Abstract
Food poisoning caused by Staphylococcus aureus (S. aureus) toxins is considered one of the foremost public health threat that usually occurs through the ingestion of raw milk contaminated with staphylococcal enterotoxins. The current study spotlights on the prevalence, antibiogram and genetic diversity of S. aureus enterotoxin genes. One hundred and fifty of raw milk (90) and ice cream (60) samples were randomly collected from local markets from Sadat city, Egypt. S. aureus was recovered from 44% of raw milk and 20% of ice cream samples. The identification for the obtained S. aureus isolates was confirmed through targeting the nuc gene. Antibiogram pattern of 32 S. aureus isolates showed high resistance to Cefoxitin, Sulpha/Trimethoprim, Tetracycline, Norfloxacin, Penicillin and Cephradine. However, high susceptibility to Gentamycin and Vancomycin were observed. Multiplex PCR was a competent practise for the recognition of Staphylococcus enterotoxin (SE) genes (SEA, SEB and SED). The phylogenetic analysis of the SED gene of enterotoxigenic S. aureus strains showed identical similarity with 100% to each other and high similarity with other international isolates in GenBank from different localities and sources. The frequency of enterotoxigenic S. aureus strains in milk products could have serious hazardous effects on humans. These results suggested possible strains transmission between different geographical areas through the food and milk product trades.
Keywords: S. aureus, Enterotoxin genes, Milk, Sequencing analysis
1. Introduction
Staphylococcus aureus (S. aureus) is classified as the third most predominant pathogen responsible for food poisoning outbreaks in humans and is considered a major contaminant for many dairy products (Rabello et al., 2007, Kadariya et al., 2014, Dittmann et al., 2017). It remains a major problematic agent in food poisoning outbreaks due to its powerful heat-stable enterotoxins (ICMSF, 1996). Moreover, Stewart, 2005, Elbehiry et al., 2017 reported that S. aureus is a common pathogenic agent in food-producing animals, causing diseases such as subclinical mastitis. Several environmental vehicles, including air, dust, and surface food products, are considered a serious source of contamination by S. aureus enterotoxins. Many surveillance studies have reported the existence and contamination of food products by S. aureus enterotoxins (De Buyser et al., 2001, Denayer et al., 2017). Milk and its products often become contaminated at the time of cheese or dairy product manufacturing, causing a serious and potential public health hazard to human consumers (Vimercati et al., 2006, Velázquez-Ordoñez et al., 2019).
The pathogenicity of S. aureus is promoted via several virulence factors, including Staphylococcus enterotoxins (SEs), haemolysins, fibronectin-binding proteins, and toxic shock syndrome toxin-1; these factors have a critical role in S. aureus pathogenicity (Puah et al., 2016). To date, approximately twenty-two SE types have been described and identified from food poisoning cases designated A-V (Argudín et al., 2010), causing vomiting, headache, abdominal colic and diarrhoea shortly after the consumption of contaminated foods (Le Loir et al., 2003). The role of SE genes as virulence determinants in S. aureus is thought to be regulated by the accessory gene regulator (agr) gene (Kornblum et al., 1990, Jenul and Horswill, 2018) in conjunction with the staphylococcal accessory regulator (sar) gene (Novick, 2000).
Due to the popular and intensive use of raw milk in Egypt by human consumers and the serious and public health importance of raw milk and its products, this investigation designed to detect the frequency, antibiogram pattern and genetic relationship among enterotoxigenic S. aureus strains recovered from milk and ice cream specimens from local Egyptian markets and their effect on public health.
2. Materials and methods
2.1. Collection of samples
One hundred and fifty milk product (90 raw milk and 60 ice cream) specimens were collected from local market/street vendors in different areas in Sadat city. Each raw milk sample weighed approximately 10 ml and each ice cream sample weighed 10 g; the samples were preserved in a sterile container and transported to the laboratory.
2.2. Phenotypic characterization of S. aureus isolates
The processed samples were inserted onto Baird-Parker agar (Oxoid Ltd.) and 7–10% defibrinated blood agar and then incubated at 37 °C for two successive days. The doubtful colonies of S. aureus were subjected to catalase, coagulase (APHA, 2004), gram staining, haemolytic activity, DNase agar (Murray et al., 2003) and Congo red medium to evaluate biofilm activity (Freeman et al., 1989).
2.3. Antibiogram profile of S. aureus isolates
The obtained S. aureus isolates were subjected to antibiotic susceptibility testing against 11 antibiotics on MHA medium according to (Finegold and Martin, 1982). Penicillin (P), Cefoxitin (FOX), Cephradine (CE), Tetracycline (TE), Erythromycin (E), Ciprofloxacin (CIP), Amoxicillin/Clavulanate acid (AX), Gentamycin (CN), Sulpha-Trimethoprim (SXT), Vancomycin (VA), CE, Norofloxacin (NOR).
2.4. Multiplex PCR for the recognition of enterotoxin genes
2.4.1. DNA extraction
From each sample, DNA was extracted by QIAamp DNA mini kits purchased from Qiagen Company, Germany. The extraction process was carried out based on the constructor’s recommendations.
