Abstract
Ultraviolet light exposure and cutaneous pigmentation are important host risk factors for cutaneous melanoma (CM), and it is well known that inherited ability to produce melanin varies in humans. The study aimed to identify single-nucleotide variants (SNVs) on pigmentation-related genes with importance in risk and clinicopathological aspects of CM. The study was conducted in two stages. In stage 1, 103 CM patients and 103 controls were analyzed using Genome-Wide Human SNV Arrays in order to identify SNVs in pigmentation-related genes, and the most important SNVs were selected for data validation in stage 2 by real-time polymerase-chain reaction in 247 CM patients and 280 controls. ADCY3 c.675+9196T>G, CREB1 c.303+373G>A, and MITF c.938-325G>A were selected for data validation among 74 SNVs. Individuals with CREB1 GA or AA genotype and allele “A” were under 1.79 and 1.47-fold increased risks of CM than others, respectively. Excesses of CREB1 AA and MITF AA genotype were seen in patients with tumors at Clark levels III to V (27.8% versus 13.7%) and at III or IV stages (46.1% versus 24.9%) compared to others, respectively. When compared to others, patients with ADCY3 TT had 1.89 more chances of presenting CM progression, and those with MITF GA or AA had 2.20 more chances of evolving to death by CM. Our data provide, for the first time, preliminary evidence that inherited abnormalities in ADCY3, CREB1, and MITF pigmentation-related genes, not only can increase the risk to CM, but also influence CM patients’ clinicopathological features.
Subject terms: Melanoma, Cancer genetics
Introduction
Cutaneous melanoma (CM) is the most deadly form of skin cancers1. Ultraviolet (UV) light exposure and individual pigmentation features are well-established host risk factors for CM1, and tumor depth and stage are the most important hallmarks of CM prognosis2. Moreover, previous studies have shown that inherited genetic variants modulate CM risk3 and outcome4.
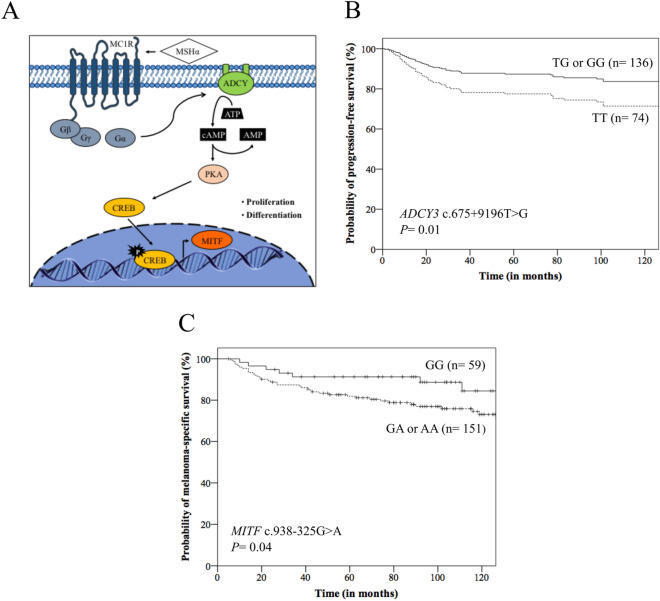
Melanocytes absorb UV radiation and survive to considerable genotoxic stress, and the main genetic mechanism involved in CM development alters the control of skin pigmentation1,5,6. Sunlight exposure induces the post-inflammatory hyperpigmentation system by the melanocortin-1 receptor (MC1R), and melanocytes express MC1R that regulates the quality and quantity of their melanin production7. The α-melanocyte stimulating hormone (MSHα) activates the membrane associated enzyme adenylate cyclase (ADCY), increasing 3′-5′-cyclic adenosine monophosphate (cAMP) levels; the increased cAMP signals activate protein kinase A (PKA) that activates cAMP responsive element binding protein (CREB)5,6,8. CREB is a transcription factor that regulates the expression of melanocyte inducing transcription factor (MITF) and consequently, proliferation and differentiation of melanocytes and melanin synthesis5,6,8 (Fig. 1A).
Figure 1.
(A) Pigmentation regulation by alpha-melanocyte stimulating hormone (MSHα) and G-proteins from melanocortin receptor 1 (MC1R): MSHα/MCR1 can trigger the activation of the adenylate cyclase (ADCY) and 3′–5′-cyclic adenosine monophosphate AMP (cAMP). The cAMP signals activate the protein kinase A (PKA) that phosphorylates and activates cAMP responsive element binding protein (CREB) transcription factor, which induces the expression of melanocyte inducing transcription factor (MITF) and induction of proliferation and differentiation of melanocytes. G-proteins: β, γ, and α; ATP: adenosine triphosphate. (B) Kaplan–Meier (K-M) curves for progression-free survival according to ADCY3 c.675+9196T>G genotypes, where patients with TT genotype presented lower survival than those with TG or GG genotype. (C) K-M curves for melanoma-specific survival according to MITF c.938-325G>A genotypes, where patients with GA or AA genotype presented lower survival than those with GG genotype.
On the other hand, melanogenesis generates mutagenic intermediates (quinones and semiquinones), neutralizes reactive oxygen species, eliminates free radicals, and modifies cell metabolism through the stimulation of aerobic glycolysis generating a hypoxic environment9,10, making melanoma cells resistant to chemo-, radio-, photo- and immunotherapy9,11. Brożyna et al. showed that nonpigmented cells were significantly more sensitive to gamma rays than pigmented cells12. Melanogenesis induction is also related to significant up-regulation of hypoxia-inducible factors (HIF) and these factors are key master regulators of cellular metabolism and therapeutic resistance11,13, contributing to the increased aggressiveness of melanoma and shorter survival time of patients with pigmented metastatic melanoma than the ones with amelanotic lesion11.
Genome-wide association studies (GWAS), conducted particularly in Caucasians, have identified single nucleotide variants (SNVs) associated with CM risk, many of which in human pigmentation genes, such as MC1R, solute carrier family 45 member 2 (SLC45A2) and tyrosinase (TYR)14–17. The most GWAS have identified SNVs located in non-coding regions of the genome which can affect gene regulatory sequences, and consequently the gene expression18. SNVs located in introns can also alter the precursor RNA messenger (pre-mRNA) splicing process19 or the binding sites for regulatory proteins splicing20, influencing the efficiency of splicing or inducing alternative splicing, and an intronic variant of poly(ADP-ribose) polymerase number 1 (PARP1) was associated with increased CM risk in Caucasians21.
The Brazilian population is highly heterogeneous, consisting of indigenous Amerindians and immigrants from Europe, Africa, and Asia22. Since other SNVs in genes with equal or even greater importance in melanogenesis may not have been selected in the previous analyzed populations, we conducted an association study in patients with CM and healthy controls from Brazil, and identified three SNVs of MC1R pathway, ADCY3 c.675+9196T>G, CREB1 c.303+373G>A and MITF c.938-325G>A in association with tumor risk and clinicopathological features.
