Abstract
Objective
The purpose of this study is to identify the biomarkers for early diagnosis of Parkinson's disease (PD) by multi-omics joint analysis, so as to identify the biomarkers for early diagnosis of PD, and to help clinicians make early diagnosis and treatment.
Methods
In this study, mice are taken as the study subjects. The model of PD mice is established, and then lymphocyte, striatum, substantia nigra protein and proteolysis are extracted. After that, the experiments of protein imprinting and 418O labeling are carried out. Mass Spectrometry (MS) analysis technology is mainly used to study proteomics and to analyze the quantitative and qualitative situation of differential proteins in striatum, substantia nigra protein and lymphocyte. By this method, biomarkers for early diagnosis of PD are analyzed and identified.
Results
The biomarkers of Parkinson's early onset are related to the same quantitative differential expression of lymphocyte, striatum, substantia nigra protein, lymphocyte and substantia nigra.
Conclusion
This experimental method can analyze and identify the biomarkers of early diagnosis of PD, help to explore the pathophysiology and pathogenesis of PD, effectively help clinicians make timely diagnosis in advance, and improve the prevention and treatment effect of the disease.
Keywords: Multi-omics, Biomarkers, Parkinson, Differential proteins
1. Introduction
Parkinson's disease (PD) is the second largest neurodegenerative disease in the world after Alzheimer's disease. Its main clinical manifestations are resting tremor, abnormal posture, muscular stiffness and dyskinesia, accompanied by cognitive impairment, depression, hypoolusive olfactory function and some mental problems (Ge et al., 2017, Hatano et al., 2016). According to epidemiological investigation and analysis, with the acceleration of population aging process, rapid economic growth and serious environmental pollution, the incidence of PD in China is increasing. The prevalence rate of people over 65 years old was 1.7% for males and 1.6% for females. Globally, the number of Parkinson's patients is expected to reach 8.7 million by 2030, more than double the increase in 2005. Among them, the number of sick people in China will account for 57% (Simonsen et al., 2005). The main pathogenesis of PD is progressive loss of dopaminergic neurons in the substantia nigra striatum system and the appearance of soma (Le et al., 2017). The brain is a highly protected organ, and the blood-brain barrier limits the exchange of fluid and substances between the brain and Lewy, making the pathological changes in the brain difficult to show in the body fluid. The particularity of human brain organs also determines that it cannot be directly sampled by routine pathological methods. Therefore, the diagnosis of neurodegenerative diseases such as PD, especially early diagnosis, has been a difficult problem in clinical medicine. The study of etiology and biomarkers of the disease has very important social and medical value (Kim, 2017, Pestova et al., 2018).
Biomarkers are indicators in blood, saliva, urine and cerebrospinal fluid (CSF), which can reflect physiological or pathological conditions directly related to clinical manifestations of the disease (Lotem et al., 2016). Tissingh et al. (2015) studied the significantly negative correlation between olfactory dysfunction and PD, and proved that there was significant olfactory impairment in early and new PD, so olfactory test may be helpful for the early diagnosis of PD (Tissingh et al., 2015). Maggio et al. (2015) studied the application of new radiology technologies such as DaT-SCAN SPECT in the early diagnosis and treatment of PD, and improved the early diagnosis and treatment of these patients (Maggio et al., 2015). Biomarkers are helpful for the early diagnosis of chronic neurodegenerative diseases, and can monitor the progress of the disease and the efficacy of drugs. Ideal biomarkers are easy to detect and quantify, have good repeatability, are not easy to mutate, and are not affected by disease factors (Chen et al., 2016). There are many methods for identification of biomarkers, including imaging, neurophysiology and some known new technologies, such as biochemistry, proteomics, metabonomics and gene probe technology.
A PD mouse model was established, and lymphocytes, striatum, substantia nigra protein and proteolysis were extracted. Then, protein imprinting experiments and 418O labelling were carried out, and mass spectrometry (MS) analysis technology was used to analyze the proteomics. By this method, biomarkers for early diagnosis of PD are analyzed and identified, in order to better understand the pathophysiology and pathogenesis of PD, and effectively help clinical medicine to make early diagnosis and treatment.
2. Materials and methods
2.1. Experimental objects and grouping
12 specific pathogen free (SPF) grade healthy male rats aged 2–3 months and weighing 180–220 g are selected as experimental subjects. Rats are fed in isolation with cages. The feeding environment are as follows: temperature 18–21 °C, humidity >40%, pressurized air supply 150 pa, natural light (12 h a day). Rats are free to take water and standard granules. Every two days, it is necessary to change the bedding and feed. The rats are divided into two groups, 12 rats in each group, sham-operated control group and mature PD model group. The difference between the sham-operated group and the PD model group is that no stereotactic injection of 6-oxidopamine (6-OHDA) (Shanghai Chenyi Biotechnology Co., Ltd., China) damaged the striatum unilaterally. This study has been approved by the ethics committee of our hospital, and all the tests were conducted in accordance with the rules and regulations formulated by the ethics committee.
