Key Points
Question
Are organism-specific International Classification of Diseases, Ninth Revision (ICD-9) administrative codes for pneumonia valid measures in identifying pneumonia etiology?
Findings
In this cross-sectional study of data from 161 529 patients hospitalized with pneumonia between 2010 and 2015, ICD-9 codes had generally low sensitivity but high specificity for pneumonia etiology identified by laboratory testing.
Meaning
In this study, ICD-9 codes appeared to underestimate prevalence of specific organisms.
Abstract
Importance
Administrative databases may offer efficient clinical data collection for studying epidemiology, outcomes, and temporal trends in health care delivery. However, such data have seldom been validated against microbiological laboratory results.
Objective
To assess the validity of International Classification of Diseases, Ninth Revision (ICD-9) organism-specific administrative codes for pneumonia using microbiological data (test results for blood or respiratory culture, urinary antigen, or polymerase chain reaction) as the criterion standard.
Design, Setting, and Participants
Cross-sectional diagnostic accuracy study conducted between February 2017 and June 2019 using data from 178 US hospitals in the Premier Healthcare Database. Patients were aged 18 years or older admitted with pneumonia and discharged between July 1, 2010, and June 30, 2015. Data were analyzed from February 14, 2017, to June 27, 2019.
Exposures
Organism-specific pneumonia identified from ICD-9 codes.
Main Outcomes and Measures
Sensitivity, specificity, positive predictive value, and negative predictive value of ICD-9 codes using microbiological data as the criterion standard.
Results
Of 161 529 patients meeting inclusion criteria (mean [SD] age, 69.5 [16.2] years; 51.2% women), 35 759 (22.1%) had an identified pathogen. ICD-9–coded organisms and laboratory findings differed notably: for example, ICD-9 codes identified only 14.2% and 17.3% of patients with laboratory-detected methicillin-sensitive Staphylococcus aureus and Escherichia coli, respectively. Although specificities and negative predictive values exceeded 95% for all codes, sensitivities ranged downward from 95.9% (95% CI, 95.3%-96.5%) for influenza virus to 14.0% (95% CI, 8.8%-20.8%) for parainfluenza virus, and positive predictive values ranged downward from 91.1% (95% CI, 89.5%-92.6%) for Staphylococcus aureus to 57.1% (95% CI, 39.4%-73.7%) for parainfluenza virus.
Conclusions and Relevance
In this study, ICD-9 codes did not reliably capture pneumonia etiology identified by laboratory testing; because of the high specificities of ICD-9 codes, however, administrative data may be useful in identifying risk factors for resistant organisms. The low sensitivities of the diagnosis codes may limit the validity of organism-specific pneumonia prevalence estimates derived from administrative data.
This cohort study assesses the validity of organism-specific International Classification of Diseases, Ninth Revision (ICD-9) administrative codes for pneumonia in adult patients using microbiological data as the criterion standard.
Introduction
Although detailed clinical data represent the criterion standard for studying epidemiology, outcomes, and temporal trends in health care delivery, such data are cumbersome and expensive to collect. It is difficult to create research data sets large enough to represent the patient mix and the variety of health care settings; medical record abstraction requires intensive review by trained professionals and is subject to interobserver variability and observer bias. The Centers for Disease Control and Prevention directs surveillance of specific health care–associated infections captured by the National Hospital Surveillance Network and engages a small number of academic centers to collect data through the Centers for Disease Control and Prevention Epicenters Program, but these data are limited in scope.1,2 In contrast, administrative data collected during routine clinical encounters for the purpose of reimbursement are copious, widely available, and generalizable. For these reasons, administrative data offer a potential alternative for some types of research. Administrative data have been used, for example, to evaluate temporal trends in pneumonia hospitalization and mortality, but there remains a paucity of efforts to validate administrative data with corresponding clinical information.3 Administrative data can be imprecise, with claims-based algorithms for some conditions demonstrating lower mortality, length of stay, and costs than independent clinical review.4
Validation studies testing the accuracy of pathogen-specific coding have been rare in hospitalizations for infectious diseases in general and in pneumonia in particular. To establish the validity of administrative data regarding pneumonia, we examined the performance of pathogen-specific administrative coding in comparison with corresponding microbiological data in the setting of community-onset pneumonia in a large multicenter US database.
