples. A study showed that miR-93-5p had more changes in AD samples105. Another study revealed that miR-93-5p was one of the effective miRNAs in the case of MDD patients106.
The miR-193b-3p was identified in a study by comparing the AD samples with the normal ones. This study examined a decrease in the value of miR-193b107. The miR-20a-5p has also been examined in several articles using the network-based method and has been regarded as a regulator in AD samples85.
The second group had only one member which was miR-106b-5p. It belonged to the overlap between MCI-AD module groups and CTL-AD module groups. The examination of this miRNA shows that many studies have named it as a miRNA which is related to Alzheimer’s disease. There is a study that showed the radiation-induced changes of miRNA-106b-5p in the blood was involved in the development of Alzheimer’s diseas108. Another study proposed that miR-106b-5p was an upregulated miRNA in Alzheimer’s disease. This claim was validated using the qRT-PCR analysis81. Another study named this miRNA as a blood-based miRNA which was related to AD in more than 34 studies109.
The third group was miR-98-5p and was observed exclusively in CTL-MCI module group. This issue has been mentioned in some studies. For example, it was proposed as a novel therapeutic target for Alzheimer’s disease because of its crucial role in the accumulation of Aβ110. It has been shown that the expression levels of this miRNA differ considerably between normal samples and Alzheimer’s disease samples111.
The fourth group included 6 miRNAs which were observed exclusively in MCI-AD module groups. They were investigated one by one similar to the previous studies. Five miRNAs in this group have not been found in clinical research studies. They include miR-4722-5p, miR-4768-3p, miR-1827, miR-940, and miR-30b-3p. miR-106a-5p was introduced in a study that considered it as an effective miRNA in AD and used it to examine the effect of Huperzine-A on β-Amyloid peptide accumulation to determine the relationship between brain damage and neuro-muscular system deficiency112. A similar study used this miRNA as an effective miRNA in Alzheimer’s disease to express the Folic acid deficiency in amyloid-β accumulation113. Finally, a study showed that miR-106a-5p was an important biomarker of Alzheimer’s disease and argued that it was a predictor variable in AD114.
Likewise, the fifth group had 6 miRNAs which belonged to the intersection of CTL-MCI and CTL-AD module groups. The miRNA group members were miR-877-3p, miR-30a-5p, miR-30c-5p, miR-181a-5p, miR-142-3p and miR-15b-5p. The first one, miR-877-3p, was indicated as a miRNA which was effective in young-onset AD96. The second one, miR-30a-5p, was indicated as the miRNA which had a considerable and high expression in affected samples which were related to the early-onset familial Alzheimer’s disease115. Another study showed the effectiveness of miR-30a-5p for the same disease in the opposite direction brain-derived neurotrophic factor116. The next miRNA, mir-30c-5p, was introduced as a differentially expressed miRNA between normal and Alzheimer’s disease samples in two separate studies81,117. It was shown that the next miRNA, miR-181a-5p, was expressed at different levels in AD in comparison with the normal samples118. In another study, mir-181a-5p was shown to be an effective miRNA in Alzheimer’s disease using SNAP-25 vesicular protein119. The fifth one was mir-142-3p. It was introduced as one of the miRNAs which had considerably different levels in AD samples and normal samples120. A different study showed that the expression level of two target genes caused a reduction in the risk of AD by reducing the expression level of mir-142-3p121. The last one was mir-15b-5p. It was examined using one of the Alzheimer’s disease cell models named swAPP695-HEK293 and revealed the upregulation in the expression of mir-15b-5p94.
Discussion
In this article, co-expression network analyses were performed for three stages of Alzheimer’s disease based on gene expressions. The experiments were performed on over 145 samples of Alzheimer stage, 80 samples of the MCI stage, and 104 samples which were at the healthy stage.
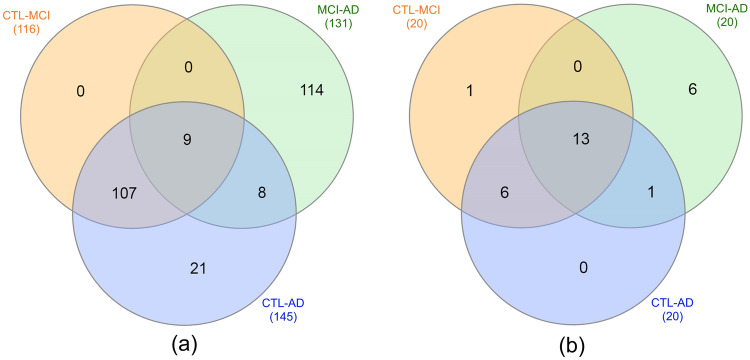
There were 6,179 genes in the total integrated dataset, which were generated by the genes with adjusted p-values smaller than 0.01. After network reconstruction, the modules were specified and merged to gain an optimal structure. Next, the target miRNAs that were related to the selected genes were extracted, and the bipartite networks were constructed for each stage. Subnetworks in the previous steps were constructed by selecting the hub miRNAs, which had pivotal roles in the regulation of the genes to reach optimal results. Then, the lists of extracted genes and miRNAs for each of the subnetworks were used to draw the Venn diagram and to indicate the intersections of three subnetworks. The related diagrams of genes and miRNAs are presented in Fig. 5. The list of the genes and miRNAs of these modules are shown separately by their different intersections and are listed in the Supplementary file as Supplementary Tables S13 and S14.