2.4.2. Oligonucleotide primers and PCR cycling conditions
As shown in Table 1, the primers used for detection of nuc and enterotoxins genes were obtained from Metabion (Germany). The PCR conditions were as follows: for nuc gene: primary denaturation was carried out for 5 min at 94 °C; 35 cycles of secondary denaturation were done for 30 secs at 94 °C; annealing was performed for 30 secs at 55 °C; then extension was done for 1 min at 72 °C and a final extension was accomplished for 10 min at 72 °C. Meanwhile for enterotoxins genes: primary denaturation for 5 min at 94 °C; 35 cycles of secondary denaturation for 30 secs at 94 °C; annealing for 45 secs at 50 °C; extension for 45 secs at 72 °C; and a final extension for 10 min at 72 °C. For multiplex PCR, the primers were added to a reaction of 50-µl comprising 25 µl of Master Mix (Takara, Japan), 7 µl of DNA template, 1 µl of each primer at 20 pmol concentrations, and 16 µl of water.
Table 1.
Primer sequences applied for nuc and enterotoxins genes in S. aureus.
| Target gene | Primer sequences (5ʹ-3ʹ) | bp | Reference |
|---|---|---|---|
| nuc | GCGATTGATGGTGATACGGTT | 270 | Louie et al. (2002) |
| AGCCAAGCCTTGACGAACTA AAGC | |||
| Sea | GGTTATCAATGTGCGGGTGG | 102 | Mehrotra et al. (2000) |
| CGGCACTTTTCTCTTCGG | |||
| Seb | CGGCACTTTTCTCTTCGG | 164 | Mehrotra et al. (2000) |
| CCAAATAGTGACGTTAGG | |||
| Sec | AGATGAAGTAGTTGATGTGTATGG | 451 | Mehrotra et al. (2000) |
| CACACTTTTAGAACCG | |||
| Sed | CCAATAATAGGAGAAAATAAAAG | 278 | Mehrotra et al. (2000) |
| ATTGGTATTTTTTTTCGTTC | |||
| See | AGGTTTTTTCACAGGTCATCC | 209 | Mehrotra et al. (2000) |
| CTTTTTTTTCTTCGGTCAATC |
2.5. Phylogenetic analysis of the S. aureus SED gene from milk
A QIAquick extraction kit was used for purification of the PCR products. A Big Dye Terminator V3.1 cycle sequencing kit purchased from the Company of Perkin-Elmer was used for the sequence reaction, and a Centrisep spin column was applied for purification of the products. The sequences of DNA were created by genetic analyser (Applied Biosystems, USA). To establish sequence identity with existing GenBank accessions, a basic local alignment search tool (BLAST®) analysis was applied (Altschul et al., 1990). The MegAlign module of Lasergene DNAStar was applied to create the phylogenetic tree (Thompson et al., 1994), and by using maximum likelihood, neighbour-joining and maximum parsimony methods in MEGA6, the phylogenetic analyses were carried out (Tamura et al., 2013). The accession numbers of enterotoxigenic S. aureus strains form raw milk and ice cream samples MF359584 and MF359585 respectively.
3. Results
3.1. Prevalence of S. aureus isolates
Out of 90 and 60 raw milk and ice cream samples, the incidence rates of S. aureus were 44% and 20%, respectively. Other staphylococci were 26% and 10%, respectively, as revealed in Table 2. The obtained isolates of S. aureus were subjected to biochemical characterization tests, including catalase, coagulase, haemolysis, and DNase testing as well as biofilm activity testing.
Table 2.
Prevalence of S. aureus isolates from raw milk and ice cream samples.
| Raw milk (90) |
Ice cream (60) |
||||||
|---|---|---|---|---|---|---|---|
|
S. aureus |
Other staphylococci |
S. aureus |
Other staphylococci |
||||
| N | % | N | % | N | % | N | % |
| 22 | 44 | 13 | 26 | 10 | 20 | 5 | 10 |
The % was estimated according to the total number of isolates (50).
3.2. Antibiogram profile of S. aureus isolates from milk products
The antibiogram pattern of 32 S. aureus isolates was determined. The results indicated that most of the S. aureus isolates exhibited high resistance to Cefoxitin, Sulpha/Trimethoprim, Tetracycline, Penicillin, Norfloxacin and Cephradine, with percentages of 78.1%, 71.9%, 65.6%, 62.5% and 59.4%, respectively. However, the same isolates, with 53.1% of each isolate, showed high sensitivity to Gentamycin and Vancomycin, as shown in Table 3.
Table 3.
