Abstract
This study used microbial rDNA sequencing for accurate detection of systemic lupus erythematosus (SLE) skin lesions. 20 lupus erythematosus dermatology patients and 20 healthy persons were selected as experimental group and control group. Feces and serum of the subjects were sampled in sterile environment. Serum samples were examined for anti dsDNA antibodies. Fecal samples were analyzed by 16 s rDNA high-throughput sequencing, using Illumina MiSeq 2 × 250 sequencing platform. The results suggested that positive rate of anti-dsDNA antibody in serum was significantly higher in the experimental group than the control group. Significant difference of intestinal microbiome was spotted between the two groups in phylum (Firmicutes, Bacteroidetes) and genus level (Lactobacillus, Allobaculum, Lachnospira, Turicibacter, Bifidobacterium). The different intestinal microbiomes existing in healthy people and patients can provide a strong tool for accurate diagnose of lupus erythematosus.
Keywords: Microorganism, rDNA, Lupus erythematosus, Skin lesion
1. Introduction
Systemic Lupus Erythematosus (SLE) is chronic autoimmune disease, dependent on T cells and induced by immune complexes. By far, it is widely believed that SLE can be triggered by pathogenic autoantibody disorder under interaction of multiple factors such as susceptibility genes, gender, environmental stimulation and so on (Agmon-Levin et al., 2012). For instance, the occurrences of SLE among females are significantly more than males. In middle 20th century, SLE was considered to be quickly lethal due to limitation of treatments. Survival rate of patients in 5 years was only 50%. In recent 50 years, thanks to the application of hormones, immunosuppressor and biological agents, the survival rate has obviously increased. The survival rate of patients in 5 years and 10 years are 94% and 98%, respectively. However, the death rate of the patients is still 2–5 times of healthy people. Most death of SLE is caused by cardiovascular disease, infection and lupus.
The skin lesions of SLE are diverse. Therefore, it is difficult to make accurate diagnose just by observation. The main symptoms of SLE are lesions on skin and mucosa tissues. Currently, there is no academic names for lesions of SLE. In clinical research, based on manifestation, sun-exposed sore, butterfly-like rash, yin-yang sore and other lesions are named. Clinical symptoms of SLE skin and mucosa lesions are various. Discoid lesion and butterfly rash are characteristic SLE skin lesions. Lupus hair and hand erythema are secondary characteristic skin lesion of SLE. Necrotizing vasculitis, erythema multiforme and Raynaud’s phenomenon are non-characteristic SLE lesions. And zygomatic dermatitis is acute SLE skin lesions. The characteristics of SLE skin lesions are strongly correlated to the development of the disease. Hence, it is important to detect and treat SLE by recognizing the characteristics of skin lesions (Fan et al., 2019).
Recently, the combination of biology, informatics, physics, mathematics, statistics and computer network is implied to analyze biological data. High throughput assay (like gene chips or DNA microarray) has been used for studying genome expression. By screening data from gene microarray, sequence comparation, statistical analysis, cluster analysis, path analysis and visualization, these technics allow us to simultaneously analyze thousands of genes and deepen our understanding of diseases from molecular level. The whole process which called bioinformatics, firstly draw theories from available data and then validate the theories in experiments. With this methodology applied in SLE research, we can further reveal the biological processes and molecular systems of the disease, such as interactions between genes and proteins, related pathways and occurrence and dynamics of SLE, which provide more possibilities to explore mechanisms of SLE occurring, healing and relapsing. Moreover, microarray technology has been used for gene expression profiling, due to its capacity of examining the expression of thousands of genes. It was first utilized in analysis of gene expression of complicated RNA communities. Microarray chips can check thousands of genes and analyzing their expression at the same time (Schrezenmeier et al., 2019).
Nowadays, nucleic acid technology has been widely used for phylogenesis, taxonomy and bacteria identification in different territories. This technology has been more and more applied in microbiome analysis in clinical research. 16S rDNA assay is an important method for nucleic acid analysis in PCR, one of the most common technics in microbiology laboratories. 16S rDNA assay includes replacing conservative and variable regions and effectively identifies microorganisms based on variable regions between different primers (Wu et al., 2019). The application of microorganism rDNA sequencing in SLE test may improve the accuracy of diagnose of SLE skin lesions. Therefore, this research combined 16S rDNA high throughput sequencing and statistical analysis to show that whether microbial rDNA assay can make more accurate diagnose of SLE skin lesion.