Methods
In this cross-sectional diagnostic accuracy study, we studied patients hospitalized with pneumonia between July 1, 2010, and June 30, 2015, using data from 178 US hospitals in the Premier Healthcare Database. Data were analyzed from February 14, 2017, to June 27, 2019. Using microbiological evidence of a pathogen as the criterion standard (test results for blood or respiratory culture, urinary antigen, or polymerase chain reaction), we derived the performance characteristics (sensitivity, specificity, positive predictive value [PPV], and negative predictive value [NPV]) of the corresponding ICD-9 organism codes as indicators of diagnosis. Because the data source was completely deidentified, the institutional review board of the Cleveland Clinic determined that this study was exempt from review and did not require informed patient consent. This study followed the Standards for Reporting of Diagnostic Accuracy (STARD) reporting guideline for diagnostic accuracy studies.
The Premier Healthcare Database is widely used for research and has been well described elsewhere.5 Between July 1, 2010, and June 30, 2015, the number of participating hospitals increased from 461 to 592. In 2015, 75% of participating hospitals were in urban settings (census block groups or blocks have a population density of at least 1000 people per square mile) and 25% were rural by the US Census Bureau definition (any territory outside urban setting),6 mirroring the membership of the American Hospital Association, although with Midwestern hospitals underrepresented and Southern hospitals overrepresented. Larger hospitals were overrepresented and teaching hospitals were underrepresented in the Premier Healthcare Database. For the current analysis, we included the 178 hospitals in the Premier Healthcare Database that reported microbiological data using the Safety Surveillor web-based tracking tool.
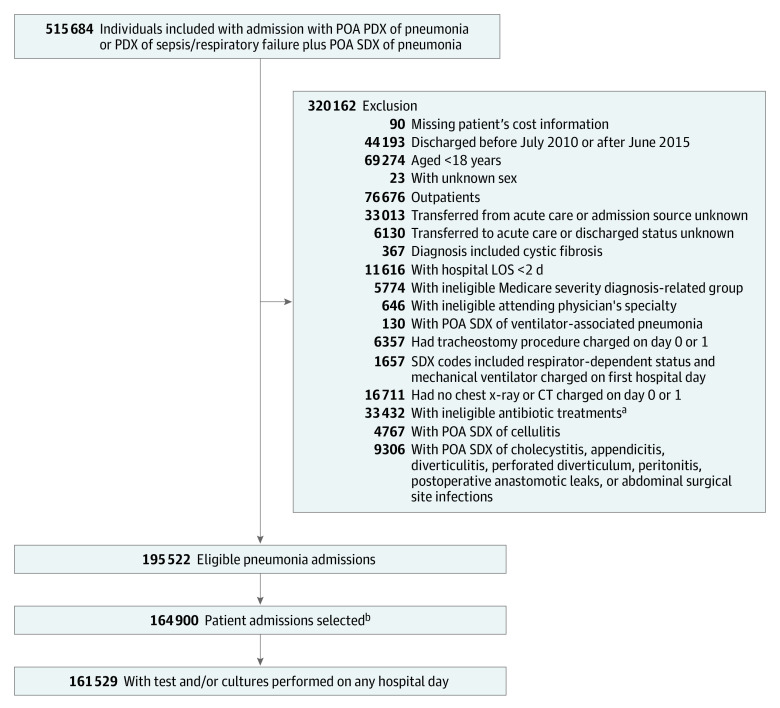
We included all patients aged 18 years or older who were discharged between July 1, 2010, and June 30, 2015, with either a principal diagnosis of pneumonia or with a principal diagnosis of respiratory failure, acute respiratory distress syndrome, respiratory arrest, sepsis, or influenza and a secondary diagnosis of pneumonia (details of the algorithm have been published previously).7 In addition, a blood culture, respiratory culture, pneumococcal urinary antigen, Legionella urinary antigen, or antibody tests or polymerase chain reaction targeting atypical bacterial pathogens (Bordetella pertussis, Chlamydophila pneumoniae, Mycoplasma pneumoniae) or viruses was required for inclusion. Included tests are listed in the eTable in the Supplement. Patients with a secondary diagnosis of cellulitis, cholecystitis, appendicitis, diverticulitis, perforated diverticulum, peritonitis, postoperative anastomotic leaks, or abdominal surgical site infections were excluded (Figure).