Material and methods
Study population
This association study was conducted in two stages. In stage 1, 103 CM patients and 103 controls were analyzed with the purpose of identifying SNVs on pigmentation-related genes with importance in CM risk, and in stage 2 the most important SNVs were selected for data validation in 247 CM patients and 280 controls.
All CM patients were diagnosed at the Clinical Oncology and Dermatology Services of University of Campinas, A.C. Camargo Cancer Center, and Barretos Cancer Hospital between April 2000 and May 2018. Patients diagnosed with the unknown primary site tumors and those with tumors located in mucous were excluded from the study. The control group was compound by blood donors seen at the Hematology and Hemotherapy Center of University of Campinas in the same period. The study was approved by the local Ethical Committees of both Institutions (numbers: 424/20016 and 1.438.601). All procedures were carried out according to the Helsinki Declaration, and appropriate informed consent was obtained.
Data and specimen collection
Clinical information of individuals (age at diagnosis, gender, skin color, skin phototype, sun exposure, type of sun exposure, and number of nevi) was obtained by specific questionnaires. Skin phototype was defined using reported criteria23. Individuals exposed to the sun for more than 2 h per day and for more ten years were considered positive for sun exposure23. Sun exposure was classified as intermittent in cases of recreational activities performed less than 50% of the week or holidays, or chronic, activities at home or work under sunlight exposure during more than 50% of the time24.
The diagnosis of CM was established by histopathological evaluation of tumor fragments embedded in paraffin and stained with hematoxylin and eosin. Pathological aspects of the tumor (tumor location, histological type, Breslow thickness, Clark level, and tumor stage) were obtained from medical records of patients25. Tumor stage was identified using the TNM classification of the American Joint Committee on Cancer, where T describes the size of tumor, N describes spread of tumor to nearby lymph nodes, and M describes distant metastasis2. Patients with desmoplastic, acro-lentiginous and amelanotic melanomas were excluded from the study.
Surgical excision (n = 217) was the primary treatment for patients with localized tumor26. Sentinel lymph node biopsy (n = 41) was recommended in patients with tumor measuring more than 1 mm (mm) and lymphadenectomy (n = 24) was performed in patients with clinically positive lymph nodes or lymph nodes with tumor infiltration on histopathological evaluation. Patients with operable single metastasis or relapse (n = 30) underwent surgical resection27. Those patients with inoperable relapse or multiple metastases (n = 30) received chemotherapy with dacarbazine28. Radiotherapy was also used in the local treatment of patients with surgical impossibility (n = 4), particularly in bleeding lesions, bone or brain metastases29.
Stage 1: screening of SNVs, candidate genes choice and SNVs selection
DNA from leukocytes of peripheral blood of CM patients and controls were genotyped for a total of 906,660 SNVs using the Affymetrix Genome-Wide Human SNV Arrays 6.0 (AFFYMETRIX, USA), according to the manufacturer’s recommended protocols. The intensities resulting from the arrays scanning process were made available via CEL files, one per DNA sample with total quality control higher than 90% (AFFYMETRIX, USA). Tools from the Bioconductor (https://www.bioconductor.org) were used to process the CEL files. The genotyping was performed applying the corrected robust linear mixture model (crlmm) algorithm30.
The genes previously reported as involved in the pigmentation pathway were selected for study. The pathway analysis was performed using the Database for Annotation, Visualization and Integrated Discovery (https://david.ncifcrf.gov31 and Kyoto Encyclopedia of Genes and Genomes pathway maps (https://www.kegg.jp)32.
Each pigmentation related-gene was analyzed using the in silico method by the Human Splicing Finder algorithm (version 3.1) (https://www.umd.be/HSF3/index.html33 in order of identifying SNVs in splicing regulatory sequences. For analysis, wild-type (ancestral) allele was taken as reference. SNVs showing deviation from the HWE and those with the minor allele frequency less than 10% were excluded from the selection34. SNVs that potentially alter expression or function of the encoding proteins13,14 were selected for further validation.
Stage 2: validation of selected SNVs in risk and characteristics of melanoma
DNA from leukocytes of peripheral blood of CM patients and controls was analyzed by real-time polymerase chain reaction with TaqMan SNV genotyping assays (APPLIED BIOSYSTEMS, USA) for ADCY3 (rs11900505, assay ID: C_7868411_20), CREB1 (rs10932201, assay ID: C_2859093_20) and MITF (rs7623610, assay ID: C_29012190_10) SNVs, following manufacturer instructions. Twenty percent of genotype determinations were carried out twice in independent experiments with 100% of concordance.
Frequencies of ADCY3 c.675+9196T>G, CREB1 c.303+373G>A and MITF c.938-325G>A genotypes, isolated and in combination, were analyzed in patients and controls, and in patients stratified by clinicopathological features.
Gene expression by quantitative PCR
Total RNA was obtained from leukocytes of peripheral blood of CM patients and controls with distinct genotypes of ADCY3 (16 and 18 with TT genotype, 16 and 19 with TG, and seven and 19 with GG, respectively), CREB1 (14 and 10 with GG genotype, 16 and 25 with GA, 9 and 18 with AA, respectively) and MITF (15 and 28 with GG genotype, 16 and 23 with GA, 6 and 23 with AA, respectively) with TRIzol reagent (LIFE TECHNOLOGIES, USA).
cDNA was generated using Maxima First Strand cDNA Synthesis kit reagents (LIFE TECHNOLOGIES, USA) following manufacturer’s instructions. The experiments were performed using SYBR Green PCR Master Mix reagents (APPLIED BIOSYSTEMS, USA) and specific primers for ADCY3 (forward: 5′-TCATCTCCGTGGTCTCCTG-3′ and reverse: 5′-CACAGGTAGAGGAA-GACGTTG-3′), CREB1 (forward 5′-CTAGTACAGCTGCCCA-ATGG-3′ and reverse: 5′-AGTTG-AAATCTGTGTTCCGG-3′), and MITF (forward: 5′-AGTCTGAAGCAAGAGCACTG-3′ and reverse: 5′-GCGCATGTCTGGATCATTTG-3′) genes, in triplicate per sample, and a control without template were included in each plate. The relative expression level of genes was normalized to that of reference housekeeping gene actin beta (forward: 5′-AGGCCAACCGCGAGAAG-3′ and reverse: 5′-ACAGCCTGGATAGCAACGTACA-3′) using 2−DDCt cycle threshold method35. Twenty percent of samples had evaluation repeated in separate experiments with 100% agreement. The results were expressed in arbitrary units (AUs).
Statistical analysis
Association between disease statuses, CM patients versus controls, and genotypes for study’s stage 1 was performed using logistic regression model, and analyses were adjusted by age at diagnosis, skin color, and sun exposure. SNVs that presented raw p-values below the 0.001 thresholds were selected for further inspection. These analyses were implemented in R software (version 3.3.0) (https://www.r-project.org).