Figure 5.
Venn diagram related to the extracted genes and miRNAs. (a) Venn diagram of genes (b) Venn diagram of miRNAs.
The listed genes and miRNAs in the above-mentioned tables were the proposed biomarkers of Alzheimer’s disease and were obtained using our proposed method. Therefore, we investigated the previous research studies and medical experiments as well as clinical studies in the field of Alzheimer’s disease.
The examination of the results of the clinical and experimental studies and the investigation of the recently published authentic articles show that almost all of our discovered genes and miRNAs have been reported in different studies of Alzheimer’s disease and neurological diseases. Most of them can be found in the latest articles. However, some of them have not been reported yet. Therefore, it can be claimed that the proposed biomarkers which were extracted using our methods can be real biomarkers that are related to Alzheimer’s disease and should be examined by experimental studies. Some of our proposed biomarkers have been reported in the aging disease that is related to Alzheimer’s disease. Eight of the discovered genes in this study, including MBOAT1, ARMC7, HNRNPUL1, LAMTOR1, PLAGL2, CREBRF, LCOR, and MRI1, have not been reported in the recent authentic Alzheimer related articles. RABL2B has been reported as a gene that is related to Phelan-McDermid Syndrome (PMS) and is not associated with Alzheimer. Therefore, these genes are the landmark finding of our study, and we propose them as the biomarkers of Alzheimer’s disease.
Moreover, we introduced Mir-615-3p as an Alzheimer-related miRNA which has been recognized as a biomarker of the Huntington disease. Furthermore, we dealt with five miRNAs, including miR-4722-5p, miR-4768-3p, miR-1827, miR-940, and miR-30b-3p, which have not been reported in the previous studies. Consequently, they can be considered as new proposed biomarkers which have to be examined by clinical experiments. Finally, we speculated that our gathered biomarkers, which were prepared in tables in the Supplementary file (Supplementary Tables S13 and S14), can be studied as potential biomarkers for the early detection of Alzheimer’s disease.
In summary, our proposed method aimed to conduct a prognostic study of Alzheimer’s disease and used the Gene co-expression Network analysis method based on the GEO database. To this end, significant modules obtained from co-expression networks were utilized to construct bipartite networks (gene-miRNA) for the three stages of the disease. Therefore, we worked with three types of samples. Each type of sample belonged to one of the normal, mild cognitive impairment, and Alzheimer’s disease stages. This study identified the hub genes. These genes have the highest connectivity degrees and are regarded to be the potential prognostic biomarkers for Alzheimer’s disease. The novel genes, including MBOAT1, ARMC7, RABL2B, HNRNPUL1, LAMTOR1, PLAGL2, CREBRF, LCOR, and MRI1 together with miRNAs comprising miR-615-3p, miR-4722-5p, miR-4768-3p, miR-1827, miR-940, and miR-30b-3p are introduced as Alzheimer-related proposed biomarkers and should be examined in the experimental studies as clinical studies. This study points out that its exploration needs further research to develop novel therapeutic approaches to drug design and drug discovery along with medical approaches to the treatment of Alzheimer’s disease.
Methods
This part explains the methods which were used in this study step by step.
Networks construction
To construct networks, we detected the outlier samples using an optimal version of hierarchical clustering, which uses distance and averaging methods to cluster the study samples. The results of the clustering approach showed that there were only two outliers at the AD stage. Moreover, they revealed that there were not any outliers at the other two stages. The WGCNA approach was utilized to perform an analysis of the gene expression data. The Co-expression networks were constructed for all of the genes at each of the stages separately. In the next step, mean connectivity and scale dependency measures were calculated to choose the proper soft power and to reconstruct the network. Lastly, soft threshold power was evaluated using network analysis functions to preserve more correlated genes based on scale-free topology.
Module extraction
The dissimilarity matrix was obtained from TOM matrix to apply the module analysis algorithm. This matrix was used to perform a hierarchical clustering to recognize the potential modules. The modules were selected by using a tree-cut algorithm and experimenting with different values for deepSplit and minimal module size parameters. The extracted modules were merged and labeled with colors. To merge the modules, we extracted eigengenes of modules. After calculating the dissimilarity of the eigengenes, the clustering method was applied to eigengenes.
At this point, module preservation analysis was performed to identify the meaningful modules at different stages of the disease. Module preservation function specified the amount of changes in the modules against the network of the next stage by calculating Zsummary. A large change was observed in the modules when the value of Zsummary was small. This issue was in line with the aim of this study since we preferred to find the modules that underwent bigger changes.
Bipartite gene-miRNA networks
In the next step, the genes of the Normal-MCI modules were merged and were experimentally validated. Moreover, the target miRNAs of these genes were extracted using the miRWalk2.0 database. The MCI-AD modules and Normal-AD modules underwent the same process. Three bipartite gene-miRNA networks were constructed by the genes and their target miRNAs by Cytoscape.3.7.0. The miRNAs with larger degrees had more connections with the selected genes and performed more regulatory roles in the network. Therefore, 20 miRNAs with the highest degree values were chosen together with their connections and were used to reconstruct the network.
