Abstract
Plasmopara viticola is the causal agent of grapevine downy mildew (DM). DM resistant varieties deploy effector-triggered immunity (ETI) to inhibit pathogen growth, which is activated by major resistance loci, the most common of which are Rpv3 and Rpv12. We previously showed that a quick metabolome response lies behind the ETI conferred by Rpv3 TIR-NB-LRR genes. Here we used a grape variety operating Rpv12-mediated ETI, which is conferred by an independent locus containing CC-NB-LRR genes, to investigate the defence response using GC/MS, UPLC, UHPLC and RNA-Seq analyses. Eighty-eight metabolites showed significantly different concentration and 432 genes showed differential expression between inoculated resistant leaves and controls. Most metabolite changes in sugars, fatty acids and phenols were similar in timing and direction to those observed in Rpv3-mediated ETI but some of them were stronger or more persistent. Activators, elicitors and signal transducers for the formation of reactive oxygen species were early observed in samples undergoing Rpv12-mediated ETI and were paralleled and followed by the upregulation of genes belonging to ontology categories associated with salicylic acid signalling, signal transduction, WRKY transcription factors and synthesis of PR-1, PR-2, PR-5 pathogenesis-related proteins.
Subject terms: Genetics, Plant sciences
Introduction
Downy mildew is one of the most destructive diseases of the grapevine, causing significant limitations on grape production in the absence of chemical protection of vineyards. Downy mildew is caused by the biotrophic oomycete Plasmopara viticola (Berk. And Curt) Berl. and Toni, which is native to North America and was introduced into Europe at the end of the nineteenth century. The European bunch grape (Vitis vinifera L.) does not normally activate the immune system in response to P. viticola with a few exceptions1,2. The introgression of resistant genes from other grape species (i.e. V. rupestris, V. amurensis, V. cinerea, V. riparia and Muscadinia rotundifolia) into the crop germplasm can alleviate the dependence of viticulture on the use of fungicides3.
The pathogen can cause serious damage to any green organ of the grapevine. P. viticola deploys a specialized structure called haustorium to establish a close interaction with grapevines and to leak nutrients from viable host cells4,5. P. viticola can feed on both susceptible and resistant grapevines and complete its life cycle on both hosts. Resistant grapevines are, however, able to detect the invading pathogen and operate defence responses. The initial phases of the infection are similar on both hosts, but mycelial growth is restricted in resistant hosts and sporangia are released at lower rates than in susceptible hosts. The similarity during early phases of infection suggests the presence of post-infection mechanisms of resistance, including callose deposition, cell wall-associated defence processes, accumulation of reactive oxygen species and hypersensitive response (HR) associated with necrosis, and production of phytoalexins and antimicrobial compounds6–12. These responses are paralleled by the up-regulation of genes coding for pathogenesis-related (PR) proteins13. The study of the defence response in resistant grape varieties can improve our understanding of the resistance mechanisms and shed light on the metabolic pathways involved. It is commonly believed that ETI requires a massive reallocation of energy and materials towards a multitude of actions against pathogen colonisation14,15, which reduce fitness in annuals16,17 and might be energetically costly in perennial crops grown under limiting conditions18,19.
We recently performed metabolomic analyses after pathogen inoculation in the grape variety ‘Bianca’ that carries a major resistance haplotype called Rpv3-1, leading to the identification of 53 metabolites associated with the defence response20. We decided to extend this investigation using another resistant grapevine that has a genetic background highly similar to ‘Bianca’. This grapevine shares a complete haploid complement with its parent ‘Bianca’ but not the resistance haplotype Rpv3-1 at the Rpv3 locus on chromosome 18 and it carries instead a different resistance gene—Rpv12—that is located on chromosome 14. We first identified a suitable genotype for this kind of experiment in an accession obtained in 2000 by P. Kozma and S. Hoffmann in Hungary from a cross between ‘Bianca’ and the breeding line SK77-4/5 and held in a germplasm repository in South Tyrol (Italy). The resistance haplotype at the Rpv12 locus was initially introgressed from the Asian species Vitis amurensis and confers higher levels of downy mildew resistance than Rpv3-1 when the resistance phenotype is quantitatively scored using the OIV452 descriptor21, but both genes trigger a similar ETI-dependent response12,21. The donor parent of the Rpv12 resistance haplotype, SK77-4/5, was bred at the University of Novi Sad, Serbia22 by crossing V. vinifera ‘Traminer’ and the resistant variety ‘Kunbarat’, which descended from the interspecific hybridization of V. amurensis and V. vinifera. In our previous study in ‘Bianca’20, we focused on Rpv3-mediated metabolome changes upon pathogen inoculation with the aim to identify biomarkers of resistance. Recently, Eisenmann and coworkers23 performed a transcriptomic analysis in ‘Regent’, a resistant variety that operates the same Rpv3-mediated ETI response as does ‘Bianca’, showing that Rpv3-mediated changes in gene expression occur early after pathogen inoculation.
Technological developments in genomics, transcriptomics, proteomics and metabolomics have provided powerful tools for systems biology studies. Metabolomics is widely used for monitoring changes in response to the developmental program, to the environment and to biotic stresses24. In grapevine, metabolomics has been used to investigate several aspects of berry development and berry ripening25–27 or to study the effects of abiotic stresses28–30. More recent studies using grape leaves that had been inoculated with P. viticola have used metabolomic analyses to monitor specific classes of compounds or a limited number of molecules31–33. In our previous work20, we extended the number of compounds that are identified and quantified in grape leaves using a single analytical run. Transcriptomics refers to the global analysis of all transcribed genes in a certain organ. Transcriptomic analyses based on RNA-Sequencing (RNA-Seq) provide a comprehensive survey of gene expression changes occurring during the response to pathogen infections. Transcriptomic analyses have been performed on grape leaves undergoing defence responses to P. viticola in native American species13 and in V. vinifera introgression lines carrying Rpv3-1, Rpv3-3, or Rpv10 resistance genes23,34,35. Using a single-omics approach it is possible to clarify only one aspect of cell metabolism. Multi-omics analyses offer a unifying view of the biological processes and have been applied to fruit crop species36 for inferring relationships between genes, transcripts, metabolites and phenotypes. Recent work has been done in grapevine by applying a multi-omics integrated approach to study drought stress, berry developmental changes, light exclusion, basal immunity and responses to Erysiphe necator infection25,26,30,37,38. Here we present two integrated omics data of Rpv12-mediated ETI responses to P. viticola in grapevine leaves and compare the results with our previous data20 and other literature reports23,31 in which the same two-omics were used separately for studying Rpv3-mediated responses.
Results
Impact of the defence response on leaf metabolome
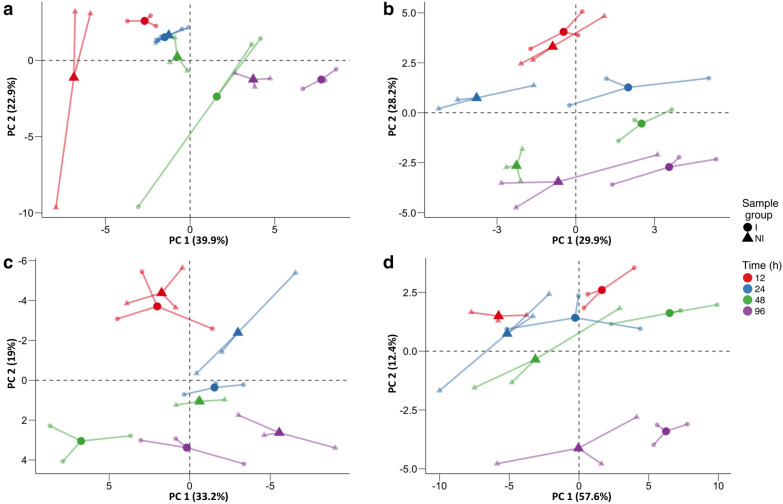
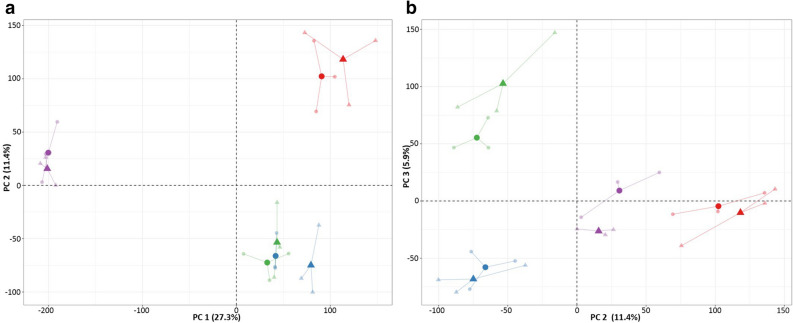
We identified and quantified 175 metabolites in inoculated and control leaves. A complete list of these compounds and their concentration is reported in Supplementary Table S1, sorted into four classes of primary metabolites, lipids, phenols, and volatile compounds. Figure 1 illustrates a lower-dimensional picture of the distribution of samples undergoing Rpv12-mediated ETI responses and controls during the course of the experiment using a Principal Component Analysis (PCA). The first two components explained 62.8, 58.1, 52.2, and 70.0% of the observed variation for primary metabolites, lipids, phenols, and volatile compounds, respectively. The first component (PC1) revealed a separation between inoculated samples and controls at 12 and 96 h post infection (hpi) for primary metabolites and volatile compounds (Fig. 1a,d), at 24 and 48 hpi for lipids (Fig. 1b), and at 48 and 96 hpi for phenols (Fig. 1d). The second component (PC2) separated the samples according to the stage of incubation.
Figure 1.
PCA of metabolite data in leaves undergoing Rpv12-mediated defence (inoculated black filled circle) and controls (black filled triangle). Three biological replicates are reported as small symbols (black filled circle, black filled triangle) and are linked to their mean values (large symbols, black filled circle, black filled triangle) by connectors (–). (a) Primary metabolites. (b) Lipids. (c) Phenols. (d) Volatile compounds. Different colours represent different time points: red filled circle, red filled triangle—12 hpi; blue filled circle, blue filled triangle—24 hpi; green filled circle, green filled triangle—48 hpi; violet filled circle, violet filled triangle—96 hpi.