The Hardy–Weinberg equilibrium (HWE) was tested using chi-square (χ2) statistics for the goodness-to-fit test, and logistic regression model served to obtain age, skin color, sun exposure, and number of nevi status-adjusted crude odds ratios (ORs) with 95% confidence intervals (CI) in comparisons evolving patients and controls for study’s stage 2. To evaluate the robustness of risk estimates, the false discovery rate (FDR) was computed, which reflects the expected ratio of false-positive findings to the total number of significant findings; the differences revealed were considered statistically significant at FDR values < 0.0525. χ2 and Fisher’s exact tests were used to evaluate associations between clinicopathological features and genotypes of selected SNVs. Bonferroni method was used to adjust values of multiple comparisons in patients stratified by tumor aspects36. For ADCY3, CREB1 and MITF expression analysis, data sets were probed for normality using Shapiro–Wilk’s test. Because data sets assume normal distribution, analysis of variance performed comparisons of groups36.
For survival analysis, the progression-free survival (PFS) was calculated from the date of surgery until the date of first recurrence, or the date of progression of disease, or the date of death by any cause, or the date of last follow-up. The melanoma-specific survival (MSS) was calculated from the date of diagnosis until the date of death by the disease or last follow-up. PFS and MSS were calculated using Kaplan–Meier estimates, and differences between survival curves were analyzed by log-rank test25. The impact of age at diagnosis, gender, tumor location, Breslow thickness37, Clark level, TNM stage and genotypes of each analyzed SNV in survival of patients were evaluated using univariate Cox proportional hazards ratio (HR) regression. At a second time, all variables with p < 0.20 were included in the multivariate Cox regression. The significant results of Cox analysis were internally validated using a bootstrap resampling study to investigate the stability of risk estimates (1,000 replications)25.
All tests were done using the SPSS 21.0 software (SPSS INCORPORATION, USA). Significance was two sided and achieved when p values were ≤ 0.05.
Results
Study population
The clinicopathological features of patients and the clinical features of controls included in stage 1 and stage 2 of the study are presented in Table 1. Controls were younger than patients, and CM patients presented more white skin color, referred more sun exposure and presented more nevi than controls, and all differences were corrected in comparisons involving patients and controls by appropriate statistical analysis. Similar clinicopathological features were observed in patients and controls analyzed in both stages of the study.
Table 1.
Clinicopathological aspects of patients with cutaneous melanoma and clinical features of controls used in screening (stage 1) and validation (stage 2) of single nucleotide variants in human pigmentation-related genes. *The numbers of individuals were not the same included in the study because no consistent information could be obtained from some individuals. NC: values were not computed. P values describe differences between patients and controls and values < 0.05 are presented in bold letters.
| Characteristics | Stage 1 | Stage 2 | ||||
|---|---|---|---|---|---|---|
| Patients n (%) |
Controls n (%) |
p value | Patients n (%) |
Controls n (%) |
p value | |
| Age (years) | ||||||
| ≤ 55 | 55 (53.4) | 93 (90.3) | < 0.0001 | 125 (50.6) | 229 (81.8) | < 0.0001 |
| > 55 | 48 (46.6) | 10 (9.7) | 122 (49.4) | 51 (18.2) | ||
| Gender | ||||||
| Male | 61 (59.2) | 55 (53.4) | 0.48 | 130 (52.6) | 145 (51.8) | 0.84 |
| Female | 42 (40.8) | 48 (46.6) | 117 (47.4) | 135 (48.2) | ||
| Skin color | ||||||
| White | 97 (94.2) | 80 (77.7) | 0.001 | 231 (93.5) | 231 (82.5) | < 0.0001 |
| Non-white | 6 (5.8) | 23 (22.3) | 16 (6.5) | 49 (17.5) | ||
| Phototype* | ||||||
| I or II | 58 (68.2) | 53 (53.0) | 0.05 | 154 (66.7) | 164 (60.1) | 0.12 |
| III to VI | 27 (31.8) | 47 (47.0) | 77 (33.3) | 109 (39.9) | ||
| Sun exposure* | ||||||
| Yes | 70 (82.4) | 52 (52.0) | < 0.0001 | 196 (83.1) | 126 (45.0) | < 0.0001 |
| No | 15 (17.6) | 48 (48.0) | 40 (16.9) | 154 (55.0) | ||
| Type of sun exposure* | ||||||
| None or intermittent | 32 (34.4) | 76 (73.8) | < 0.0001 | 96 (44.2) | 210 (75.0) | < 0.0001 |
| Chronic | 61 (65.6) | 27 (26.2) | 121 (55.8) | 70 (25.0) | ||
| Number of nevi* | ||||||
| ≤ 50 | 68 (80.0) | 91 (97.8) | 0.0001 | 183 (75.9) | 260 (98.1) | < 0.0001 |
| > 50 | 17 (20.0) | 2 (2.2) | 58 (24.1) | 5 (1.9) | ||
| Tumor location | ||||||
| Head | 16 (18.4) | NA | NC | 46 (18.6) | NC | NC |
| Trunk | 41 (47.1) | NA | NC | 118 (47.8) | NC | NC |
| Upper limb | 13 (14.9) | NA | NC | 40 (16.2) | NC | NC |
| Lower limb | 17 (19.6) | NA | NC | 43 (17.4) | NC | NC |
| Histological type* | ||||||
| Superficial spreading | 37 (50.7) | NA | NC | 119 (57.2) | NC | NC |
| Lentigo malign | 9 (12.3) | NA | NC | 26 (12.5) | NC | NC |
| Nodular | 27 (37.0) | NA | NC | 63 (30.3) | NC | NC |
| Breslow thickness (mm)* | ||||||
| ≤ 1.5 | 44 (46.8) | NA | NC | 123 (52.8) | NC | NC |
| > 1.5 | 50 (53.2) | NA | NC | 110 (47.2) | NC | NC |
| Clark level* | ||||||
| I | 14 (16.5) | NA | NC | 31 (13.2) | NC | NC |
| II | 12 (14.1) | NA | NC | 42 (17.9) | NC | NC |
| III | 27 (31.8) | NA | NC | 60 (25.5) | NC | NC |
| IV | 30 (35.3) | NA | NC | 94 (40.0) | NC | NC |
| V | 2 (2.3) | NA | NC | 8 (3.4) | NC | NC |
| TNM stage* | ||||||
| 0 | 12 (13.8) | NA | NC | 30 (12.3) | NC | NC |
| I | 23 (26.4) | NA | NC | 92 (37.7) | NC | NC |
| II | 27 (31.0) | NA | NC | 83 (34.0) | NC | NC |
| III | 22 (25.3) | NA | NC | 27 (11.1) | NC | NC |
| IV | 3 (3.5) | NA | NC | 12 (4.9) | NC | NC |
Screening of SNVs, candidate genes choice and SNVs selection
We found 12,495 new SNVs associated with CM risk; 6,497 (52.0%) of them were in introns, 5,928 (47.4%) in gene regulatory regions, and 70 (0.6%) in coding regions. The genome association data were deposited at Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo) with accession number GSE129890.
The most significant melanoma associated SNVs identified in stage 1 (p < 0.0001) are presented in Table S1 Supplement. Seventy-four SNVs in 28 human pigmentation-related genes were found to be involved with CM risk (Table S2 Supplement).