Figure. Participant Selection Flow Diagram.
Abbreviations: CT, computed tomography; LOS, length of stay (in days); PDX, principal diagnosis; POA, present on admission; SDX, secondary diagnosis.
aPatients with viral pneumonia as principal diagnosis but without initial antibiotic treatments were included.
bOn a 1-patient 1-admission basis, with a single eligible admission randomly selected from each patient's eligible admissions (178 hospitals).
Statistical Analysis
Baseline characteristics of patients with pneumonia were summarized by frequency distributions for categorical variables and mean (SD) or quartiles for continuous variables. We cross-classified the presence vs absence of an ICD-9 code for each of the 12 common organisms identified as causing pneumonia in hospitalized patients with the presence or absence of a laboratory sample identifying that organism. From these cross-classifications, we calculated 4 measures of ICD-9 code performance in designating a laboratory-confirmed organism: (1) sensitivity, the fraction of patients with a positive laboratory finding of an organism for whom the ICD-9 code of that organism was present; (2) specificity, the fraction of patients without a positive laboratory finding for an organism for whom the ICD-9 code of that organism was also not present; (3) PPV, the fraction of patients with an ICD-9 code of an organism for whom a corresponding laboratory finding was present; and (4) NPV, the fraction of patients without an ICD-9 code for an organism who were also without a laboratory finding for the organism. For the 4 most common organisms, sensitivity, specificity, PPVs, and NPVs were also tracked across year and by whether the diagnosis was primary or secondary. For any particular laboratory test, patients without a result were assumed not to have had that test. Data management and analysis were performed with SAS statistical software version 9.4 (SAS Institute).
Results
The database included 515 684 patients before exclusions (Figure); among the 164 900 patients admitted with pneumonia who met inclusion criteria, cultures were obtained from 161 529 (98.0%) (mean [SD] age, 69.5 [16.2] years; 51.2% women) (Table 1), including blood cultures from 154 034 (93.4%) patients. Most patients (71.8%) were insured by Medicare, 87.8% were admitted through the emergency department, and most had a principal diagnosis of pneumonia (61.9%, including 9.3% by aspiration) or sepsis (32.1%). One-quarter of patients were treated in the intensive care unit, and 8.4% received invasive mechanical ventilation. The in-hospital mortality rate was 9.2%, and median length of stay was 5 days (interquartile range, 3-8 days). Median cost was $8356.41 (interquartile range, $5035.31-$14 928.49). Of the entire eligible cohort, 35 759 (22.1%) had a positive test result.
Table 1. Characteristics of 161 529 Patients Hospitalized With Pneumonia in the 2010-2015 Premier Healthcare Database Sample.