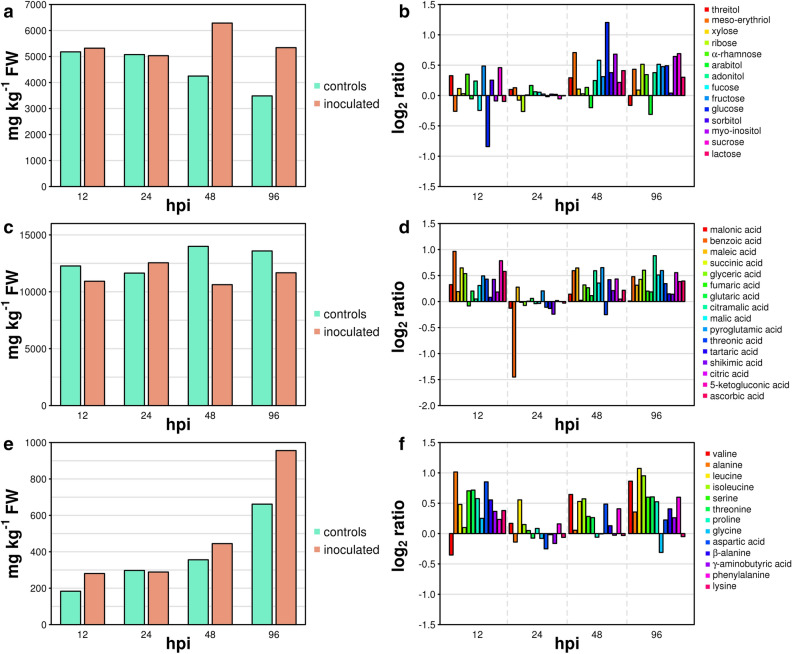
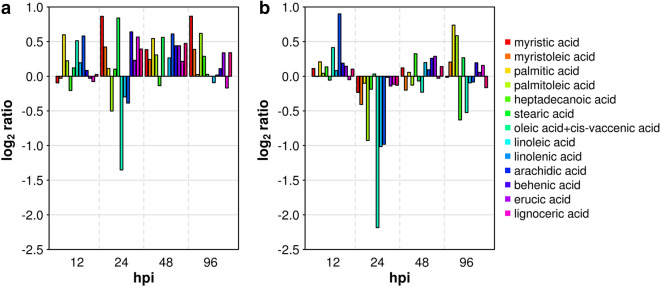
With regard to primary metabolism, we observed two distinct temporal patterns of activation. A rapid and transient increase in the concentration of some sugars and most of organic and amino acids was detected at 12 hpi (Fig. 2). A delayed and more sustained increase of almost all sugars, organic acids and amino acids occurred from 48 to 96 hpi (Fig. 2). We observed the least differences in primary metabolites between inoculated samples and controls at the stage of 24 hpi with the notable exception of benzoic acid that was greatly reduced in samples undergoing Rpv12-mediated defence (Fig. 2d). Benzoic acid is the precursor of salicylic acid (SA) in the phenylalanine ammonia-lyase (PAL) pathway39. We therefore assume that the benzoic acid pool is depleted at 24 hpi by a rapid conversion of this substrate to SA. The concentration of fatty acids also increased in samples undergoing Rpv12-mediated defence (Fig. 3a). Linolenic acid (18:3) was among the fatty acids with elevated concentration in samples undergoing Rpv12-mediated defence early at 12 hpi compared to controls (Fig. 3a) and this behaviour was in common with samples undergoing Rpv3-mediated defence (Fig. 3b) and with literature reports40. Linolenic acid is reported to activate NADPH–oxidase—an enzyme responsible for the production of reactive oxygen species (ROS)—and is important for the modulation of the strength of the HR41 if not responsible itself for the onset of the oxidative burst in programmed cell death (PCD)42. In a context of an elevated lipid metabolism (Fig. 3a), we observed a marked decrease in palmitoleic acic (16:1), linoleic acid (18:2), linolenic acid (18:3), and especially in oleic and vaccenic acids (18:1) at the stage of 24 hpi. It is noteworthy that the concentration of the same five fatty acids showed the most remarkable decreases in samples undergoing Rpv3-mediated defence at the same stage of incubation (Fig. 3b). Low levels of 18:1 fatty acids in Arabidopsis mutants, which are a consequence of an inactivating mutation in the SSI2 gene encoding an isoform of stearoyl-desaturase for the conversion of stearic acid (18:0) to oleic acid (18:1), cause constitutive SA-signalling and expression of PR proteins43. We also observed a general increase in the concentration of volatile compounds in samples undergoing Rpv12-mediated defence (Supplementary Table S1) and a monotonic pattern of increased levels of all terpenoids at all stages of incubation (Supplementary Fig. S1). We detected a clear and consistent pattern of increasingly higher levels of benzaldehyde in samples undergoing the Rpv12-mediated defence compared to controls during the entire course of the defence response (Fig. 4). We previously proposed benzaldehyde, a promoter of SA-mediated defence44, as a biomarker for the defence response in Rpv3-mediated resistance because this compound markedly increased at 48 and 96 hpi20. In Rpv12-mediated ETI, the rapid accumulation of this biomarker from 12 to 96 hpi suggests a more prompt and prolonged activation of the defence response compared to Rpv3-mediated ETI.
Figure 2.
Major classes of primary metabolites in leaves undergoing Rpv12-mediated defence (inoculated) and controls. Cumulative concentration of (a) sugars, (c) organic and (e) amino acids. (b,d,f) Report the log2 ratio of the concentration of each compound in inoculated leaves versus controls.
Figure 3.
Differences in fatty acids between leaves undergoing (a) Rpv12- or (b) Rpv3-mediated defence. On the y-axis the log2 ratio of the concentration of each compound in inoculated leaves versus controls is shown. Graph in (b) was plotted using raw data obtained from our previous paper20.
Figure 4.
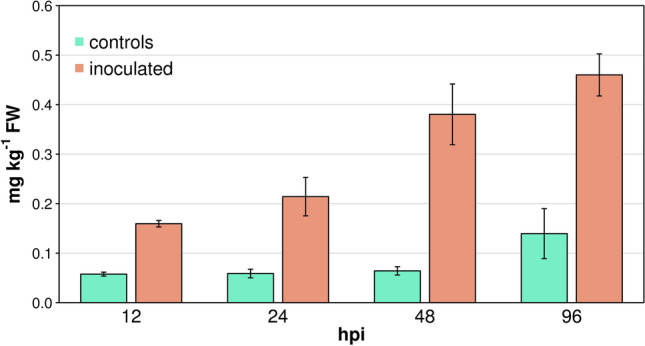
Concentration of benzaldehyde in leaves undergoing Rpv12-mediated defence (inoculated) and controls during the course of infection. Bars represent ± standard errors.
In order to identify the metabolites that better explain the differences between inoculated samples and controls, we selected 88 metabolites that showed an absolute value of the t statistic greater than 3 (|t|> 3) in one time point at least (Supplementary Fig. S2a, Supplementary Table S2). We summarized this information in the network represented in Supplementary Fig. S2b. We identified the largest number of nodes in the network at 48 hpi, indicating that the effects of metabolic reprogramming in samples undergoing Rpv12-mediated defence were maximal between 24 and 48 hpi and these changes involved mainly secondary metabolites.
Key metabolite changes in secondary metabolism
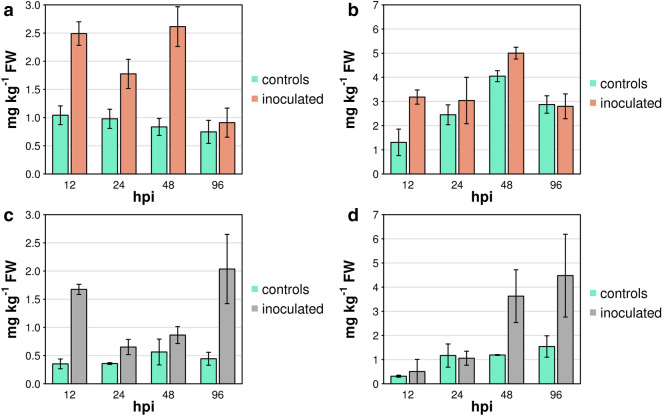
It has been previously shown that Rpv3-mediated defence is characterised by the two following early events: PCD occurs in the mesophyll cells underneath the infected stomata and non-toxic stilbenes, mainly trans-resveratrol, accumulate in leaves undergoing PCD. Elevated trans-resveratrol synthesis and PCD are operated between 12 and 24 hpi and their outcomes become effective, visible, and measurable between 24 and 48 hpi12,20,23. The conversion of non–toxic trans-resveratrol into fungi–toxic derivatives, mainly ε-viniferin, reached a peak between 48 and 96 hpi20,23. In the current experiment, the amount of trans-resveratrol was consistently two to threefold higher from 12 to 48 hpi in leaves undergoing Rpv12-mediated resistance than in controls (Fig. 5a), whereas this increase occurred more transiently from 12 to 24 hpi in our previous experiment with Rpv3-mediated resistance (Fig. 5c). As a result of this, the concentration of trans-resveratrol stayed within an higher range of variation (between 1.8 and 3.1 mg kg−1 of fresh weight (FW)) during the first 48 hpi in leaves undergoing Rpv12-mediated resistance, whereas the concentration remained within a lower range (between 0.7 and 1.7 mg kg−1 of FW) in Rpv3-mediated resistance20. This strong increase of trans-resveratrol in Rpv12-mediated resistance compared to controls was not paralleled or followed by a commensurate increase in ε-viniferin, which originates from oxidative dimerization of trans-resveratrol. The concentration of ε-viniferin was higher in inoculated samples undergoing Rpv12-mediated resistance than in controls only at 12 hpi (Fig. 5b), whereas it markedly increased at 48 and 96 hpi in Rpv3-mediated resistance (Fig. 5d).