In accord with results of the in silico analysis, the variant allele “G” of ADCY3 c.675+9196T>G may abolish a potential branch point site and an exonic splicing enhancer (ESE), and this variation may create a site of ligation for SRp55 and 9G8 splicing proteins. Besides, new sites for an exon-identity element (EIE) and an intron-identity element (IIE) may be created. The variant allele “A” of CREB1 c.303+373G>A may abolish a splice donor site (5′ end of the intron), an exonic splicing silencer (ESS), an IIE site, and a binding site for the hnRNP A1, and this variation may create a new branch point, an EIE site, and a putative exonic splicing enhancer. The variant allele “A” of MITF c.938-325G>A may create a splice donor site, an ESS, and a binding site for the hnRNP A1, and this variation may break a potential branch point site, an EIE, and silencer motifs and an IIE site (Table S3 Supplement). ADCY3 c.675+9196T>G, CREB1 c.303+373G>A, and MITF c.938-325G>A SNVs were selected for analysis in stage 2 of the study due to their potential effects on encoding proteins13,14.
Selected SNVs in risk and clinicopathological features of melanoma
Patient and control samples included in the stage 2 were in HWE at ADCY3 c.675+9196T>G (χ2 = 0.58, p = 0.44; χ2 = 0.70, p = 0.40), and CREB1 c.303+373G>A (χ2 = 0.08, p = 0.77; χ2 = 1.55, P = 0.21) loci, respectively. Controls’ samples (χ2 = 0.92, p = 0.33) but not patients’ samples (χ2 = 4.40, p = 0.03) confirmed the HWE at MITF rs7623610 locus.
CREB1 GA or AA genotype and allele “A” were more common in patients than in controls; carriers of the above genotypes and allele were under 1.79 and 1.47-fold increased risks for CM than those with the GG genotype and allele “G”, respectively (Table 2). No associations between ADCY3, CREB1 and MITF SNVs combined genotypes were seen in CM patients and controls (Table S4 Supplement).
Table 2.
ADCY3 c.675+9196T>G, CREB1 c.303+373G>A, and MITF c.938-325G>A genotypes and alleles in 247 patients with cutaneous melanoma and 280 controls. OR: odds ratio adjusted by age, skin color, sun exposure, and number of nevi by multiple regression analysis; CI confidence interval; Pc values are p values corrected for multiple testing by the false discovery rate test. P and pc values < 0.05 are presented in bold letters.
| Genotype or allele | Patients N (%) |
Controls N (%) |
P value (pc value) | OR (95% CI) |
|---|---|---|---|---|
| ADCY3 c.675+9196T>G | ||||
| TT | 84 (34.0) | 86 (30.7) | 0.84 (0.84) | 1.04 (0.66–1.64) |
| TG or GG | 163 (66.0) | 194 (69.3) | Reference | |
| TT or TG | 209 (84.6) | 218 (77.9) | 0.07 (0.12) | 1.67 (0.95–2.93) |
| GG | 38 (15.4) | 62 (22.1) | Reference | |
| Allele T | 294 (59.5) | 304 (54.3) | 0.35 (0.35) | 1.15 (0.85–1.56) |
| Allele G | 200 (40.5) | 256 (45.7) | Reference | |
| CREB1 c.303+373G>A | ||||
| GG | 68 (27.5) | 110 (39.3) | 0.01 (0.04) | Reference |
| GA or AA | 179 (72.5) | 170 (60.7) | 1.79 (1.14–2.82) | |
| GG or GA | 189 (76.5) | 233 (83.2) | 0.19 (0.28) | Reference |
| AA | 58 (23.5) | 47 (16.8) | 1.43 (0.83–2.46) | |
| Allele G | 257 (52.0) | 344 (61.4) | 0.01 (0.04) | Reference |
| Allele A | 237 (48.0) | 216 (38.6) | 1.47 (1.08–2.00) | |
| MITF c.938-325G>A | ||||
| GG | 71 (28.7) | 92 (32.9) | 0.49 (0.55) | Reference |
| GA or AA | 176 (71.3) | 188 (67.1) | 1.17 (0.74–1.85) | |
| GG or GA | 178 (72.1) | 222 (79.3) | 0.02 (0.06) | Reference |
| AA | 69 (27.9) | 58 (20.7) | 1.76 (1.07–2.89) | |
| Allele G | 249 (52.0) | 344 (61.4) | 0.06 (0.12) | Reference |
| Allele A | 237 (48.0) | 216 (38.6) | 1.28 (0.84–1.94) | |
No associations of studied SNVs genotypes were seen in CM patients stratified by age, gender, and skin color (Table S5), phototype, sun exposure, type of sun exposure, and number of nevi (Table S6). However, CREB1 AA genotype was more common in patients with tumors located in limbs than in head or trunk (31.7% versus 15.9%, p = 0.009) and tumors with Clark levels III to V than in those with tumors of I or II Clark levels (27.8% versus 13.7%, p = 0.012), and MITF AA genotype was more common in patients with III or IV tumor stage than in those with tumors at 0 to II stages (46.1% versus 24.9%, p = 0.007). These results were significant even after Bonferroni correction (corrected p value: 0.0125) (Table 3).
Table 3.
ADCY3 c.675 + 9196 T > G, CREB1 c.303 + 373G > A, and MITF c.938-325A > G genotypes in 247 patients with cutaneous melanoma stratified by tumor features. Values are expressed as number and percentage. *The numbers of patients were not the same included in the study (n = 247) because no consistent information could be obtained from some individuals. P values < 0.05 are presented in bold letters. **Significant even after Bonferroni correction for multiple comparisons (corrected p value = 0.0125).