2. Methods and materials
2.1. Experiment subjects and groups
According to the 1997 ARA modified criteria for SLE diagnose, a SLE patient should meet at least 4 out of 11 criteria after the possibilities of external infections, tumors or other diseases in connective tissues are eliminated. For example, SLE can often be confused with: (1) rheumatoid arthritis; (2) polymyositis or dermatomyositis; (3) tuberous polyarteritis; (4) mixed connective tissue disease; (5) others, such as systemic sclerosis, rheumatic fever, leukaemia, serum disease, primary hemolytic anemia, primary thrombocytopenic purpura, Hodgkin's disease, primary nephrotic syndrome, chronic urticaria, etc.
Besides, factors mentioned in Yue’s (2019) paper that may affect the experimental outcomes were also excluded, along with the following factors: 1. Females in pregnancy and lactation; 2. Patients with neuropsychic symptoms caused by infections, electrolyte disorder, metabolism, uremia and drugs; 3. Patients with past history of primary cerebral diseases, like stroke, aneurysm, cerebral vasculitis and Vascular Dementia (VD); 4. Patients with brain injury history, diabetes, hypertensive encephalopathy and anti-NMDA receptor antibody encephalitis; 5. Patients intaking aspirin for the last six months; 6. Patient used to have hyperthyroidism; 7, Patient used to have syphilis (Zhang et al., 2019, Yue, 2019).
The data used in this research came from multiple databases and practical surveys. Along with searching and referring to GEO PubMed databases of NCBI, EXALT database and 2019 After-healing Long-term SLE Tracking Database in Jiangsu Province, we gathered primary data by independent research. Based on the selecting criteria mentioned above, 20 patients from the database of Jiangsu Province were selected as experimental group, including 18 females and 2 males (Li et al., 2018). The average age of the patients was 31.52 ± 8.76. Average duration of SLE was 4.85 years, with longest duration of 6.97 and shortest duration of two months. 20 healthy people were selected as control group. All subjects had not taken any immunosuppressor, nor had other skin lesions, cardio-cerebrovascular diseases, renal or hepatic diseases or related infections. Subjects without complete experimental data or records were also excluded. This experiment was approved by the ethics committee in Jiangsu Province and corresponding informed consent sheet was signed by all the patients.
2.2. Instruments and reagents
Reagents and instruments used in this research include: enzyme-substrate chromogenic reagent from SeebioR (Shanghai); Marker antibody from ABIOCENTER; Total fecal DNA extraction kit, BeaverBeads, from Xiyan Instrument; Antigen dsDNA from New England Biolab Inc.; Illumina MiSeq 2 × 250 sequencing platform provided by Novogene (Tianjing).
2.3. Sampling
The serum and feces of the subjects were sampled in sterile environment. Samples were reserved in centrifuge tubes in −70 °C.
2.4. DNA extraction and sequencing
Total microbiome DNA from fecal samples was extracted by the extraction kit and the concentration and purity of DNA were checked with agarose electrophoresis. Then the extracted DNA was diluted to 1.5 ng/μL as template. High-variable region V3-V4 of 16S rRNA was amplified by PCR. The qualified sequences from PCR were mixed to establish DNA library (López Suárez et al., 2016, Katz-Agranov and Zandman-Goddard, 2017, Mu et al., 2015). After checking the quality of the DNA library, 16S rDNA sequencing was performed by Illumina MiSeq 2 × 250 platform.
2.5. Data analysis
Primary sequences were excluded by base removal. Validated sequences were obtained after the elimination of chimera sequences. Based on the similarity of the validated sequences, microbial communities in each sample were classified. The sequences with similarity over 96% were gathered as one operational taxonomic unit (OTU) (Agmon-Levin et al., 2012). According to OTUs, bioinformatical analysis were implemented to reveal microbial communities, such as species identification, cluster analysis, component difference assay and diversity analysis.
2.6. Statistical analysis
In past studies, statistical analysis software 19.0 SPSS was used, the results was shown by x ± s. Correlation analysis was performed after Q-Q normal distribution detection, and P < 0.05 was considered statistically significant. After Q-Q analysis indicating that the data did not conform to normal distribution. Kendall and Spearman correlation analysis was conducted. Different from the existing literature, the data and calculation of Shannon were processed with QIIME 1.8.0 Software. Difference between groups of Beta and Alpha diversity of UniFrac and Simpson index were analysis by R studio. The data from serum was analyzed by GraphPad Prism 6 Software. The significance of difference was showed by t-test. The difference between group was considered significant when P < 0.05.