2.2. Establishment of rat model PD model
6-OHDA is the first chemical to be found to have neurotoxic effects on catechol pathways. Usually, 6-OHDA is injected into medial forebrain tract, substantia nigra or striatum by unilateral stereotaxic method to construct animal models of PD Rat models are established by stereotactic injection of 6-OHDA, which is the key to unilateral striatal lesion and then to establish the model. At the 4th week of stereotactic injection, rats' rotational behavior is tested once a week for three consecutive weeks. The experimental steps are as follows. At the dose of 0.5 mg/kg, rats are intraperitoneally injected with 0.05% apomorphine solution. After 5–10 min, the rats are counted and the number of rotation cycles of the head and tail are recorded for 40 min. Rats are regarded as successful models of PD rats with constant rotation to the healthy side and rotation cycles (≥7 r/min).
2.3. Extraction of rat striatum and substantia nigra protein
After cardiac perfusion with normal saline, rat brain is removed. After stripping the striatum and excising the substantia nigra of the midbrain, tissue lysate is added at the ratio of 1:10 of mass to volume, respectively. A 1.5 ml tissue grinder is used to grind 15 times in ice bath at the same intensity to break up tissues and cells. When no tissue is observed, the grinding suspension is broken by ultrasound again. Ultrasound breaking conditions are as follows: pause 2 s after 1 s, ultrasonic amplitude 28%, ultrasonic time of 1 min, total time of 3 min. After that, centrifugation is carried out at 17,000g and 4 °C for 30 min. The supernatant is absorbed and the protein concentration is determined according to 2.2:5.1. The supernatant is stored at −80 °C. The substantia nigra proteins of PD group and sham-operated control group are mixed by enzymolysis, labeling and 1:1 equivalence, respectively. After desalination of C18 column by solid phase extraction, peptide segments are separated by off-line strong cation exchange (SCX) high performance liquid chromatography to reduce the complexity of biological samples.
2.4. Extraction of rat lymphocyte protein
After the rat lymph cells are separated and extracted, 200 μL lysate is added and broken by ultrasound. After that, centrifugation is carried out at 17,000g and 4 °C for 30 min. The supernatant is absorbed and the protein concentration is determined according to 2.2:5.1. The supernatant is stored at −80 °C.
2.5. Proteolysis
10.0 mg BSA (Beijing solabo Technology Co., Ltd., China) is weighed and dissolved in 1 ml denatured solution. Without adding urea, it is directly denatured at 37 °C for 4 h. A solution of 100 mM iodoacetamide (IAA) (Beijing solabo Technology Co., Ltd., China) of 15 ml is added and alkylated in the dark for 1 h. Afterwards, 195 μL 50 mM ammonium bicarbonate (NH4HCO3) solution (Beijing solabo Technology Co., Ltd., China) is added to dilute the urea concentration in the sample, reducing the urea concentration to 1 M. Then, according to the ratio of protein to trypsin 50:1, 6 μg trypsin (Beijing solabo Technology Co., Ltd., China) (trypsin dissolved in 100 μL 50 mM NH4HCO3 solution in advance, i.e. 0.2 μg/ml) is added and enzymatic hydrolysis is carried out in water bath at 37 degrees centigrade for 20 h. After enzymatic hydrolysis, the samples are bathed in boiling water for 10 min, to be used.
2.6. 18O labeling
The PD model group is labeled with 18O, while the sham-operated control group is labeled with 16O. After the labeling is completed, the trypsin is inactivated by boiling water for 10 min and adding 5% formic acid (Taixing Jiuxin Biotechnology Co., Ltd., China).
2.7. Separation by strong cation exchange chromatography (SCX)
The peptide mixtures of substantia nigra and lymphocytes are separated by off-line strong cation exchange chromatography (SCX) (Tianjin Beisile Chromatographic Technology Development Center). The chromatographic system is Agilent 1100 series high performance liquid chromatography (Agilent 1100 HPLC). The chromatographic conditions are as follows: Agilent Zorbax column (2.1 mm * 150 mm, 5 um, 300 A), mobile phase A is 25% acetonitrile solution containing 10 mM ammonium formate (PH 3.0), mobile phase B is 25% acetonitrile containing 500 mM ammonium formate (PH 6.6), flow rate is 0.2 ml/min. The detection wavelength is 280 nm. A tube of fractions is collected every minute. The fractions are merged according to the chromatographic peaks and concentrated in vacuum for drying.