Antibiogram results of S. aureus isolates from milk products (raw milk and ice cream).
| Antibiotics/Abbreviation | Resistant |
Intermediate |
Sensitive |
|||
|---|---|---|---|---|---|---|
| No. | % | No. | % | No. | % | |
| Penicillin (P) | 20 | 62.5 | 5 | 15.6 | 7 | 21.9 |
| Cefoxitin (FOX) | 25 | 78.1 | 7 | 21.9 | – | – |
| Tetracycline (TE) | 21 | 65.6 | 5 | 15.6 | 6 | 18.8 |
| Erythromycin (E) | 15 | 46.9 | 9 | 28.1 | 8 | 25 |
| Ciprofloxacin (CIP) | 18 | 56.3 | 11 | 34.3 | 3 | 9.4 |
| Amoxicillin/clavulanate acid (AX) | 11 | 34.3 | 6 | 18.8 | 15 | 46.9 |
| Gentamycin (CN) | 15 | 46.9 | – | – | 17 | 53.1 |
| Sulpha/Trimethoprim (SXT) | 23 | 71.9 | – | – | 9 | 28.1 |
| Vancomycin (VA) | 4 | 12.5 | 11 | 34.4 | 17 | 53.1 |
| Cephradine CE | 19 | 59.4 | 8 | 25 | 5 | 15.6 |
| Norfloxacin (NOR) | 20 | 62.5 | 8 | 25 | 4 | 12.5 |
3.3. Molecular identification of isolated S. aureus by nuc gene
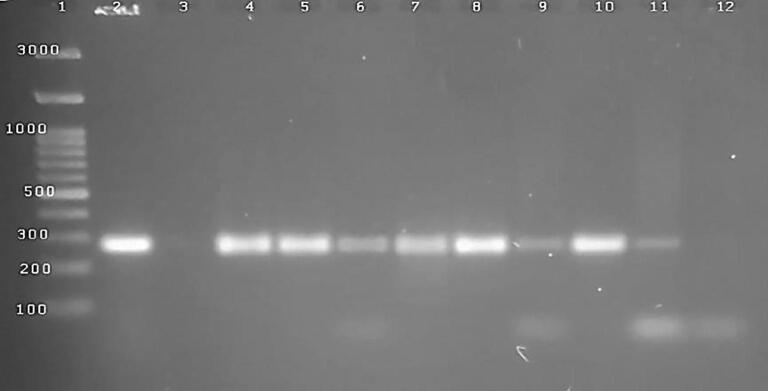
Successful amplification and targeting the nuc gene was carried out using specific primer sets at 270 bp. Among tested ten isolates of S. aureus, the nuc gene was recorded in seven strains (7∕10) as illustrated in Fig. 1.
Fig. 1.
1.5% agrose gel electrophoresis of PCR product for 10 S. aureus isolates (3–7 isolated from raw milk; 8–12 from ice cream), the nuc genes were positive in 8 samples (4–11) at 27 bp., 2 control positive, 3,12 were negative samples.
3.4. Multiplex PCR for the recognition of enterotoxin genes in S. aureus strains
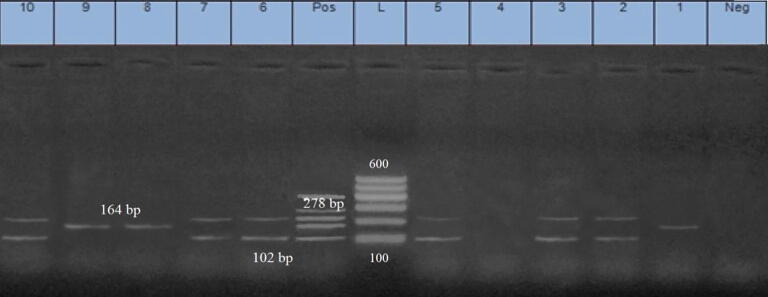
Ten strains of S. aureus were carefully chosen for the recognition of five classical enterotoxin genes (SEA, SEB, SEC, SED, and SEE). The successful amplification of the SEA and SED genes, at 102 bp and 278 bp, correspondingly, was performed; there was a 60% prevalence rate among the tested S. aureus isolates. The successful amplification of the SEB gene, at 160 bp, was performed, with a prevalence rate of 30%; there was no recognition of the SEC and SEE genes in any of the tested samples, as shown in Fig. 2.
Fig. 2.
Multiplex PCR for 10 strains of S. aureus (1–5 recovered from fresh milk; 5–10 from ice cream), 2, 3, 5, 6, 7 and 10 samples were positive to sea genes at 102 bp; 1, 8 & 9 samples, the seb genes was recognized at 164 bp and in samples number 2, 3, 5, 6, 7 & 10, the sed was identified at 278 bp.