| Characteristic | No. (%) |
|---|---|
| Age, mean (SD), y | 69.5 (16.2) |
| Male | 78 786 (48.8) |
| Race/ethnicity | |
| White | 124 574 (77.1) |
| Black | 20 348 (12.6) |
| Other | 16 607 (10.3) |
| Admission source | |
| Emergency department | 141 861 (87.8) |
| SNF/ICF | 12 431 (7.7) |
| Clinic | 7100 (4.4) |
| Other | 137 (0.08) |
| Discharge status | |
| Home | 95 782 (59.3) |
| Hospice | 8534 (5.3) |
| Died | 14 798 (9.2) |
| SNF | 38 957 (24.1) |
| Other | 3458 (2.1) |
| Insurance | |
| Medicare | 116 020 (71.8) |
| Medicaid | 14 059 (8.7) |
| Commercial/managed care | 22 312 (13.8) |
| Other | 9138 (5.7) |
| Principal diagnosis code | |
| Viral pneumonia | 63 (0.04) |
| Pneumonia | 81 359 (50.4) |
| Aspiration pneumonia | 15 021 (9.3) |
| Influenza pneumonia | 3499 (2.2) |
| Sepsis | 51 823 (32.1) |
| Respiratory failure | 9764 (6.0) |
| Teaching hospital | 66 317 (41.1) |
| Urban setting (US Census Bureau definition)a | 142 356 (88.1) |
| Beds, No. | |
| ≤200 | 33 145 (20.5) |
| 201-400 | 69 184 (42.8) |
| ≥401 | 59 200 (36.6) |
| Region (US Census Bureau classification)8 | |
| Midwest | 40 664 (25.2) |
| Northeast | 30 183 (18.7) |
| South | 69 041 (42.7) |
| West | 21 641 (13.4) |
| Combined comorbidity score, mean (SD)b | 3.2 (2.7) |
| Hypertension | 106 492 (65.9) |
| Fluid and electrolyte disorders | 80 617 (49.9) |
| Chronic pulmonary disease | 75 327 (46.6) |
| Anemia | 53 072 (32.9) |
| Diabetes | 52 827 (32.7) |
| Congestive heart failure | 45 295 (28.0) |
| Other neurological disorders | 27 301 (16.9) |
| Hypothyroidism | 27 452 (17.0) |
| Depression | 25 379 (15.7) |
| Weight loss | 20 720 (12.8) |
| Obesity | 21 797 (13.5) |
| Valvular disease | 15 392 (9.5) |
| Coagulopathy | 15 309 (9.5) |
| Peripheral vascular disease | 13 204 (8.2) |
| Pulmonary circulation disease | 12 820 (7.9) |
| Psychoses | 10 130 (6.3) |
| Admit from SNF | 12 431 (7.7) |
| Previous admission (within 6 mo) | 17 395 (10.8) |
| Immunosuppressed | 21 638 (13.4) |
| ICU | 41 226 (25.5) |
| IMV | 13 572 (8.4) |
| Vasopressor | 11 413 (7.1) |
| In-hospital mortality | 14 798 (9.2) |
| Length of stay, median (IQR), d | 5.0 (3.0-8.0) |
| Cost, median (IQR), $ | 8356.41 (5035.31-14 928.49) |
Abbreviations: ICF, intermediate care facility; ICU, intensive care unit; IMV, invasive mechanical ventilation; IQR, interquartile range; SNF, skilled nursing facility.
Urban setting indicates census block groups or blocks with a population density of at least 1000 people per square mile.6
Theoretical range, −2 (lowest mortality risk) to 20 (highest mortality risk); range in these data, −1 to 19.
Most patients (110 360 [68.3%]) had an ICD-9 code for pneumonia, organism unspecified (486). The organisms most frequently specified were influenza (5891 [3.6%]), S pneumoniae (4090 [2.5%]), and methicillin-resistant Staphylococcus aureus (MRSA) (3747 [2.3%]). Overall, 35 759 (22.1%) patients had a laboratory-identified etiology (19.4% bacterial, 3.2% viral, and 0.1% fungi). Table 2 lists ICD-9 codes and laboratory results. The proportions of patients with positive laboratory findings and with organism-specific positive ICD-9 codes were, respectively, 5.4% and 0.8% for methicillin-susceptible S aureus (MSSA), 3.6% and 2.3% for MRSA, 2.0% and 0.4% for Escherichia coli, 1.3% and 0.6% for Klebsiella pneumoniae, 3.6% and 3.0% for S pneumoniae, and 2.7% and 1.6% for Pseudomonas species.