| Genotypes | Histological type* | Breslow thickness (mm)* | Clark level* | TNM stage* | ||||
|---|---|---|---|---|---|---|---|---|
| Head/trunk | Upper/lower limb | ≤ 1.5 | > 1.5 | I or II | III to V | 0 to II | III or IV | |
| ADCY3 | ||||||||
| TT | 53 (32.3) | 31 (37.4) | 42 (34.1) | 37 (33.6) | 23 (31.5) | 57 (35.2) | 71 (34.6) | 12 (30.8) |
| TG or GG | 111 (67.7) | 52 (62.6) | 81 (65.9) | 73 (66.4) | 50 (68.5) | 105 (64.8) | 134 (65.4) | 27 (69.2) |
| P value | 0.18 | 0.93 | 0.58 | 0.64 | ||||
| TT or TG | 121 (83.4) | 55 (87.3) | 104 (84.5) | 95 (86.4) | 65 (89.0) | 134 (82.7) | 173 (84.4) | 34 (87.2) |
| GG | 24 (16.6) | 8 (12.7) | 19 (15.5) | 15 (13.6) | 8 (11.0) | 28 (17.3) | 32 (15.6) | 5 (12.8) |
| P value | 0.53 | 0.69 | 0.24 | 0.39 | ||||
| CREB1 | ||||||||
| GG | 48 (33.1) | 13 (20.6) | 36 (29.3) | 28 (25.4) | 23 (31.5) | 41 (25.3) | 57 (27.8) | 11 (28.2) |
| GA or AA | 97 (66.9) | 50 (79.4) | 87 (70.7) | 82 (74.6) | 50 (68.5) | 121 (74.7) | 148 (72.2) | 28 (71.8) |
| P value | 0.07 | 0.51 | 0.32 | 0.95 | ||||
| GG or GA | 122 (84.1) | 43 (68.3) | 100 (81.3) | 79 (71.8) | 63 (86.3) | 117 (72.2) | 156 (76.1) | 32 (82.0) |
| AA | 23 (15.9) | 20 (31.7) | 23 (18.7) | 31 (28.2) | 10 (13.7) | 45 (27.8) | 49 (23.9) | 7 (18.0) |
| P value | 0.009** | 0.08 | 0.012** | 0.53 | ||||
| MITF | ||||||||
| GG | 43 (29.6) | 14 (32.6) | 36 (29.3) | 30 (22.7) | 25 (34.7) | 41 (25.3) | 62 (30.2) | 6 (15.4) |
| GA or AA | 102 (70.4) | 49 (67.4) | 87 (70.7) | 80 (77.3) | 48 (65.3) | 121 (74.7) | 143 (69.8) | 33 (84.6) |
| P value | 0.26 | 0.15 | 0.73 | 0.07 | ||||
| GG or GA | 106 (73.1) | 42 (66.7) | 93 (75.6) | 76 (69.1) | 52 (71.2) | 119 (73.5) | 154 (75.1) | 21 (53.9) |
| AA | 39 (26.9) | 21 (33.3) | 30 (24.4) | 34 (30.9) | 21 (28.8) | 43 (26.5) | 51 (24.9) | 18 (46.1) |
| P value | 0.34 | 0.72 | 0.26 | 0.007** | ||||
ADCY3, CREB1 and MITF expression
Similar mRNA expressions (in arbitrary units ± standard deviation) were seen in CM patients with distinct genotypes of ADCY3 (TT: 1.13 ± 0.55, TG: 0.90 ± 0.66, GG: 1.16 ± 0.85; p = 0.52) (Figure S1A Supplement), CREB1 (GG: 1.21 ± 0.76, GA: 1.19 ± 0.79, AA: 1.14 ± 0.63; p = 0.98) (Figure S1B Supplement), and MITF (GG: 1.08 ± 0.47, GA: 0.98 ± 0.74, AA: 1.03 ± 0.61; p = 0.91) (Figure S1C Supplement). Expressions of mRNA were also similar in controls with distinct genotypes of ADCY3 (TT: 1.06 ± 0.34, TG: 1.06 ± 0.44, GG: 1.08 ± 0.74; p = 0.98) (Figure S1D Supplement), CREB1 (GG: 1.06 ± 0.42, GA: 1.24 ± 0.78, AA: 1.48 ± 0.70; p = 0.29) (Figure S1E Supplement), and MITF (GG: 1.15 ± 0.69, GA: 1.24 ± 0.45, AA: 0.99 ± 0.46; p = 0.29) (Figure S1F Supplement).
Association of clinicopathological aspects and genotypes with patients’ survival
We obtained consisted survival data from 210 CM patients. The median follow-up time of patients enrolled in the survival analysis was 97 months (range 5–228 months). The patient’s final status was established on January 2020, when 136 patients were alive (132 without disease, 4 with disease) and 74 patients had died (46 due to disease, 28 of unrelated causes).
At 60 months of follow-up, the PFS was lower in males (68.5% versus 81.3%, p = 0.02), patients with tumors located in head or trunk (70.9% versus 82.0%, p = 0.03), patients with tumor with Breslow index higher 1.5 mm (54.9% versus 94.4%, p < 0.0001), Clark levels III to V (66.3% versus 94.1%, p < 0.0001) and III or IV stage (32.3% versus 82.2%, p < 0.0001) (Kaplan–Meier estimates). Differences among groups remained the same in univariate analysis. In multivariate analysis, CM located in head or trunk (HR: 2.38), thicker tumors (HR: 4.93), stage III or IV tumors (HR: 3.30), and ADCY3 TT genotype (HR: 1.89) (Fig. 1B) were predictors of poor PFS. At 60 months of follow-up, the MSS was lower in males (76.4% versus 93.7%, p < 0.0001), patients with tumors with Breslow index higher 1.5 mm (72.8% versus 97.1%, p < 0.0001), Clark levels III to V (79.1% versus 98.5%, p < 0.0001) and stage III or IV (45.2% versus 91.3%, p < 0.0001), and MITF GA or AA genotype (81.9% versus 91.3%, p = 0.04) (Fig. 1C) (Kaplan–Meier estimates). Differences among groups remained the same in univariate analysis; patients with MITF GA or AA genotype had 2.20 more chances of evolving to death by CM than others. In multivariate analysis, males (HR: 3.12), thicker tumors (HR: 4.86) and III or IV tumor stage (HR: 4.01) were predictors of poor MMS (Table 4).
Table 4.
Clinicopathological aspects and genotypes in survival of 210 cutaneous melanoma patients. HR, hazard ratio; CI, confidence interval; NC, characteristic not computed in multivariate analysis. *The total numbers of individuals differed from the total quoted because it was not possible to obtain consistent information about characteristics in some individuals. aPbootstrap = 0.01; bPbootstrap = 0.001. cPbootstrap = 0.001. dPbootstrap = 0.02; ePbootstrap = 0.002. fPbootstrap = 0.006. gPbootstrap < 0.0001 in multivariate analysis. Significant differences between groups are presented in bold letters.