Figure 5.
Concentration of trans-resveratrol (a,c) and ε-viniferin (b,d) in samples undergoing either (a,b) Rpv12- or (c,d) Rpv3-mediated defence response (inoculated) and controls. Graphs in (c,d) were plotted using raw data obtained from our previous paper20.
Impact of the defence response on the leaf transcriptome and global changes in metabolic pathways
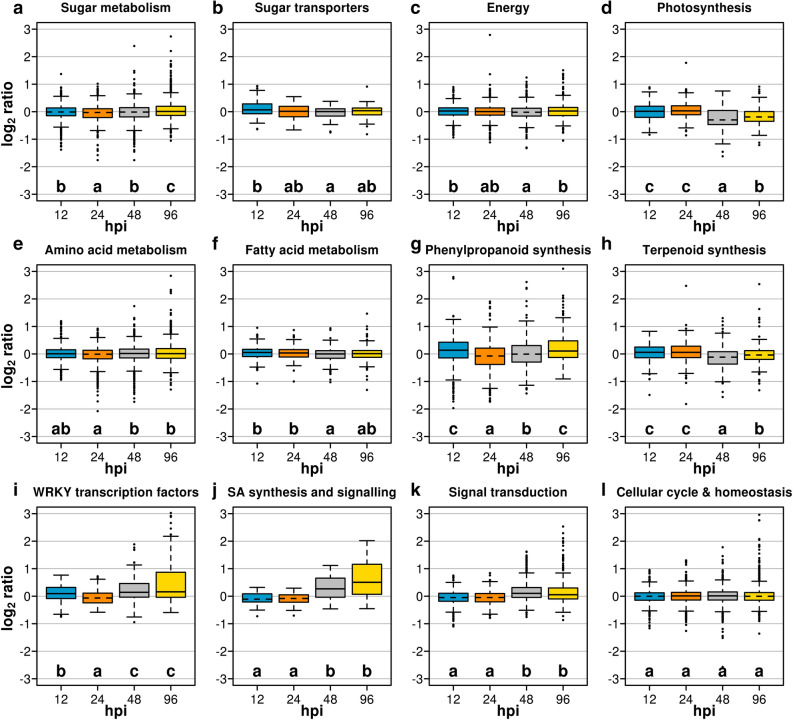
Figure 6 illustrates a lower-dimensional picture of the distribution of samples undergoing Rpv12-mediated defence response and controls during the course of the experiment based on a PCA using gene expression values for all transcribed genes. Replicates of samples undergoing Rpv12-mediated defence responses at 24 hpi grouped separately from replicated controls along the first component (PC1, Fig. 6a). Replicates of samples undergoing Rpv12-mediated defence responses grouped separately from replicated controls at 48 and 96 hpi along the third component (PC3, Fig. 6b). Figure 7 illustrates the distributions of FPKM log2 ratios between samples undergoing Rpv12-mediated ETI and controls at each stage of incubation for 12 groups of genes sorted by gene ontology (GO) category and/or metabolic pathway. Genes that are involved in sugar metabolism and generation of energy showed small global shifts in expression between samples undergoing Rpv12-mediated ETI and controls. The distributions of FPKM log2 ratios for sugar metabolism genes (Fig. 7a) were shifted towards negative median values from 12 to 48 hpi and were significantly shifted towards higher values and with a positive median value only at 96 hpi, suggesting that sugar metabolism pathways were slightly modulated at early stages in samples undergoing Rpv12-mediated ETI response. However, categories including genes that encode sugar transporters (Fig. 7b) and are involved in fatty acid metabolism (Fig. 7f) showed the most remarkable global shift in FPKM log2 ratios towards higher and positive values at 12 and 24 hpi. This is consistent with the notion that the activation of sugar signaling and fatty acid modulators of signal transduction pathways relies on mobilization and deployment of existing resources at the initial stages of the ETI response. These metabolic activities were sustained by the activation of pathways involved in generation of energy as we observed slight global shifts of distributions of FPKM log2 ratios for genes assigned to energy-related gene categories towards positive median values at 12, 24, and 96 hpi (Fig. 7c). Although the ETI response seems to require substantial amounts of carbon and energy, the global expression of genes involved in photosynthesis (Fig. 7d) suggested that these resources were not contributed by an activation of the photosynthetic pathway that was on the contrary strongly reduced at 48 and 96 hpi. We observed in samples undergoing Rpv12-mediated ETI two distinct temporal patterns of elevated expression of the gene family encoding WRKY transcription factors (Fig. 7i). A rapid and transient increase was detected at 12 hpi and was followed by a more sustained increase from 48 to 96 hpi. WRKY transcription factors are known to activate defence response in a number of ways through the up-regulation of genes involved in the phenylpropanoid pathway and in SA synthesis and signalling. Indeed, we found that structural genes of the phenylpropanoid and downstream pathways—including those required for stilbene synthesis—showed global shifts in the distributions of FPKM log2 ratios towards positive median values at 12 and 96 hpi (Fig. 7g). This observation recalls the notion that compounds in this class activate early signalling pathways and, at later stages of the incompatible interaction, sustain the production of stilbenes. We also observed statistically significant shifts towards higher FPKM log2 ratios with positive median values in the distribution for genes involved in SA synthesis and signalling at 48 and 96 hpi (Fig. 7j), concomitant with similar increases in the expression of genes involved in calcium and mitogen activated protein (MAP) kinase signalling (Fig. 7k). We finally used the set of genes assigned to the GO categories of cell cycle and cell homeostasis that are not expected to be recruited in defence responses as a negative control for this analysis. We indeed observed that these GO categories showed no statistically significant difference in their global distributions of FPKM log2 ratios between stages and their median log2 ratios pointed to zero at each stage of incubation (Fig. 7l).
Figure 6.
PCA of transcript data in leaves undergoing Rpv12-mediated defence (inoculated black filled circle) and controls (black filled triangle). Three biological replicates are reported as small symbols (black filled circle, black filled triangle) and are linked to their mean values (large symbols, black filled circle, black filled triangle) by connectors (–) for the first and second component in (a) and the second and third component in (b). Different colours represent different time points: red filled circle, red filled triangle 12 hpi; blue filled circle, blue filled triangle 24 hpi; green filled circle, green filled triangle 48 hpi; violet filled circle, violet filled triangle 96 hpi.
Figure 7.
Global differences of gene expression in leaves undergoing Rpv12-mediated defence (inoculated) and controls. Genes were grouped by metabolic pathways (a–l). Box–plot distributions of FPKM log2 ratios at four stages of incubation. Each letter identifies distributions that are not significantly different using a Wilcoxon–Mann–Whitney test (P < 0.05). Alphabetical order indicates increasing median values. The line pointing to the median value in each box is plotted as a solid line for positive median values and as a dotted line for negative median values.
Statistically significant and major differences in individual gene expression
We then defined a set of differentially expressed genes (DEGs) between inoculated samples and controls by selecting genes with an absolute value of log2 Fold Change (FC) in expression greater than 1 and adjusted p value P < 0.05. We found 431 DEGs including alternative transcripts with differential expression for 35 genes (Supplementary Table S3). We validated by quantitative PCR (qPCR) a sample of DEGs representative of different GOs and found consistency both in the log2 ratios (Supplementary Fig. S3) and in the statistical significance of the difference (Supplementary Table S4) between samples undergoing Rpv12-mediated ETI and controls. Only two DEGs were identified at 12 hpi, which were both up-regulated in samples undergoing Rpv12-mediated defence responses. These two genes are VIT_207s0129g00660 encoding an Indole-3-acetic acid (IAA)-amido synthetase (IAA) and VIT_209s0002g06890 encoding a blue copper protein. IAA-amido synthetases, also known as GH3 proteins, dampen free-IAA levels by catalysing the conjugation of IAA to amino acids. GH3-overexpressing plants in Arabidopsis and rice showed enhanced resistance to Pseudomonas syringae45 and Magnaporthe oryzae46, respectively, and it is generally assumed that suppression of IAA signalling is an integral part of SA-mediated resistance against biotrophic pathogens47,48. The second up-regulated gene (VIT_209s0002g06890) showed elevated expression levels also at 24 and 48 hpi. We found 70 DEGs at 24 hpi (17 up-regulated; 53 down-regulated in samples undergoing Rpv12-mediated defence responses); 50 DEGs at 48 hpi (41 up-regulated; 9 down-regulated in samples undergoing Rpv12-mediated defence responses) and 340 genes at 96 hpi (326 up-regulated; 14 down-regulated in samples undergoing Rpv12-mediated defence responses). The expression of a fraction of genes was statistically different between samples undergoing Rpv12-mediated defence responses and controls at more than one stage of incubation as reported in the Venn diagram of Supplementary Fig. S4. We focused our attention on the top 100 most significantly enriched GO categories for biological processes represented in the list of DEGs and the 20 GO categories with lowest p values are shown in Supplementary Table S5. At 48 and 96 hpi, we found up-regulation of DEGs belonging to the “plant hypersensitive response” (GO:0009626). We also found up-regulation for DEGs belonging to GO categories such as the “salicylic acid biosynthetic process” (GO:0009697), “systemic acquired resistance, salicylic acid mediated signalling pathway” (GO:0009862) and “response to salicylic acid stimulus” (GO:0009751). It was worth noting that at the same stages of incubation we found a significant up-regulation of the gene VIT_208s0058g01390 encoding a WRKY70 transcription factor that acts in Arabidopsis as an activator of SA-induced PR genes and is required for RPP4-mediated disease resistance against Hyaloperonospora parasitica49. It is known that resistance genes activate ETI responses through the SA-dependent signalling pathway and through signalling cascades that amplify the spectra of defence responses. Indeed, we found up-regulation of DEGs belonging to the “MAP kinase cascade” category (GO:0000165). For instance, the gene VIT_209s0002g04560 encoding a calmodulin-binding protein was significantly up-regulated at 48 and 96 hpi in samples undergoing Rpv12-mediated defence responses. Calmodulin-binding proteins play crucial roles in cellular signalling cascades through the regulation of numerous target proteins50. We also observed up-regulation of the gene VIT_203s0088g00910 encoding a class 1 pathogenesis related protein (PR-1) at 24, 48 and 96 hpi and of the gene VIT_202s0025g04330 encoding a thaumatin-like class 5 protein (TLP or PR-5) at 48 and 96 hpi in samples undergoing Rpv12-mediated defence responses (Supplementary Table S4). At 24 and 48 hpi we also found up-regulation of DEGs VIT_208s0007g06040 and VIT_208s0007g06060 encoding beta-glucanases that operate cell wall lysis of invading pathogens as a part of the inducible defence responses. DEGs with predicted functions related to amino acid, carbohydrate and lipid metabolism were up-regulated at 96 hpi together with genes belonging to secondary metabolism pathways within the GOs “coumarin biosynthetic process” (GO:0009805), and “stilbene biosynthetic process” (GO:0009811). At 96 dpi, we also found significant down-regulation of individual genes involved in photosynthesis and photosynthetic electron transport chain processes, such as VIT_210s0003g02900, VIT_217s0000g06350, VIT_210s0003g02890 encoding different chlorophyll a-b binding proteins.