Table 2. Cross-Classification of Laboratory Findings by ICD-9 Diagnosis Code for 12 Specific Pneumonia Microbial Agents.
| Organism | No. (%) | |||
|---|---|---|---|---|
| ICD-9+/laboratory+ | ICD-9−/ laboratory− | ICD-9+/ laboratory− | ICD-9−/laboratory+ | |
| MSSA | 1233 (0.8) | 152 722 (94.5) | 120 (0.1) | 7454 (4.6) |
| MRSA | 2849 (1.8) | 154 379 (95.6) | 898 (0.6) | 2936 (1.8) |
| Streptococcus pneumoniae | 3525 (2.2) | 154 668 (95.8) | 1280 (0.8) | 2345 (1.5) |
| Pseudomonas spp | 2052 (1.3) | 156 651 (97.0) | 505 (0.3) | 2321 (1.4) |
| Escherichia coli | 557 (0.3) | 158 240 (98.0) | 71 (0.0) | 2661 (1.6) |
| Klebsiella pneumoniae | 774 (0.5) | 159 154 (98.5) | 205 (0.1) | 1396 (0.9) |
| Haemophilus influenzae | 708 (0.4) | 159 747 (98.9) | 128 (0.1) | 946 (0.6) |
| Mycoplasma pneumoniae | 564 (0.3) | 160 126 (99.1) | 348 (0.2) | 491 (0.3) |
| Legionella spp | 486 (0.3) | 160 806 (99.6) | 103 (0.1) | 134 (0.1) |
| Influenza virus | 4168 (2.6) | 155 460 (96.2) | 1723 (1.1) | 178 (0.1) |
| Respiratory syncytial virus | 41 (0.0) | 161 339 (99.9) | 20 (0.0) | 129 (0.1) |
| Parainfluenza virus | 20 (0.0) | 161 371 (99.9) | 15 (0.0) | 123 (0.1) |
Abbreviations: ICD-9, International Classification of Diseases, Ninth Revision; ICD-9+, presence of an ICD-9 code; ICD-9−, absence of an ICD-9 code; laboratory+, presence of a laboratory sample identifying an organism; laboratory−, absence of a laboratory sample identifying an organism; MSSA, methicillin-sensitive Staphylococcus aureus; MRSA, methicillin-resistant Staphylococcus aureus; spp, several species.
Among 34 263 (21.2%) patients with either an organism-specific code or laboratory evidence, concordance between the 2 existed in fewer than half (eFigure in the Supplement). Table 3 shows the characteristics of ICD-9 coding against the microbiology criterion standard for certain common organisms. In general, specificities were high, with 98.9% for influenza virus and 99.9% for MSSA. Sensitivities were substantially lower for most organisms, ranging from 95.9% (95% CI, 95.3%-96.5%) for influenza virus to 14.0% (95% CI, 8.8%-20.8%) for parainfluenza virus. Although both NPV and PPV values were higher than 75% for most bacterial organisms, owing to the low prevalence, the NPVs were substantially higher than the PPVs. The PPVs varied widely, from as low as 57.1% (95% CI, 39.4%-73.7%) for parainfluenza virus to as high as 91.1% (95% CI, 89.5%-92.6%) for MSSA, and were notably lower (57.1%-70.8%) for mycoplasma (61.8%), influenza (70.8%), respiratory syncytial virus (67.2%), and parainfluenza virus (57.1%) than for most other organisms, eg, MRSA (76.0%), E coli (88.7%), and Legionella species (82.5%).
Table 3. Performance Measures of Organism-Specific ICD-9 Pneumonia Diagnosis Codes for Identifying Organism-Positive Laboratory Findings.