| Variable | Progression-free survival | Melanoma-specific survival | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| N total/N events | Univariate Cox analysis | Multivariate Cox analysis | N total/ N events |
Univariate Cox analysis | Multivariate Cox analysis | |||||
| HR (95% CI) | p | HR (95% CI) | p | HR (95% CI) | P | HR (95% CI) | p | |||
| Median age | ||||||||||
| ≤ 55 years | 102/31 | Reference | 0.50 | Reference | 0.17 | 102/21 | Reference | 0.50 | NA | NA |
| > 55 years | 108/36 | 1.18 (0.72–1.91) | 1.40 (0.86–2.29) | 108/25 | 1.22 (0.68–2.18) | |||||
| Gender | ||||||||||
| Male | 113/43 | 1.78 (1.07–2.94) | 0.02 | 1.09 (0.63–1.88) | 0.74 | 113/36 | 3.61 (1.79–7.29) | < 0.001 | 3.12 (1.52–6.41) | 0.002e |
| Female | 97/24 | Reference | Reference | 97/10 | Reference | Reference | ||||
| Tumor location | ||||||||||
| Head or trunk | 143/52 | 1.83 (1.03–3.26) | 0.03 | 2.38 (1.22–4.62) | 0.01a | 143/35 | 1.54 (0.78–3.03) | 0.21 | NA | NA |
| Upper/lower limb | 67/15 | Reference | Reference | 67/11 | Reference | |||||
| Breslow thickness* | ||||||||||
| ≤ 1.5 mm | 109/12 | Reference | < 0.001 | Reference | < 0.001b | 109/7 | Reference | < 0.001 | Reference | < 0.001f |
| > 1.5 mm | 90/47 | 6.51 (3.44–12.29) | 4.93 (2.53–9.57) | 90/33 | 6.64 (2.93–15.04) | 4.86 (2.09–11.31) | ||||
| Clark level* | ||||||||||
| I or II | 69/6 | Reference | < 0.001 | Reference | 0.20 | 69/3 | Reference | 0.001 | Reference | 0.29 |
| III-V | 132/55 | 5.71 (2.45–13.29) | 1.89 (0.70–5.05) | 132/38 | 7.39 (2.28–23.96) | 2.08 (0.53–8.12) | ||||
| TNM stage* | ||||||||||
| 0-II | 177/43 | Reference | < 0.001 | Reference | < 0.001c | 177/26 | Reference | < 0.001 | Reference | < 0.001 g |
| III or IV | 31/23 | 5.18 (3.08–8.71) | 3.30 (1.85–5.89) | 31/20 | 7.51 (4.17–13.56) | 4.01 (2.07–7.74) | ||||
| ADCY3 | ||||||||||
| TT | 74/29 | 1.41 (0.87–2.29) | 0.16 | 1.89 (1.11–3.21) | 0.01d | 74/21 | 1.58 (0.88–2.83) | 0.12 | 1.49 (0.79–2.82) | 0.21 |
| TG or GG | 136/38 | Reference | Reference | 136/25 | Reference | Reference | ||||
| TT or TG | 180/58 | 1.04 (0.51–2.12) | 0.89 | NC | NC | 180/40 | 1.06 (0.42–2.37) | 0.99 | NC | NA |
| GG | 30/9 | Reference | 30/6 | Reference | ||||||
| CREB1 | ||||||||||
| GG | 60/15 | Reference | 0.19 | Reference | 0.23 | 60/10 | Reference | 0.32 | NC | NA |
| GA or AA | 150/52 | 1.51 (0.87–2.63) | 1.45 (0.78–2.70) | 150/36 | 1.42 (0.70–2.87) | |||||
| GG or GA | 164/49 | Reference | 0.19 | Reference | 0.28 | 164/34 | Reference | 0.50 | NC | NA |
| AA | 46/18 | 1.43 (0.83–2.46) | 1.37 (0.76–2.47) | 46/12 | 1.24 (0.64–2.41) | |||||
| MITF | ||||||||||
| GG | 59/14 | Reference | 0.22 | NC | NC | 59/7 | Reference | 0.05 | Reference | 0.19 |
| GA or AA | 151/53 | 1.44 (0.80–2.60) | 151/39 | 2.20 (1.00–4.93) | 1.79 (0.74–4.34) | |||||
| GG or GA | 150/45 | Reference | 0.25 | NC | NC | 150/30 | Reference | 0.22 | NC | NC |
| AA | 60/22 | 1.34 (0.80–2.23) | 60/16 | 1.45 (0.79–2.66) | ||||||
Discussion
In this study, we investigated and identified intronic SNVs ADCY3 c.675+9196T>G, CREB1 c.303+373G>A, and MITF c.938-325G>A in pigmentation-related genes in association with CM risk and clinicopathological features.
After screening SNVs (stage 1), we found more than 6,000 SNVs associated with CM risk in introns of genes, according to previous studies14–18, and we selected three SNVs involved in the splicing regulatory sequences of pigmentation-related genes for data validation, due to their potential roles in determining abnormalities in production and/or function of the respective encoded proteins13,14.
In fact, previous GWAS have shown that the majority of disease-associated variants reside in the non-coding regions of the genome, suggesting that gene regulatory changes contribute to disease risk18. On the other hand, splicing comprises a two-step reaction of intron removal and exon ligation and is essential for gene expression: pre-mRNA splicing is catalyzed by the spliceosome, a large complex of ribonucleoproteins (RNPs), and this complex recognizes the target sequences and assembles on pre-mRNA20.
After SNVs validation (stage 2), we observed that CREB1 GA or AA genotype and allele “A” were more common in CM patients than in controls, and that individuals with referred genotypes and allele were under 1.79 and 1.47-fold increased risks of CM than others, respectively.
CREB1 was highly expressed in tumor cells, such as human gastric cell lines and knockdown of CREB1 inhibited human gastric cancer cells growth38. CREB1 has also been seen as an important gene in CM development8, and analysis of common network from cancer type-specific RNA-Seq co-expression data showed CREB1 as a melanoma-associated gene39. To the best of our knowledge, there are no previous studies focusing the roles of ADCY3 c.675+9196T>G, CREB1 c.303+373G>A, and MITF c.938-325G>A SNVs in risk of CM, and therefore the association of CREB1 GA or AA genotype and allele “A” with CM risk seen in the present study is a new finding. The search for potential splicing regulatory elements using in silico algorithm in this study indicated that gene variants induce the creation or abrogation of binding sites33. The allele “A” of CREB1 c.303+373G>A may alter binding sites for splicing elements, such as the hnRNP A1, and possibly increases CREB1 activity due to altered efficiency of splicing19,20,33. Since CREB1 is a transcription factor that stimulates the MITF activity, the increase of its activity may in turn increase MITF activity, having proliferation of abnormal melanocytes and increased risk for CM as consequence8.
When genotypes were analyzed in patients stratified by clinicopathological aspects, we noted that CREB1 AA variant genotype was more common in patients with tumors located in limbs than in patients with tumors located in head or trunk and with tumors at Clark level III to IV than in patients with tumors at I or II level. In addition, an excess of MITF AA genotype was found in patients with tumors at stage III or IV than in those with stage I or II tumors.
It was already described that acquisition of metastatic phenotype in CM involved the gain in expression of CREB/activating transcription factor-1 (CREB/ATF-1)40 and MITF amplification41. However, how far our knowledge reaches, this study is the first to describe the influence of CREB1 c.303+373G>A and MITF c.938-325G>A SNVs on clinicopathological features of CM. Indeed, CREB1 promotes tumorigenesis by increasing cell migration, proliferation, and invasiveness, through its effects on the MITF pathway8. The in silico analysis showed that the variant allele “A” of MITF c.938-325G>A may create a site of ligation for splicing factors, including the hnRNP A1, possibly determining increase in gene expression33. Thus, we postulate that CREB1 AA and MITF AA genotypes may increase abnormal melanocytes proliferation and consequently improve aggressiveness of CM.
We also noted that ADCY3 c.675+9196 TT genotype was associated with shorter PFS while MITF GA or AA genotype was associated with shorter MSS in CM patients, when compared to the remaining genotypes.
Up-regulation of ADCY3 increased the tumorigenic potential of gastric cells42 and predicted shorter overall survival in patients with pancreatic cancer43. Overexpression of ADCY2 was previously associated with aggressive behavior of CM44, and MITF amplification predicted worst survival of CM patients41. The in silico analysis showed that the ADCY3 c.675+9196T>G variant may alter sites of ligation for splicing factors, including the SRp55 and 9G8, with a possible increase in the efficiency of splicing and gene expression19,20,33. Since ADCYs participate in CREB activation, and CREB regulates the expression of MITF8, the increase in ADCY3 activity in CM patients with the TT genotype may favor proliferation of abnormal melanocytes leading to relapse or death by CM effects. Again, the possible increased activity of MITF in patients with GA or AA genotype may have contributed to this clinical unfavorable outcome.