Functional enrichment analysis
The Annotation Visualization and Integrated Discovery (DAVID) database was used to study the biological mechanism and gene ontology of the selected genes122,123. The biological processes of the selected genes were listed, and the nodes (p-value < 0.01) were reported as important processes. Kyoto Encyclopedia Gene and Genomes (KEGG) database124 was used to perform the pathway enrichment analysis and the significant genes (p-value < 0.05) were selected.
Supplementary information
Author contributions
S.P. and H.M. designed and directed the project. N.S.S. wrote the first draft of the manuscript and prepared all figures. N.S.S. conducted the experiments and analyzed the obtained results. All authors reviewed and approved the final manuscript. All authors contributed to the manuscript.
Data availability
The datasets which were processed in the present study can be provided by the corresponding author on reasonable request. The raw dataset is available on Information Gene expression Omnibus (GEO) with GSE63063 accession number ( https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE63063).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-69249-8.
References
- 1.Kerepesi C, Daróczy B, Sturm Á, Vellai T, Benczúr A. Prediction and characterization of human ageing-related proteins by using machine learning. Sci. Rep. 2018;8:4094. doi: 10.1038/s41598-018-22240-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Association AS. 2017 Alzheimer's disease facts and figures. Alzheimer Dement. 2017;13:325–373. [Google Scholar]
- 3.Association AS. 2018 Alzheimer's disease facts and figures. Alzheimer Dement. 2018;14:367–429. [Google Scholar]
- 4.Kochanek KD, Xu J, Murphy SL, Minino AM, Kung H-C. National vital statistics reports. Natl Vital Stat Rep. 2011;59:1. [PubMed] [Google Scholar]
- 5.Rollo JL, et al. Unraveling the mechanistic complexity of Alzheimer's disease through systems biology. Alzheimer Dement. 2016;12:708–718. doi: 10.1016/j.jalz.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 6.Meng G, Zhong X, Mei H. A systematic investigation into aging related genes in brain and their relationship with Alzheimer’s disease. PLoS ONE. 2016;11:e0150624. doi: 10.1371/journal.pone.0150624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Motieghader H, Najafi A, Sadeghi B, Masoudi-Nejad A. A hybrid gene selection algorithm for microarray cancer classification using genetic algorithm and learning automata. Inform. Med. Unlocked. 2017;9:246–254. [Google Scholar]
- 8.MotieGhader H, Gharaghani S, Masoudi-Sobhanzadeh Y, Masoudi-Nejad A. Sequential and mixed genetic algorithm and learning automata (SGALA, MGALA) for feature selection in QSAR. Iran. J. Pharm. Res. IJPR. 2017;16:533. [PMC free article] [PubMed] [Google Scholar]
- 9.Li Q, et al. Multi-modal discriminative dictionary learning for Alzheimer's disease and mild cognitive impairment. Comput. Methods Programs Biomed. 2017;150:1–8. doi: 10.1016/j.cmpb.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 10.Biju K, Alfa S, Lal K, Antony A, Akhil MK. Alzheimer’s detection based on segmentation of MRI image. Procedia Comput. Sci. 2017;115:474–481. [Google Scholar]
- 11.Ali EM, Seddik AF, Haggag MH. Automatic detection and classification of Alzheimer's disease from MRI using TANNN. Int. J. Comput. Appl. 2016;148:9. [Google Scholar]
- 12.Lama, R. K., Gwak, J., Park, J.-S. & Lee, S.-W. Diagnosis of Alzheimer’s disease based on structural mri images using a regularized extreme learning machine and PCA features. J. Healthc. Eng.2017 (2017). [DOI] [PMC free article] [PubMed]
- 13.Golkov, V. et al. q-Space deep learning for Alzheimer’s disease diagnosis: global prediction and weakly-supervised localization.
