Abstract
Melanopsin, an atypical vertebrate visual pigment, mediates non-image-forming light responses including circadian photoentrainment and pupillary light reflexes and contrast detection for image formation. Melanopsin-expressing intrinsically photosensitive retinal ganglion cells are characterized by sluggish activation and deactivation of their light responses. The molecular determinants of mouse melanopsin’s deactivation have been characterized (i.e., C-terminal phosphorylation and β-arrestin binding), but a detailed analysis of melanopsin’s activation is lacking. We propose that an extended third cytoplasmic loop is adjacent to the proximal C-terminal region of mouse melanopsin in the inactive conformation, which is stabilized by the ionic interaction of these two regions. This model is supported by site-directed spin labeling and electron paramagnetic resonance spectroscopy of melanopsin, the results of which suggests a high degree of steric freedom at the third cytoplasmic loop, which is increased upon C-terminus truncation, supporting the idea that these two regions are close in three-dimensional space in wild-type melanopsin. To test for a functionally critical C-terminal conformation, calcium imaging of melanopsin mutants including a proximal C-terminus truncation (at residue 365) and proline mutation of this proximal region (H377P, L380P, Y382P) delayed melanopsin’s activation rate. Mutation of all potential phosphorylation sites, including a highly conserved tyrosine residue (Y382), into alanines also delayed the activation rate. A comparison of mouse melanopsin with armadillo melanopsin—which has substitutions of various potential phosphorylation sites and a substitution of the conserved tyrosine—indicates that substitution of these potential phosphorylation sites and the tyrosine residue result in dramatically slower activation kinetics, a finding that also supports the role of phosphorylation in signaling activation. We therefore propose that melanopsin’s C-terminus is proximal to intracellular loop 3, and C-terminal phosphorylation permits the ionic interaction between these two regions, thus forming a stable structural conformation that is critical for initiating G-protein signaling.
Significance
Melanopsin is an important visual pigment in the mammalian retina that mediates non-image-forming responses such as circadian photoentrainment and pupil constriction and supports contrast detection for image formation. In this study, we detail two critical structural features of mouse melanopsin—its third cytoplasmic loop and C-terminus—that are important in the activation of melanopsin’s light responses. Furthermore, we propose that these two regions directly participate in coupling mouse melanopsin to its G-protein. These findings contribute to further understanding of GPCR-G-protein coupling, and given recent findings suggesting flexibility of melanopsin signal transduction in the retina (possibly by coupling to more than one G-protein type), these findings provide insight into the molecular basis of melanopsin function in the retina.
Introduction
G-protein-coupled receptors (GPCRs) make up the largest family of integral membrane receptors and are activated by a variety of biological stimuli, including hormones, odorants, small peptides, neurotransmitters, and photons (1,2). Upon activation, GPCRs undergo a series of conformational changes that facilitate the binding and activation of intracellular heterotrimeric G-proteins, which initiate a variety of signaling responses (3,4). Visual pigments are specialized light-detecting GPCRs that are comprised of an opsin protein covalently attached to a chromophore, typically 11-cis-retinal in the mammalian retina. The absorption of a photon by 11-cis-retinal results in its isomerization to all-trans-retinal. This conformational change in the chromophore results in the activation of visual pigment and the phototransduction cascade. In the mammalian retina, three distinct classes of photoreceptor cells detect light. Rod and cone photoreceptor cells found in the outer retina express distinct specialized visual pigments that mediate photon absorption for image-forming vision. A less well-known third photoreceptor class in the inner retina is composed of a small subset of intrinsically photosensitive ganglion cells (ipRGCs) that express melanopsin, a rhabdomeric-type opsin (5,6). In the mouse, ipRGCs are divided into six subtypes (M1–M6) that are unique in their morphology, projections into the inner plexiform layer of the retina, amount of melanopsin expressed, transcription factors, and their projections to the brain, and thus their functions (7,8). For example, M1-ipRGCs exhibit the highest level of melanopsin expression and project to the suprachiasmatic nucleus (among other brain nuclei) and thus are implicated in circadian photoentrainment (9), whereas M4-ipRGCs possess the largest soma of all ipRGC subtypes, project to the dorsal lateral geniculate nucleus, and are implicated in image formation (10). Light detected by melanopsin regulates non-image-forming functions such as circadian photoentrainment, pupillary light reflex, sleep, and melatonin synthesis (9,11, 12, 13, 14). Unlike rods and cones, melanopsin in M1-type ipRGCs signal through a Gαq-mediated pathway that leads to the opening of TRPC6/7 channels (15, 16, 17, 18). Recent studies analyzing the M4 ipRGC subtype suggest that melanopsin in these cells signals either through the Gαq transduction cascade (19) or through a cyclic-nucleotide cascade and the opening of HCN-channels (20).
Structural and molecular determinants of GPCR-G-protein complex formation have been described in both a receptor specific (for review of rhodopsin activation, see (21)) and systematic manner (22). However, it remains difficult to attribute specific receptor properties (e.g., amino acid sequence, polarity, charge, steric properties) to their selectivity for their cognate G-protein. The notion of a singular cognate G-protein for each receptor is also one that is challenged by evidence of receptor promiscuity to several Gα classes (23,24), including melanopsin promiscuously activating transducin in vitro (25). GPCR regions important for G-protein binding selectivity include the second and third intracellular loops and cytoplasmic extensions of transmembrane helices 5 and 6, which form critical contacts with helices 4 and 5 on Gα (26). GPCR C-termini can regulate G-protein activation as well as serve as substrates for GPCR kinase to mediate arrestin binding (for melanopsin C-terminal phosphorylation and deactivation, see (27, 28, 29, 30, 31, 32, 33)). Structural analysis of rhodopsin-G-protein complexes reveals contacts between helix 8 on rhodopsin’s C-terminus and the C-terminal helix 5 on Gαi (34) or transducin (35), and contacts have been found on rhodopsin’s C-terminus with Gβ when in complex with Gαi (36). Additionally, rhodopsin C-terminal peptides bind and facilitate transducin signaling by enhancing phosphodiesterase activity and inhibiting transducin’s GTPase activity (37). The role of melanopsin’s cytoplasmic loops or C-terminus in stimulating G-protein activity remain to be determined.
In this study, we aimed to determine the critical cytoplasmic domains on mouse melanopsin that contribute to phototransduction activation. Melanopsin-mediated behaviors are diverse and are both image and non-image forming. Additionally, ipRGC light responses are sluggish (single flash responses can persist across minutes) (38) compared to rapid photoresponses observed in rod and cone photoreceptors (these cells can respond in a precise manner to millisecond-scale light flashes). Melanopsin Gαq signal transduction has been shown to be repurposed to modulate leak potassium channels (rather than activating phospholipase C) or could possibly couple to another G-protein to stimulate cyclic-nucleotide signal transduction in M4-type ipRGCs (19,20). Within the M1-type population, there is a striking heterogeneity in the light responses, in which there is a mixture of cells that optimally respond to certain irradiances or respond in a linear fashion in response to increasing irradiance, thus allowing for this M1 population to encode for a breadth of light intensities (39). Additionally, melanopsin phosphorylation by Protein Kinase A, likely on the intracellular loops, can attenuate light responses in a dopamine and cyclic AMP-dependent manner (28). It is therefore critical to examine melanopsin-G-protein complex formation at a structural level to shed light on the molecular determinants underlying the functional diversity observed in melanopsin signal transduction. Based on amino acid sequence analysis and homology modeling using Todarodes pacificus rhodopsin as a template, melanopsin is predicted to have extended fifth and sixth transmembrane helices and also a uniquely long C-terminus (Fig. 1 for two-dimensional (2D) schematic of amino acids, Fig. 2 for three-dimensional figures), which is 171 amino acids long (residues T350–L521). Given the functional significance of these regions in other GPCRs, we synthesized a series of mutants designed to test if these regions contribute to phototransduction activation. Our findings provide a detailed examination of the structural basis of melanopsin activation and, furthermore, provide a more robust foundation to examine ipRGC activation and structure-function relationships of other related visual pigments.