3. Results
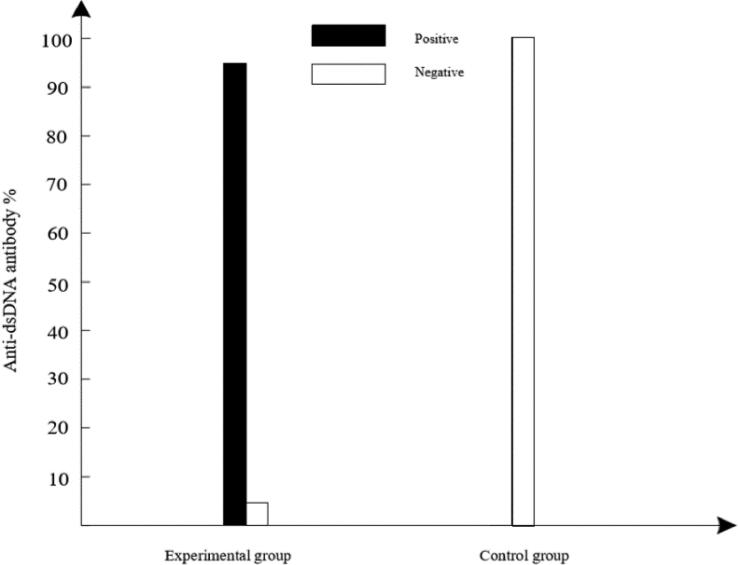
The level of anti-dsDNA antibody in serum of experimental and control groups was showed in Fig. 1.
Fig. 1.
Anti-dsDNA antibody level in experimental and control groups.
Fig. 1 illustrates that there was significant difference between experimental group and control group (p < 0.0001). In experimental group, dsDNA-positive ratio was 95% while dsDNA-negative ratio was only 5%. In control group, dsDNA- negative ratio of all subjects was 100%. These results suggested that anti-dsDNA antibody level can effectively detect SLE.
After sequencing of the fecal samples from experimental and control groups. Sequences with high similarity (>97%) were gathered as one OUT and microbial abundance in fecal samples was analyzed. The results suggested that microbial community in fecal samples mostly consisted of Bacteroidetes, Firmicutes, Actinobacteria and Proteobacteria. The dominating phyla were Bacteroidetes and Firmicutes.
The abundance of different taxonomic levels was obtained to further reveal the content in each sample. First ten abundant organisms on phylum and genus level were summarized. Table 1 shows the relative abundance on phylum level in different groups.
Table 1.
Relative abundance on phylum level in different groups.
| Phylum | Experimental group (%) | Control group (%) |
|---|---|---|
| Firmicutes | 57.37 | 48.82 |
| Bacteroidetes | 34.54 | 43.87 |
| Actinobacteria | 5.06 | 4.92 |
| Proteobacteria | 1.48 | 1.16 |
| Acidobacteria | 0.47 | 0.43 |
| Verrucomicrobia | 0.43 | 0.18 |
| Chloroflexi | 0.18 | 0.12 |
| Helotiales | 0.16 | 0.12 |
| Cyanobacteria | 0.11 | 0.08 |
| Deferribacteres | 0.04 | 0.01 |
Table 1 suggests that among intestinal microbiome, abundance of Firmicutes was obviously higher while abundance of Bacteroidetes was lower in experimental group than control group. These results indicted that there was significant difference of abundance of dominating phyla in two groups and the difference of Firmicutes and Bacteroidetes was strongly correlated with SLE skin lesion.
Table 2 shows the relative abundance on genus level in different groups.
Table 2.
Relative abundance on genus level in different groups.
| Genus | Experimental group (%) | Control group (%) |
|---|---|---|
| Lactobacillus | 16.85 | 11.23 |
| Allobaculum | 12.32 | 20.13 |
| Lachnospiraceae | 5.91 | 3.06 |
| Turicibacter | 5.91 | 3.49 |
| Bifidobacterium | 5.13 | 4.98 |
| Roseburia | 2.03 | 1.02 |
| Prevotella | 1.43 | 0.16 |
| Alistipes | 1.34 | 1.71 |
| Parabacteroides | 1.26 | 0.63 |
| Alloprevotella | 0.35 | 0.93 |
The results showed in Table 2 suggested that in experimental group, species with high abundance were Lactobacillus (16.85%), Allobaculum (12.32%), Lachnospiraceae (5.91%), Turicibacter (5.91%) and Bifidobacterium (5.13%). In control group, the dominating genera were Allobaculum (20.13%), Lactobacillus (11.23%), Bifidobacterium (4.98%), Turicibacter (3.49%) and Lachnospiraceae (3.06%). The component and ratio of dominating genera were significantly different in two groups and therefore can be utilized as reference for diagnose of SLE skin lesion.