MCIA of metabolites and transcripts
We used Multiple Co-Inertia Analysis (MCIA), a method developed by Meng et al.51, to identify co-relationships between the two-omics datasets using a multivariate dimension reduction approach. MCIA rescales the values from heterogeneous datasets using a covariance criterion and projects simultaneously multiple datasets onto the same plane. The results of MCIA analysis are reported in Fig. 8. Individual transcripts and metabolites are represented by grey filled squares and black filled dots, respectively, regardless of the sample type, of the stage of incubation and of the replicate. The colored dots, which are differentiated by the stage of incubation, recapitulate the global distribution of each omics dataset on the same bidimensional space, sorted by sample type and replicate. Large colored triangles and circles recapitulate the integration of the two-omics datasets, sorted by sample type, stage of incubation and replicate. The length of the connectors between large symbols and the corresponding small coloured dots are inversely correlated with the extent to which the two-omics datasets are coherent in the separation by sample type, stage of incubation and replicate on the bidimensional space. The MCIA plot showed that the strongest separation between samples undergoing Rpv12-mediated defence responses and controls occurred at 96 hpi.
Figure 8.
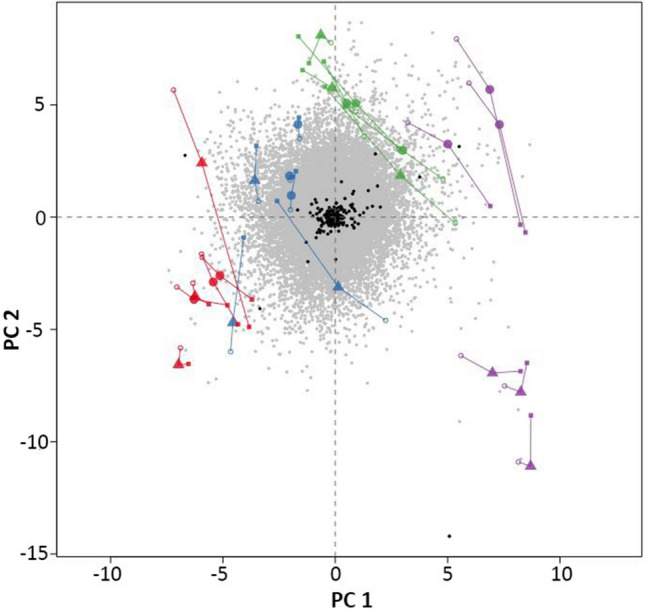
Multiple Co-Inertia Analysis of two-omics datasets including all transcript levels (grey filled square) and metabolite concentrations (black filled circle). The two axes report the first two principal components (PC). Small coloured symbols summarise transcript or metabolite data separately for each replicated sample and stage (red filled square, red open circle 12 hpi; blue filled square, blue open circle 24 hpi; green filled square, green open circle 48 hpi; violet filled square, violet open circle 96 hpi, respectively) and are linked by connectors (–) to their integrated value for samples undergoing Rpv12-mediated defence (red filled circle; blue filled circle; green filled circle; violet filled circle) and controls (red filled triangle; blue filled triangle; green filled triangle; violet filled triangle).
Discussion
In grapevine leaf discs that are operating Rpv12-mediated resistance against the infection of P. viticola, the host tissues are characterised by wider and faster metabolome changes than those observed in a similar experiment performed on leaf discs that were operating Rvp3-mediated resistance20. Using the same analytical and statistical methods and the same cut-off thresholds, we identified a higher number of metabolites (88 versus 53) that were significantly modulated over a 96 h period after the inoculation with the pathogen. We also found that substantial metabolite changes occurred earlier in Rpv12-mediated resistance compared to Rpv3-mediated resistance with the highest number of modulated metabolites at 48 hpi instead of 96 hpi20. Although we paid particular attention to the choice of an appropriate Rpv12-carrying genotype in order to minimise the confounding effects of the genetic background, we cannot exclude that part of the faster and stronger defence responses of the Rpv12-carrier compared to the Rpv3-carrier was due to more effective signal transduction components downstream of the R-gene in the genotype under study. We indeed observed substantial variation in field resistance against natural infections of P. viticola among dozens of grape varieties that all carry the same Rpv3-1 resistance haplotype in an ample variation of genetic backgrounds52. While for choosing the Rpv3-carrier20 we had background information available to select an average variety, such as ‘Bianca’, as a representative of a known distribution52, similar data are still not available for a substantial number of Rpv12-carriers, due to a more recent origin of this lineage. While Rpv3-carrying varieties had been introduced in cultivation since the late 1800’s and since then used for further breeding, it was only in 1960 that Koleda and Tamássy backcrossed V. amurensis × V. vinifera hybrids to generate the first two resistant varieties (‘Kunleany’ and ‘Kunbarat’53) carrying a new resistance that later has become known as Rpv12-dependent21. It took another 15 years after Koleda and Tamássy, before Cindrić and Korać had the intuition that those two varieties could become excellent progenitors for a new lineage of resistant varieties, performing in 1977 the backcross of ‘Kunbarat’ that has led to origin of the individual SK77-4/554, a parent of the individual of this study. It is therefore possible that we used by chance, among the few available, an Rpv12-carrier with a particularly favourable genetic background, although several reports converge on a consensus that, based on OIV452 scores, Rpv12-dependent resistance is stronger across varieties than Rpv3-dependent resistance55,56. Bove and Rossi57 also showed that three varieties operating Rpv12-dependent defence clustered together and separately from varieties carrying other Rpv genes for five defence components (i.e. proportion of inoculated sites that develop disease symptoms, length of the latent period, number of waves of sporangial release during the infectious period upon extended incubation of sporulating lesions, sporangial density per lesion, infectivity of the released sporangia), suggesting that the outcome of the defence response is still more dependent on the type of R-gene than on the genetic background of the variety. Despite the inherent limitations of this kind of comparative experiments, we observed that changes leading to a common metabolic reprogramming necessary for triggering and operating cell death at the infection sites and for SA-dependent synthesis of PR-proteins have occurred in Rpv12-mediated resistance with similar timing and in the same direction as observed for Rpv3-mediated resistance.
While the causal factor that triggers Rpv3-mediated HR has been identified in the gene products of two nearby TIR-NB-LRRs58, the causal gene that would explain the observed Mendelian inheritance of Rpv12-mediated resistance has not been isolated yet21. The sequence of the reference grapevine genome that has been assembled from a DM sensitive genotype predicts the presence of allelic forms of CC-NB-LRR genes in the Rpv12 locus21,59. Although the Rpv12-resistance haplotype has yet to be sequenced, the Rpv12-mediated response analysed in this paper shows the hallmarks of a ROS- and SA-dependent HR triggered by a NB-LRR gene product. Here, we provided evidence that gene expression profiling and changes in transcription levels of gene categories grouped by metabolic pathways lend support to the observed metabolite changes over a period of 96 hpi and highlight striking similarities with the canonical model of ETI-mediated defence responses to biotrophic pathogens in plants60. Indeed, cumulative evidence from metabolomic and transcriptomic data suggests that the Rpv12-mediated resistance relies on a rapid activation of a broad set of inducible responses that take place within 12 hpi. At 12 hpi, we detected elevated expression of gene categories encoding sugar transporters, which suggest a mobilisation of existing carbon resources, and an increase of sugar concentration. Sugar sensing and sugar-induced signalling have been shown to activate pathways leading to the synthesis of PR-proteins, to elicit PCD61 and to induce a metabolic swift from source to sink through a negative feedback loop62. At 12 hpi, samples undergoing Rpv12-mediated ETI also showed elevated concentration of linolenic acid that is reported to directly activate NADPH-oxidase41, an enzyme required for oxidative burst. An increase in linolenic acid has also been observed at the early stages of the Rpv3-mediated defence response in ‘Regent’40. Elevated levels of trans-resveratrol were observed at 12 hpi in both Rpv12- and Rpv3-mediated defence responses. In contrast with metabolite and transcript analyses of Rpv3-mediated20,23 and of Rpv10-mediated defence35, we did not find statistically significant differences between samples undergoing Rpv12-mediated defence response and controls in the expression of the gene VIT_212s0028g01840, which encodes a peroxidase possibly involved in the dimerization of trans-resveratrol into the fungi-toxic ε-viniferin. Nor did we find evidence for a significant increase of ε-viniferin concomitant with or subsequent to the increase of trans-resveratrol in samples undergoing Rpv12-mediated defence response. Although the two roles are not mutually exclusive, these lines of evidence in Rpv12-mediated resistance point to a primary role of trans-resveratrol as a signaling molecule in ROS formation and initiation of programmed cell death, as proposed by63, rather than as a precursor of toxic stilbenes, as it occurs in Rpv3- and in Rpv10-mediated defence. An augmented and prolonged accumulation of resveratrol, without a substantial conversion into ε-viniferin, has also been observed in otherwise sensitive grapevines after they were administered a specific sulphated derivative of laminarin that primed H2O2 production and suppressed the development of P. viticola in inoculated leaf discs64.