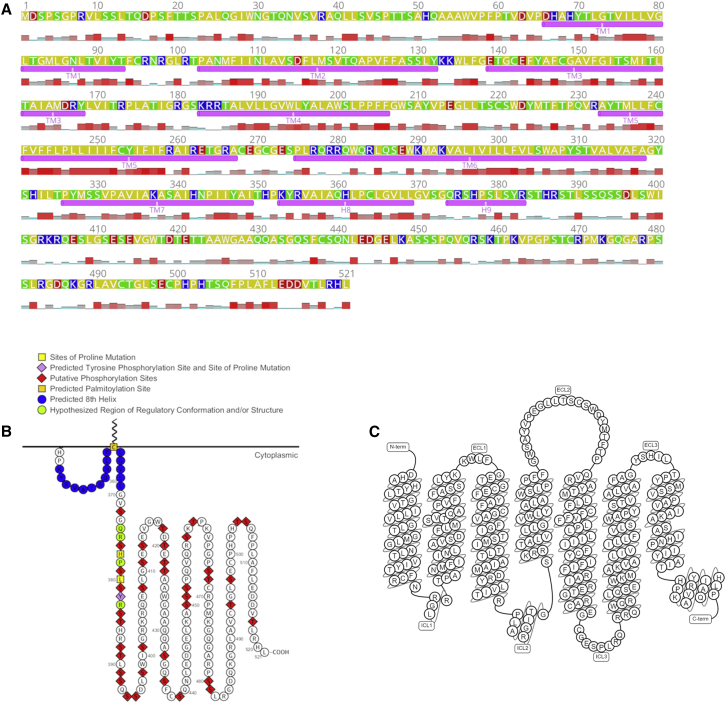
Figure 1.
Mouse melanopsin’s amino acid sequence. Mouse melanopsin is 521 amino acids long, with a long C-terminus domain that is 171 amino acids long. (A) Transmembrane and cytoplasmic helices are annotated below the amino acid sequence. Amino acid property is denoted by the color of the residue: yellow, nonpolar; green, polar and noncharged; blue, positively charged; and red, negatively charged. Hydrophobicity at each residue is plotted below the amino acid sequence; taller and red bars indicate a high level of hydrophobicity at that position. Identities of residues possessing secondary structures are based on prediction from the homology mode generated (see Fig. 2). Figure was generated using Geneious software (63). (B) 2D schematic of mouse melanopsin C terminus depicting functionally significant residues. Figure was made using Protter software (64). (C) 2D snakeplot of mouse melanopsin’s secondary structure. Figure is from GPCRdb, using Swiss-Prot for secondary structure prediction (65). Note that the amino acids labeled as transmembrane helices in (A), not (C), correspond to identities of the amino acids in the homology model in Fig. 2.
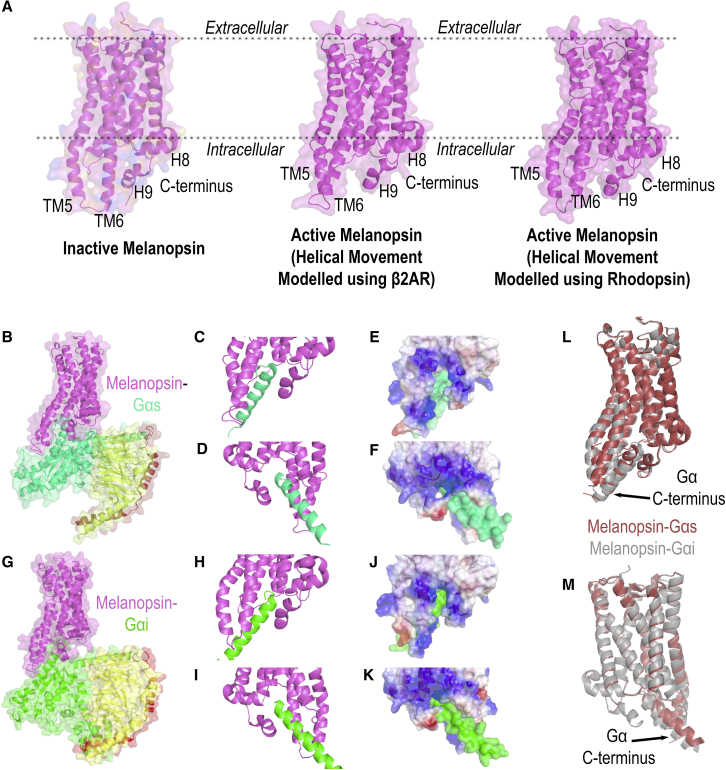
Figure 2.
Homology modeling of mouse melanopsin. (A) Inactive and active conformations of mouse melanopsin, generated using squid (T. pacificus) rhodopsin as a template and using β2 adrenergic receptor or rhodopsin to model helical movements in the active melanopsin models. (B) Melanopsin in complex with Gαs: purple, melanopsin; green, Gα; yellow, Gβ; and dark red, Gγ. (C and D) Zoomed-in views of mouse melanopsin’s (purple) cytoplasmic domains when in complex with Gαs C-terminal peptide (green). (E and F) Zoomed-in views are depicted in (C) and (D) and depicted as a space-filling model with melanopsin’s electrostatic potentials plotted on the surface (blue: positively charged; red: negatively charged; white: neutral). (G) Melanopsin in complex with Gαi: purple, melanopsin; green, Gα; yellow, Gβ; and dark red, Gγ. (H and I) Zoomed-in views of mouse melanopsin’s (purple) cytoplasmic domains when in complex with Gαi C-terminal peptide (green). (J and K) Zoomed-in views depicted in (H) and (I) and depicted as a space-filling model with melanopsin’s electrostatic potentials plotted on the surface (blue: positively charged; red: negatively charged; white: neutral). (L and M) Overlay of Gαs and Gαi complexed melanopsin structures.
Materials and Methods
Note that certain commercial equipment, instruments, and materials are identified in this article to specify an experimental procedure as completely as possible. In no case does the identification of particular equipment or materials imply a recommendation or endorsement by National Institute of Standards and Technology, nor does it imply that the materials, instruments, or equipment are necessarily the best available for the purpose. The opinions expressed in this article are the authors’ own and do not necessarily represent the views of National Institute of Standards and Technology.
Homology modeling of mouse melanopsin
Mouse melanopsin amino acid sequence (UniProt: Q9QXZ9-1; OPN4L) was used to produce a homology model using the online protein structure service LOMETS, which ranks models derived from 11 servers using a Z-score as a metric of confidence for each model (40). The melanopsin structural model that was generated used T. pacificus rhodopsin (Protein Data Bank, PDB: 2ZIY) (41) as the template, which was the highest ranked model and was produced using the HHpred threading program (42). To model the active conformation of mouse melanopsin, the LOMETS-generated homology model was aligned to the crystal structure of the β2 adrenergic receptor in complex with Gαs (PDB: 3SN6) (43) and cryo-electron microscopy structure of rhodopsin in complex with Gαi (PDB: 6CMO) (34) in PyMol (Schrödinger, New York, NY) using the cealign tool, and predicted melanopsin helical movements were modeled using the positions of the transmembrane helices of the β2 adrenergic receptor or rhodopsin structures. Surface electrostatic potential of melanopsin was calculated using vacuum electrostatics function on PyMol.
Synthesis of melanopsin mutants
All mutant genes were constructed using the mouse melanopsin coding sequence (National Center for Biotechnology Information, NCBI: NM_013887.2) cloned in the mammalian expression plasmid PMT3 (44). Melanopsin Δ365, H377A L380A Y382A, phosphonull + Y382S, melanopsin ICL3 Null, melanopsin 280-285 alanine, armadillo melanopsin constructs, and mouse 1D4-Gαq (Mus musculus Gnaq with N-terminal 1D4 tag: amino acid sequence TETSQVAPA, corresponding to the last 9 amino acids of bovine rhodopsin, NCBI: NM_008139.5) were synthesized by cassette synthesis using synthetically synthesized gene fragments. Melanopsin C268 was constructed using Gibson assembly (45) of synthetically synthesized gene fragments to generate mutagenesis of residues C95, C271, C438, C469, C493, and C499 to alanine residues to eliminate nonspecific spin label attachment. These constructs for electron paramagnetic resonance (EPR) spectroscopy were also C-terminally tagged with the 1D4 epitope to facilitate immunoprecipitation when expressed heterologously. Melanopsin C268 Δ365 was synthesized through cassette synthesis using melanopsin C268 as a PCR template and with a reverse primer to amplify from the start of the gene to the end of the codon encoding for L365. Melanopsin H377P L380P Y382P, phosphonull + Y382A and phosphonull + Y382F were generated using QuikChange-based site-directed mutagenesis (46). All plasmids were sequence verified through Sanger sequencing.