| Organism | ICD-9 code | ICD-9 performance vs laboratory criterion standard, %a | |||
|---|---|---|---|---|---|
| Test performance | Predictive value | ||||
| Sensitivity | Specificity | Positive | Negative | ||
| MSSA | 482.41 | 14.2 | 99.9 | 91.1 | 95.4 |
| MRSA | 482.42 | 49.3 | 99.4 | 76.0 | 98.1 |
| Streptococcus pneumoniae | 481, 482.30 | 60.1 | 99.2 | 73.4 | 98.5 |
| Pseudomonas spp | 482.1 | 46.9 | 99.7 | 80.3 | 98.5 |
| Escherichia coli | 482.82 | 17.3 | 100 | 88.7 | 98.4 |
| Klebsiella pneumoniae | 482 | 35.7 | 99.9 | 79.1 | 99.1 |
| Haemophilus influenzae | 482.2 | 42.8 | 99.9 | 84.7 | 99.4 |
| Mycoplasma pneumoniae | 483 | 53.5 | 99.8 | 61.8 | 99.7 |
| Legionella spp | 482.84 | 78.4 | 99.9 | 82.5 | 99.9 |
| Influenza virus | 487.x, 488.x | 95.9 | 98.9 | 70.8 | 99.9 |
| Respiratory syncytial virus | 480.1 | 24.1 | 100 | 67.2 | 99.9 |
| Parainfluenza virus | 480.2 | 14.0 | 100 | 57.1 | 99.9 |
Abbreviations: ICD-9, International Classification of Diseases, Ninth Revision; MRSA, methicillin-resistant Staphylococcus aureus; MSSA, methicillin-sensitive Staphylococcus aureus; spp several species.
Microbiological data (test results for blood or respiratory culture, urinary antigen, or polymerase chain reaction) are considered as the criterion standard.
Temporal trends for 5 selected organisms are given in Table 4. Despite year-to-year variance, there do not appear to be consistent trends in sensitivity, specificity, or PPVs.
Table 4. Temporal Trends of ICD-9 and Laboratory Data for Selected Organisms.
| Organism | Performance | % | |||||
|---|---|---|---|---|---|---|---|
| Overall (n = 161 529) | July 2010-June 2011 (n = 37 259) | July 2011-June 2012 (n = 31 302) | July 2012-June 2013 (n = 33 803) | July 2013-June 2014 (n = 32 388) | July 2014-June 2015 (n = 30 148) | ||
| MSSA | ICD-9+ | 0.8 | 0.8 | 0.8 | 0.8 | 0.9 | 0.9 |
| Laboratory+ | 5.4 | 5.8 | 5.4 | 5.4 | 5.3 | 4.8 | |
| Sensitivity | 14.2 | 13.4 | 12.6 | 13.4 | 15.7 | 16.4 | |
| Specificity | 99.9 | 99.9 | 99.9 | 99.9 | 99.9 | 99.9 | |
| PPV | 91.1 | 92.8 | 91.3 | 89.3 | 90.7 | 91.4 | |
| NPV | 95.4 | 94.9 | 95.2 | 95.3 | 95.5 | 95.9 | |
| MRSA | ICD-9+ | 2.3 | 2.4 | 2.4 | 2.4 | 2.4 | 2.1 |
| Laboratory+ | 3.6 | 4.0 | 3.7 | 3.5 | 3.4 | 3.2 | |
| Sensitivity | 49.3 | 47.1 | 48.9 | 50.3 | 50.7 | 49.9 | |
| Specificity | 99.4 | 99.5 | 99.4 | 99.4 | 99.3 | 99.5 | |
| PPV | 76.0 | 80.3 | 75.7 | 75.6 | 72.4 | 75.5 | |
| NPV | 98.1 | 97.8 | 98.1 | 98.2 | 98.3 | 98.4 | |
| Streptococcus pneumoniae | ICD-9+ | 3.0 | 3.2 | 2.8 | 3.3 | 2.9 | 2.6 |
| Laboratory+ | 3.6 | 3.9 | 3.5 | 4.0 | 3.5 | 3.1 | |
| Sensitivity | 60.1 | 61.8 | 60.2 | 61.3 | 58.7 | 56.9 | |
| Specificity | 99.2 | 99.2 | 99.3 | 99.1 | 99.1 | 99.2 | |
| PPV | 73.4 | 75.5 | 74.9 | 74.3 | 70.8 | 70.1 | |
| NPV | 98.5 | 98.5 | 98.6 | 98.4 | 98.5 | 98.6 | |
| Pseudomonas spp | ICD-9+ | 1.6 | 1.8 | 1.7 | 1.6 | 1.4 | 1.3 |
| Laboratory+ | 2.7 | 3.0 | 3.0 | 2.7 | 2.5 | 2.3 | |
| Sensitivity | 46.9 | 49.5 | 44.5 | 48.3 | 45.9 | 45.5 | |
| Specificity | 99.7 | 99.6 | 99.7 | 99.7 | 99.7 | 99.7 | |
| PPV | 80.3 | 81.0 | 79.4 | 79.8 | 80.8 | 80.1 | |
| NPV | 98.5 | 98.5 | 98.3 | 98.6 | 98.6 | 98.7 | |
| Influenza virus | ICD-9+ | 3.7 | 1.9 | 0.9 | 4.1 | 4.7 | 6.9 |
| Laboratory+ | 2.7 | 1.3 | 0.6 | 3.1 | 3.3 | 5.4 | |
| Sensitivity | 95.9 | 91.2 | 92.9 | 96.0 | 96.9 | 96.9 | |
| Specificity | 98.9 | 99.3 | 99.7 | 98.8 | 98.4 | 98.3 | |