This ample set of early changes that had already occurred by 12 hpi in samples undergoing Rpv12-mediated defence suggests that the metabolism is timely reconfigured shortly after pathogen perception and the entire defence machinery comes immediately into operation as a part of a crisis management program. However, the full deployment of the activated defence weaponry remains on standby until a point of no return that is surpassed when ROS and SA synergistically potentiate the response and drive cells into an auto-toxic and energetically costly process. At 24 hpi, we observed strong reduction in 18:1 fatty acids that suggests initiation of ROS production, as it occurs in the Rpv3-mediated defence response with a timing that coincides in that case with the execution of PCD23. In Arabidopsis, it is reported that lower levels of 18:1 fatty acids directly stabilise the NITRIC OXIDE ASSOCIATED1 enzyme, increasing nitric oxide concentration65, a signalling gaseous radical that potentiates ROS-dependent PCD and up-regulates gene expression in the phenylpropanoid and SA pathways. It is possible that we only detected early transcriptional and metabolic signals associated with the onset of PCD at 24 hpi. Light exclusion—an experimental condition imposed for minimising the effect of light-dependent circadian fluctuations on the transcriptome at different sampling stages—might have prevented us from observing the occurrence and consequences of light-dependent oxidation, including lipid peroxidation40, and ROS production that are known to regulate the strength of PCD-dependent HR. However, we observed a similar trend of changes in fatty acid composition in response to Rpv12-mediated defence response as that reported by40 in Rpv3-mediated defence response in ‘Regent’, which did not occur in sensitive host controls40. These authors reported a transient and slight increase of oleic, linoleic and linolenic acids at the earliest stages of the defence response (6 hpi), followed by a rapid decrease of their concentrations at 12 hpi40. We observed a delay in this trend, with slight increases at 12 hpi and decreases at 24 hpi, but the drop at 24 hpi was much more pronounced and consistent with what we previously observed in Rpv3-mediated defence response in ‘Bianca’20.
We observed at 48 hpi full deployment of important components of the defence weaponry, such as SA-signalling, calcium and MAP kinase signal transduction, PR gene expression, and strong repression of photosynthesis that marked the complete source-to-sink reconfiguration of the metabolism to shuttle resources into energy-generating pathways and to prioritize the synthesis of defence-related compounds. The hypothesis of a central role of SA-signalling in eliciting some components of the Rpv12-mediated response was corroborated by the observation of a general up-regulation of the WRKY family of transcription factors. Noteworthy was the up-regulation of grapevine homologs of WRKY28 that binds to the promoter of isochorismate synthase66, the rate-limiting enzyme in SA biosynthetic, and of WRKY70, a component of gene-for-gene resistance against the oomycete Hyaloperonospora parasitica in Arabidopsis that causes induction of genes encoding PR-1, PR-2, and PR-5 proteins49,67. The grapevine VvWRKY28 has also been proposed as a positional candidate residing in a QTL confidence interval that explained 10.8% of the phenotypic variation for cis-resveratrol content in a segregating population68. In the same work, some stilbenoids showed a general trend of augmented concentration in individuals with higher OIV-452 scores and OIV-452 scores were higher in full-sibling individuals that carried an Rpv3-3 haplotype than in those who did not. However, no significant QTL for stilbenoid concentration was found to colocalise with the QTL for OIV-452 scores, which corresponded to the Rpv3 locus and explained 19.7% of the observed variation in P. viticola resistance68. Another transcription factor, VvWRKY33, was found by69 to be induced in ‘Regent’ during the Rpv3-mediated response from 2 to 120 hpi, with highest differences in expression levels compared to mock-inoculated controls between 4 and 10 hpi and correlation with increased expression of a pathogenesis related protein 10.1 (PR-10.1) gene. VIT_208s0058g00690 is the most similar reference gene for the WRKY33 sequence isolated from ‘Regent’ (GenBank accession no. KF800706.169). We found elevated levels of VIT_208s0058g00690 expression at 48 and 96 hpi in leaf discs undergoing the Rpv12-mediated response compared to controls (log2 Fold Change 0.70 and 0.88, respectively for the primary transcripts), although these difference remained below the threshold of statistical significance. In a similar way, we found a trend of augmented levels, yet not statistically significant, of three PR-10.1 gene copies (VIT_05s0077g01530, VIT_05s0077g01580, VIT_05s0077g01560) within a tandemly arrayed gene cluster at 48 hpi (log2 Fold Change 0.19, 0.16, 0.16, respectively) and 96 hpi (log2 Fold Change 0.19, 0.23, 0.23, respectively) in leaf discs undergoing the Rpv12-mediated response. In the Rpv12-mediated response, we vice versa found statistically significant induction of a thaumatin-like protein gene similar to a V. amurensis ortholog that impaired mycelial growth, causing hyphal malformation, in transgenic lines of sensitive controls70. An increasing number of yet genetically uncharacterized sources of P. viticola resistance have emerged in the past years from Asian species, which include V. pseudoreticulata71 and Vitis quinquangularis72. Further studies will be needed to clarify if these novel downy mildew resistances share the same type of ETI and follow the same lines of defence reactions as those observed in Rpv12-mediated resistance.
Materials and methods
Plant material and artificial inoculation
Dormant canes for the experiment were obtained from mother plants held in a germplasm repository in the South Tyrol region, Italy. Two bud cuttings (n = 45) were rooted in potted soil and forced to sprout in isolation within a greenhouse free from sources of inoculum. A single shoot was raised from each rooted cutting until the stage of 12 expanded leaves. Water was supplied by capillary action by placing the pots, containing the rooted cuttings, onto a layer of capillary matting that was kept constantly wet. We then sorted the plants into three homogenous groups. Each group of plants (n = 15) formed a biological replicate. At the time of the inoculation the plants were healthy, with no symptoms of any foliar disease. In parallel, shoots were raised from three cuttings of the variety ‘Pinot Noir’ in order to prepare sensitive controls. The three distal fully expanded leaves were detached from all plants of a biological replicate, bulked, and rinsed with ultrapure water. Discs were excised from each leaf lamina using a 1.1-cm diameter cork borer and plated randomly with the abaxial surface upside on wet paper in Petri dishes, collectively representing the biological replicate. Leaf discs were acclimated at 21 °C in the dark for 12 h in order to dispel wounding-induced jasmonic acid signalling73. Petri dishes from each biological replicate were divided randomly into two groups, one used for pathogen inoculation and one used for mock-inoculation, hereafter referred to as controls. Inoculated samples were sprayed with a P. viticola suspension of 106 sporangia ml−1, while controls were sprayed with ultrapure water. P. viticola was originally collected as a natural mixture of sporangia from diverse V. vinifera cultivars in an unsprayed vineyard in San Michele all’Adige, Northern Italy. Sporangia were stored at − 20 °C prior to use for the experiment of this study and for the experiment reported in20, which was conducted two weeks earlier during the same growing season. The inoculum was propagated on plants of V. vinifera ‘Pinot Gris’ grown in potted soil in a greenhouse by spraying a 5 × 105 ml−1 suspension of the frozen sporangia in distilled water. Plants were maintained at > 90% relative humidity at 21 °C until sporulation. Fresh sporangia were collected from ‘Pinot Gris’ plants by soaking sporulating leaves in refrigerated (4 °C) distilled water. The suspension was diluted to the final concentration using a Burker chamber and sprayed immediately on the experimental leaf discs. All Petri dishes were incubated at 21 °C in the dark until sampling. Samples were collected at 12, 24, 48 and 96 hpi and immediately frozen in liquid nitrogen. A set of five Petri dishes was available per sampling stage, per treatment and per biological replicate. Four Petri dishes in each set collectively containing a minimum of 100 leaf discs were used for metabolomic analyses by pooling and grinding together all discs of the biological replicate. The remaining Petri dish of each set containing a minimum of 25 leaf discs was used for transcriptomic analyses by sampling two discs per time point that were pooled and ground together for RNA extraction. The remaining leaf discs were incubated until sporulation to confirm P. viticola infection on inoculated samples and to exclude P. viticola contamination prior to sample collection on mock-inoculated controls. Sporulation was not observed on inoculated Rpv12-resistant leaf discs during the course of sampling and until 120 hpi. The resistance phenotype scored a value of 9 on the OIV452 descriptor scale12, due to concomitant absence of sporulation and presence of barely visible necrotic spots. Sporulation was already visible at the last stage of sampling (96 hpi) in inoculated control leaf discs of the sensitive ‘Pinot Noir’ and was confirmed after the conclusion of the experiment at 120 hpi. Sporulation was not observed on mock-inoculated control leaf discs of ‘Pinot Noir’ until 120 hpi. Gene expression analyses were performed at the genomics facility of the Fondazione Edmund Mach. Three biological replicates per treatment per time point were used for metabolite and gene expression analyses.
Metabolite analysis
Primary metabolites were determined following the methodology published by Chitarrini and coworkers20. GC/MS analysis was performed by injecting 1 µL of derivatised extract into a Trace GC Ultra instrument combined with a TSQ Quantum GC mass spectrometer (Thermo Electron Corporation, MA). Compounds were separated using an RXI-5-Sil MS w/Integra-Guard column. Data acquisition was run in full scan mode from 50 to 700 m/z. Data were processed using the XCALIBUR 2.2 software.
Lipids were determined using the method developed by Della Corte et al.74. Chromatographic analysis was carried out using a UHPLC Dionex 3000 instrument (Thermo Fischer Scientific, DE) combined with an API 5500 triple quadrupole mass spectrometer (Applied Biosystems/MDS Sciex, CA). Compounds were separated using an RP Ascentis Express column.
Phenols were determined with slight modifications of the method developed by Vrhovsek et al.75. Chromatographic analysis was carried out using a Waters Acquity UPLC instrument combined with a Waters Xevo triple quadrupole mass spectrometer detector. Compounds were separated using a Waters Acquity HSS T3 column. Data processing was performed using Waters MassLynx V4.1 software.