Cell culture and transfection of HEK293 cells
Human embryonic kidney HEK293 cells were grown in a monolayer on 10-cm culture dishes in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with fetal bovine serum (to a final total protein concentration of 3.6 g/L), 0.006 g/L penicillin, 0.01 g/L streptomycin, and 2.5 × 10−5 g/L amphotericin B in a humid CO2 incubator at 37°C. Cells were passaged by disassociation using 2.5 g/L trypsin-EDTA and seeded to a density of up to 5 million cells per 10-cm dish and 240,000 cells per well in a six-well dish. Transfections were done using TurboFect Transfection Reagent (Thermo Fisher Scientific, Waltham, MA) as per manufacturer’s protocol. Briefly, 4 or 10 μg DNA (six-well and 10-cm dish, respectively) was diluted in nonsupplemented DMEM, and then the transfection reagent was added to the mixture. After a 20-min incubation at room temperature, the transfection mixture was added to the cells, and the cells were incubated in a humid CO2 incubator for 24–48 h.
Calcium imaging of melanopsin-transfected HEK293 cells
24 h after transfection, HEK293 cells were trypsin disassociated from the plates and seeded to 96-well plates at a density of 100,000 cells per well. The 96-well plate is incubated overnight in a humid CO2 incubator in a dark room for dark adaptation. The cells are then incubated with an equivalent volume of Fluo-4 AM Direct Calcium Dye supplemented with 20 mmol/L probenecid and 20 μmol/L 9-cis-retinal and are incubated for 1 h in a humid CO2 incubator. Calcium kinetics were then measured on a commercial fluorescence plate reader by exciting the sample at 487 nm and recording the emission fluorescence at 516 nm at a rate of 1 Hz. The plate reader monochromator has a bandwidth of <9 nm for excitation light. Every melanopsin construct in each transfection had six replicates.
Protein expression and preparation for EPR spectroscopy
Melanopsin C268 and C268 Δ365 constructs were transfected in HEK293 cells in 60–80 10-cm plate batches, harvested 48 h later, and stored at −80°C. Frozen cell pellets were then thawed on ice, then immediately resuspended in PBS with 40 μmol/L 9-cis-retinal, and incubated for at least an hour at 4°C to reconstitute the visual pigment. The sample was centrifuged at maximal speed on a clinical centrifuge, and the cell pellet was then resuspended in solubilization buffer composed of 1 g/L n-dodecyl-β-D-maltoside (DM), 3 mmol/L MgCl2, 140 mmol/L NaCl, 1 mmol/L phenylmethylsulfonyl fluoride, and 50 mmol/L HEPES (pH 7.5). After a minimum of 1 h on a tube rotator at 4°C, the solubilized cell lysate was centrifuged at maximal speed on a clinical centrifuge, and the supernatant was collected. The solubilized protein was then incubated with Sepharose 4B resin conjugated with monoclonal α-1D4 antibody (provided generously by Prof. Daniel Oprian) for at least an hour at 4°C. The 1D4 resin was then washed by centrifuging 30 s at 500 × gn, removing the supernatant, and resuspending the resin with solubilization buffer at least 10 times. After the final wash, the 1D4 resin was resuspended in solubilization buffer containing 2 mmol/L 2,2,5,5-tetramethyl-1-oxyl-3-methyl methanethiosulfonate (MTSL) to attach the spin label at C268 for EPR spectroscopy and was incubated overnight on a tube rotator at 4°C. The sample was then washed by centrifuging 30 s at 500 × gn, removing the supernatant, and resuspending the resin with solubilization buffer at least five times. MTSL spin-labeled melanopsin was then eluted by incubating the 1D4 resin in solubilization buffer containing 50 μmol/L 1D4 peptide (amino acid sequence TETSQVAPA). Eluate was centrifuged using 10,000 g/mol (10 kDa) size-exclusion centrifugal filters to concentrate the sample and filter out the eluting peptide.
EPR spectroscopy of single spin-labeled melanopsin
Continuous wave (CW) EPR spectra at X-band (∼9.8 GHz) were recorded using a commercial EPR spectrometer equipped with a commercial microwave cavity in perpendicular mode. Samples were drawn, by capillary action, into calibrated 25-μL disposable borosilicate glass micropipettes sealed with a hematocrit tube sealing putty and subsequently inserted into a critically coupled EPR cavity at ambient temperature. Spectra (88 averages) were recorded using 0.1 mT modulation amplitude at 100 kHz, with an incident microwave power of 1.5 mW. Simulations of EPR data were done using EasySpin (47).
Calculation of melanopsin activation rates
The activation phases of calcium imaging assay data (corresponding to the part of the data before the peak fluorescence level) for all melanopsin constructs was fitted to a one-phase association function using GraphPad Prism software (GraphPad Software, San Diego, CA). The following function was used:
where Y(0) is the value at t = 0, Plateau is the peak activation value, and k is the rate constant. Data across multiple transfections (six replicates per transfection) were pooled together and averaged, and the activation rate of the averaged data was calculated. Standard error of the mean of the activation rates of all melanopsin constructs and statistical analysis were calculated using GraphPad Software. Statistical significance was determined by performing unpaired t-tests of all mutant melanopsin constructs with respect to wild-type melanopsin or between specific melanopsin constructs of interest.
Results
Three-dimensional homology modeling of mouse melanopsin predicts that an extended third cytoplasmic loop and the proximal C-terminus mediate G-protein binding
The structural basis of melanopsin activation is not well understood; specifically, the unique molecular mechanisms underlying its capability to couple its cognate G-protein have not been elucidated in detail. To shed light on this problem, we constructed a homology model of mouse melanopsin using squid rhodopsin as a template. The generated homology model had 68% of melanopsin’s amino acid sequence aligned to the template (Fig. S1), and among these amino acids, they shared 37% sequence identity to the squid (T. pacificus) rhodopsin template. The sequence coverage of our model includes the entirety of the seven-transmembrane helical domain and the proximal portion of the C-terminus, up to S398. In the inactive state (Fig. 2 A), the model predicts that melanopsin forms a seven-transmembrane visual pigment with an extended third intracellular loop, which is comprised of cytoplasmic extensions of transmembrane helices 5 and 6. In the active state, which we modeled based on helical movements of β2 adrenergic receptor (43) and rhodopsin (34), melanopsin’s fifth and sixth transmembrane helices are predicted to swing away from the C-terminus (Fig. 2 A), thereby making way for the attachment of G-protein to the newly formed binding pocket (Fig. 2, B–K). The generated homology model also predicts a C-terminal ninth helix (Fig. 2 A), but further experimental data are needed to test the existence of this structure in melanopsin. However, the data presented in this study strongly support the predicted C-terminal conformation shown in the homology model.
The crystal structures obtained for squid rhodopsin reveal contacts between intracellular loop 3 (specifically, the cytoplasmic extension of transmembrane helix 6) and its C-terminal ninth helix (41,48). Additionally, the presence of positively charged residues on squid rhodopsin’s third intracellular loop and negatively charged residues on the C-terminus may facilitate the formation of contacts between these regions or potentially to the Gαq C-terminus. Additionally, although most GPCRs display promiscuous binding to several Gα subtypes, several molecular features on the GPCR determine the selectivity for cognate G-proteins—the cognate G-protein being Gαq for melanopsin. Systematic analysis of GPCRs suggest that selectivity to the Gαq/11 family is determined primarily by the amino acid properties of the receptor’s C-terminus, specifically through enrichment of nonaromatic residues (22). Interestingly, neither the length of either the C-terminus nor the length of the third intracellular loop are positively correlated with selectivity to Gαq/11 subtypes and are the only selectivity features in the binding of Gα12/13 (22). Surface electrostatic analysis of our homology model of melanopsin suggests an enrichment of positively charged residues on the intracellular loops, particularly intracellular loop 3 (Fig. 2, E and J) and 2 (Fig. 2, F and K). The proximal region of melanopsin’s C-terminus is also positively charged (Fig. 2, E and J), and we predict it will make contacts with positively charged residues on transmembrane helix 6 in the inactive conformation (Fig. 2 A). Thus, the ionic contacts between the C-terminus and third intracellular loop would require amino acid modification (namely, C-terminal phosphorylation) to provide the necessary charge for these ionic contacts. Comparison of both our active-state melanopsin models suggests that melanopsin would have to undergo different helical movements, specifically a greater outward swing of transmembrane helix 6 when melanopsin is modeled with Gαs (Fig. 2, L and M). Systematic analysis of GPCRs suggests no positive correlation with the negative surface charge at these critical cytoplasmic domains on the receptor and their capability to couple Gαq (22). However, we propose that the amino acid ionic charges and the conformation of melanopsin’s cytoplasmic domains play critical roles in its ability to couple to G-protein(s) and initiate ipRGC phototransduction.