2.8. Protein imprinting experiment
Samples of rats in sham-operated group and PD group are taken separately and mixed with 4 x sample buffer according to the volume ratio of 3:1. Samples are bathed in boiling water for 5 min and centrifuged for 5 min before supernatant is taken. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) is performed using Bio-Rad vertical electrophoresis cell (Beijing Bingyang Technology Co., Ltd., China). The concentration of gel for electrophoresis separation is 12% and that of gel concentrate is 5%. The voltage used for gel concentrate is set at 80 V during electrophoresis. When the electrophoretic strips reach the interface of separating and concentrating gels, the voltage is adjusted to 110 V and the electrophoresis time is 1.5 h. After electrophoresis, protein transmembrane is carried out using Bio-Rad standard wet transmembrane device. The polyvinylidene fluoride (PVDF) membrane (0.22 um) is immersed in methanol for 30 s, and the filter paper and sponge are also immersed in the transfer buffer. When the electrophoresis is finished, the gel is unloaded (the gel concentrate is removed). According to the order of negative electrode-sponge-filter paper-glue-PVDF membrane filter paper sponge positive electrode, the rotating membrane core is installed. The electrophoresis cell is placed in the ice-water mixture to cool down. The voltage of the transfer membrane is set to 110 V, and the transfer time is 1.5 h. After transmembrane, PVDF membrane is sealed at room temperature for 90 min (except goat-derived antibodies are sealed with 5% rabbit serum, other species-derived antibodies are sealed with 5% skimmed milk powder). Tris Buffered Saline Tween (TBST) (Beijing Baileibo Technology Co., Ltd., China) is used to rinse the membrane 4 times for 10 min after overnight incubation with 1 antibody (beta-actin 1:8000, caspase-31:1000, syntaxin-1:1000, serum albumin 1:100, V/V). It is then incubated at room temperature for 1 h with horseradish peroxidase labelled antibodies (except goat-derived antibodies using rabbit-derived antibodies, and goat-derived antibodies using goat-derived antibodies, 1:5000, V/V). The membrane is rinsed four times with TBST for 10 min each time. Finally, ECL (enhanced chemiluminescence) is performed.
2.9. Data analysis
The data collected by mass spectrometry are retrieved by MS/MS through PEAKS Studio software. The database is derived from Swissport protein database. Rattus norvigicus is selected as species, fixed as Carbamidomethy1 (C), modified as oxidation (M) and O-labelling (C-term), which allowed 2 leakage sites of trypsinase and MS mass number. The error is 50 ppm, the mass error is 0.1 Da, and the false discovery rate (FDR) is 1%. The full O-labelling method is selected for quantitative analysis. The differential protein GO is clustered by DAVID (database for annotation, visualization, and integrated discovery).
3. Results
3.1. Quantitative and qualitative analysis of striatal differential proteins
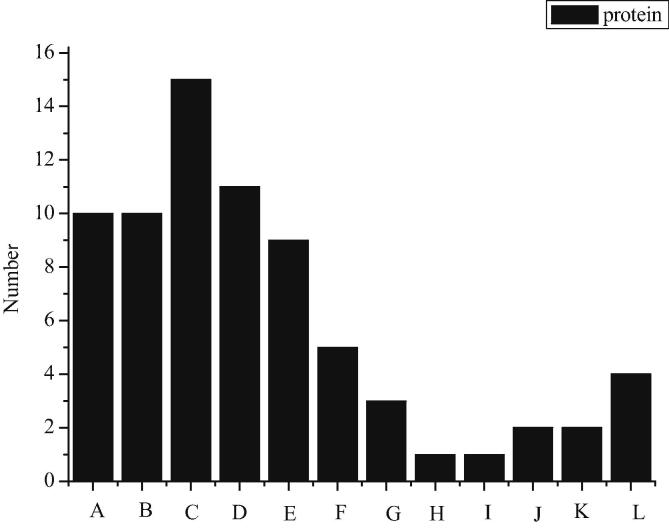
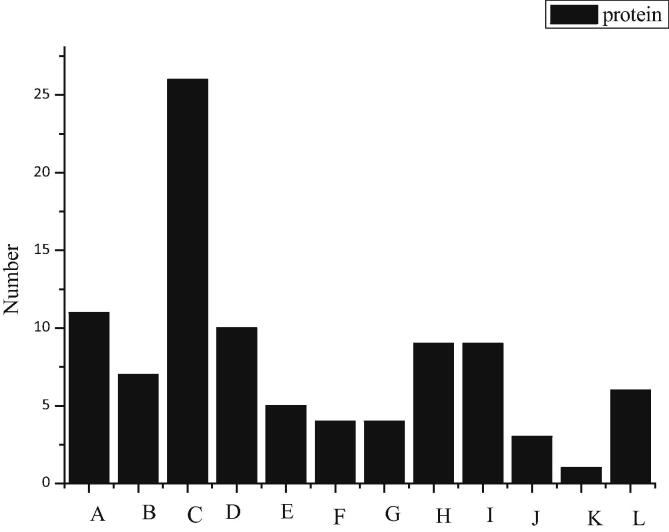
In this study, the whole striatal protein is labeled by 18O after trypsinase hydrolysis, detected by online two-dimensional liquid chromatography-mass spectrometry platform, and the mass spectrometry data are retrieved and analyzed by PEAKS. When the FDR of the peptide segment is less than 1.0%, there are 1232 MS matching maps of the peptide segment and the qualitative and quantitative results of 370 proteins, as shown in Fig. 1. The results are as follows. Among them, A, B, C, D, E, F, G, H, I, J, K and L represent structural proteins, signal transduction proteins, regulatory proteins, transporters, energy metabolic proteins, carbohydrate metabolic proteins, amino acid metabolic proteins, nucleotide metabolic proteins, transcriptional proteins, proteasomes, antioxidant proteins, and other proteins with undetermined functions. According to the descriptive information of proteins, these differential proteins are divided into 12 groups. Among them, 10 proteins (13.3%) are structural proteins, 10 (13.3%) are signal transduction proteins, 15 (20%) are regulatory proteins, 11 (14.7%) are transporters (including 7 synaptic related proteins), 9 (12.0%) are energy metabolism related proteins, 5 (6.7%) are involved in carbohydrate metabolism, and 3 (4.7%) are energy metabolism related proteins. 1 (1.3%) protein participates in amino acid metabolism, 1 (1.3%) protein participates in nucleotide metabolism, the rest proteins play a role in transcription (1, 1.3%), proteasome (2, 2.7%) and antioxidant process (2, 2.7%). Four (5.3%) proteins have not yet been identified.