3.5. Phylogenetic analysis of the SED gene in field S. aureus
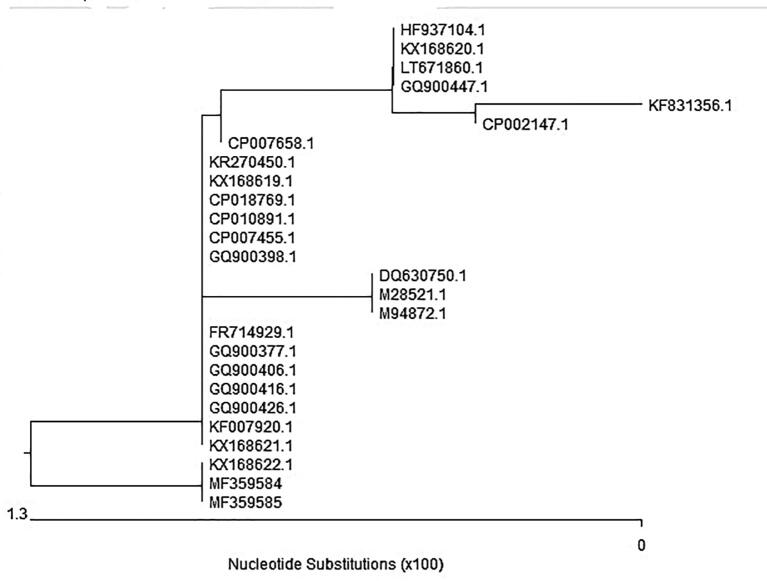
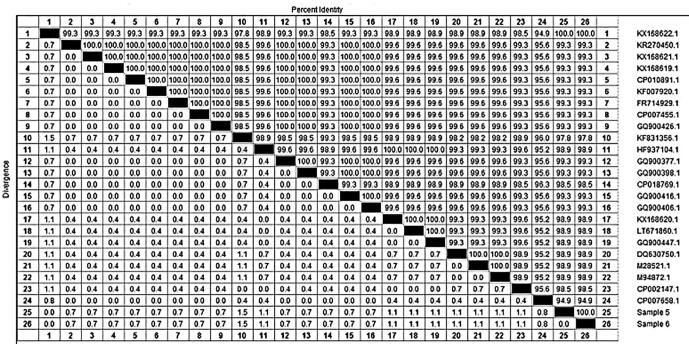
The sequencing of the SED gene was carried out to analyse the similarity amongst the S. aureus isolates from the raw milk and ice cream samples. Fig. 3, Fig. 4 showed an identical similarity between the two analysed isolates accession numbers (MF359584) and (MF359585) from the raw milk and ice cream samples, respectively. The phylogenetic tree is classified into two main clusters. The first cluster has several isolates. The second cluster included our two isolates (MF359584 and MF359585) which showed 100% similarity with each other and with the (KX168622.1) isolate of human origin from Switzerland. Additionally, high homology and similarity (99.3%) to numerous isolates of rabbit origin from Switzerland (KX168621.1), from a patient’s wound in Iran (KF007920.1), and from the United Kingdom (GQ900426.1) were observed. Furthermore, there was high genetic similarity to three clinical isolates from the USA (GQ900416.1; GQ900406.1; GQ900377.1) and one isolate from Sweden (FR714929.1). However, a distinct segregation from (DQ630750.1), isolated from food poisoning cases in India, was noted.
Fig. 3.
Dendrogram showed the genetic homogeneity of two S. aureus isolates (MF359584) and (MF359585) from raw milk and ice cream respectively with other related international isolates.
Fig. 4.
Show the identical similarity between our isolates of accession number sample 5 (MF359584) and sample 6 (MF359585) within the other international isolates on Gene bank.
4. Discussion
The normal compositions of milk products enhance the growth of several microorganisms, resulting in severe serious food poisoning hazards to humans (Soomro et al., 2003, Velázquez-Ordoñez et al., 2019). S. aureus is a public health pathogen implicated in food-borne illness in humans. For example, approximately 13,420 food-borne cases were reported in Japan (Hennekinne et al., 2012). In the current study, S. aureus was recovered from 44% and 20% of 90 raw milk and 60 ice cream specimens, correspondingly. Similar results in Egypt were reported by (Saly et al., 2019), who detected S. aureus in 46% of the examined milk and milk products.
Moreover, (Wang et al., 2017) isolated S. aureus from 46.2% of raw milk samples recovered from two dairy farms in China. Furthermore, (Liu et al., 2017) stated that the frequency rate of S. aureus was 22% in the raw milk of cows. In Brazil, a comparative study conducted by Rall et al., (2008) recovered S. aureus from 20.4% of 162 pasteurized and raw milk samples. In contrast, a lower incidence of 3.9% was reported in pasteurized milk samples in China (Dai et al., 2019). However, an increased prevalence was obtained by (Gündogan et al., 2006), who recorded the frequency rate of 56.6% from 180 pasteurized, raw milk, and ice cream samples. The variation between studies in the frequency of S. aureus recovered from milk and their products may be attributed to differences in sample sizes, origins, and geographic locations and may reflect the degree of applicable sanitary measures.
In this study, the amplification of nuc gene was carried out using specific primer sets for confirmatory identification of S. aureus isolates. This result was previously supported by several surveillances (Hu et al., 2013, Sahebnasagh et al., 2014, Sudhaharan et al., 2015, Elbehiry et al., 2019) they discussed the actual role of nuc gene as potential and gold standard marker gene in detection of S. aureus isolates.