EPR spectroscopy of single spin-labeled mouse melanopsin supports the proposed C-terminus-intracellular loop 3 arrangement
To test our model that predicted the C-terminus and cytoplasmic loop, we synthesized a site-directed spin labeling mutant melanopsin for EPR spectroscopy analysis, Melanopsin C268. All solvent-accessible, nonspecific cysteines were mutated to alanine residues with the exception of C364, a predicted palmitoylation site, and residues C142 and C220, which are predicted sites of a disulfide bridge, a common molecular feature of opsins necessary for proper folding and retention of covalently linked retinal molecule (49,50). This melanopsin C268 mutant was designed for single labeling using the nitroxide spin label MTSL at C268 on intracellular loop 3 to analyze the steric freedom at this position in the presence (Melanopsin C268) or absence (Melanopsin C268 Δ365) of the C-terminus.
The CW EPR spectrum of single spin-labeled, dark-adapted melanopsin C268 (Fig. 3 A) in DM presents a sharper spectral lineshape compared to similar EPR measurements on rhodopsin (51,52), a finding that indicates a high degree of rotational mobility of the MTSL spin label at this position. In addition to the qualitative observation of a sharper spectral lineshape, mathematical fitting of the EPR spectrum of single spin-labeled melanopsin C268 to extract a rotational correlation time (τcorr) of the spin label supports the higher degree of steric freedom at this region compared to rhodopsin. The calculated τcorr of melanopsin C268 was 9.36 ns (Fig. S2), which is lower than τcorr of both inactive and active forms of rhodopsin (77 and 26 ns, respectively) when spin labeled at the cytoplasmic end of transmembrane helix 6 (V250C) (53). One possible explanation for the increased spin label rotational mobility in the C268-labeled melanopsin is that the third intracellular loop is extended into the cytoplasm, as predicted by the structural model. The broader lineshape and longer rotational correlation times for rhodopsin (51,52,54) suggest that the spin label’s rotational mobility (MTSL attached at the cytoplasmic face of transmembrane helices) is hindered because of closer proximity to the DM micelle compared to melanopsin’s spin label. The CW EPR spectrum of single spin-labeled, dark-adapted C-terminal truncated melanopsin (Melanopsin C268 Δ365) in DM (Fig. 3 B) possesses an even sharper spectral lineshape than full-length melanopsin and resembles the lineshape of the spectrum observed when measuring free MTSL in DM solution without protein (Fig. 3 C). Together, these data indicate a higher degree of MTSL rotational mobility at the third intracellular loop, which we attribute to the elimination of any steric hinderance caused by the C-terminus consistent with the C-terminal conformation proposed in Fig. 2.
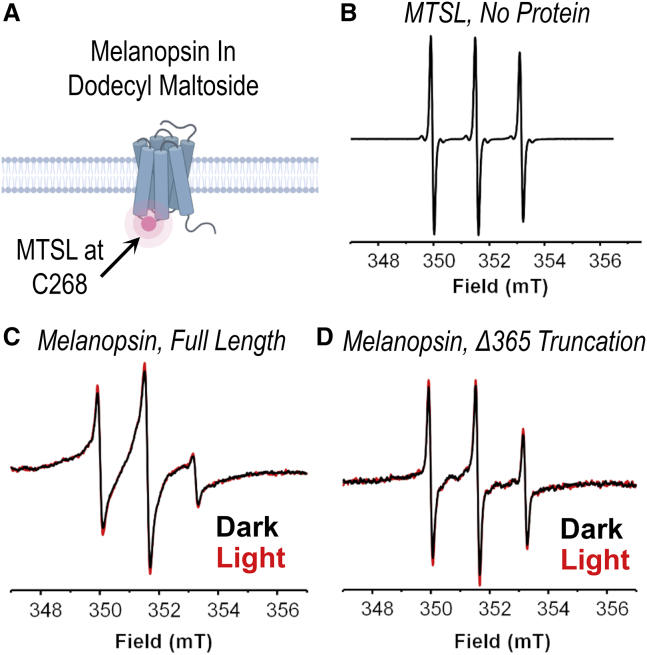
Figure 3.
Electron paramagnetic resonance (EPR) spectroscopy of site-directed spin-labeled mouse melanopsin in DM. (A) Cartoon schematic of the EPR measurement of melanopsin. Affinity-purified melanopsin in dodecyl maltoside solution was site directed and spin labeled at C268 on the third cytoplasmic loop. (B) EPR spectrum of MTSL in solution with no protein. (C and D) EPR spectra of mouse melanopsin, MTSL-conjugated at C268. EPR spectra were recorded in darkness under dim red-light illumination at room temperature. (C) EPR spectrum of full-length melanopsin C268 in darkness (black trace) and after 30-s light illumination (red trace). (D) EPR spectrum of C-terminally truncated melanopsin C268 in darkness (black trace) and after 30-s light illumination (red trace).
After measurement of the EPR spectra of full-length and truncated Melanopsin C268, the samples were exposed to 30 s of 300 W white light to test for differences in the spectra upon light-induced conformational change of the visual pigment. Comparison of the dark and light EPR spectra indicates minimal differences between the two spectra of each sample (Fig. 3, A and B), indicating that the rotational mobility of the spin label at position C268 is minimally affected by light-induced conformational change.
The proximal region of mouse melanopsin’s C-terminus is necessary for rapid phototransduction activation
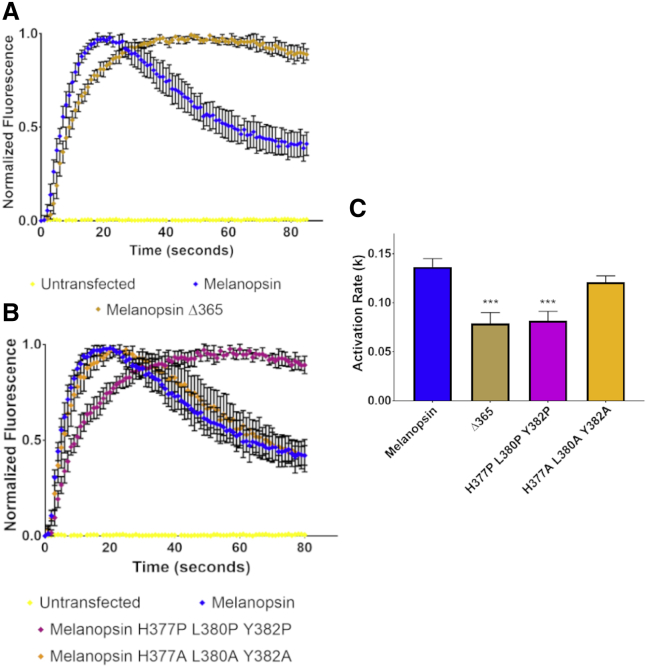
To test the hypothesis that melanopsin’s C-terminus regulates signaling initiation, we synthesized melanopsin C-terminal mutants (positions in Fig. 1 B) aimed to disrupt or ablate C-terminal structure or configuration in its proximal region (before residue D396). To test for potential C-terminal regulation of phototransduction activation, we synthesized a mouse melanopsin C-terminus truncation mutant, Melanopsin Δ365, which is truncated at residue L365 and eliminates the C-terminal region predicted to form part of the G-protein binding pocket in our structural model. Calcium imaging of this mutant reveals delayed activation kinetics (Fig. 4 A) compared to wild-type melanopsin, supporting a functional role of this C-terminal region in mediating signaling activation. Previous studies that examined melanopsin C-terminal truncations (29) suggest that the distal C-terminus (after D396) does not contribute to the production of rapid signaling activation and can potentially sterically hinder the G-protein from accessing the cytoplasmic side of the receptor (32). To disrupt the predicted proximal C-terminal conformation (C-terminus near cytoplasmic loop 3, depicted in Fig. 2), we introduced proline mutations (H377P L380P Y382P) in the critical region predicted to work synergistically with the cytoplasmic loops to regulate phototransduction activation. These proline mutations introduced backbone rigidity and thus will disrupt any important conformations or potential secondary structures found in the wild-type protein (55). Calcium imaging of this triple proline mutant (Fig. 4 B) reveals that it has delayed activation kinetics compared to wild-type melanopsin, indicating that this region has a functionally significant conformation, as suggested by the structural modeling. Mutation of those three residues produces the most pronounced reduction in activation rate as pairwise proline melanopsin mutants H377P L380P and L380P Y382P show slight but not statistically significant reductions in activation rate, whereas the H377P Y382P and H377P L380P Y382P show significant activation rate reduction (Fig. S3). To test that the activation defect observed in the proline mutant is attributable to a proline-induced kink or disruption in structure, we synthesized and tested another mutant of melanopsin with alanine mutations at the same sites of interest in the proline mutant (H377A L380A Y382A). We predicted that these mutations would not disturb any functionally important C-terminal structure or conformation, unlike the proline mutations. Calcium imaging of this mutant shows no reduction in the activation rate, suggesting that the proline mutant’s defect is indeed due to a disruption of C-terminal structure or conformation.