Fig. 1.
6 Differential protein classification map of striatum expression in 6-OHDA unilateral damaged rat model.
In addition, based on GO annotation, cell components and biological processes are clustered according to DAVID. The clustering results with a clustering score greater than 1.3 are shown in Figs. 2 and 3. In Fig. 2A–E represent mitochondrial components, cytoskeleton components, neuron projection components, organelle membrane components and vesicle components, respectively. A, B, C, D, E, F, G, H and I in Fig. 3 represent energy and metabolite production, exocytosis regulation, stimulation response, neurodifferentiation, proton transport, synaptic conduction regulation, ATP biosynthesis, vesicle-mediated transport, respectively. It can be seen that most of the proteins are related to vesicle components, accounting for 26.5%. Because of the special function of striatum, 14 differentially expressed proteins are also found to belong to neuronal projection components. According to the biological process clustering, there are 8 groups whose clustering score is more than 1.3. Among them, 29.4% are involved in the regulation of exocytosis and vesicle-mediated transport. The protein expression level is down-regulated. It can be known that the biological process of endocytosis in PD rats is affected by the disease.
Fig. 2.
Cluster analysis based on cell component enrichment.
Fig. 3.
Cluster analysis of enrichment according to biological processes.
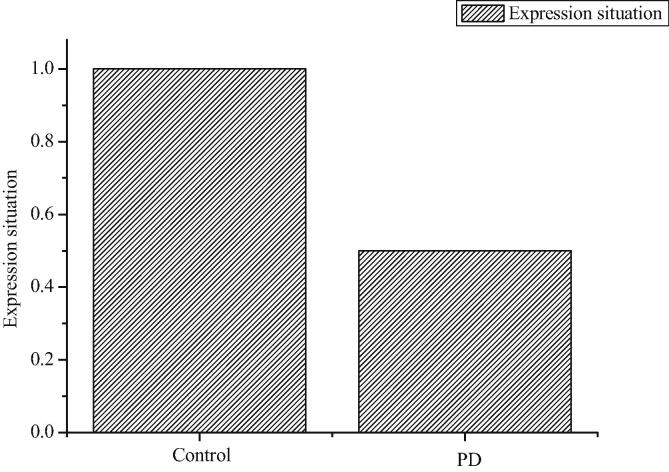
According to Fig. 4, compared with the sham-operated control group, the expression of syntaxin-1A in PD mice decreases, but the results are the same as those of mass spectrometry.
Fig. 4.
Expression of syntaxin-1A in sham-operated control group and PD group.
3.2. Quantitative and qualitative analysis of differentially expressed proteins in substantia nigra
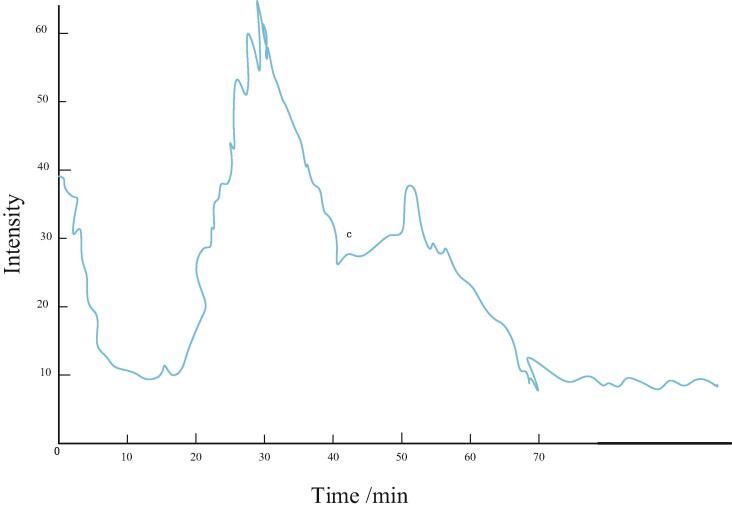
Fig. 5 is an ion exchange chromatogram of the peptide segment after enzymatic hydrolysis of substantia nigra. After pre-separation by SCX, the mixed peptide fragments are distributed in different fractions according to the retention time, thus achieving the effect of reducing sample complexity. In subsequent ESI mass spectrometry, it can reduce ion suppression and facilitate detection of peptides. After ion exchange separation, fractions are merged according to the intensity of peak response, fractions with high response intensity are not merged or merged less, while fractions with low response intensity are merged in multiple tubes.