The misuse of antibiotics in dairy animals may have a serious hazard for the broadcast of resistant strains of bacteria to humans and the environment (Sharma et al., 2017). Our findings showed that S. aureus isolates exhibited high resistance to Cefoxitin, Sulphate/Trimethoprim, Tetracycline, Norfloxacin, Penicillin and Cephradine, at 78.1%, 71.9%, 65.6%, 62.5%, 62.5% and 59.4%, respectively. However, high susceptibility to Gentamycin and Vancomycin, at 53.1%, was recorded. Similar findings were reported in a recent study in Egypt (El Faramaway et al., 2019), who illustrated increased resistance in S. aureus clinical mastitis isolates to Penicillin (95.65%), Oxacillin (67.39%), Tetracycline (56.52%) and Erythromycin (52.17%); the highest sensitivity was to Gentamycin (71.74%) and Vancomycin (69.57%).
Moreover, (Abraha et al., 2017) recovered S. aureus isolates from raw milk that were highly susceptible to Gentamicin (100%) in Ethiopia. Furthermore, a study from Bangladesh (Islam et al., 2016) reported the tolerance of S. aureus strains from uncooked milk samples to Tetracycline (73.33%) and susceptibility to Tentamicin (100%). In contrast, (Akanbi et al., 2017) demonstrated high susceptibility to Tetracycline and Sulfamethoxazole-Trimethoprim (56.7%), Ciprofloxacin (66.7%) and Cefoxitin (76.7%). In addition, (Okpo et al., 2018) indicated that all tested S. aureus strains were sensitive to Ciprofloxacin, although 50% of these strains were unaffected by Tetracycline. Additionally, (Aliyu et al., 2018) tested 12 S. aureus isolates obtained from pasteurized milk in Nigeria that showed high susceptibility to Ciprofloxacin and high resistance to Amoxicillin, Erythromycin, and Norfloxacin.
The pathogenic and toxigenic effects of S. aureus mainly rely on the existence of some virulence determinants. Among these determinants, SE genes in S. aureus isolates in different food and milk products have an important and emerging role; these genes have been screening identified in many surveillance studies and represent a serious public health hazard (Wang et al., 2017). In our study, the SEA and SED genes were the predominant SE genes (60%) amongst the examined strains, followed by the SEB gene (30%). Similar to our results (Zhang et al., 2015), the predominance of the SEA and SED genes among S. aureus isolates from retail foods in China has been reported. Moreover, the predominance of the SED gene among S. aureus isolates of milk origin was previously demonstrated in several findings in Poland (McMillan et al., 2016, Liu et al., 2017). Furthermore, (Wang et al., 2017) demonstrated that more than 90% of S. aureus isolates enclosed the SEA and SEB genes, and 80% contained the SED gene. In a similar finding, the SEA gene was described as the major aetiological agent of S. aureus food poisoning eruptions (Argudín et al., 2010, Gholamzad et al., 2015). Nevertheless, (Veras et al., 2008) found that each of the SEs (SEB, SEC or SED) alone or in combination with other genes were concerned in food poisoning eruptions all over the world. On the other hand, the results of (Gholamzad et al., 2015, Chang et al., 2016) disagreed with our findings; these studies found that the SEB gene was the most common and prevalent S. aureus enterotoxin gene.
The phylogenetic analysis of the SED gene demonstrated 100% similarity between the two analysed isolates (accession numbers MF359584 and MF359585), and with other international isolates. Interestingly, (Patel, 2001) used the rrs gene for genotyping and the genetic analysis of S. aureus isolates of milk origin that were not easily phenotypically identified. Also, (Herron-Olson et al., 2007) performed genetic characterization of the tst and sec genes of bovine strain RF122. Moreover, (Ikawaty et al., 2008) declared that the multiple-locus variable-number tandem-repeat analysis (MLVA) technique was effective for the detection of the close genetic relationship among human S. aureus isolates. The clear diversity between S. aureus enterotoxin genotypes from different farms and countries may be attributed to differences in geographical areas and isolate origins (Larsen et al., 2002, Monistero et al., 2018).
5. Conclusions
The current results spot highlights on the high prevalence of S. aureus enterotoxins among fresh milk and ice cream samples that contribute a serious public health hazardous to humans. In addition to, the close relationship between the circulating S. aureus strains recovered from these products in Egypt and other strains from the USA may reflect the possible transmission of these strains through the importation of dairy cows and milk products from these countries and highlight the important role of animal transport and milk product trading in spreading these circulating strains.
Author contributions
All authors contributed to the reagents/materials/analysis tools, collected the material, analysed the data and wrote and revised the manuscript.
Acknowledgment
The authors extend their appreciation to the Deanship of Scientific Research at King Saud University for funding the work through the research group project No.: (RG-162)
Footnotes
Peer review under responsibility of King Saud University.
References
- APHA, 2004. Standard Methods for the Examination of Dairy Products, 17th ed., American Public Health Association, Washington, DC., USA.