4. Discussion
Environmental and genetic factors greatly affect the occurrence of SLE. Age, race and ambient viruses can also correlate to the development of SLE. Intestinal microbiome is “the second genome” of human. The disturbance of intestinal microorganisms can directly influence immune system, especially inflammatory diseases and autoimmune diseases (Li et al., 2019). Therefore, although genetic factors are proved to be vital to the occurrence of SLE, environmental factors are also highly correlated to SLE. Previous studies speculated that intestinal microbes probably relate to anti-dsDNA antibody level as an environmental factor in TC mice in SLE model. Relative reports suggested that inducements of SLE included usage of antibiotics, antiparasitic drugs and drugs for gastrointestinal diseases, which is the evidence of correlation of intestinal microbes and SLE.
More and more evidences suggest that gastrointestinal microbes affect the process of autoimmune diseases in model rodents, possibly partly through mediation of intestinal microbes. Unsaturated fatty acid, vitamin A, vitamin D, vitamin E and plant estrogen promote lupus condition in SLE model animals, mostly reducing proteinuria and glomerulonephritis (Gao et al., 2017). Moreover, Reifen et al. pointed out that n-3 polyunsaturated fatty acids could prevent fetus loss and other clinical symptoms caused by antiphospholipid syndrome (APS). Similar research showed diet influenced SLE and corresponding APS in great extent, probably by regulating the structure of intestinal microbial communities (Ye, 2014).
The coevolution of host and internal microbes shapes the important dependence between human and microorganisms. Millions of microorganisms living inside our bodies and most of them are not pathogenic. These microbes exist on the surface of mucosa barrier of skin, gastrointestinal tract, genital tract and respiratory tract and reach highest density in downstream of gastrointestinal tract. Every part of mucosa and skin directly exposed to the environment has its unique and complicated combination of microbial species, which is called microbial community. Intestine has most abundant and diverse microbial communities which function in metabolism and regulation of nutrient and immunity. The subtle balance of intestinal microbial communities is the key to maintain intestinal immunity and homeostasis of body. Any disturbance on the balance may cause severe pathological and physiological consequences. Hence, the disturbance on intestinal microbial communities is also called microbial dysbiosis. Microbial dysbiosis is correlated to many autoimmune and chronic inflammatory diseases, such as rheumatoid arthritis, type 1 diabetes, inflammatory intestinal disease and SLE. Due to the direct interaction between intestinal microbes and mucosa, microbial communities may influence permeability of mucosa of intestine, and further participate in partial or general immune inflammation (Cox et al., 2015).
The study of microbial communities on phylum and genus level indicates great difference of intestinal microbial composition in healthy people and patients with various phenotypes of SLE. The structure of intestinal microbial communities is strongly correlated with anti-dsDNA antibody level which can accurately detect SLE. Regulation of intestinal microbial communities in the patients by diet and probiotics may regain the balance of intestinal microbes and cure SLE.
Bacteroidetes and Firmicutes are the two dominating phyla in intestinal microbial communities, accounting for 90% of the total microbes in intestine. During clinical practice, the ratio of the two phyla, F/B ratio, is an effective criterion to determine the balance of intestinal microbial communities. Relative research showed F/B ratio in SLE patients is lower than which in healthy people. Our study confirmed this conclusion.
Some studies suggested that severity of SLE may be affected by increased Lachnospiraceae and consumed Lactobacillus at certain extent. Their results showed that Lachnospiraceae was significantly higher in SLE patients than in healthy people. There was significant discrepancy of Proteobacteria between two groups of people. Although most of Proteobacteria are pathogenic and their content in intestinal microbial communities is low, the discrepancy suggested that Proteobacteria directly affect the outcome of SLE test. These results may provide strong tool for the diagnose of SLE.
5. Conclusion
SLE is a spectrum disease. Without timely diagnose and treatment, it may cause multiple systematic damage to our bodies, even complications like renal failure. Many researchers have proved the direct relationship of intestinal microbes and SLE. The result of 16S rDNA high throughput sequencing of fecal samples indicated huge difference of intestinal microbial communities on phylum and genus level between patient and healthy people. The results suggested that positive rate of anti-dsDNA antibody in serum was significantly higher in the experimental group than the control group. Significant difference of intestinal microbiome was spotted between the two groups in phylum (Firmicutes, Bacteroidetes) and genus level (Lactobacillus, Allobaculum, Lachnospira, Turicibacter, Bifidobacterium). The different intestinal microbiomes existing in healthy people and patients can provide a strong tool for accurate diagnose of lupus erythematosus. Our result can provide helpful evidence for accurate diagnose of SLE.