Figure 4.
Calcium imaging of melanopsin C-terminal mutants. (A) Calcium imaging of HEK293 cells expressing melanopsin Δ365, a C-terminus mutant truncated at residue L365, and (B) calcium imaging of melanopsin H377P L380P Y382P, a C-terminus mutant with point mutations designed to disrupt the critical conformation at this region. (C) Calculated activation rates of melanopsin constructs tested in these experiments. All error bars represent standard error of the mean (S.E.M.) of three independent transfections. Statistical significance tested by using Student’s t-test: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. All constructs were compared to wild-type melanopsin’s rate, and statistical significance is indicated over individual bars.
C-terminal palmitoylation anchors and stabilizes GPCR C-termini (56), and in rhodopsin, it regulates its proper expression and stability in the membrane (57,58). Mutation of mouse melanopsin’s predicted palmitoylation site from a cysteine to serine residue and calcium imaging of this mutant, melanopsin C364S, reveals a very modest increase in this mutant’s time to reach peak activation and a slower deactivation compared to wild-type melanopsin (Fig. S4). These data suggest that C364, if palmitoylated, might stabilize the proximal C-terminus for signal transduction, possibly in a cooperative manner with the critical C-terminal conformation downstream of this site. Taken together, we propose that the proximal region of the C-terminus, between residues L365 and D396, holds functional significance that is dependent on an important structural conformation of these residues.
Proximal C-terminal phosphorylation regulates melanopsin phototransduction activation kinetics
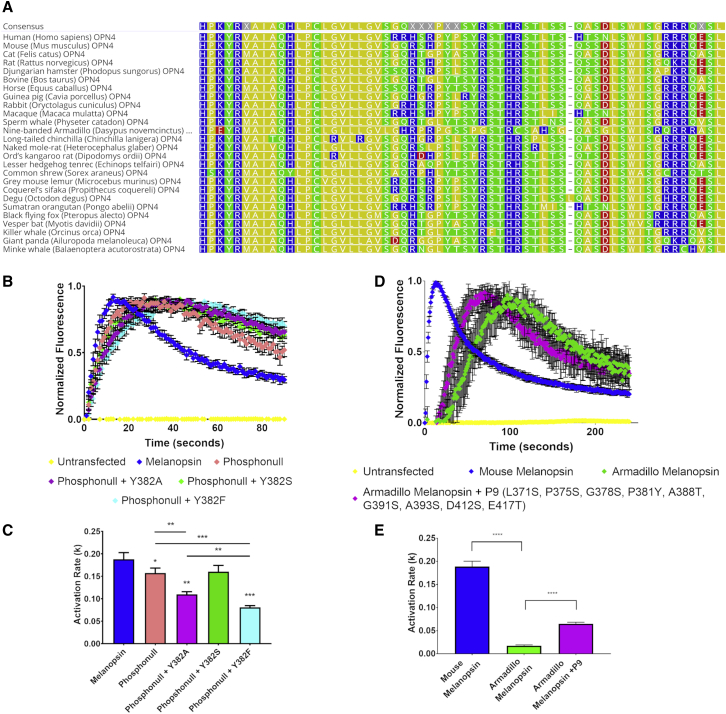
Relative to rod and cone opsins, mouse melanopsin has a uniquely long C-terminus that possesses 38 serine and threonine residues (Fig. 1 B) that may serve as potential phosphorylation sites. It is well established that melanopsin C-terminal phosphorylation is critical for signaling deactivation and the lifetime of melanopsin-driven behaviors such as pupil constriction and jet-lag photoentrainment (28,29,31,32). Given the enrichment of serine and threonine residues at the proximal region (from residues H351 to T385) of mouse melanopsin’s C-terminus, we tested if phosphorylation of these sites contributes to the regulation of phototransduction activation, in addition to deactivation. To test this idea, we performed calcium imaging of transiently transfected HEK293 cells expressing melanopsin C-terminal phosphorylation mutants: phosphonull melanopsin (28,31), a mutant with all 38 C-terminal serine and threonine residues mutated to alanine residues, and phosphonull + Y382A melanopsin, a mutant with an evolutionarily conserved tyrosine residue (Fig. 5 A) mutated to an alanine residue in addition to the phosphonull C-terminus mutations. Calcium imaging of these mutants (Fig. 5 B) suggests that melanopsin C-terminal phosphorylation contributes to signaling activation because of both mutants displaying slower rates of activation (Fig. 5 C) compared to wild-type melanopsin. Phosphonull + Y382A melanopsin displays a slower rate of activation than phosphonull melanopsin, similar to melanopsin Δ365 and melanopsin H377P L380P Y382P mutants. This implies that Y382, an evolutionarily conserved residue in mammalian melanopsins, holds key significance for regulating the kinetics of signaling activation.
Figure 5.
Calcium imaging of melanopsin C-terminal phosphorylation mutants. (A) Amino acid alignment of mammalian melanopsins focused on the proximal C-termini. Arrow depicts the conserved tyrosine Y382 in mouse melanopsin. Alignment generated using Geneious software (Kearse et al. (63)). (B) Calcium imaging of HEK293 cells expressing melanopsin C-terminal phosphorylation mutants. (C) Calculated activation rates of melanopsin constructs depicted in (B). (D) Calcium imaging of HEK293 cells expressing mouse and armadillo melanopsins. (E) Calculated activation rates of melanopsin constructs depicted in (D). All error bars represent standard error of the mean (S.E.M.) of three independent transfections. Statistical significance tested by using Student’s t-test: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. All constructs were compared to wild-type melanopsin’s rate, and statistical significance is indicated over individual bars. Additional statistical analyses between constructs are indicated with the lines above the appropriate bars.
To elucidate the mechanism of this tyrosine residue, specifically its potential phosphorylation to facilitate ionic interaction with intracellular loop 3, we synthesized additional point mutations at that position, specifically phosphonull + Y382S and phosphonull + Y382F (Fig. 5 B). By mutating Y382 to serine and phenylalanine residues, we aimed to test if potential phosphorylation at this position (by means of the phosphorylatable residue serine) contributes to activation kinetics. Specifically, if the elimination of phosphorylation but retention of the atomic mass (phenylalanine has a similar aromatic structure as tyrosine but lacks the hydroxyl group for phosphorylation modification) produces a similar defect as phosphonull + Y382A, then we can conclude that the addition of a phosphate group is important. Calcium imaging of these mutants reveals that phosphonull + Y382S has a higher rate of activation compared to phosphonull + Y382A, has a similar rate as phosphonull, and is not statistically lower than wild-type melanopsin’s rate of activation (Fig. 5 C). This supports the importance of potential phosphorylation at this position. Analysis of the activation rate of phosphonull + Y382F shows that it has slow activation kinetics, slower than phosphonull + Y382A (Fig. 5 C), supporting the conclusion that charge and not atomic size is important in this region of melanopsin.
To further test the evolutionary basis of C-terminal phosphorylation and the proposed functional tyrosine Y382 in signaling activation, we transiently transfected armadillo melanopsin in HEK293 cells and tested its signaling kinetics using calcium imaging. Armadillo melanopsin possesses a shorter C-terminus than mouse melanopsin, and it has fewer C-terminal serine and threonine residues than mouse melanopsin (Armadillo melanopsin has 14 potential phosphorylation sites compared to 38 in mouse melanopsin, Fig. S5). Additionally, armadillo melanopsin has a substitution of the evolutionarily conserved tyrosine residue (P381 in armadillo melanopsin, Fig. 5 A), which may potentially directly impact the activation kinetics. Calcium imaging of mouse and armadillo melanopsins (Fig. 5 D) indicates a striking difference in signaling kinetics, most notably armadillo melanopsin’s activation kinetics are significantly slower than mouse melanopsin (Fig. 5 E) and displays a brief time period of inactivity before phototransduction onset. This suggests that the lack of C-terminal serine and threonine residues, particularly the proximally located (before residue D396) residues, and the lack of the tyrosine residue in armadillo melanopsin directly affects its capability to activate phototransduction. Furthermore, point mutations of armadillo melanopsin to mimic mouse melanopsin residues (L371S P375S G378S P381Y A388T G391S A393S D412S E417T) resulted in faster activation kinetics (Fig. 5, D and E) than wild-type armadillo melanopsin, further supporting the importance of C-terminal phosphorylation in producing rapid signaling activation.