- Abraha H., Hadish G., Aligaz B., Eyas G., Workelule K. Antimicrobial resistance profile of Staphylococcus aureus isolated from raw cow milk and fresh fruit juice in Mekelle, Tigray, Ethiopia. J. Vet. Med. Anim. Heath. 2017;10(4):106–113. [Google Scholar]
- Akanbi O.E., Njom H.A., Fri J., Otigbu C.A., Clarke M.A. Antimicrobial Susceptibility of Staphylococcus aureus Isolated from Recreational Waters and Beach Sand in Eastern Cape Province of South Africa. Int. J. Environ. Res. Public Health. 2017;14:2–15. doi: 10.3390/ijerph14091001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aliyu Y., Jibril U.Y., Jibrin S.M., Salawu E.M. Occurrence and Antibiotic Resistant Phenotypes of Staphylococcus aureus Isolated from some Hospital Environments in Nasarawa town, Nasarawa State, Nigeria. Eur. J. Pharm. Med. Res. 2018;5(11):20–26. [Google Scholar]
- Altschul S.F., Gish W.J., Miller W. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Argudín M.Á., Mendoza M.C., Rodicio M.R. Food poisoning and Staphylococcus aureus enterotoxins. Toxins. 2010;2:1751–1773. doi: 10.3390/toxins2071751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y., Gao H., Zhu Z., Ye S., Yang Y., Shen X. High prevalence and properties of enterotoxin-producing Staphylococcus aureus ST5 strains of good sources in China. Foodborne Pathog. Dis. 2016;13:386–390. doi: 10.1089/fpd.2015.2085. [DOI] [PubMed] [Google Scholar]
- Dai J., Wu S., Huang J., Wu Q., Zhang F., Zhang J. Prevalence and Characterization of Staphylococcus aureus Isolated From Pasteurized Milk in China. Front Microbiol. 2019;10:641. doi: 10.3389/fmicb.2019.00641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Buyser M.L., Dufour B., Maire M., Lafarge V. Implication of milk and milk products in food-borne disease in France and in different industrialized countries. Int. J. Food Microbiol. 2001;67:1–17. doi: 10.1016/s0168-1605(01)00443-3. [DOI] [PubMed] [Google Scholar]
- Denayer S., Delbrassinne L., Nia Y., Botteldoorn N. Food-borne outbreak investigation and molecular typing: high diversity of Staphylococcus aureus strains and importance of toxin detection. Toxins (Basel) 2017;9(12):407. doi: 10.3390/toxins9120407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmann K.K., Chaul L.T., Lee S.H.I., Corassin C.H., Fernandes de Oliveira C.A., Pereira De Martinis E.C., Alves V.F., Gram L., Oxaran V. Staphylococcus aureus in some Brazilian dairy industries: changes of contamination and diversity. Front Microbiol. 2017;8:2049. doi: 10.3389/fmicb.2017.02049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbehiry A., Al-Dubaib M., Marzouk E., Moussa I. Antibacterial effects and resistance induction of silver and gold nanoparticles against Staphylococcus aureus-induced mastitis and the potential toxicity in rats. Microbiologyopen. 2019;8(4) doi: 10.1002/mbo3.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbehiry A., Marzouk M., Hamada M., Al-Dubaib M., Alyamani E., Moussa I.M., AlRowaidhan A., Hemeg H.A. Application of MALDI-TOF MS fingerprinting as a quick tool for identification and clustering of foodborne pathogens isolated from food products. New Microbiol. 2017;40(4):269–278. [PubMed] [Google Scholar]
- El Faramaway R.T., Abdeen E.E., Ashraf A.A., Mousa W.S. Antibiogram profile and molecular characterization of Coa and spa genes of methicillin resistant Staphylococcus aureus (MRSA) from clinical mastitis. Alex. J. Vet. Sci. 2019;61(1):32–38. [Google Scholar]
- Finegold S.M., Martin W.J. 16th Ed. Th C.V. Mosby Co.; St. Louis Toronto, London: 1982. Diagnosis Microbiology. [Google Scholar]
- Freeman D.J., Falkiner F.R., Keane C.T. New method for detecting slime production by coagulase negative staphylococci. J. Clin. Pathol. 1989;42:872–874. doi: 10.1136/jcp.42.8.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gholamzad M., Khatami M.R., Ghassemi S., Vaise Malekshahi Z., Shooshtari M.B. Detection of Staphylococcus Enterotoxin B (SEB) using an immune chromatographic test strip. Jundishapur J. Microbiol. 2015;8(9) doi: 10.5812/jjm.26793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gündogan N., Citak S., Turan E. Slime production, DNAse activity and antibiotic resistance of Staphylococcus aureus isolated from raw milk, pasteurized milk and ice cream samples. Food Control. 2006;17:389–392. [Google Scholar]
- Hennekinne J.A., De Buyser M.L., Dragacci S. Staphylococcus aureus and its food poisoning toxins: characterization and outbreak investigation. FEMS Microbiol. 2012;36:815–836. doi: 10.1111/j.1574-6976.2011.00311.x. [DOI] [PubMed] [Google Scholar]
- Herron-Olson L., Fitzgerald J.R., Musser J.M., Kapur V. Molecular correlates of host specialization in Staphylococcus aureus. PLoS ONE. 2007;2 doi: 10.1371/journal.pone.0001120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Meng J., Shi C., Hervin K., Fratamico P.M., Shi X. Characterization and comparative analysis of a second thermonuclease from Staphylococcus aureus. Microbiol. Res. 2013;168(3):174–182. doi: 10.1016/j.micres.2012.09.003. [DOI] [PubMed] [Google Scholar]
- ICMSF, 1996. Microorganisms in Foods 5: Characteristics of Microbial Pathogens, Roberts, T. A., Baird-Parker, A. C., Tompkin, R. B. (eds.), Blackie Academic & Professional, London (ISBN 0 412 47350 X).