| PPV | 70.8 | 61.3 | 63.8 | 72.2 | 67.4 | 76.4 | |
| NPV | 99.9 | 99.9 | 100 | 99.9 | 99.9 | 99.8 | |
Abbreviations: ICD-9, International Classification of Diseases, Ninth Revision; ICD-9+, presence of an ICD-9 code; laboratory+, presence of a laboratory sample identifying an organism; MRSA, methicillin-resistant Staphylococcus aureus; MSSA, methicillin-sensitive Staphylococcus aureus; PPV, positive predictive value; NPV, negative predictive value; spp, several species.
Discussion
In this cross-sectional diagnostic study of more than 160 000 patients undergoing culture or antigen testing for pneumonia infection in 178 US hospitals, we found that just 35 759 (22.1%) had an identified pathogen. ICD-9-coded organisms and laboratory findings differed notably. Although specificities and NPVs exceeded 95% for all codes, sensitivities ranged from 95.9% for influenza virus to 14.0% for parainfluenza virus, and PPVs were as high as 91.1% (95% CI, 89.5%-92.6%) for S aureus and as low as 57.1% (95% CI, 39.4%-73.7%) for parainfluenza virus. Because of the high specificities, for most diagnoses an ICD-9 code was a reliable marker of a positive culture, but because of the low sensitivities, use of only administrative codes may potentially undercount almost all diagnoses.
Previous studies have examined the concordance between administrative and clinical data in various infectious syndromes, including pneumonia. Guevara and colleagues9 reported sensitivity of 58.3% for the pneumococcal pneumonia code (481.0), similar to what we observed. Schweizer et al10 questioned the validity of the ICD-9 code for identifying incident MRSA infection ([V09] not limited to pneumonia), and found sensitivity of 24% with a PPV of 31%, lower than our findings but using a different coding approach. In a study focused on multidrug-resistant organisms, Burnham and colleagues11 found that a higher rate of coding for MRSA was associated with infectious disease consultation, and counseled against using that ICD-9 code to estimate rates of multidrug-resistant organism infection in hospitals. The present study expands on this body of validation work by increasing the pool of common pneumonia pathogens beyond S pneumoniae and MRSA.
Understanding the epidemiology of pneumonia is important for resource allocation and risk prediction based on demographic characteristics and geospatial location.12 The low sensitivity of administrative data with regard to specific microorganisms has implications for interpreting epidemiological studies. Smith and colleagues,13 for example, used the Nationwide Inpatient Sample to explore the association between introduction of pneumococcal vaccine and distribution of pathogens among admissions of patients with pneumonia. They reported a reduction in S pneumoniae among pneumonia codes following the year 2000, suggesting that the pneumococcal vaccination was conferring its desired benefits. Our findings that the S pneumoniae ICD-9 code identifies only 54% of culture-confirmed infections might call such an assertion into question. However, if coding practices remained constant throughout the study time frame, relative reductions in longitudinal trends would be unaffected by such discrepancies. We found that over a 5-year period sensitivity of the S pneumoniae code declined slightly, while specificity remained constant.