Primary metabolites, lipids, and phenols. Compounds were identified based on their reference standard, retention time and qualifier and quantifier ion. Compounds were quantified using linear calibration curves and expressed as mg kg−1 of FW.
Volatile compounds were extracted with slight modifications of the methods developed by Matarese et al.76 and Salvagnin et al.77 as described by Chitarrini et al.20. Compounds were captured from the headspace using a Supelco 2-cm DVB/CAR/PDMS 50/30 μm fibre (Supelco, PA). Chromatographic analysis was carried out using a Trace GC Ultra gas chromatograph combined with a Quantum XLS mass spectrometer (Thermo Scientific, Electron Corporation, MA). Compounds were separated using a Stabilwax-DA column (Restek Corporation, PA). Data were processed using the XCALIBUR 2.2 software. Identification of volatile compounds was carried out by injecting pure reference standards when available or comparing retention index and mass spectra using the NIST MS Search 2.0 database. Results were expressed in µg kg−1 of FW with semi-quantification using 1-heptanol as internal standard.
RNA extraction and gene expression analysis
Approximately, 100 mg of leaf discs were ground to powder in liquid nitrogen. RNA was isolated according to the manufacturer’s instructions using the Spectrum Plant Total RNA Kit (Sigma-Aldrich, DE). Total RNA quality was checked on RNA ScreenTape using an Agilent 2200 TapeStation (Agilent Technologies, CA).
RNA-Seq and gene expression analysis
cDNA libraries were constructed using 1 µg of total RNA and the KAPA Stranded mRNA-Seq Kit (Kapa Biosystems, MA). Each library was barcoded using the SeqCap Adapter Kit A and B (NimbleGen, Roche, CA) and the target range of fragment sizes (250–280 bp) was confirmed on High Sensitivity D1000 ScreenTape using the Tapestation 2200 (Agilent Technologies, CA,). All the libraries were quantified with a KAPA Library Quantification Kit (Kapa Biosystems, MA) using a LightCycler 480 (Roche, DE) and multiplexed in four equimolar pools. Each library pool was sequenced on an Illumina HiSeq 2500 sequencer generating 2 × 50 bp paired-end reads. An average of 54.4 million paired-end reads was generated per sample. Base calling and quality control were performed using the Illumina RTA sequence analysis pipeline. Reads were aligned to the grapevine PN40024 reference transcriptome59,78 using Bowtie279, obtaining an average rate of read alignment of 84.7%.
Use of RNA-Seq data for the description of the genetic makeup of the genotype under-study
We also used RNA-Seq reads from all samples and stages to perform allele-specific read alignment against a modified version of the grapevine genome reference that included the reference Rpv3-PN40024 haplotype and the alternative resistance Rpv3-1 haplotype and called variant sites using the procedure described in58. RNA reads revealed a substantial number of homozygous sites in transcribed genes of the Rpv3-1 sequence, which allowed us to exclude that the genotype analysed in this study had inherited from its parent ‘Bianca’ the resistance haplotype at the Rpv3 locus. To corroborate this analysis and to widen the description of the genetic constitution of the genotype under study, we realigned RNA-Seq reads from all samples and stages against the canonical grapevine genome reference59 with TopHat2 and called variants sites in transcribed genes with GATK. We filtered all SNPs present in a comprehensive diversity-panel of vinifera as described in58 and plotted the density of residual (non-vinifera) SNPs in 100-Kb genomic windows. We generated evidence that the introgression in the upper arm of chromosome 14 extends from the upper telomere down to a genomic window whose distal edge is located at the chromosomal coordinate chr14:11,683,740 (Supplementary Fig. S7), approximately 2 Mb downstream of the reported location of the Rpv12 locus21. A closer inspection into the introgression around the Rpv12 locus was conducted using available DNA sequencing reads of ‘Merlot Khorus’, a variety known to carry the Rpv12 resistance haplotype, which derived from the backcross to vinifera of a full-sibling of the genotype under study (uncle–nephew relationship), and UD-2107658, which derived from the selfing of ‘Bianca’, the second parent of the genotype under study (Supplementary Fig. S8). We could extract evidence from the nearly complete sharing of non–vinifera SNPs between ‘Merlot Khorus’ and the genotype under study (Supplementary Fig. S8e) that they indeed share the same Asian introgression across a large part of the upper arm, including the Rpv12 locus (Supplementary Fig. 8a,c–e). The resistance chromosome recombined with the vinifera counterpart at 11,7 Mbp in the megaspore mother cell of SK77–4/5 that formed the gamete transmitted to the genotype under study. We could also confirm that the homologous chromosome was donated to the genotype under study by ‘Bianca’, because this homolog shared with UD–21,076 exactly the same recombination break points at two introgressed regions (Supplementary Fig. S8b–e). One of these introgressions extended from 5.8 to 11 Mbp. As a result of this, the individual under study is heterozygous for an Asian (Rpv12 resistance haplotype) and an American introgression across approximately 5 Mbp that also include the Rpv12 locus (Supplementary Fig. 8c). Since we did not detect any significant QTL effect on P. viticola resistance on chromosome 14 in ‘Bianca’12, which is heterozygous for the American introgression and a vinifera haplotype across this locus, we infer that this particular haplotypic combination occurred by chance and was not a target of intentional selection in this individual. We also confirmed that the genotype under study carries two vinifera homologouos copies along the entirety of chromosomes 18 and 9 (Supplementary Fig. S7) and it did not show any signal of introgression in or around the reported locations of the Rpv3 locus58 and the Rpv1080 locus on those chromosomes.
qPCR validation
cDNA was generated using Invitrogen SuperScript IV VILO Master Mix (ThermoFisher Scientific, DE) and 1.5 µg of total RNA. A set of 9 DEGs representative of different GO categories were selected for primer design and validation (Supplementary Table S3). Quantitative PCR reactions were performed using a LightCycler480 thermocycler (Roche, CH) and a 2 × qPCRBIO SyGreen Kit (PCR Biosystems, UK) following the manufacturer’s instructions. Transcripts were quantified using the LC480 software (Roche, CH) and the 2–∆∆CT method using the glyceraldehyde-3-phosphate dehydrogenase (GADPH) housekeeping gene VIT_17s0000g10430 for normalisation with the following primer sequences (forward 5′ → 3′ TTCTCGTTGAGGGCTATTCCA; reverse 5′ → 3′ CCACAGACTTCATCGGTGACA).
Data analysis and graphical representation
Missing data points were replaced with a random value between zero and the limit of quantification of each metabolite. A log10-transformation of metabolite concentration was performed prior to statistical analysis81. The t-statistic was computed using the R package Stats82. Metabolite networks were generated using ggraph83. DEG analysis was performed with DeSeq284. Gene enrichment analysis was performed using blast2GO85. Gene functional annotation was retrieved from VitisNet86, VitisCyc87, and V2.1 annotation78. A two-sided Wilcoxon–Mann–Whitney test was used for assessing significant differences between distributions of values. Multivariate data analysis was performed using the R package FactoMineR88 and the related graphs in Figs. 1 and 6 were generated using the R package Factoextra89. The R package omicade451 was used to perform MCIA and to generate the plot reported in Fig. 8. Graphs in all other figures were generated with R (version 3.3)82. Metabolite concentrations of Rpv3-mediated defence response were obtained from raw data of our previous paper20.
Supplementary information
Acknowledgements
We thank Domenico Masuero and Cesare Lotti for the technical help. This research was supported by the FIRST International PhD program and ADP 2015/2016 project funded by the Autonomous Province of Trento.
Author contributions
G.C., L.Z., A.V., M.S., G.D.G. and U.V. designed the experiment. M.S. provided the plant material. G.C., G.D.G., L.Z., and A.V. performed inoculation and sampling. G.C. performed metabolomic analyses. M.P., G.M., S.F. and S.L. performed RNA-Seq analysis and qPCR validation. S.R., A.C. and P.F. carried out statistical analyses. G.M., S.F., M.M., and G.D.G performed genotypic and haplotypic analyses on the genetic constitution of the genotype under study. G.C., S.F., M.M., G.D.G., and U.V. wrote the manuscript. All authors interpreted the results and commented on the manuscript.
Data availability
RNA-Seq reads were deposited in the European Nucleotide Archive repository under the Unique Name Experiment ERX2989847. Metabolite data are provided as Supplementary Table S1. ‘Merlot Khorus’ FASTQ files of the reads aligning to chromosome 14 are publicly available at figshare under the https://doi.org/10.6084/m9.figshare.12240881.
Competing interests
G.D.G. and M.M. are inventors of five patented varieties carrying Rpv12-mediated resistance. All other authors declare no potential conflict of interest. Funding bodies had no role in the design of the study and in the collection, analysis and interpretation of data, or writing the manuscript.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Gabriele Di Gaspero, Email: digaspero@appliedgenomics.org.
Urska Vrhovsek, Email: urska.vrhovsek@fmach.it.
Supplementary information
is available for this paper at 10.1038/s41598-020-69051-6.