It is also worth to comment that pigmentation-related genes have been seen as potential therapeutic targets. Previous studies showed that increased ADCY expression generated resistance to MAPK inhibitions and up regulates MITF in melanoma cells8, and the suppression of MITF expression by the CH6868398 agent caused melanoma cell growth inhibition45. Inhibition of p300 acetyltransferase transcriptional coactivator of MITF by p300/CBP complex had growth inhibitory effects in melanoma cells expressing MITF46,47, and Kazinol U reduced melanogenesis by inhibition of MITF in melanoma cells48. Since response to new agents depends on ADCY3 and MITF expressions, it is possible that patients with distinct genotypes of these genes present differentiated responses to therapies.
At this time, we draw attention to the fact that no differences in ADCY3, CREB1 and MITF expressions were identified in leukocytes of peripheral blood of individuals with the distinct genotypes of ADCY3, CREB1 and MITF SNVs. It is possible that the sample size evaluated was not enough to identify differences in gene expression among individuals or, alternatively, these variants may determine gene expression abnormalities only in tumor tissue or only protein functional changes.
In summary, we described for the first time the potential importance of ADCY3 c.675+9196T>G, CREB1 c.303+373G>A, and MITF c.938-325G>A SNVs in the pigmentation-related genes in CM risk and clinicopathological features in Brazilian individuals. We recognize that the present study has limitations: it was conducted on a relatively small number of individuals and only quantitative analysis of gene expression in normal leukocytes was performed. Thus, we believe that our results will require confirmation in a further larger epidemiological study in our population and others, and quantitative and functional analyses of ADCY3, CREB1 and MITF SNVs in melanoma cells. If these findings are confirmed, they might help to identify individuals with high risk for CM who deserves to receive additional recommendations for CM prevention and early tumor detection and/or differentiated treatment, perhaps including the targeting lineage specific MC1R signalizing pathway agents.
Supplementary information
Acknowledgements
We would like to thank Barretos Cancer Hospital Biobank and Research Support Department (NAP) for the clinical data and specimen collection, and “Fundação de Amparo à Pesquisa do Estado de São Paulo” (FAPESP) (Grants: 2012/15880-3, 2012/16617-4, 2016/02193-9), “Conselho Nacional de Desenvolvimento Científico e Tecnológico” (CNPq) (Grant: 402873/2016-5), and “Coordenação de Aperfeiçoamento de Pessoal de Nível Superior” (CAPES) for the financial support.
Author contributions
G.J.L. and C.O. were responsible for the study design, experiments, acquisition, analysis and interpretation of data, and drafting the manuscript. C.T., J.K.S., G.V.B.G., and W.L.O. made relevant contributions in genotyping and analysis of gene expression. G.J.L. and B.S.C. analyzed the data. J.A.R., V.L.V., S.S., and A.M.M. conducted the clinical treatment of patients. C.S.P.L. was responsible for the concept and study design, acquisition and interpretation of data, and critical revision of the manuscript. All authors approved the final version of the manuscript.
Data availability
The authors declare that all data of the present study are available for the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Gustavo Jacob Lourenço and Cristiane Oliveira.
Supplementary information
is available for this paper at 10.1038/s41598-020-68945-9.
References
- 1.Schadendorf D, et al. Melanoma. Nat. Rev. Dis. Primers. 2015;1:15003. doi: 10.1038/nrdp.2015.3. [DOI] [PubMed] [Google Scholar]
- 2.Balch CM, et al. Final version of 2009 AJCC melanoma staging and classification. J. Clin. Oncol. 2009;27:6199–6206. doi: 10.1200/JCO.2009.23.4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Potrony M, et al. Update in genetic susceptibility in melanoma. Ann. Transl. Med. 2015;3:210. doi: 10.3978/j.issn.2305-5839.2015.08.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Potrony M, et al. IRF4 rs12203592 functional variant and melanoma survival. Int. J. Cancer. 2017;140:1845–1849. doi: 10.1002/ijc.30605. [DOI] [PubMed] [Google Scholar]
- 5.D'Mello SA, Finlay GJ, Baguley BC, Askarian-Amiri ME. Signaling pathways in melanogenesis. Int. J. Mol. Sci. 2016 doi: 10.3390/ijms17071144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin JY, Fisher DE. Melanocyte biology and skin pigmentation. Nature. 2007;445:843–850. doi: 10.1038/nature05660. [DOI] [PubMed] [Google Scholar]
- 7.Plonka PM, et al. What are melanocytes really doing all day long…? Exp. Dermatol. 2009;18:799–819. doi: 10.1111/j.1600-0625.2009.00912.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodríguez CI, Setaluri V. Cyclic AMP (cAMP) signaling in melanocytes and melanoma. Arch. Biochem. Biophys. 2014;563:22–27. doi: 10.1016/j.abb.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 9.Slominski A, Tobin DJ, Shibahara S, Wortsman J. Melanin pigmentation in mammalian skin and its hormonal regulation. Physiol. Rev. 2004;84:1155–1228. doi: 10.1152/physrev.00044.2003. [DOI] [PubMed] [Google Scholar]
- 10.Slominski A, Zbytek B, Slominski R. Inhibitors of melanogenesis increase toxicity of cyclophosphamide and lymphocytes against melanoma cells. Int. J. Cancer. 2009;124:1470–1477. doi: 10.1002/ijc.24005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brożyna AA, Jóźwicki W, Roszkowski K, Filipiak J, Slominski AT. Melanin content in melanoma metastases affects the outcome of radiotherapy. Oncotarget. 2016;7:17844–17853. doi: 10.18632/oncotarget.7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brozyna AA, VanMiddlesworth L, Slominski AT. Inhibition of melanogenesis as a radiation sensitizer for melanoma therapy. Int. J. Cancer. 2008;123:1448–1456. doi: 10.1002/ijc.23664. [DOI] [PubMed] [Google Scholar]