Positively charged residues on mouse melanopsin’s third intracellular loop play a critical role in G-protein activation
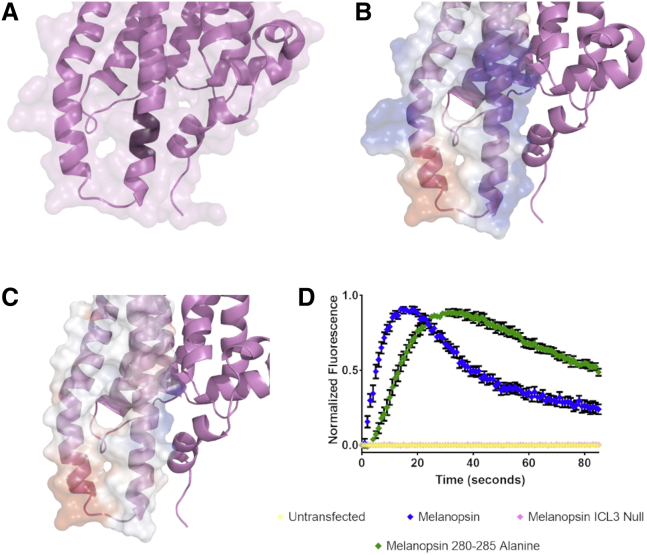
The functional importance of GPCR intracellular loop domains in the coupling and activation of its cognate G-protein has been established in many receptors. We wanted to test what molecular features of melanopsin’s cytoplasmic loops confer its capability to activate G-protein signaling. We focused on the third intracellular loop because of its extended nature and predicted interaction with the C-terminus. In addition, melanopsin’s third intracellular loop is rich with positively charged residues, and we wanted to test if this property has any role in phototransduction activation. First, we synthesized a melanopsin mutant in which amino acids 280–285 (R280 Q281 W282 Q283 R284 L285) on the cytoplasmic extension of transmembrane helix 6 were mutated to alanine residues (Melanopsin 280–285 Alanine, Fig. 6 A) to disrupt any hydrogen bonding or ionic contacts with the C-terminus, as predicted by our modeling. Calcium imaging of this mutant reveals sluggish activation kinetics (Fig. 6 D), similar to our C-terminal activation mutants (Fig. 4). This data, when analyzed with the structural model, suggests that this region of the third cytoplasmic loop potentially forms functionally significant contacts between intracellular loop 3 and the C-terminus. Furthermore, when these data are taken together with the data in Fig. 5, we propose that C-terminal phosphorylation could add the requisite negative charge to allow ionic contacts to form with these positively charged intracellular loop 3 residues. Although we propose this functionally important cytoplasmic conformation and a mechanism for its stabilization (i.e., intracellular loop 3-C-terminus ionic interaction facilitated by C-terminal phosphorylation), further point-mutation analyses would be required to validate this interaction and to identify the required residues for this interaction to occur.
Figure 6.
Calcium imaging of intracellular loop 3 mutants. (A) Homology model of melanopsin depicting the region of mutations (darker purple color on the cytoplasmic extension of transmembrane helix 6) in the mutant melanopsin 280–285 Alanine, in which amino acids 280–285 are mutated to alanine residues. (B) Homology model depicting the electrostatic charge of the third cytoplasmic loop in wild-type melanopsin and (C) the mutant melanopsin ICL3 Null, in which all positively charged residues were mutated to alanines to neutralize the charge of this region (blue: positively charged; red: negatively charged; white: neutral). (D) Calcium imaging of melanopsin and the mutants depicted in (A) and (C). Error bars represent SEM of three independent transfections.
To test for any functional significance of the positive charge localized at the third cytoplasmic loop, we mutated all positively charged residues on the third intracellular loop (Fig. 6 B) to alanine residues (Melanopsin R259A R262A R266A R277A R279A R280A R284A K390A K293A or Melanopsin ICL3 Null, Fig. 6 C) to alter the loop’s surface charge closer to neutral. Calcium imaging of this mutant shows a striking inability to activate phototransduction (Fig. 6 D), suggesting that the positive charge at the third intracellular loop is critical for melanopsin to couple to G-protein in any capacity; thus, we propose this feature as a contributor to melanopsin’s capability to couple to its cognate G-protein.
Discussion
The diversity of molecular features and amino acid properties in the cytoplasmic regions of GPCRs complicates the prediction of receptor-G-protein binding properties in an unstudied receptor. It is evident in many class A GPCRs, including the prototypical receptor and visual pigment rhodopsin, that cytoplasmic domains, including the cytoplasmic loops and the C-terminus, are critical for G-protein binding and selectivity. Melanopsin-expressing ipRGCs in the mammalian retina regulate a diversity of nonvisual and visual functions, and functional heterogeneity is observed across ipRGC subtypes and even within the same subtypes. In this study, we propose our model of melanopsin, which consists of a functionally critical cytoplasmic conformation consisting of extended transmembrane helices 5 and 6 that form a third cytoplasmic loop domain that protrudes far into the cytoplasm. Additionally, in the dark inactive state, the proximal region of the C-terminus (before residue D396, Fig. 4 A) takes on a conformation in which, after the predicted eighth helix, it is adjacent to the cytoplasmic extension of transmembrane helix 6 and close enough to form contacts. These contacts could potentially be ionic in nature, and our data support the importance of C-terminal phosphorylation sites and Y382—which may also be phosphorylated—in phototransduction activation. Thus, C-terminal phosphorylation could provide the negative charge that is needed for the C-terminus to ionically interact with the arginine and lysine residue-rich third cytoplasmic loop in the inactive state before light-induced activation. Based on our homology modeling, we propose that upon photon-mediated conformational change to active-state melanopsin, the fifth and sixth transmembrane helices undergo an outward swing, moving the cytoplasmic ends of these helices—and the third cytoplasmic loop—away from the C-terminus. We also propose that the C-terminus, together with the third cytoplasmic loop, forms a critical part of the G-protein binding pocket and may play a role in G-protein specificity or stabilization of the melanopsin-G-protein complex. Previous studies have also generated homology models of melanopsin utilizing squid rhodopsin (59,60) or bovine rhodopsin (61), but these studies focused primarily on making computational predictions of melanopsin’s chromophore chemistry and did not focus on structural determinants of melanopsin-G-protein binding. However, these previous models also generated a predicted structure containing the features we highlight here (i.e., extended fifth and sixth transmembrane helices/third intracellular loop and C-terminal conformation).
Our EPR spectroscopy measurements on melanopsin support the cytoplasmic extension of the third cytoplasmic loop because of the spectral lineshape that is indicative of high spin label rotational mobility at that region. Interestingly, although a clear difference was observed in full-length and truncated melanopsin C268 EPR spectra, we did not observe dramatic differences in the spectral lineshape after light exposure in both samples. This result was unexpected as we did predict seeing light-induced conformational change reflected in the EPR spectra. Given the steric freedom at C268 and in the third intracellular loop region, it is possible that this region is unaffected by helical movement or conformational change because of its predicted distance from the membrane and from other structures. However, our model and data—including our EPR data of full-length and C-terminal truncations—support the idea that the proximal C-terminus is adjacent to the third intracellular loop. Our model also predicts that upon light exposure, if melanopsin adopts a similar active conformation as the β2 adrenergic receptor, another family A GPCR, then we would expect these regions to separate and form the G-protein binding pocket. This would lead to the prediction that the EPR spectrum of the light-exposed full-length melanopsin C268 sample should differ from the dark spectrum. It is possible that this movement happens as predicted by our modeling but does not produce a significant difference in the EPR spectral lineshape. Alternatively, it is also possible that the predicted interactions between these two regions, the C-terminus and the third cytoplasmic loop, remain intact even after light-induced activation. We observed no differences in the dark and light EPR spectra of the Δ365 truncated melanopsin C268 because this mutant lacks this C-terminal region. We therefore propose that our model of melanopsin activation is robust, but identification of structural and conformational changes of melanopsin after light exposure remains an important question for further study.