Fig. 5.
Ion exchange chromatogram of peptide segment after proteolysis of substantia nigra.
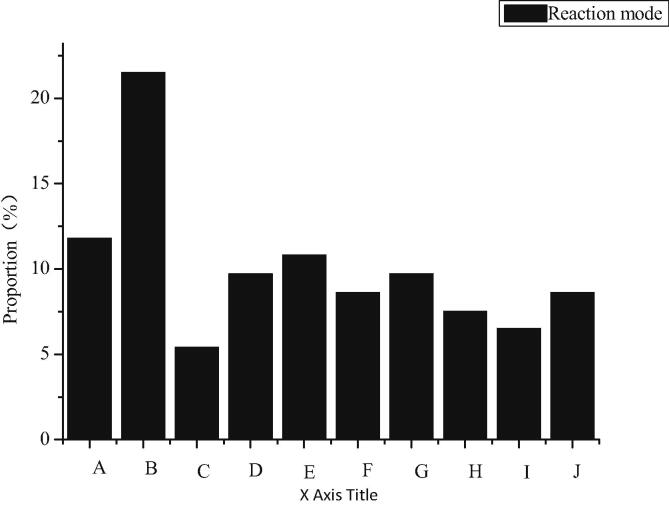
The peptide fragments separated by SCX are detected by nano-ESI-Q-TOF. Mass spectrometry data are retrieved and analyzed. When the FDR of the peptide segment is less than 1.0%, there are 8349 MS matching maps of the peptide segment and the qualitative and quantitative results of 1344 proteins, as shown in Fig. 6. The results are as follows. Among them, A, B, C, D, E, F, G, H, I, J, K and L represent structural proteins, signal transduction proteins, regulatory proteins, transporters, energy metabolic proteins, carbohydrate metabolic proteins, amino acid metabolic proteins, nucleotide metabolic proteins, transcriptional proteins, proteasomes, antioxidant proteins, and other proteins with undetermined functions. According to the descriptive information of protein function, these differential proteins are divided into 12 groups. Among them, 11 proteins (12.0%) are structural proteins, constituting cytoskeleton and microtubules. Seven (7.6%) are signal transduction proteins. 26 (28.3%) are regulatory proteins. Ten (10.9%) are transporters responsible for binding ions, transporters, molecules and proteins. Five (5.4%) proteins are energy metabolism related proteins. Four (4.3%) proteins are involved in carbohydrate metabolism. Four (4.3%) proteins have homeostasis function. Nine (9.8%) proteins are proteases, which participate in biosynthesis and redox reactions. The rest includes 9 (9.8%) proteins that play a role in RNA binding, processing and maturation, 3 (3.3%) proteins and proteasomes associate with transcription (1, 1.1%) and 6 (6.5%) proteins that have not yet been identified.
Fig. 6.
Differential protein classification map of sex expression in 6-OHDA unilateral damage rat model.
In addition, based on GO annotation, differential proteins were clustered according to DAVID using biological processes. The clustering results with a clustering score greater than 1.3 are shown in Fig. 7. Among them, A, B, C, D, E, F, G, H, I and J represent protein complexes, assembly energy metabolism, cytoskeleton and cell morphology regulation, cell adhesion and transport processes, macromolecule metabolism, transcription and translation, proteolysis and protein folding, other stress responses, immune response, and other responses. It can be seen that the largest proportion of differential proteins is involved in the biological process of stress response, a total of 20. Among them, 13 proteins are associated with immune response.
Fig. 7.
Cluster analysis of differential proteins enriched by biological processes.
According to Fig. 8, compared with the sham-operated control group, the expression level of serum albumin in PD mice decreased, but the result was the same as that of mass spectrometry.
Fig. 8.
Serum albumin expression in sham-operated control group and model group.
3.3. Quantitative and qualitative analysis of lymphocyte differential proteins
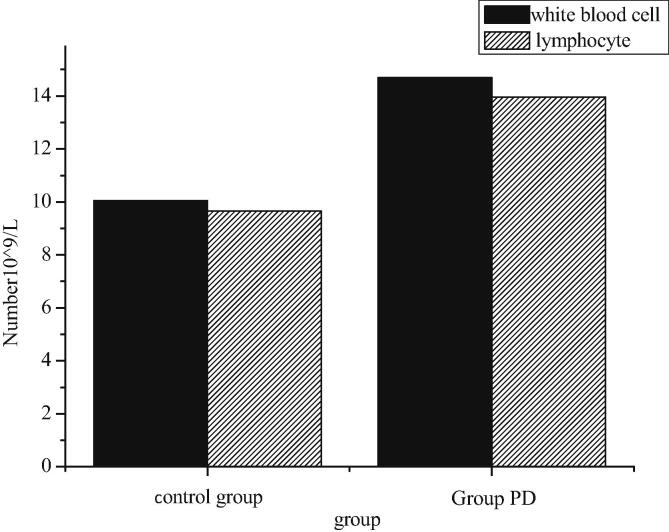
In this study, it is found that the number of white blood cells and lymphocyte in 6-OHDA unilateral lesion model is significantly higher than that in sham-operated control group (P < 0.01), as shown in Fig. 9. The number of leukocytes and lymphocytes in sham-operated control group are 10.05 ± 0.42 (10 ^ 9/L) and 9.65 ± 0.31 (10 ^ 9/L) respectively. The number of white blood cells and lymphocyte in PD group is 14.70 ± 1.58 (10 ^ 9/L) and 13.96 ± 1.64 (10 ^ 9/L), respectively.