- 13.Slominski A, et al. The role of melanogenesis in regulation of melanoma behavior: melanogenesis leads to stimulation of HIF-1α expression and HIF-dependent attendant pathways. Arch. Biochem. Biophys. 2014;563:79–93. doi: 10.1016/j.abb.2014.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duffy DL, et al. Novel pleiotropic risk loci for melanoma and nevus density implicate multiple biological pathways. Nat. Commun. 2018;9:4774. doi: 10.1038/s41467-018-06649-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Law MH, et al. Genome-wide meta-analysis identifies five new susceptibility loci for cutaneous malignant melanoma. Nat. Genet. 2015;47:987–995. doi: 10.1038/ng.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ransohoff KJ, et al. Two-stage genome-wide association study identifies a novel susceptibility locus associated with melanoma. Oncotarget. 2017;8:17586–17592. doi: 10.18632/oncotarget.15230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Visconti A, et al. Genome-wide association study in 176,678 Europeans reveals genetic loci for tanning response to sun exposure. Nat. Commun. 2018;9:1684. doi: 10.1038/s41467-018-04086-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu Y, Tazearslan C, Suh Y. Challenges and progress in interpretation of non-coding genetic variants associated with human disease. Exp. Biol. Med. (Maywood) 2017;242:1325–1334. doi: 10.1177/1535370217713750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baralle D, Baralle M. Splicing in action: assessing disease causing sequence changes. J. Med. Genet. 2005;42:737–748. doi: 10.1136/jmg.2004.029538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Will CL, Lührmann R. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 2011 doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi J, et al. A common intronic variant of PARP1 confers melanoma risk and mediates melanocyte growth via regulation of MITF. Nat. Genet. 2017;49:1326–1335. doi: 10.1038/ng.3927. [DOI] [PubMed] [Google Scholar]
- 22.Pena SD, et al. The genomic ancestry of individuals from different geographical regions of Brazil is more uniform than expected. PLoS ONE. 2011;6:e17063. doi: 10.1371/journal.pone.0017063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fitzpatrick TB. The validity and practicality of sun-reactive skin types I through VI. Arch. Dermatol. 1988;124:869–871. doi: 10.1001/archderm.1988.01670060015008. [DOI] [PubMed] [Google Scholar]
- 24.Rigel DS, Friedman RJ, Levenstein MJ, Greenwald DI. Relationship of fluorescent lights to malignant melanoma: another view. J. Dermatol. Surg. Oncol. 1983;9:836–838. doi: 10.1111/j.1524-4725.1983.tb00741.x. [DOI] [PubMed] [Google Scholar]
- 25.Gomez GVB, et al. PDCD1 gene polymorphisms as regulators of T-lymphocyte activity in cutaneous melanoma risk and prognosis. Pigment Cell Melanoma Res. 2018;31:308–317. doi: 10.1111/pcmr.12665. [DOI] [PubMed] [Google Scholar]
- 26.Gillgren P, et al. 2-cm versus 4-cm surgical excision margins for primary cutaneous melanoma thicker than 2 mm: a randomised, multicentre trial. Lancet. 2011;378:1635–1642. doi: 10.1016/S0140-6736(11)61546-8. [DOI] [PubMed] [Google Scholar]
- 27.Sondak VK, Gibney GT. Indications and options for systemic therapy in melanoma. Surg. Clin. North Am. 2014;94:1049–1058, viii. doi: 10.1016/j.suc.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 28.Sasse AD, Sasse EC, Clark LG, Ulloa L, Clark OA. Chemoimmunotherapy versus chemotherapy for metastatic malignant melanoma. Cochrane Database Syst. Rev. 2007 doi: 10.1002/14651858.CD005413.pub2. [DOI] [PubMed] [Google Scholar]
- 29.Burmeister BH, et al. Adjuvant radiotherapy versus observation alone for patients at risk of lymph-node field relapse after therapeutic lymphadenectomy for melanoma: a randomised trial. Lancet Oncol. 2012;13:589–597. doi: 10.1016/S1470-2045(12)70138-9. [DOI] [PubMed] [Google Scholar]
- 30.Carvalho BS, Louis TA, Irizarry RA. Quantifying uncertainty in genotype calls. Bioinformatics. 2010;26:242–249. doi: 10.1093/bioinformatics/btp624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang DW, et al. Extracting biological meaning from large gene lists with DAVID. Curr. Protoc. Bioinform. 2009;Chapter 3:Unit 13.11. doi: 10.1002/0471250953.bi1311s27. [DOI] [PubMed] [Google Scholar]
- 32.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Desmet FO, et al. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walters R, Laurin C, Lubke GH. An integrated approach to reduce the impact of minor allele frequency and linkage disequilibrium on variable importance measures for genome-wide data. Bioinformatics. 2012;28:2615–2623. doi: 10.1093/bioinformatics/bts483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 36.Tortorelli GA, et al. CASP8 (rs3834129) and CASP3 (rs4647601) polymorphisms in oropharynx cancer risk, tumor cell differentiation, and prognosis in a cohort of the Brazilian population. Mol. Biol. Rep. 2019;46:6557–6563. doi: 10.1007/s11033-019-05107-9. [DOI] [PubMed] [Google Scholar]
- 37.Davis LE, Shalin SC, Tackett AJ. Current state of melanoma diagnosis and treatment. Cancer Biol. Ther. 2019;20:1366–1379. doi: 10.1080/15384047.2019.1640032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rao M, Zhu Y, Cong X, Li Q. Knockdown of CREB1 inhibits tumor growth of human gastric cancer in vitro and in vivo. Oncol. Rep. 2017;37:3361–3368. doi: 10.3892/or.2017.5636. [DOI] [PubMed] [Google Scholar]
- 39.Wu K, et al. Integration of protein interaction and gene co-expression information for identification of melanoma candidate genes. Melanoma Res. 2019;29:126–133. doi: 10.1097/CMR.0000000000000525. [DOI] [PubMed] [Google Scholar]
- 40.Melnikova VO, Bar-Eli M. Transcriptional control of the melanoma malignant phenotype. Cancer Biol. Ther. 2008;7:997–1003. doi: 10.4161/cbt.7.7.6535. [DOI] [PubMed] [Google Scholar]
- 41.Garraway LA, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 42.Hong SH, et al. Upregulation of adenylate cyclase 3 (ADCY3) increases the tumorigenic potential of cells by activating the CREB pathway. Oncotarget. 2013;4:1791–1803. doi: 10.18632/oncotarget.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fukuhisa H, et al. Gene regulation by antitumor miR-130b-5p in pancreatic ductal adenocarcinoma: the clinical significance of oncogenic EPS8. J. Hum. Genet. 2019 doi: 10.1038/s10038-019-0584-6. [DOI] [PubMed] [Google Scholar]
- 44.Masugi Y, et al. Overexpression of adenylate cyclase-associated protein 2 is a novel prognostic marker in malignant melanoma. Pathol. Int. 2015;65:627–634. doi: 10.1111/pin.12351. [DOI] [PubMed] [Google Scholar]
- 45.Aida S, et al. MITF suppression improves the sensitivity of melanoma cells to a BRAF inhibitor. Cancer Lett. 2017;409:116–124. doi: 10.1016/j.canlet.2017.09.008. [DOI] [PubMed] [Google Scholar]
- 46.Wang R, et al. Targeting lineage-specific MITF pathway in human melanoma cell lines by A-485, the selective small-molecule inhibitor of p300/CBP. Mol. Cancer Ther. 2018;17:2543–2550. doi: 10.1158/1535-7163.MCT-18-0511. [DOI] [PubMed] [Google Scholar]
- 47.Kim E, et al. MITF expression predicts therapeutic vulnerability to p300 inhibition in human melanoma. Cancer Res. 2019 doi: 10.1158/0008-5472.CAN-18-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lim J, et al. Kazinol U inhibits melanogenesis through the inhibition of tyrosinase-related proteins via AMP kinase activation. Br. J. Pharmacol. 2019;176:737–750. doi: 10.1111/bph.14560. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data of the present study are available for the corresponding author upon reasonable request.