- 14.Miao Y, Jiang H, Liu H, Yao Y-D. An Alzheimers disease related genes identification method based on multiple classifier integration. Comput. Methods Programs Biomed. 2017;150:107–115. doi: 10.1016/j.cmpb.2017.08.006. [DOI] [PubMed] [Google Scholar]
- 15.Ahmed OB, et al. Recognition of Alzheimer's disease and mild cognitive impairment with multimodal image-derived biomarkers and multiple kernel learning. Neurocomputing. 2017;220:98–110. [Google Scholar]
- 16.Er F, et al. Distinguishing age-related cognitive decline from dementias: a study based on machine learning algorithms. J. Clin. Neurosci. 2017;42:186–192. doi: 10.1016/j.jocn.2017.03.021. [DOI] [PubMed] [Google Scholar]
- 17.Xu L, Wu X, Chen K, Yao L. Multi-modality sparse representation-based classification for Alzheimer's disease and mild cognitive impairment. Comput. Methods Programs Biomed. 2015;122:182–190. doi: 10.1016/j.cmpb.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 18.Kyrtsos CR, Baras JS. A systems biology approach to understanding Alzheimer's disease. Biophys. J. 2010;98:738a. [Google Scholar]
- 19.Park C, Yoon Y, Oh M, Yu SJ, Ahn J. Systematic identification of differential gene network to elucidate Alzheimer's disease. Expert Syst. Appl. 2017;85:249–260. [Google Scholar]
- 20.Hao W, Friedman A. Mathematical model on Alzheimer’s disease. BMC Syst. Biol. 2016;10:108. doi: 10.1186/s12918-016-0348-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martínez-Ballesteros M, García-Heredia JM, Nepomuceno-Chamorro IA, Riquelme-Santos JC. Machine learning techniques to discover genes with potential prognosis role in Alzheimer’s disease using different biological sources. Inf. Fusion. 2017;36:114–129. [Google Scholar]
- 22.Zhang R, Simon G, Yu F. Advancing Alzheimer’s research: a review of big data promises. Int. J. Med. Inform. 2017;106:48–56. doi: 10.1016/j.ijmedinf.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Land WH, Schaffer JD. A Machine intelligence designed bayesian network applied to Alzheimer's detection using demographics and speech Data. Procedia Comput. Sci. 2016;95:168–174. [Google Scholar]
- 24.Barati M, Ebrahimi M. Identification of genes involved in the early stages of Alzheimer disease using a neural network algorithm. Gene Cell Tissue. 2016;3:1–10. [Google Scholar]
- 25.Zafeiris D, Rutella S, Ball GR. An artificial neural network integrated pipeline for biomarker discovery using Alzheimer's disease as a case study. Comput. Struct. Biotechnol. J. 2018;16:77–87. doi: 10.1016/j.csbj.2018.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Y, Li Y, Narayan R, Subramanian A, Xie X. Gene expression inference with deep learning. Bioinformatics. 2016;32:1832–1839. doi: 10.1093/bioinformatics/btw074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Danaee P, Ghaeini R, Hendrix DA. Pacific Symposium on Biocomputing. Singapore: World Scientific; 2017. pp. 219–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu J, Wang X, Cheng Y, Zhang L. Tumor gene expression data classification via sample expansion-based deep learning. Oncotarget. 2017;8:109646. doi: 10.18632/oncotarget.22762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moradi E, Marttinen M, Häkkinen T, Hiltunen M, Nykter M. Supervised pathway analysis of blood gene expression profiles in Alzheimer's disease. Neurobiol. Aging. 2019;84:98–108. doi: 10.1016/j.neurobiolaging.2019.07.004. [DOI] [PubMed] [Google Scholar]
- 30.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinform. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Motieghader H, Kouhsar M, Najafi A, Sadeghi B, Masoudi-Nejad A. mRNA–miRNA bipartite network reconstruction to predict prognostic module biomarkers in colorectal cancer stage differentiation. Mol. BioSyst. 2017;13:2168–2180. doi: 10.1039/c7mb00400a. [DOI] [PubMed] [Google Scholar]
- 32.32Adhami, M., MotieGhader, H., Haghdoost, A. A., Afshar, R. M. & Sadeghi, B. Gene co-expression network approach for predicting prognostic microRNA biomarkers in different subtypes of breast cancer. Genomics (2019). [DOI] [PubMed]
- 33.Liang J-W, et al. Application of weighted gene co-expression network analysis to explore the key genes in Alzheimer’s disease. J. Alzheimer Dis. 2018;1:1–12. doi: 10.3233/JAD-180400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo HM, et al. Gene expression analysis reveals novel gene signatures between young and old adults in human prefrontal cortex. Front. Aging Neurosci. 2018;10:259. doi: 10.3389/fnagi.2018.00259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li J, et al. TAM 2.0: tool for microRNA set analysis. Nucleic Acids Res. 2018;46:W180–W185. doi: 10.1093/nar/gky509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loerch PM, et al. Evolution of the aging brain transcriptome and synaptic regulation. PLoS ONE. 2008;3:e3329. doi: 10.1371/journal.pone.0003329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roses AD. The medical and economic roles of pipeline pharmacogenetics: Alzheimer's disease as a model of efficacy and HLA-B* 5701 as a model of safety. Neuropsychopharmacology. 2009;34:6. doi: 10.1038/npp.2008.153. [DOI] [PubMed] [Google Scholar]