References
- 1.Toffolatti SL, Venturini G, Maffi D, Vercesi A. Phenotypic and histochemical traits of the interaction between Plasmopara viticola and resistant or susceptible grapevine varieties. BMC Plant Biol. 2012 doi: 10.1186/1471-2229-12-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schröder S, et al. Crop wild relatives as genetic resources—The case of the European wild grape. Can. J. Plant Sci. 2015;95:905–912. [Google Scholar]
- 3.Fischer BM, et al. Quantitative trait locus analysis of fungal disease resistance factors on a molecular map of grapevine. Theor. Appl. Genet. 2004;108:501–515. doi: 10.1007/s00122-003-1445-3. [DOI] [PubMed] [Google Scholar]
- 4.Whisson SC, et al. A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature. 2007;450:115–118. doi: 10.1038/nature06203. [DOI] [PubMed] [Google Scholar]
- 5.Fawke S, Doumane M, Schornack S. Oomycete interactions with plants: Infection strategies and resistance principles. Microbiol. Mol. Biol. Rev. 2015;79:263–280. doi: 10.1128/MMBR.00010-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dercks W, Creasy LL. Influence of fosetyl-Al on phytoalexin accumulation in the Plasmopara viticola-grapevine interaction. Physiol. Mol. Plant Pathol. 1989;34:203–213. [Google Scholar]
- 7.Dai G, Andary C, Mondolot-Cosson L, Boubals D. Histochemical studies on the interaction between three species of grapevine, Vitis vinifera, V. rupestris and V. rotundifolia and the downy mildew fungus, Plasmopara viticola. Physiol. Mol. Plant Pathol. 1995;46:177–188. [Google Scholar]
- 8.Gindro K, Pezet R, Viret O. Histological study of the responses of two Vitis vinifera cultivars (resistant and susceptible) to Plasmopara viticola infections. Plant Physiol. Biochem. 2003;41:846–853. [Google Scholar]
- 9.Kortekamp A, Zyprian E. Characterization of Plasmopara-resistance in grapevine using in vitro plants. J. Plant Physiol. 2003;160:1393–1400. doi: 10.1078/0176-1617-01021. [DOI] [PubMed] [Google Scholar]
- 10.Pezet R, Gindro K, Viret O, Richter H. Effects of resveratrol, viniferins and pterostilbene on Plasmopara viticola zoospore mobility and disease development. Vitis. 2004;43:145–148. [Google Scholar]
- 11.Díez-Navajas AM, Wiedemann-Merdinoglu S, Greif C, Merdinoglu D. Nonhost versus host resistance to the grapevine downy mildew, Plasmopara viticola, studied at the tissue level. Phytopathology. 2008;98:776–780. doi: 10.1094/PHYTO-98-7-0776. [DOI] [PubMed] [Google Scholar]
- 12.Bellin D, et al. Resistance to Plasmopara viticola in grapevine ‘Bianca’ is controlled by a major dominant gene causing localised necrosis at the infection site. Theor. Appl. Genet. 2009;120:163–176. doi: 10.1007/s00122-009-1167-2. [DOI] [PubMed] [Google Scholar]
- 13.Polesani M, et al. General and species-specific transcriptional responses to downy mildew infection in a susceptible (Vitis vinifera) and a resistant (V. riparia) grapevine species. BMC Genomics. 2010;11:117. doi: 10.1186/1471-2164-11-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bolton MD. Current review: Primary metabolism and plant defense-fuel for the fire. Mol. Plant Microbe Interact. 2009;22:487–497. doi: 10.1094/MPMI-22-5-0487. [DOI] [PubMed] [Google Scholar]
- 15.Rojas CM, Senthil-Kumar M, Tzin V, Mysore KS. Regulation of primary plant metabolism during plant-pathogen interactions and its contribution to plant defense. Front. Plant Sci. 2014;5:17. doi: 10.3389/fpls.2014.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heil M, Baldwin IT. Fitness costs of induced resistance: Emerging experimental support for a slippery concept. Trends Plant Sci. 2002;7:61–67. doi: 10.1016/s1360-1385(01)02186-0. [DOI] [PubMed] [Google Scholar]
- 17.Smedegaard-Petersen V. The effect of defence reactions on the energy balance and yield of resistant plants. In: Wood RKS, editor. Active Defense Mechanisms in Plants. New York: Springer; 1982. pp. 299–315. [Google Scholar]
- 18.Sampedro L, Moreira X, Zas R. Costs of constitutive and herbivore-induced chemical defences in pine trees emerge only under low nutrient availability. J. Ecol. 2011;99:818–827. [Google Scholar]
- 19.Stevens MT, Waller DM, Lindroth RL. Resistance and tolerance in Populus tremuloides: Genetic variation, costs, and environmental dependency. Evol. Ecol. 2007;21:829–847. [Google Scholar]
- 20.Chitarrini G, et al. Identification of biomarkers for defense response to Plasmopara viticola in a resistant grape variety. Front. Plant Sci. 2017;8:1524. doi: 10.3389/fpls.2017.01524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Venuti S, et al. Historical introgression of the downy mildew resistance gene Rpv12 from the Asian species Vitis amurensis into grapevine varieties. PLoS ONE. 2013;8:e61228. doi: 10.1371/journal.pone.0061228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cindric P, Korac N, Kovac V. Grape breeding in the vojvodina province. Acta Hortic. 2000;528:499–504. [Google Scholar]
- 23.Eisenmann B, et al. Rpv3–1 mediated resistance to grapevine downy mildew is associated with specific host transcriptional responses and the accumulation of stilbenes. BMC Plant Biol. 2019;19:343. doi: 10.1186/s12870-019-1935-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernie AR, Trethewey RN, Krotzky AJ, Willmitzer L. Metabolite profiling: From diagnostics to systems biology. Nat. Rev. Mol. Cell Biol. 2004;5:763–769. doi: 10.1038/nrm1451. [DOI] [PubMed] [Google Scholar]
- 25.Zamboni A, et al. Identification of putative stage-specific grapevine berry biomarkers and omics data integration into networks. Plant Physiol. 2010;154:1439–1459. doi: 10.1104/pp.110.160275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fortes AM, et al. Transcript and metabolite analysis in Trincadeira cultivar reveals novel information regarding the dynamics of grape ripening. BMC Plant Biol. 2011;11:149. doi: 10.1186/1471-2229-11-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cuadros-Inostroza A, et al. GC–MS metabolic profiling of Cabernet Sauvignon and Merlot cultivars during grapevine berry development and network analysis reveals a stage- and cultivar-dependent connectivity of primary metabolites. Metabolomics. 2016;12:1–17. doi: 10.1007/s11306-015-0927-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grimplet J, et al. Proteomic and selected metabolite analysis of grape berry tissues under well-watered and water-deficit stress conditions. Proteomics. 2009;9:2503–2528. doi: 10.1002/pmic.200800158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Savoi S, et al. Transcriptome and metabolite profiling reveals that prolonged drought modulates the phenylpropanoid and terpenoid pathway in white grapes (Vitis vinifera L.) BMC Plant Biol. 2016;16:1–17. doi: 10.1186/s12870-016-0760-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Savoi S, et al. Multi-omics and integrated network analyses reveal new insights into the systems relationships between metabolites, structural genes, and transcriptional regulators in developing grape berries (Vitis vinifera L.) exposed to water deficit. Front. Plant Sci. 2017;8:1124. doi: 10.3389/fpls.2017.01124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ali K, et al. Alterations in grapevine leaf metabolism upon inoculation with Plasmopara viticola in different time-points. Plant Sci. 2012;191–192:100–107. doi: 10.1016/j.plantsci.2012.04.014. [DOI] [PubMed] [Google Scholar]
- 32.Algarra Alarcon A, et al. Emission of volatile sesquiterpenes and monoterpenes in grapevine genotypes following Plasmopara viticola inoculation in vitro. J. Mass Spectrom. 2015;50:1013–1022. doi: 10.1002/jms.3615. [DOI] [PubMed] [Google Scholar]
- 33.Viret O, Spring JL, Gindro K. Stilbenes: Biomarkers of grapevine resistance to fungal diseases. OENO ONE. 2018;52:235. [Google Scholar]
- 34.Malacarne G, et al. Resistance to Plasmopara viticola in a grapevine segregating population is associated with stilbenoid accumulation and with specific host transcriptional responses. BMC Plant Biol. 2011;11:1–13. doi: 10.1186/1471-2229-11-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fröbel S, Dudenhöffer J, Töpfer R, Zyprian E. Transcriptome analysis of early downy mildew (Plasmopara viticola) defense in grapevines carrying the Asian resistance locus Rpv10. Euphytica. 2019;215:28. [Google Scholar]
- 36.Shiratake K, Suzuki M. Omics studies of citrus, grape and rosaceae fruit trees. Breed. Sci. 2016;66:122–138. doi: 10.1270/jsbbs.66.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Agudelo-Romero P, et al. Transcriptome and metabolome reprogramming in Vitis vinifera cv. Trincadeira berries upon infection with Botrytis cinerea. J. Exp. Bot. 2015;66:1769–1785. doi: 10.1093/jxb/eru517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Figueiredo A, et al. Transcriptional and metabolic profiling of grape (Vitis vinifera L.) leaves unravel possible innate resistance against pathogenic fungi. J. Exp. Bot. 2008;59:3371–3381. doi: 10.1093/jxb/ern187. [DOI] [PubMed] [Google Scholar]
- 39.Dempsey DA, Vlot AC, Wildermuth MC, Klessig DF. Salicylic acid biosynthesis and metabolism. Arab. B. 2011;9:e0156. doi: 10.1199/tab.0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laureano G, et al. The interplay between membrane lipids and phospholipase A family members in grapevine resistance against Plasmopara viticola. Sci. Rep. 2018;8:1–15. doi: 10.1038/s41598-018-32559-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yaeno T, Matsuda O, Iba K. Role of chloroplast trienoic fatty acids in plaint disease defense responses. Plant J. 2004;40:931–941. doi: 10.1111/j.1365-313X.2004.02260.x. [DOI] [PubMed] [Google Scholar]
- 42.Overmyer K, Brosché M, Kangasjärvi J. Reactive oxygen species and hormonal control of cell death. Trends Plant Sci. 2003;8:335–342. doi: 10.1016/S1360-1385(03)00135-3. [DOI] [PubMed] [Google Scholar]
- 43.Shah J, Kachroo P, Nandi A, Klessig DF. A recessive mutation in the Arabidopsis SSI2 gene confers SA- and NPR1-independent expression of PR genes and resistance against bacterial and oomycete pathogens. Plant J. 2001;25:563–574. doi: 10.1046/j.1365-313x.2001.00992.x. [DOI] [PubMed] [Google Scholar]