Fig. 9.
Number of leukocyte and lymphocyte in rat peripheral blood.
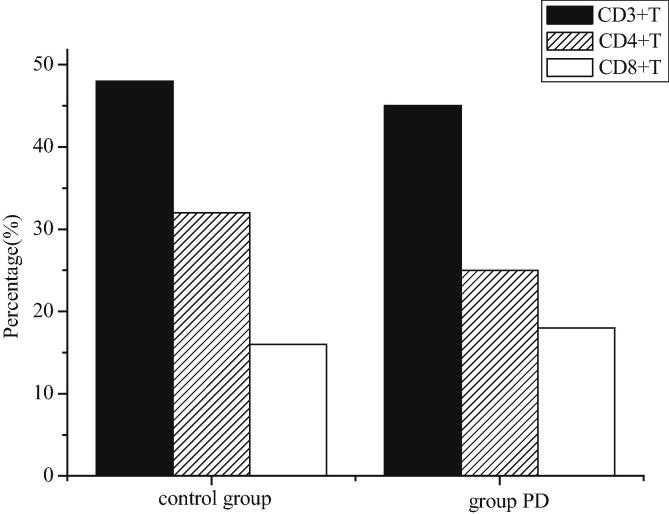
According to Fig. 10, there is no significant difference in lymphocyte percentage between model group and sham-operated control group (P > 0.05). The percentage of CD4+T cells in the model group is significantly lower than that in the sham-operated control group (P < 0.05), but the percentage of CD8+T cells in the model group is not significantly different from that in the sham-operated control group (P > 0.05). Therefore, the ratio of CD4+T to CD8+T also decreases (P < 0.01). It can be known that the decrease of the proportion of helper cells will affect the function of transforming helper cells into effector cells and promoting the production of antibodies by cells, which will lead to the disorder of immune function in the body.
Fig. 10.
Percentage of peripheral blood lymphocyte subsets (CD3+T, CD4+T, CD8+T) in rats.
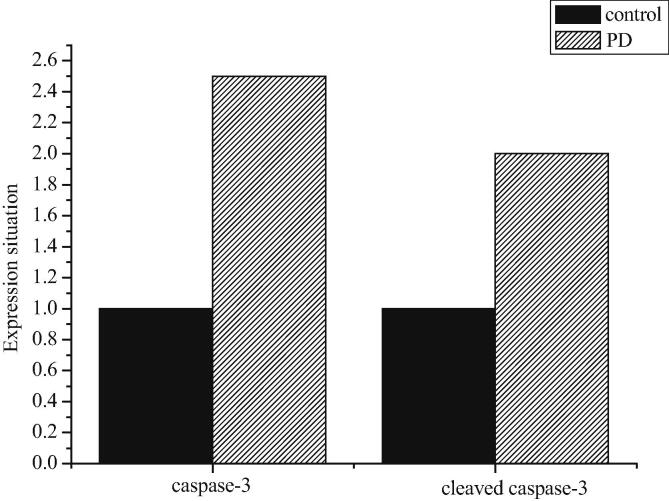
According to Fig. 11, the expression levels of caspase-3 and cleaved caspase-3 in the model group were higher than those in the control group.
Fig. 11.
Expression of caspase-3 and cleaved caspase-3 in sham-operated control group and model group.
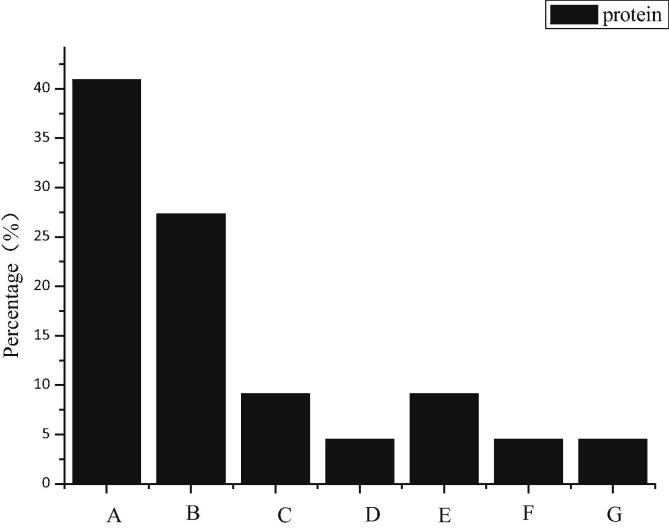
According to Fig. 12A–G represent carbohydrate proteins, signal transduction regulatory proteins, cell homeostasis proteins, transporters, translation proteins, catalytic proteins and proteins with undetermined functions. When the FDR of the peptide segment is less than 0.1%, there are 367 matched mass spectra of the peptide segment, and 136 qualitative and quantitative results of the protein are obtained. According to the descriptive information of protein function, these differential proteins are divided into seven groups. Among them, 6 proteins (27.3%) play carbohydrate metabolic functions, 9 proteins (40.9%) have signal transduction and regulation functions, and the rest proteins include maintaining cell homeostasis (1, 4.5%), transporting function (1, 4.5%), translating process (2, 9.1%) and catalytic function (1, 4.5%). 5% of proteases and 2 (9.1%) proteins have not yet been identified. It can be seen that differentially expressed proteins play important roles in carbohydrate metabolism, cell homeostasis, signal transduction and regulation.