- Ikawaty R., Brouwer E.C., Jansen M.D., Van Duijkeren E., Mevius D., Verhoef J., Fluit A.C. Characterization of Dutch Staphylococcus aureus from bovine mastitis using a multiple locus variable number tandem repeat analysis. Vet. Microbiol. 2008;136:277–284. doi: 10.1016/j.vetmic.2008.10.034. [DOI] [PubMed] [Google Scholar]
- Islam A., Nath A.D., Islam K., Islam S., Chakma S., Hossain M.B. Isolation, identification and antimicrobial resistance profile of Staphylococcus aureus in cockroaches (Periplaneta americana) J. Adv. Vet. Anim. Res. 2016;3:221–228. [Google Scholar]
- Jenul C., Horswill A.R. Regulation of Staphylococcus aureus virulence. Microbiol. Spectr. 2018 doi: 10.1128/microbiolspec.GPP3-0031-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadariya J., Smith T.C., Thapaliya D. Staphylococcus aureus and staphylococcal food-borne disease: an ongoing challenge in public health. Biomed Res Int. 2014;2014 doi: 10.1155/2014/827965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblum, J., Kreiswirth, B., Projan, S.J., Ross, H., Novick, R.P., 1990. Agr: a polycistronic locus regulating exoprotein synthesis in Staphylococcus aureus, in Molecular Biology of the Staphylococci, ed Novick R. P., editor. (New York, NY: VCH Publishers. 373–401.
- Larsen H.D., Aarestrup F.M., Jensen N.E. Geographical variation in the presence of genes encoding superantigenic exotoxins and β-hemolysin among Staphylococcus aureus isolated from bovine mastitis in Europe and USA. Vet. Microbiol. 2002;85(1):61. doi: 10.1016/s0378-1135(01)00478-3. [DOI] [PubMed] [Google Scholar]
- Le Loir Y., Baron F., Gautier M. Staphylococcus aureus and food poisoning. Genet. Mol. Res. 2003;2:63–76. [PubMed] [Google Scholar]
- Liu H., Archer N.K., Dillen C.A., Wang Y., Ashbaugh A.G., Ortines R.V. Staphylococcus aureus epicutaneous exposure drives skin inflammation via IL-36-mediated T cell responses. Cell Host Microbe. 2017;22:653–666. doi: 10.1016/j.chom.2017.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louie L., Goodfellow J., Mathieu P., Glatt A., Louie M., Simor A.E. Rapid detection of methicillin-resistant staphylococci from blood culture bottles by using a multiplex PCR assay. J. Clin. Microbiol. 2002;40:2786–2790. doi: 10.1128/JCM.40.8.2786-2790.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan K., Moore C.S., McAuley C.M., Fegan N., Fox E.M. Characterization of Staphylococcus aureus isolates from raw milk sources in Victoria. BMC Microbiol. 2016;16:1–12. doi: 10.1186/s12866-016-0789-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrotra M., Wang G., Johnson M. Multiplex PCR for detection of genes for Staphylococcus aureus enterotoxins exfoliative toxins toxic shock syndrome toxin1 and methicillin resistance. J. Clin. Microbiol. 2000;38:1032–1035. doi: 10.1128/jcm.38.3.1032-1035.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monistero V., Graber H.U., Pollera C. Staphylococcus aureus isolates from bovine mastitis in eight countries: genotypes, detection of genes encoding different toxins and other virulence genes. Toxins (Basel) 2018;10(6):247. doi: 10.3390/toxins10060247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray P.R., Baron E.J., Jorgensen J.H. Manual of Clinical Microbiology. Clin. Infec. Dis. 2003;38:1199–1200. [Google Scholar]
- Novick R.P. Pathogenicity factors and their regulation. In: Fischetti V.A., Novick R.P., Feretti J.J., Portnoy D.A., Rood J.I., editors. Gram Positive Pathogens. ASM Press; Washington, D.C., USA: 2000. pp. 392–407. [Google Scholar]
- Okpo N.O., Abdullahi I.O., Whong C.M.Z., Ameh J.B. Occurrence and Antibiogram of Staphylococcus aureus in some Dairy Products sold in parts of Kaduna State, Nigeria. UMYM J. Microbiol. Res. 2018;1(1):56–60. [Google Scholar]
- Patel J.B. 16S rRNA gene sequencing for bacterial pathogen identification in the clinical laboratory. Mol. Diag. 2001;6(4):313–321. doi: 10.1054/modi.2001.29158. [DOI] [PubMed] [Google Scholar]