- 44.Ribnicky DM, Shulaev V, Raskin I. Intermediates of salicylic acid biosynthesis in tobacco. Plant Physiol. 1998;118:565–572. doi: 10.1104/pp.118.2.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Z, et al. Dual regulation role of GH3.5 in salicylic acid and auxin signaling during arabidopsis-Pseudomonas syringae interaction. Plant Physiol. 2007;145:450–464. doi: 10.1104/pp.107.106021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Domingo C, Andrés F, Tharreau D, Iglesias DJ, Talón M. Constitutive expression of OsGH3.1 reduces auxin content and enhances defense response and resistance to a fungal pathogen in rice. Mol. Plant-Microbe Interact. 2009;22:201–210. doi: 10.1094/MPMI-22-2-0201. [DOI] [PubMed] [Google Scholar]
- 47.Wang D, Pajerowska-Mukhtar K, Culler AH, Dong X. Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr. Biol. 2007;17:1784–1790. doi: 10.1016/j.cub.2007.09.025. [DOI] [PubMed] [Google Scholar]
- 48.Fu J, Wang S. Insights into auxin signaling in plant-pathogen interactions. Front. Plant Sci. 2011;2:74. doi: 10.3389/fpls.2011.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Knoth C, Ringler J, Dangl JL, Eulgem T. Arabidopsis WRKY70 is required for full RPP4-mediated disease resistance and basal defense against Hyaloperonospora parasitica. Mol. Plant-Microbe Interact. 2007;20:120–128. doi: 10.1094/MPMI-20-2-0120. [DOI] [PubMed] [Google Scholar]
- 50.Ranty B, Aldon D, Galaud JP. Plant calmodulins and calmodulin-related proteins: Multifaceted relays to decode calcium signals. Plant Signal. Behav. 2006;1:96–104. doi: 10.4161/psb.1.3.2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meng C, Kuster B, Culhane AC, Gholami AM. A multivariate approach to the integration of multi-omics datasets. BMC Bioinform. 2014;15:162. doi: 10.1186/1471-2105-15-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Foria S, Magris G, Morgante M, Di Gaspero G. The genetic background modulates the intensity of Rpv3-dependent downy mildew resistance in grapevine. Plant Breed. 2018;137:220–228. [Google Scholar]
- 53.Koleda I. Ergebnisse von Kreuzungen zwischen Vitis amurensis und Vitis vinifera in der Züchtung frostwiderstandsfähiger Reben. Vitis. 1975;14:1–5. [Google Scholar]
- 54.Cindric P, Korac N. Grape breeding for disease resistance at Sremski Karlovci. Acta Hortic. 1998;473:105–112. [Google Scholar]
- 55.Zini E, et al. R-loci arrangement versus downy and powdery mildew resistance level: A vitis hybrid survey. Int. J. Mol. Sci. 2019;20:3526. doi: 10.3390/ijms20143526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kosev K, Simeonov I, Ivanov M, Nakov Z, Hvarleva T. Phenotypic and molecular characterization of 18 Bulgarian newly bred grapevine varieties in relation to their resistance to downy mildew. Biotechnol. Biotechnol. Equip. 2017;31:68–74. [Google Scholar]
- 57.Bove F, Rossi V. Components of partial resistance to Plasmopara viticola enable complete phenotypic characterization of grapevine varieties. Sci. Rep. 2020;10:1–12. doi: 10.1038/s41598-020-57482-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Foria S, et al. Gene duplication and transposition of mobile elements drive evolution of the Rpv3 resistance locus in grapevine. Plant J. 2020;101:529–542. doi: 10.1111/tpj.14551. [DOI] [PubMed] [Google Scholar]
- 59.Jaillon O, et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- 60.Wang Y, Tyler BM, Wang Y. Defense and counterdefense during plant-pathogenic oomycete infection. Annu. Rev. Microbiol. 2019;73:667–696. doi: 10.1146/annurev-micro-020518-120022. [DOI] [PubMed] [Google Scholar]
- 61.Moghaddam MRB, Van Den Ende W. Sugars and plant innate immunity. J. Exp. Bot. 2012;63:3989–3998. doi: 10.1093/jxb/ers129. [DOI] [PubMed] [Google Scholar]
- 62.Scharte J, Schon H, Weis E. Photosynthesis and carbohydrate metabolism in tobacco leaves during an incompatible interaction with Phytophthora nicotianae. Plant Cell Environ. 2005;28:1421–1435. [Google Scholar]
- 63.Chang X, Heene E, Qiao F, Nick P. The phytoalexin resveratrol regulates the initiation of hypersensitive cell death in Vitis cell. PLoS ONE. 2011;6:e26405. doi: 10.1371/journal.pone.0026405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Adrian M, et al. Metabolic fingerprint of PS3-induced resistance of grapevine leaves against Plasmopara viticola revealed differences in elicitor-triggered defenses. Front. Plant Sci. 2017;08:101. doi: 10.3389/fpls.2017.00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mandal MK, et al. Oleic acid-dependent modulation of nitric oxide ASSOCIATED1 protein levels regulates nitric oxide-mediated defense signaling in arabidopsis. Plant Cell. 2012;24:1654–1674. doi: 10.1105/tpc.112.096768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van Verk MC, Bol JF, Linthorst HJM. WRKY transcription factors involved in activation of SA biosynthesis genes. BMC Plant Biol. 2011;11:1–12. doi: 10.1186/1471-2229-11-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Eulgem T. Dissecting the WRKY web of plant defense regulators. PLoS Pathog. 2006;2:e126. doi: 10.1371/journal.ppat.0020126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vezzulli S, et al. The Rpv3-3 haplotype and stilbenoid induction mediate downy mildew resistance in a grapevine interspecific population. Front. Plant Sci. 2019;10:1–23. doi: 10.3389/fpls.2019.00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Merz PR, et al. The transcription factor VvWRKY33 is involved in the regulation of grapevine (Vitis vinifera) defense against the oomycete pathogen Plasmopara viticola. Physiol. Plant. 2015;153:365–380. doi: 10.1111/ppl.12251. [DOI] [PubMed] [Google Scholar]
- 70.He R, et al. Overexpression of a thaumatin-like protein gene from Vitis amurensis improves downy mildew resistance in Vitis vinifera grapevine. Protoplasma. 2017;254:1579–1589. doi: 10.1007/s00709-016-1047-y. [DOI] [PubMed] [Google Scholar]
- 71.Liu R, et al. Transcriptomic analysis of Chinese wild Vitis pseudoreticulata in response to Plasmopara viticola. Protoplasma. 2019;256:1409–1424. doi: 10.1007/s00709-019-01387-x. [DOI] [PubMed] [Google Scholar]
- 72.Luo Y, et al. The effect of transcription factor MYB14 on defense mechanisms in Vitis quinquangularis-pingyi. Int. J. Mol. Sci. 2020;21:706. doi: 10.3390/ijms21030706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Casagrande K, Falginella L, Castellarin SD, Testolin R, Di Gaspero G. Defence responses in Rpv3-dependent resistance to grapevine downy mildew. Planta. 2011;234:1097–1109. doi: 10.1007/s00425-011-1461-5. [DOI] [PubMed] [Google Scholar]
- 74.Della Corte A, et al. A rapid LC-MS/MS method for quantitative profiling of fatty acids, sterols, glycerolipids, glycerophospholipids and sphingolipids in grapes. Talanta. 2015;140:52–61. doi: 10.1016/j.talanta.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 75.Vrhovsek U, et al. A versatile targeted metabolomics method for the rapid quantification of multiple classes of phenolics in fruits and beverages. J. Agric. Food Chem. 2012;60:8831–8840. doi: 10.1021/jf2051569. [DOI] [PubMed] [Google Scholar]
- 76.Matarese F, Cuzzola A, Scalabrelli G, D’Onofrio C. Expression of terpene synthase genes associated with the formation of volatiles in different organs of Vitis vinifera. Phytochemistry. 2014;105:12–24. doi: 10.1016/j.phytochem.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 77.Salvagnin U, et al. Homologous and heterologous expression of grapevine E-(β)-caryophyllene synthase (VvGwECar2) Phytochemistry. 2016;131:76–83. doi: 10.1016/j.phytochem.2016.08.002. [DOI] [PubMed] [Google Scholar]
- 78.Vitulo N, et al. A deep survey of alternative splicing in grape reveals changes in the splicing machinery related to tissue, stress condition and genotype. BMC Plant Biol. 2014;14:99. doi: 10.1186/1471-2229-14-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Langmead B. Aligning short sequencing reads with Bowtie. Curr. Protoc. Bioinform. 2010 doi: 10.1002/0471250953.bi1107s32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schwander F, et al. Rpv10: A new locus from the Asian Vitis gene pool for pyramiding downy mildew resistance loci in grapevine. Theor. Appl. Genet. 2012;124:163–176. doi: 10.1007/s00122-011-1695-4. [DOI] [PubMed] [Google Scholar]
- 81.van den Berg RA, Hoefsloot HCJ, Westerhuis JA, Smilde AK, van der Werf MJ. Centering, scaling, and transformations: Improving the biological information content of metabolomics data. BMC Genomics. 2006;7:142. doi: 10.1186/1471-2164-7-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing , Vienna, Austria (2017). https://www.r-project.org/. Accessed 31 Dec 2019.
- 83.Pedersen, T. ggraph: An implementation of grammar of graphics for graphs and networks version 2.0.0 from CRAN. (2017). https://rdrr.io/cran/ggraph/. Accessed 30 Dec 2019.
- 84.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Conesa A, et al. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 86.Grimplet J, et al. Comparative analysis of grapevine whole-genome gene predictions, functional annotation, categorization and integration of the predicted gene sequences. BMC Res. Notes. 2012;5:213. doi: 10.1186/1756-0500-5-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Naithani S, et al. VitisCyc: A metabolic pathway knowledgebase for grapevine (Vitis vinifera) Front. Plant Sci. 2014;5:644. doi: 10.3389/fpls.2014.00644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Edgar RC. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kassambara, A. & Mundt, F. Factoextra: Extract and visualize the results of multivariate data analyses.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-Seq reads were deposited in the European Nucleotide Archive repository under the Unique Name Experiment ERX2989847. Metabolite data are provided as Supplementary Table S1. ‘Merlot Khorus’ FASTQ files of the reads aligning to chromosome 14 are publicly available at figshare under the https://doi.org/10.6084/m9.figshare.12240881.