Fig. 12.
Differential protein classification map of lymphocyte expression in 6-OHDA unilateral damage rat model.
3.4. Quantitative and qualitative analysis of the same protein in substantia nigra and lymphocyte
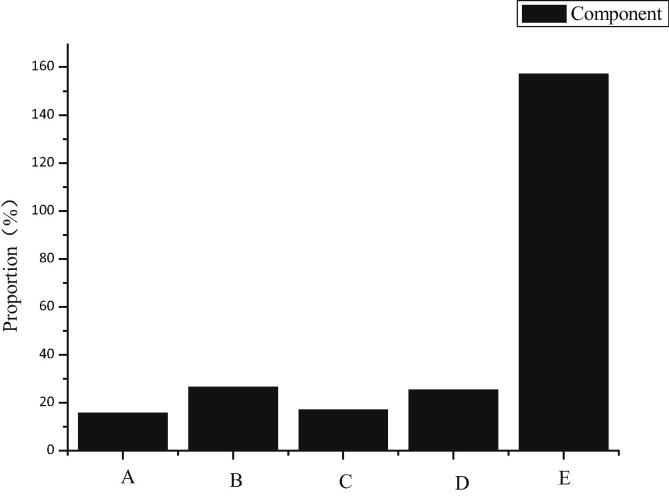
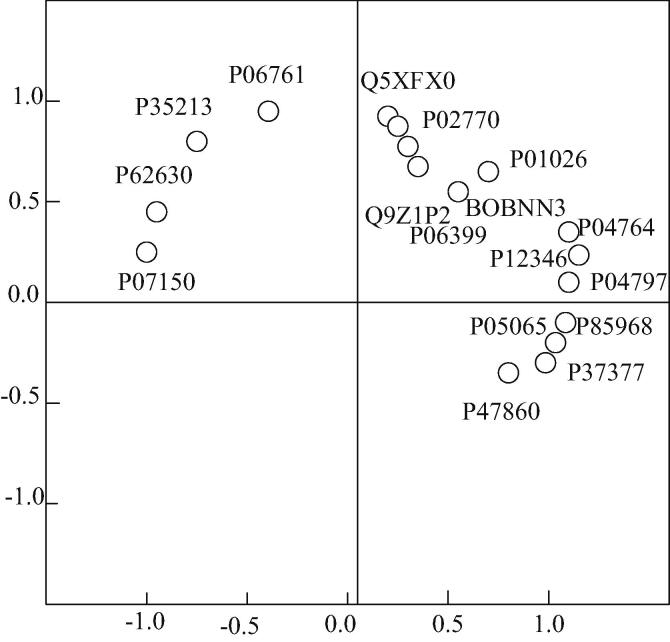
In this study, 18O-labeled differential proteomics is used to obtain differential proteins in substantia nigra and lymphocytes of unilaterally damaged PD rats. Differential proteins in substantia nigra are up-regulated in the expression of immune response-related proteins, suggesting that immune function plays a role in the development of disease. Differentially expressed proteins in lymphocytes are mainly involved in the processes of glucose metabolism, apoptosis and immune response. They are necessary biological processes for lymphocyte activation and proliferation, maintaining cell homeostasis and performing the main functions of cells. In order to find the relationship between peripheral and brain, the differentially expressed proteins in the substantia nigra and lymphocyte are compared with those in the substantia nigra. The expression of the same proteins is shown in Fig. 13.
Fig. 13.
Principal component analysis of differentially expressed proteins.
According to Fig. 13, the protein in component 1 is up-regulated in peripheral lymphocyte, but not in substantia nigra. The protein concentration in component 2 is up-regulated in both peripheral lymphocytes and substantia nigra, with the same trend.