- 38.Middleton D, et al. Frequency of HLA-A and B alleles in early and late-onset Alzheimer's disease. Neurosci. Lett. 1999;262:140–142. doi: 10.1016/s0304-3940(99)00045-2. [DOI] [PubMed] [Google Scholar]
- 39.Phelan K, McDermid H. The 22q133 deletion syndrome (Phelan-McDermid syndrome) Mol. Syndromology. 2011;2:186–201. doi: 10.1159/000334260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Costales JL, Kolevzon A. Phelan–McDermid syndrome and SHANK3: implications for treatment. Neurotherapeutics. 2015;12:620–630. doi: 10.1007/s13311-015-0352-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gatta V, D’aurora M, Granzotto A, Stuppia L, Sensi S. Early and sustained altered expression of aging-related genes in young 3xTg-AD mice. Cell Death Dis. 2014;5:e1054. doi: 10.1038/cddis.2014.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lunnon K, et al. A blood gene expression marker of early Alzheimer's disease. J. Alzheimer Dis. 2013;33:737–753. doi: 10.3233/JAD-2012-121363. [DOI] [PubMed] [Google Scholar]
- 43.Climer LK, Dobretsov M, Lupashin V. Defects in the COG complex and COG-related trafficking regulators affect neuronal Golgi function. Front. Neurosci. 2015;9:405. doi: 10.3389/fnins.2015.00405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Browne F, Wang H, Zheng H. Investigating the impact human protein–protein interaction networks have on disease-gene analysis. Int. J. Mach. Learn. Cybern. 2018;9:455–464. [Google Scholar]
- 45.Wang G, et al. Mutation and association analyses of dementia-causal genes in Han Chinese patients with early-onset and familial Alzheimer's disease. J. Psychiatr. Res. 2019;113:141–147. doi: 10.1016/j.jpsychires.2019.03.026. [DOI] [PubMed] [Google Scholar]
- 46.Hang Wong T, et al. Three VCP mutations in patients with frontotemporal dementia. J. Alzheimer Dis. 2018;1:1–8. doi: 10.3233/JAD-180301. [DOI] [PubMed] [Google Scholar]
- 47.Cruts M, Van Broeckhoven C. Progranulin. New York: Springer; 2018. pp. 81–92. [DOI] [PubMed] [Google Scholar]
- 48.Al-Obeidi E, et al. Genotype-phenotype study in patients with valosin-containing protein mutations associated with multisystem proteinopathy. Clin. Genet. 2018;93:119–125. doi: 10.1111/cge.13095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moya-Alvarado G, Gershoni-Emek N, Perlson E, Bronfman FC. Neurodegeneration and Alzheimer's disease (AD). What can proteomics tell us about the Alzheimer's brain? Mol. Cell. Proteomics. 2016;15:409–425. doi: 10.1074/mcp.R115.053330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kajiwara Y, et al. GJA1 (connexin43) is a key regulator of Alzheimer’s disease pathogenesis. Acta Neuropathol. Commun. 2018;6:144. doi: 10.1186/s40478-018-0642-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tanahashi H, Tabira T. Genomic organization of the human X11L2 gene (APBA3), a third member of the X11 protein family interacting with Alzheimer's β-amyloid precursor protein. NeuroReport. 1999;10:2575–2578. doi: 10.1097/00001756-199908200-00025. [DOI] [PubMed] [Google Scholar]
- 52.Jiang S, et al. Trafficking regulation of proteins in Alzheimer’s disease. Mol. Neurodegener. 2014;9:6. doi: 10.1186/1750-1326-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Q, et al. Impaired cognitive function and altered hippocampal synaptic plasticity in mice lacking dermatan sulfotransferase Chst14/D4st1. Front. Mol. Neurosci. 2019;12:26. doi: 10.3389/fnmol.2019.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu Z, Wu C, Pan W, ASDN Initiative Imaging-wide association study: Integrating imaging endophenotypes in GWAS. Neuroimage. 2017;159:159–169. doi: 10.1016/j.neuroimage.2017.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boada M, et al. ATP5H/KCTD2 locus is associated with Alzheimer's disease risk. Mol. Psychiatry. 2014;19:682–687. doi: 10.1038/mp.2013.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Traylor M, et al. Shared genetic contribution to ischemic stroke and Alzheimer's disease. Ann. Neurol. 2016;79:739–747. doi: 10.1002/ana.24621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jensen HLB, et al. Expression of nucleotide excision repair in Alzheimer’s disease is higher in brain tissue than in blood. Neurosci. Lett. 2018;672:53–58. doi: 10.1016/j.neulet.2018.02.043. [DOI] [PubMed] [Google Scholar]