- Puah S.M., Chua K.H., Tan J.A. Virulence factors and antibiotic susceptibility of Staphylococcus aureus isolates in ready-to-eat foods: detection of S. aureus contamination and a high prevalence of virulence genes. Int. J. Environ. Res. Public Health. 2016;13(2):199. doi: 10.3390/ijerph13020199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabello R.F., Moreira B.M., Lopes R.M.M., Teixeira L.M., Riley L.W., Castro A.C.D. Multilocus sequence typing of Staphylococcus aureus isolates recovered from cows with mastitis in Brazilian dairy herds. J. Med. Microbiol. 2007;56:1505–1511. doi: 10.1099/jmm.0.47357-0. [DOI] [PubMed] [Google Scholar]
- Rall V.L.M., Vieira F.P., Rall R., Vieitis R.L., Fernandes A.J.R., Candeias J.M. PCR for detection of staphylococcus enterotoxin genes in staphylococcus aureus strains isolated from raw and pasteurized milk. Vet. Microbiol. 2008;132(3–4):408–413. doi: 10.1016/j.vetmic.2008.05.011. [DOI] [PubMed] [Google Scholar]
- Sahebnasagh R., Saderi H., Owlia P. The prevalence of resistance to methicillin in Staphylococcus aureus strains isolated from patients by PCR method for detection of mecA and nuc genes. Iran. J. Public Health. 2014;43:84–92. [PMC free article] [PubMed] [Google Scholar]
- Saly M.E.T., Elbialy A.A., Zaky M.M.M., El-Shafey A.S. Prevalence of Staphylococcus aureus in raw milk and some dairy products in port said governorate. Am. J. Zool. 2019;1(2):40–46. [Google Scholar]
- Sharma V., Sharma S., Dahiya D.K., Khan A., Mathur M., Sharma A. Coagulase gene polymorphism, enterotoxigenecity, biofilm production, and antibiotic resistance in Staphylococcus aureus isolated from bovine raw milk in North West India. Ann. Clin. Microbiol. Antimicrob. 2017;16(1):65. doi: 10.1186/s12941-017-0242-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soomro A.H., Arain M.A., Khaskheli M., Bhutto B. Isolation of Staphylococcus aureus from milk products sold at sweet meat shops of Hyderabad. Online J. Biol. Sci. 2003;3:91–94. [Google Scholar]
- Stewart G.C. Staphylococcus aureus. In: Fratamico P.M., Bhunia A.K., Smith J.L., editors. Foodborne Pathogens. Microbiology and Molecular Biology. Caister Academic Press; Norfolk: 2005. [Google Scholar]
- Sudhaharan S., Vanjari L., Mamidi N., Ede N., Vemu L. Evaluation of LAMP assay using phenotypic tests and conventional PCR for detection of nuc and mecA genes among clinical isolates of Staphylococcus spp. J. Clin. Diagn. Res. 2015;9:6–9. doi: 10.7860/JCDR/2015/13962.6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson D., Higgins G., Gibson J. Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nuc. Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velázquez-Ordoñez, V., Valladares-Carranza, B., Tenorio-Borroto, E., Talavera-Rojas, M., Varela-Guerrero, J.A., Acosta-Dibarrat, J., Puigvert, F., Grille, L., Revello, Á.G., Pareja, L., 2019. Microbial contamination in milk quality and health risk of the consumers of raw milk and dairy products. http://dx.doi.org/10.5772/intechopen.86182.
- Veras, J.F., do Carmo, L.S., Tong, L.C., Shupp, J.W., Cummings, C., Dos Santos, D.A., 2008. A study of the enterotoxigenicity of coagulase-negative and coagulase-positive staphylococcal isolates from food poisoning outbreaks in Minas Gerais, Brazil. Int. J. Infect. Dis. 12, 410–415. [DOI] [PubMed]
- Vimercati C., Cremonesi P., Castiglioni B., Pisoni G., Boettcher P.J., Stella A. Molecular typing of Staphylococcus aureus isolated from cows, goats and sheep with intramammary infections on the basis of gene polymorphisms and toxins genes. J. Veterinary Med. B. Infect. Dis. Vet. Public Health. 2006;53(9):423–428. doi: 10.1111/j.1439-0450.2006.00980.x. [DOI] [PubMed] [Google Scholar]
- Wang X., Thompso C.D., Weidenmaier C., Lee J.C. Release of Staphylococcus aureus extracellular vesicles and their application as a vaccine platform. Nat. Commun. 2017;9:1379. doi: 10.1038/s41467-018-03847-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., Xiao, M., Kong, F., O'sullivan, M.V., Mao, L.L., Zhao, H.R., 2015. A multicentre study of meticillin-resistant Staphylococcus aureus in acute bacterial skin and skin-structure infections in China: susceptibility to ceftaroline and molecular epidemiology. Int. J. Antimicrob. Agents 45, 347–350. [DOI] [PubMed]