Precise structural and molecular determinants of GPCR selectivity to its cognate G-protein(s) are difficult to isolate because of the promiscuity of GPCRs for multiple G-proteins. Although it would be erroneous to suggest that, like melanopsin, all Gαq-binding GPCRs require similar structural features (i.e., extended fifth and sixth transmembrane helices/third cytoplasmic loop, positively charged cytoplasmic loops, and C-terminus contacting G-protein) for their selectivity, we propose that similar features are shared among rhabdomeric-type visual pigments. Of the available resolved protein structures of GPCRs, two are of rhabdomeric visual pigments: two squid rhodopsin structures and jumping spider rhodopsin (41,48,62). Similar structural features are observed in melanopsin’s predicted structure and the structures of the resolved rhabdomeric-type visual pigments. Specifically, the extended fifth and sixth transmembrane helices are a common feature among melanopsin and the invertebrate visual pigments, and like squid rhodopsin, melanopsin also has an extended C-terminus, much longer than vertebrate, ciliary-type image-forming visual pigments like rhodopsin. Melanopsin’s intracellular loops are predicted to possess a more uniform positive charge compared to the surface charges observed on the loops of squid and jumping spider rhodopsin, which are also positively charged but do possess regions of negative charge, particularly on the fifth transmembrane portion of the third cytoplasmic loop. It is evident that this positive charge holds significance in melanopsin, but it remains unclear as to the extent of this feature’s significance for Gαq interaction. Given the enrichment of potential phosphorylation sites on melanopsin’s C-terminus and intracellular loops, this provides an interesting mechanism to modify the electrostatic potential of these regions and thereby provides an interesting way to alter G-protein binding and potentially specificity. Thus, with these findings, we identified critical regions that regulate melanopsin’s phototransduction activation to shed light onto ipRGC function and provide targets for protein engineering for use as an optogenetic tool or further study of its phototransduction.
Author Contributions
J.C.V.-L., S.T.P., M.P.D., M.G., and R.J.B. conducted experiments. J.C.V.-L., E.G.C., J.B.W., V.A.S., and P.R.R. intellectually contributed and conceived experiments. J.C.V.-L. wrote the manuscript. P.R.R. edited the manuscript.
Acknowledgments
We thank Prof. Daniel D. Oprian for generously providing 1D4-antibody. J.C.V.-L also wishes to acknowledge and thank Charles Thomas and Ramses Long for assistance in optimizing protein expression.
We thank and acknowledge National Institutes of Health Training Grant T32 GM066706 awarded to J.C.V.-L., generously made possible by Prof. Katherine Seley-Radtke, and National Institutes of Health R01 EY027202-01A1 awarded to P.R.R. M.P.D. acknowledges support under the Cooperative Research Agreement between the University of Maryland Baltimore County and the National Institute of Standards and Technology Center for Nanoscale Science and Technology Award 70NANB10H193 through the University of Maryland.
Editor: Elizabeth Rhoades.
Footnotes
Stephen T. Petr’s present address is Eastern Virginia Medical School, Norfolk, Virginia.
Matthew P. Donohue’s present address is Salubris Biotherapeutics, Gaithersburg, Maryland.
Meheret Gebreeziabher’s present address is National Human Genome Research Institute, National Institutes of Health, Bethesda, Maryland.
Evan G. Cameron’s present address is Department of Ophthalmology, Byers Eye Institute, Mary M. and Sash A. Spencer Center for Vision Research, Stanford University School of Medicine, Palo Alto, California.
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2020.06.013.
Supporting Material
References
- 1.Fredriksson R., Lagerström M.C., Schiöth H.B. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol. Pharmacol. 2003;63:1256–1272. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- 2.Takeda S., Kadowaki S., Mitaku S. Identification of G protein-coupled receptor genes from the human genome sequence. FEBS Lett. 2002;520:97–101. doi: 10.1016/s0014-5793(02)02775-8. [DOI] [PubMed] [Google Scholar]
- 3.Flock T., Ravarani C.N.J., Babu M.M. Universal allosteric mechanism for Gα activation by GPCRs. Nature. 2015;524:173–179. doi: 10.1038/nature14663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hofmann K.P., Scheerer P., Ernst O.P. A G protein-coupled receptor at work: the rhodopsin model. Trends Biochem. Sci. 2009;34:540–552. doi: 10.1016/j.tibs.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 5.Provencio I., Jiang G., Rollag M.D. Melanopsin: an opsin in melanophores, brain, and eye. Proc. Natl. Acad. Sci. USA. 1998;95:340–345. doi: 10.1073/pnas.95.1.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Provencio I., Rodriguez I.R., Rollag M.D. A novel human opsin in the inner retina. J. Neurosci. 2000;20:600–605. doi: 10.1523/JNEUROSCI.20-02-00600.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmidt T.M., Chen S.K., Hattar S. Intrinsically photosensitive retinal ganglion cells: many subtypes, diverse functions. Trends Neurosci. 2011;34:572–580. doi: 10.1016/j.tins.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quattrochi L.E., Stabio M.E., Berson D.M. The M6 cell: a small-field bistratified photosensitive retinal ganglion cell. J. Comp. Neurol. 2019;527:297–311. doi: 10.1002/cne.24556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hattar S., Liao H.W., Yau K.W. Melanopsin-containing retinal ganglion cells: architecture, projections, and intrinsic photosensitivity. Science. 2002;295:1065–1070. doi: 10.1126/science.1069609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Estevez M.E., Fogerson P.M., Berson D.M. Form and function of the M4 cell, an intrinsically photosensitive retinal ganglion cell type contributing to geniculocortical vision. J. Neurosci. 2012;32:13608–13620. doi: 10.1523/JNEUROSCI.1422-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hattar S., Lucas R.J., Yau K.W. Melanopsin and rod-cone photoreceptive systems account for all major accessory visual functions in mice. Nature. 2003;424:76–81. doi: 10.1038/nature01761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peirson S.N., Halford S., Foster R.G. The evolution of irradiance detection: melanopsin and the non-visual opsins. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2009;364:2849–2865. doi: 10.1098/rstb.2009.0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hattar S., Kumar M., Berson D.M. Central projections of melanopsin-expressing retinal ganglion cells in the mouse. J. Comp. Neurol. 2006;497:326–349. doi: 10.1002/cne.20970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rupp A.C., Ren M., Schmidt T.M. Distinct ipRGC subpopulations mediate light’s acute and circadian effects on body temperature and sleep. eLife. 2019;8:e44358. doi: 10.7554/eLife.44358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lucas R.J. Mammalian inner retinal photoreception. Curr. Biol. 2013;23:R125–R133. doi: 10.1016/j.cub.2012.12.029. [DOI] [PubMed] [Google Scholar]
- 16.Do M.T., Yau K.W. Intrinsically photosensitive retinal ganglion cells. Physiol. Rev. 2010;90:1547–1581. doi: 10.1152/physrev.00013.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graham D.M., Wong K.Y., Berson D.M. Melanopsin ganglion cells use a membrane-associated rhabdomeric phototransduction cascade. J. Neurophysiol. 2008;99:2522–2532. doi: 10.1152/jn.01066.2007. [DOI] [PubMed] [Google Scholar]
- 18.Panda S., Nayak S.K., Jegla T. Illumination of the melanopsin signaling pathway. Science. 2005;307:600–604. doi: 10.1126/science.1105121. [DOI] [PubMed] [Google Scholar]
- 19.Sonoda T., Lee S.K., Schmidt T.M. Melanopsin phototransduction is repurposed by ipRGC subtypes to shape the function of distinct visual circuits. Neuron. 2018;99:754–767.e4. doi: 10.1016/j.neuron.2018.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang Z., Yue W.W.S., Yau K.W. Cyclic-nucleotide- and HCN-channel-mediated phototransduction in intrinsically photosensitive retinal ganglion cells. Cell. 2018;175:652–664.e12. doi: 10.1016/j.cell.2018.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou X.E., Melcher K., Xu H.E. Structure and activation of rhodopsin. Acta Pharmacol. Sin. 2012;33:291–299. doi: 10.1038/aps.2011.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inoue A., Raimondi F., Russell R.B. Illuminating G-protein-coupling selectivity of GPCRs. Cell. 2019;177:1933–1947.e25. doi: 10.1016/j.cell.2019.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masuho I., Ostrovskaya O., Martemyanov K.A. Distinct profiles of functional discrimination among G proteins determine the actions of G protein-coupled receptors. Sci. Signal. 2015;8:ra123. doi: 10.1126/scisignal.aab4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okashah N., Wan Q., Lambert N.A. Variable G protein determinants of GPCR coupling selectivity. Proc. Natl. Acad. Sci. USA. 2019;116:12054–12059. doi: 10.1073/pnas.1905993116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Newman L.A., Walker M.T., Robinson P.R. Melanopsin forms a functional short-wavelength photopigment. Biochemistry. 2003;42:12734–12738. doi: 10.1021/bi035418z. [DOI] [PubMed] [Google Scholar]