- 58.Henriques AG, et al. Altered protein phosphorylation as a resource for potential AD biomarkers. Sci. Rep. 2016;6:1–12. doi: 10.1038/srep30319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dayon L, et al. Alzheimer disease pathology and the cerebrospinal fluid proteome. Alzheimer's Res. Therapy. 2018;10:66. doi: 10.1186/s13195-018-0397-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Waninger S, et al. Event-related potentials during sustained attention and memory tasks: utility as biomarkers for mild cognitive impairment. Alzheimer Dement. 2018;10:452–460. doi: 10.1016/j.dadm.2018.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kong W, et al. Differences of immune disorders between Alzheimer’s disease and breast cancer based on transcriptional regulation. PLoS ONE. 2017;12:1. doi: 10.1371/journal.pone.0180337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pearl JR, et al. Genome-scale transcriptional regulatory network models of psychiatric and neurodegenerative disorders. Cell Syst. 2019;8:122–135. doi: 10.1016/j.cels.2019.01.002. [DOI] [PubMed] [Google Scholar]
- 63.Sun LL, Yang SL, Sun H, Li WD, Duan SR. Molecular differences in Alzheimer's disease between male and female patients determined by integrative network analysis. J. Cell Mol. Med. 2019;23:47–58. doi: 10.1111/jcmm.13852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Montibeller L, de Belleroche J. Amyotrophic lateral sclerosis (ALS) and Alzheimer’s disease (AD) are characterised by differential activation of ER stress pathways: focus on UPR target genes. Cell Stress Chaperones. 2018;23:897–912. doi: 10.1007/s12192-018-0897-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Twine, N. A., Janitz, K., Wilkins, M. R. & Janitz, M. Whole transcriptome sequencing reveals gene expression and splicing differences in brain regions affected by Alzheimer's disease. PloS ONE6 (2011). [DOI] [PMC free article] [PubMed]
- 66.Bennett J, Keeney P. Micro RNA’s (mirna’s) may help explain expression of multiple genes in Alzheimer’s Frontal Cortex. J. Syst. Integrat. Neurosci. 2017;3:1–9. [Google Scholar]
- 67.Liang WS, et al. Neuronal gene expression in non-demented individuals with intermediate Alzheimer's disease neuropathology. Neurobiol. Aging. 2010;31:549–566. doi: 10.1016/j.neurobiolaging.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yamada D, et al. Inhibition of the glutamine transporter SNAT1 confers neuroprotection in mice by modulating the mTOR-autophagy system. Commun. Biol. 2019;2:1–11. doi: 10.1038/s42003-019-0582-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bailey JA, Maloney B, Ge Y-W, Lahiri DK. Functional activity of the novel Alzheimer's amyloid β-peptide interacting domain (AβID) in the APP and BACE1 promoter sequences and implications in activating apoptotic genes and in amyloidogenesis. Gene. 2011;488:13–22. doi: 10.1016/j.gene.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen J, et al. MicroRNA expression data analysis to identify key miRNAs associated with Alzheimer's disease. J. Gene Med. 2018;20:e3014. doi: 10.1002/jgm.3014. [DOI] [PubMed] [Google Scholar]
- 71.Leidinger P, et al. A blood based 12-miRNA signature of Alzheimer disease patients. Genome Biol. 2013;14:R78. doi: 10.1186/gb-2013-14-7-r78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chang W-S, Wang Y-H, Zhu X-T, Wu C-J. Genome-wide profiling of miRNA and mRNA expression in Alzheimer’s disease. Med. Sci. Monit. 2017;23:2721. doi: 10.12659/MSM.905064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Denk J, et al. Specific serum and CSF microRNA profiles distinguish sporadic behavioural variant of frontotemporal dementia compared with Alzheimer patients and cognitively healthy controls. PLoS ONE. 2018;13:e0197329. doi: 10.1371/journal.pone.0197329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Erdal ME, et al. miRNA expression profile is altered differentially in the rat brain compared to blood after experimental exposure to 50 Hz and 1 mT electromagnetic field. Prog. Biophys. Mol. Biol. 2018;132:35–42. doi: 10.1016/j.pbiomolbio.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 75.Mendes-Silva AP, et al. Shared biologic pathways between Alzheimer disease and major depression: a systematic review of microRNA expression studies. Am. J. Geriatric Psychiatry. 2016;24:903–912. doi: 10.1016/j.jagp.2016.07.017. [DOI] [PubMed] [Google Scholar]
- 76.Hara N, et al. Serum microRNA miR-501-3p as a potential biomarker related to the progression of Alzheimer’s disease. Acta Neuropathol. Commun. 2017;5:10. doi: 10.1186/s40478-017-0414-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Martinez B, Peplow PV. MicroRNAs as diagnostic and therapeutic tools for Alzheimer’s disease: advances and limitations. Neural Regener. Res. 2019;14:242. doi: 10.4103/1673-5374.244784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rahman, M. R. et al. Blood-based molecular biomarker signatures in Alzheimer’s disease: Insights from systems biomedicine analyses. BioRxiv, 481879 (2018).