COVID-19 have caused tremendous pressure on the current world health, there is a lack of research in this paper, the relevant research of past studies have found that SLE patients are more likely to suffer from severe pneumonia (SP), but the susceptible factors is not clear, while past research has suggested that lymphocytes reduce disease activity to reduce the use of the hormone and immune inhibitors complement viscera damage is the risk factor of lupus co-infection, however there is no related research focus on the risk factors of severe pneumonia in lupus. Our team will apply the conclusions of this paper in the future to increase research in this area.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Peer review under responsibility of King Saud University.
References
- Agmon-Levin N., Mosca M., Petri M., Shoenfeld Y. Systemic lupus erythematosus one disease or many? Autoimmun. Rev. 2012;11(8):593–595. doi: 10.1016/j.autrev.2011.10.020. [DOI] [PubMed] [Google Scholar]
- Cox A.J., West N.P., Cripps A.W. Obesity, inflammation, and the gut microbiota. The Lancet Diabetes Endocrinol. 2015;3(3):207–215. doi: 10.1016/S2213-8587(14)70134-2. [DOI] [PubMed] [Google Scholar]
- Fan Z., Xue M., Jin L. Significance of elevated peripheral blood il-17 +Foxp3+ regulatory T cells in patients with active systemic lupus erythematosus. Chin. J. Microbiol. Immunol. 2019;39(3):174–179. [Google Scholar]
- Gao W., Wang Y., Wang W., Shi L. The first multiplication atom-bond connectivity index of molecular structures in drugs. Saudi Pharmaceut. J. 2017;25(4):548–555. doi: 10.1016/j.jsps.2017.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz-Agranov N., Zandman-Goddard G. The microbiome and systemic lupus erythematosus. Immunol. Res. 2017;65(2):432–437. doi: 10.1007/s12026-017-8906-2. [DOI] [PubMed] [Google Scholar]
- Li W., Jia M., Deng J., Wang J., Lin Q., Liu C., Wang S., Tang J., Zeng X., Ma L., Su W., Liu X. Isolation, genetic identification and degradation characteristics of COD-degrading bacterial strain in slaughter wastewater. Saudi J. Biol. Sci. 2018;12(25):1800–1805. doi: 10.1016/j.sjbs.2018.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Jia M., Wang J., Lu J., Deng J., Tang J. Association of MMP9-1562C/T and MMP13-77A/G polymorphisms with non-small cell lung cancer in Southern Chinese population. Biomolecules. 2019;9(3):107. doi: 10.3390/biom9030107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López Suárez, P., Paz Cazón, B. D., Rodríguez Carrio, J., Hevia, A., Sánchez García, B., Margolles Barros, A., & Suárez, A. (2016). Th17 responses and natural IgM antibodies are related to gut microbiota composition in systemic lupus erythematosus patients. Scientific Reports. [DOI] [PMC free article] [PubMed]
- Mu Q., Zhang H., Luo X.M. SLE: Another autoimmune disorder influenced by microbes and diet? Front. Immunol. 2015;6:608. doi: 10.3389/fimmu.2015.00608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrezenmeier E., Weißenberg S.Y., Stefanski A.L., Szelinski F., Wiedemann A., Lino A.C., Dörner T. Postactivated B cells in systemic lupus erythematosus: Update on translational aspects and therapeutic considerations. Curr. Opin. Rheumatol. 2019;31(2):175–184. doi: 10.1097/BOR.0000000000000576. [DOI] [PubMed] [Google Scholar]
- Wu J., Wei W., Zhang L., Wang J., Ius R.D.E., Li J., Wang H., Wang G., Zhang X., Yuan J., Niak M.W. Risk assessment of hypertension in steel workers based on LVQ and Fisher-SVM deep excavation. IEEE Access. 2019;7(1):23109–23119. [Google Scholar]
- Ye D.Q. Environmental factors in the pathogenesis of systemic lupus erythematosus and their interactions with several chemokine genes. Chin. J. Epidemiol. 2014;11:35–39. [Google Scholar]
- Yue R. Southern Medical University; 2019. Analysis of cognitive dysfunction and its related factors in patients with systemic lupus erythematosus. [Google Scholar]
- Zhang S., Tang Y., Liang J. Curcumin reverses the impaired phagocytic function induced by exosomes in patients with systemic lupus erythematosus. Chin. J. Dermatol. 2019;52(6):378–382. [Google Scholar]
Further Reading
- Falagas M.E., Manta K.G., Betsi G.I., Pappas G. Infection-related morbidity and mortality in patients with connective tissue diseases: a systematic review. Clin. Rheumatol. 2007;26(5):663–670. doi: 10.1007/s10067-006-0441-9. [DOI] [PubMed] [Google Scholar]