4. Discussion
At present, PD has become the most common disease among the elderly in China. As a chronic disease, it is not fatal enough to cause death, but it has brought great inconvenience and difficulties to many elderly patients and families. It will bring great obstacles to people's daily activities. The impairment and decline of muscle coordination ability will cause a series of problems such as poor balance, swaying gait and so on during walking, and even cause problems such as decreased reactivity, hearing loss, finger tremor and so on. Because the early onset of PD is not obvious, it is often unable to make timely diagnosis and treatment in advance. There are many causes of PD, such as genetic problems, exposure to toxic chemicals, loss of dopamine, degradation of Lewis bodies, age and gender, which are the causes of PD. However, in the early diagnosis, it is difficult to make a timely and accurate judgment. Therefore, it is of great significance to analyze the pathophysiology of the disease and the markers for the diagnosis of the disease.
In this study, proteomic methods are used to diagnose early biomarkers of PD. By establishing a mouse model of PD, lymphocyte, striatum, substantia nigra protein and proteolysis are extracted. Then, protein imprinting experiments and 418O labeling are carried out. MS analysis technology is mainly used for the study of map proteomics, the quantitative and qualitative analysis of striatum, substantia nigra and lymphocyte differential proteins. The results showed that biomarkers of early PD were related to the same quantitative differential expression of lymphocytes, striatum, substantia nigra protein, lymphocytes, and substantia nigra. Chen et al. (2019) showed that the identification of plasma α-synuclein oligomer level combined with enhanced T2 star weighted angiography (ESWAN) had certain value in the early diagnosis of PD (Chen et al., 2019). Chen et al. (2017) suggested that the abnormal up regulation of MicroRNA-4639-5p, a PD related gene, would lead to the down regulation of DJ-1 protein level, leading to serious oxidative stress and neuronal death, so MicroRNA-4639-5p may become a peripheral diagnostic biomarker and therapeutic target for early PD (Chen et al., 2017), which is basically consistent with the results of this study. Thus, using this method to analyze and identify the biomarkers for early diagnosis of PD can better understand the pathophysiology and pathogenesis of PD, effectively help clinicians to make diagnosis in advance and timely, and contribute to the prevention and treatment of the disease.
Declaration of Competing Interest
The authors declared that there is no conflict of interest.
Footnotes
Peer review under responsibility of King Saud University.
References
- Ge W.H., Yong L., Li S. Identification of biomarkers for early diagnosis of acute myocardial infarction. J. Cell. Biochem. 2017;119(1):650. doi: 10.1002/jcb.26226. [DOI] [PubMed] [Google Scholar]
- Hatano T., Saiki S., Okuzumi A. Identification of novel biomarkers for Parkinson's disease by metabolomic technologies. J. Neurol. Neurosurg. Psychiatry. 2016;87(3):295. doi: 10.1136/jnnp-2014-309676. [DOI] [PubMed] [Google Scholar]
- Simonsen A.H., Mcguire J., Podust V.N. O1–4 identification of a novel panel of CSF biomarkers for the early diagnosis of Alzheimer's disease. Revue Neurol. 2005;161(12):59–60. [Google Scholar]
- Le W., Dong J., Li S. Can biomarkers help the early diagnosis of Parkinson’s disease. Neurosci. Bull. 2017;33(5):535–542. doi: 10.1007/s12264-017-0174-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.W. Mild cognitive impairment in Parkinson’s disease. Curr. Neurol. Neurosci. Reports. 2017;37(02):167–175. [Google Scholar]
- Pestova K., Koch A.J., Quesenberry C.P. Identification of fluorescence in situ hybridization assay markers for prediction of disease progression in prostate cancer patients on active surveillance. BMC Cancer. 2018;18(1):2. doi: 10.1186/s12885-017-3910-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- M. Lotem, S. Merims, S. Frank et al., Adjuvant autologous melanoma vaccine for macroscopic stage III disease: survival, biomarkers, and improved response to CTLA-4 blockade, J. Immunol. Res. (1) (2016) 1-12 (2016-5-18). [DOI] [PMC free article] [PubMed]
- Tissingh G., Berendse H.W., Bergmans P. Loss of olfaction in de novo and treated Parkinson's disease: possible implications for early diagnosis. Mov. Disord. 2015;16(1):41–46. doi: 10.1002/1531-8257(200101)16:1<41::aid-mds1017>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Maggio M., Ceda G.P., Nardelli A. Utilization of the DaT-SCAN SPECT in the diagnosis of Parkinson's disease in older subjects. Lett. Drug Des. Discovery. 2015:12(8). [Google Scholar]
- Chen H.L., Wang G., Ma C. An efficient hybrid kernel extreme learning machine approach for early diagnosis of Parkinson׳s disease. Neurocomputing. 2016;184:131–144. [Google Scholar]
- Chen X.Q., Niu J.P., Peng R.Q. The early diagnosis of Parkinson's disease through combined biomarkers. Acta Neurol. Scand. 2019;140(4):268–273. doi: 10.1111/ane.13140. [DOI] [PubMed] [Google Scholar]
- Chen Y., Gao C., Sun Q. MicroRNA-4639 is a regulator of DJ-1 expression and a potential early diagnostic marker for Parkinson’s disease. Front. Aging Neurosci. 2017;9:232. doi: 10.3389/fnagi.2017.00232. [DOI] [PMC free article] [PubMed] [Google Scholar]