- 79.Patil KS, et al. Combinatory microRNA serum signatures as classifiers of Parkinson's disease. Parkinsonism Relat. Disord. 2019;64:202–210. doi: 10.1016/j.parkreldis.2019.04.010. [DOI] [PubMed] [Google Scholar]
- 80.Satoh J-I, Kino Y, Niida S. MicroRNA-Seq data analysis pipeline to identify blood biomarkers for Alzheimer's disease from public data. Biomark. Insights. 2015;10:S25132. doi: 10.4137/BMI.S25132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kumar S, Reddy PH. Are circulating microRNAs peripheral biomarkers for Alzheimer's disease? Biochim. Biophys. Acta. 2016;1862:1617–1627. doi: 10.1016/j.bbadis.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cheng À, et al. Prognostic serum miRNA biomarkers associated with Alzheimer’s disease shows concordance with neuropsychological and neuroimaging assessment. Mol. Psychiatry. 2015;20:1188. doi: 10.1038/mp.2014.127. [DOI] [PubMed] [Google Scholar]
- 83.Capelli, E. et al. Low-frequency pulsed electromagnetic field is able to modulate miRNAs in an experimental cell model of Alzheimer’s disease. J. Healthc. Eng.2017 (2017). [DOI] [PMC free article] [PubMed]
- 84.Huynh RA, Mohan C. Alzheimer’s disease: biomarkers in the genome, blood, and cerebrospinal fluid. Front. Neurol. 2017;8:102. doi: 10.3389/fneur.2017.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rahman MR, et al. Network-based approach to identify molecular signatures and therapeutic agents in Alzheimer’s disease. Comput. Biol. Chem. 2019;78:431–439. doi: 10.1016/j.compbiolchem.2018.12.011. [DOI] [PubMed] [Google Scholar]
- 86.Siedlecki-Wullich D, et al. Altered microRNAs related to synaptic function as potential plasma biomarkers for Alzheimer’s disease. Alzheimer Res. Ther. 2019;11:46. doi: 10.1186/s13195-019-0501-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kumar S, Vijayan M, Reddy PH. MicroRNA-455-3p as a potential peripheral biomarker for Alzheimer’s disease. Hum. Mol. Genet. 2017;26:3808–3822. doi: 10.1093/hmg/ddx267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sørensen SS, Nygaard A-B, Christensen T. miRNA expression profiles in cerebrospinal fluid and blood of patients with Alzheimer’s disease and other types of dementia: an exploratory study. Transl. Neurodegener. 2016;5:6. doi: 10.1186/s40035-016-0053-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hoss AG, et al. MicroRNAs located in the Hox gene clusters are implicated in huntington's disease pathogenesis. PLoS Genet. 2014;10:e1004188. doi: 10.1371/journal.pgen.1004188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hoss AG, et al. miR-10b-5p expression in Huntington’s disease brain relates to age of onset and the extent of striatal involvement. BMC Med. Genomics. 2015;8:10. doi: 10.1186/s12920-015-0083-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Karnati HK, Panigrahi MK, Gutti RK, Greig NH, Tamargo IA. miRNAs: key players in neurodegenerative disorders and epilepsy. J. Alzheimer Dis. 2015;48:563–580. doi: 10.3233/JAD-150395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Regev K, et al. Identification of MS-specific serum miRNAs in an international multicenter study. Neurol. Neuroimmunol. Neuroinflamm. 2018;5:e491. doi: 10.1212/NXI.0000000000000491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Herrera-Espejo S, Santos-Zorrozua B, Álvarez-González P, Lopez-Lopez E, Garcia-Orad Á. A systematic review of microRNA expression as biomarker of late-onset Alzheimer’s disease. Mol. Neurobiol. 2019;1:1–16. doi: 10.1007/s12035-019-01676-9. [DOI] [PubMed] [Google Scholar]
- 94.Liu H-Y, et al. miR-15b-5p targeting amyloid precursor protein is involved in the anti-amyloid eflect of curcumin in swAPP695-HEK293 cells. Neural Regener. Res. 2019;14:1603. doi: 10.4103/1673-5374.255979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Piscopo P, et al. Circulating miR-127-3p as a potential biomarker for differential diagnosis in frontotemporal dementia. J. Alzheimer Dis. 2018;1:1–11. doi: 10.3233/JAD-180364. [DOI] [PubMed] [Google Scholar]
- 96.McKeever PM, et al. MicroRNA expression levels are altered in the cerebrospinal fluid of patients with young-onset Alzheimer’s disease. Mol. Neurobiol. 2018;55:8826–8841. doi: 10.1007/s12035-018-1032-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Spencer P, Fry RC, Kisby GE. Unraveling 50-year-old clues linking neurodegeneration and cancer to cycad toxins: are microRNAs common mediators? Front. Genet. 2012;3:192. doi: 10.3389/fgene.2012.00192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang M, Qin L, Tang B. MicroRNAs in Alzheimer’s disease. Front. Genet. 2019;10:153. doi: 10.3389/fgene.2019.00153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hu Y-B, et al. Diagnostic value of microRNA for Alzheimer’s disease: a systematic review and meta-analysis. Front. Aging Neurosci. 2016;8:13. doi: 10.3389/fnagi.2016.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dellago H, Bobbili MR, Grillari J. MicroRNA-17-5p: at the crossroads of cancer and aging-a mini-review. Gerontology. 2017;63:20–28. doi: 10.1159/000447773. [DOI] [PubMed] [Google Scholar]
- 101.Mendes-Silva AP, et al. MiRNAs and their role in the correlation between major depressive disorder, mild cognitive impairment and Alzheimer’s disease. Alzheimer Dement. 2017;13:P1017–P1018. [Google Scholar]