- 26.Flock T., Hauser A.S., Babu M.M. Selectivity determinants of GPCR-G-protein binding. Nature. 2017;545:317–322. doi: 10.1038/nature22070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blasic J.R., Jr., Lane Brown R., Robinson P.R. Light-dependent phosphorylation of the carboxy tail of mouse melanopsin. Cell. Mol. Life Sci. 2012;69:1551–1562. doi: 10.1007/s00018-011-0891-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blasic J.R., Jr., Brown R.L., Robinson P.R. Phosphorylation of mouse melanopsin by protein kinase A. PLoS One. 2012;7:e45387. doi: 10.1371/journal.pone.0045387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blasic J.R., Jr., Matos-Cruz V., Robinson P.R. Identification of critical phosphorylation sites on the carboxy tail of melanopsin. Biochemistry. 2014;53:2644–2649. doi: 10.1021/bi401724r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cameron E.G., Robinson P.R. β-Arrestin-dependent deactivation of mouse melanopsin. PLoS One. 2014;9:e113138. doi: 10.1371/journal.pone.0113138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Somasundaram P., Wyrick G.R., Robinson P.R. C-terminal phosphorylation regulates the kinetics of a subset of melanopsin-mediated behaviors in mice. Proc. Natl. Acad. Sci. USA. 2017;114:2741–2746. doi: 10.1073/pnas.1611893114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mure L.S., Hatori M., Panda S. Melanopsin-encoded response properties of intrinsically photosensitive retinal ganglion cells. Neuron. 2016;90:1016–1027. doi: 10.1016/j.neuron.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mure L.S., Hatori M., Panda S. Sustained melanopsin photoresponse is supported by specific roles of β-Arrestin 1 and 2 in deactivation and regeneration of photopigment. Cell Rep. 2018;25:2497–2509.e4. doi: 10.1016/j.celrep.2018.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang Y., Kuybeda O., Xu H.E. Cryo-EM structure of human rhodopsin bound to an inhibitory G protein. Nature. 2018;558:553–558. doi: 10.1038/s41586-018-0215-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao Y., Hu H., Skiniotis G. Structures of the rhodopsin-transducin complex: insights into G-protein activation. Mol. Cell. 2019;75:781–790.e3. doi: 10.1016/j.molcel.2019.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsai C.J., Marino J., Schertler G. Cryo-EM structure of the rhodopsin-Gαi-βγ complex reveals binding of the rhodopsin C-terminal tail to the gβ subunit. eLife. 2019;8:e46041. doi: 10.7554/eLife.46041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Phillips W.J., Cerione R.A. A C-terminal peptide of bovine rhodopsin binds to the transducin alpha-subunit and facilitates its activation. Biochem. J. 1994;299:351–357. doi: 10.1042/bj2990351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emanuel A.J., Do M.T. Melanopsin tristability for sustained and broadband phototransduction. Neuron. 2015;85:1043–1055. doi: 10.1016/j.neuron.2015.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Milner E.S., Do M.T.H. A population representation of absolute light intensity in the mammalian retina. Cell. 2017;171:865–876.e16. doi: 10.1016/j.cell.2017.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu S., Zhang Y. LOMETS: a local meta-threading-server for protein structure prediction. Nucleic Acids Res. 2007;35:3375–3382. doi: 10.1093/nar/gkm251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shimamura T., Hiraki K., Miyano M. Crystal structure of squid rhodopsin with intracellularly extended cytoplasmic region. J. Biol. Chem. 2008;283:17753–17756. doi: 10.1074/jbc.C800040200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zimmermann L., Stephens A., Alva V. A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J. Mol. Biol. 2018;430:2237–2243. doi: 10.1016/j.jmb.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 43.Rasmussen S.G.F., DeVree B.T., Kobilka B.K. Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Franke R.R., Sakmar T.P., Khorana H.G. A single amino acid substitution in rhodopsin (lysine 248----leucine) prevents activation of transducin. J. Biol. Chem. 1988;263:2119–2122. [PubMed] [Google Scholar]
- 45.Gibson D.G., Young L., Smith H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 46.Wang W., Malcolm B.A. Two-stage PCR protocol allowing introduction of multiple mutations, deletions and insertions using QuikChange site-directed mutagenesis. Biotechniques. 1999;26:680–682. doi: 10.2144/99264st03. [DOI] [PubMed] [Google Scholar]
- 47.Stoll S., Schweiger A. EasySpin, a comprehensive software package for spectral simulation and analysis in EPR. J. Magn. Reson. 2006;178:42–55. doi: 10.1016/j.jmr.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 48.Murakami M., Kouyama T. Crystal structure of squid rhodopsin. Nature. 2008;453:363–367. doi: 10.1038/nature06925. [DOI] [PubMed] [Google Scholar]
- 49.Karnik S.S., Khorana H.G. Assembly of functional rhodopsin requires a disulfide bond between cysteine residues 110 and 187. J. Biol. Chem. 1990;265:17520–17524. [PubMed] [Google Scholar]
- 50.Hwa J., Reeves P.J., Khorana H.G. Structure and function in rhodopsin: further elucidation of the role of the intradiscal cysteines, Cys-110, -185, and -187, in rhodopsin folding and function. Proc. Natl. Acad. Sci. USA. 1999;96:1932–1935. doi: 10.1073/pnas.96.5.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim J.M., Altenbach C., Hubbell W.L. Structure and function in rhodopsin: rhodopsin mutants with a neutral amino acid at E134 have a partially activated conformation in the dark state. Proc. Natl. Acad. Sci. USA. 1997;94:14273–14278. doi: 10.1073/pnas.94.26.14273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van Eps N., Caro L.N., Hubbell W.L. Conformational equilibria of light-activated rhodopsin in nanodiscs. Proc. Natl. Acad. Sci. USA. 2017;114:E3268–E3275. doi: 10.1073/pnas.1620405114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kusnetzow A.K., Altenbach C., Hubbell W.L. Conformational states and dynamics of rhodopsin in micelles and bilayers. Biochemistry. 2006;45:5538–5550. doi: 10.1021/bi060101v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Resek J.F., Farahbakhsh Z.T., Khorana H.G. Formation of the meta II photointermediate is accompanied by conformational changes in the cytoplasmic surface of rhodopsin. Biochemistry. 1993;32:12025–12032. doi: 10.1021/bi00096a012. [DOI] [PubMed] [Google Scholar]
- 55.Li S.C., Goto N.K., Deber C.M. α-helical, but not β-sheet, propensity of proline is determined by peptide environment. Proc. Natl. Acad. Sci. USA. 1996;93:6676–6681. doi: 10.1073/pnas.93.13.6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chini B., Parenti M. G-protein-coupled receptors, cholesterol and palmitoylation: facts about fats. J. Mol. Endocrinol. 2009;42:371–379. doi: 10.1677/JME-08-0114. [DOI] [PubMed] [Google Scholar]
- 57.Seno K., Hayashi F. Palmitoylation is a prerequisite for dimerization-dependent raftophilicity of rhodopsin. J. Biol. Chem. 2017;292:15321–15328. doi: 10.1074/jbc.M117.804880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maeda A., Okano K., Palczewski K. Palmitoylation stabilizes unliganded rod opsin. Proc. Natl. Acad. Sci. USA. 2010;107:8428–8433. doi: 10.1073/pnas.1000640107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rinaldi S., Melaccio F., Olivucci M. Comparison of the isomerization mechanisms of human melanopsin and invertebrate and vertebrate rhodopsins. Proc. Natl. Acad. Sci. USA. 2014;111:1714–1719. doi: 10.1073/pnas.1309508111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sekharan S., Wei J.N., Batista V.S. The active site of melanopsin: the biological clock photoreceptor. J. Am. Chem. Soc. 2012;134:19536–19539. doi: 10.1021/ja308763b. [DOI] [PubMed] [Google Scholar]
- 61.Hermann R., Poppe L., Lerchl A. Predicted 3D-structure of melanopsin, the non-rod, non-cone photopigment of the mammalian circadian clock, from Djungarian hamsters (Phodopus sungorus) Neurosci. Lett. 2005;376:76–80. doi: 10.1016/j.neulet.2004.11.029. [DOI] [PubMed] [Google Scholar]
- 62.Varma N., Mutt E., Lesca E. Crystal structure of jumping spider rhodopsin-1 as a light sensitive GPCR. Proc. Natl. Acad. Sci. USA. 2019;116:14547–14556. doi: 10.1073/pnas.1902192116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kearse M., Moir R., Drummond A. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Omasits U., Ahrens C.H., Wollscheid B. Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics. 2014;30:884–886. doi: 10.1093/bioinformatics/btt607. [DOI] [PubMed] [Google Scholar]
- 65.Pándy-Szekeres G., Munk C., Gloriam D.E. GPCRdb in 2018: adding GPCR structure models and ligands. Nucleic Acids Res. 2018;46:D440–D446. doi: 10.1093/nar/gkx1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.