- 102.Lugli G, et al. Plasma exosomal miRNAs in persons with and without Alzheimer disease: altered expression and prospects for biomarkers. PLoS ONE. 2015;10:e0139233. doi: 10.1371/journal.pone.0139233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kang Q, et al. MiR-124-3p attenuates hyperphosphorylation of Tau protein-induced apoptosis via caveolin-1-PI3K/Akt/GSK3β pathway in N2a/APP695swe cells. Oncotarget. 2017;8:24314. doi: 10.18632/oncotarget.15149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Llorens F, et al. MicroRNA expression in the locus coeruleus, entorhinal cortex, and hippocampus at early and middle stages of Braak neurofibrillary tangle pathology. J. Mol. Neurosci. 2017;63:206–215. doi: 10.1007/s12031-017-0971-4. [DOI] [PubMed] [Google Scholar]
- 105.Wu Y, et al. Lower serum levels of miR-29c-3p and miR-19b-3p as biomarkers for Alzheimer’s disease. Tohoku J. Exp. Med. 2017;242:129–136. doi: 10.1620/tjem.242.129. [DOI] [PubMed] [Google Scholar]
- 106.Liu X, et al. Identification of suitable plasma-based reference genes for miRNAome analysis of major depressive disorder. J. Affect. Disord. 2014;163:133–139. doi: 10.1016/j.jad.2013.12.035. [DOI] [PubMed] [Google Scholar]
- 107.Meng F, et al. MicroRNA-193b-3p regulates chondrogenesis and chondrocyte metabolism by targeting HDAC3. Theranostics. 2018;8:2862. doi: 10.7150/thno.23547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Segaran, R. C., Chan, L. Y., Wang, H., Sethi, G. & Tang, F. R. Neuronal Development-related miRNAs as biomarkers for Alzheimer's disease, depression, schizophrenia and ionizing radiation exposure. Curr. Med. Chem. (2020). [DOI] [PubMed]
- 109.Nagaraj S, Zoltowska KM, Laskowska-Kaszub K, Wojda U. microRNA diagnostic panel for Alzheimer’s disease and epigenetic trade-off between neurodegeneration and cancer. Ageing Res. Rev. 2019;49:125–143. doi: 10.1016/j.arr.2018.10.008. [DOI] [PubMed] [Google Scholar]
- 110.Li Q, Li X, Wang L, Zhang Y, Chen L. miR-98-5p acts as a target for Alzheimer’s disease by regulating Aβ production through modulating SNX6 expression. J. Mol. Neurosci. 2016;60:413–420. doi: 10.1007/s12031-016-0815-7. [DOI] [PubMed] [Google Scholar]
- 111.Tan L, et al. Genome-wide serum microRNA expression profiling identifies serum biomarkers for Alzheimer's disease. J. Alzheimer Dis. 2014;40:1017–1027. doi: 10.3233/JAD-132144. [DOI] [PubMed] [Google Scholar]
- 112.Turkseven CH, et al. Effects of Huperzin-A on the Beta-amyloid accumulation in the brain and skeletal muscle cells of a rat model for Alzheimer's disease. Life Sci. 2017;184:47–57. doi: 10.1016/j.lfs.2017.07.012. [DOI] [PubMed] [Google Scholar]
- 113.Liu H, et al. Folic acid deficiency enhances abeta accumulation in APP/PS1 mice brain and decreases amyloid-associated miRNAs expression. J. Nutr. Biochem. 2015;26:1502–1508. doi: 10.1016/j.jnutbio.2015.07.020. [DOI] [PubMed] [Google Scholar]
- 114.Yılmaz ŞG, Erdal ME, Özge AA, Sungur MA. Can peripheral MicroRNA expression data serve as epigenomic (upstream) biomarkers of Alzheimer's disease? Omics. 2016;20:456–461. doi: 10.1089/omi.2016.0099. [DOI] [PubMed] [Google Scholar]
- 115.Lv Z, et al. Comparative study of microRNA profiling in one Chinese Family with PSEN1 G378E mutation. Metab. Brain Dis. 2018;33:1711–1720. doi: 10.1007/s11011-018-0279-2. [DOI] [PubMed] [Google Scholar]
- 116.Croce N, et al. NPY modulates miR-30a-5p and BDNF in opposite direction in an in vitro model of Alzheimer disease: a possible role in neuroprotection? Mol. Cell. Biochem. 2013;376:189–195. doi: 10.1007/s11010-013-1567-0. [DOI] [PubMed] [Google Scholar]
- 117.Nagaraj S, et al. Profile of 6 microRNA in blood plasma distinguish early stage Alzheimer’s disease patients from non-demented subjects. Oncotarget. 2017;8:16122. doi: 10.18632/oncotarget.15109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Burgos K, et al. Profiles of extracellular miRNA in cerebrospinal fluid and serum from patients with Alzheimer's and Parkinson's diseases correlate with disease status and features of pathology. PLoS ONE. 2014;9:e94839. doi: 10.1371/journal.pone.0094839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Agostini S, et al. Serum miRNAs expression and SNAP-25 genotype in Alzheimer’s disease. Front. Aging Neurosci. 2019;11:52. doi: 10.3389/fnagi.2019.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cosín-Tomás M, et al. Plasma miR-34a-5p and miR-545-3p as early biomarkers of Alzheimer’s disease: potential and limitations. Mol. Neurobiol. 2017;54:5550–5562. doi: 10.1007/s12035-016-0088-8. [DOI] [PubMed] [Google Scholar]
- 121.Ghanbari M, et al. A functional variant in the miR-142 promoter modulating its expression and conferring risk of Alzheimer disease. Hum. Mutat. 2019;40:2131–2145. doi: 10.1002/humu.23872. [DOI] [PubMed] [Google Scholar]
- 122.Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 123.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2008;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ogata H, et al. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets which were processed in the present study can be provided by the corresponding author on reasonable request. The raw dataset is available on Information Gene expression Omnibus (GEO) with GSE63063 accession number ( https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE63063).