The high specificities of administrative codes for individual uncommon organisms make administrative data well suited for deriving predictive models, because specificity is a primary component of PPV when prevalence is low. PPV exceeded 70% for influenza and all bacterial species except mycoplasma, suggesting that characteristics of patients who have codes for specific organisms may be representative of patients who actually have infection with those organisms. At least 9 models have been created to predict drug-resistant organisms, such MRSA and Pseudomonas species, in pneumonia. Nearly all models perform better than the health care–associated pneumonia criteria, but no single model is yet accurate enough to guide antibiotic stewardship.14,15 More sophisticated models would be particularly useful as decision aids to help clinicians optimize empirical treatment while avoiding overuse of broad-spectrum agents. High specificity is critical to ensure the accuracy of such predictive instruments. Although missing cases may have minimal implications for the model’s discrimination, low sensitivity of ICD-9 data could result in miscalibration, consistently underpredicting risk.
Similarly, changing resistance patterns over time are of concern to clinicians prescribing empirical antibiotic therapy. The Centers for Disease Control and Prevention National Healthcare Safety Network provides robust data describing trends in resistance, for example, changing prevalence of MRSA and the emergence of multidrug-resistant gram-negative bacteria.16 Our temporal analysis shows that the association between administrative and laboratory data for MSSA and MRSA has been stable between 2010 and 2014, with a slight uptick in sensitivity in 2015 for both organisms. This finding suggests utility for efforts at more generalizable infection surveillance using administrative data.
Limitations
This study has limitations. The case selection algorithm may have insufficiently discriminated pneumonia from other infection diagnoses, and identified pathogens may represent colonization rather than infection. For example, many patients had culture growth but no corresponding ICD-9 coding event. This subset of patients tended to have more comorbid conditions. Although all patients had a diagnosis of pneumonia, it is possible that the specific pneumonia code was missed owing to truncation of diagnoses. MRSA and MSSA may also be coded as present based on the result of a nasal swab, which we did not include because nasal passages may not accurately represent lung flora. The presence of an ICD-9 code indicating infection without culture results could represent coding based on clinical suspicion, available data from a transferring institution, or late reporting of growth after hospital discharge. We also excluded patients who did not have any cultures. Although this number was small, it may have introduced a bias in detection rates.
Of note, International Statistical Classification of Diseases and Related Health Problems, Tenth Revision (ICD-10) coding was implemented in the US on October 1, 2015, and is far more detailed than ICD-9, making it difficult to study a population across the transition. Thus, our analysis is limited to 2010-2015. Nonetheless, the process by which codes are chosen has not changed, and it is possible to crosswalk from ICD-10 to ICD-9. In addition, it is not known how our results may be applicable to other large administrative data sets. Because this was a national sample, coding is likely to be similar, but it is possible that hospitals outside of the Premier Healthcare Database have different coding patterns. Validation in additional data sets would be welcome.
Conclusions
In this study, organism-specific administrative codes in hospitalized patients undergoing laboratory testing for infection appear to have limited sensitivities in the setting of pneumonia, although specificities and NPVs are high, and PPVs are reasonable considering the low pretest probabilities and consequent challenges of ruling in specific organisms. This finding may have important implications for the reliability of research conducted in administrative databases. Although the high specificity is conducive to predictive modeling, low sensitivities may limit the utility of organism-specific administrative codes for surveillance purposes, as organism-specific prevalence estimates based on administrative codes may underestimate true organism-specific burden. Future studies may need to examine whether microbiology trends indicated by ICD-9 codes represent actual pathogen shifts or are consequences of alterations in coding practices.
eFigure. Venn Diagram of Etiology and Laboratory Result
eTable. Tests Required for Study Inclusion
References
- 1.CDC/NHSN Surveillance of Healthcare-Associated Infections Accessed August 1, 2019. https://www.cdc.gov/nhsn/acute-care-hospital/index.html
- 2.CDC’s Prevention Epicenters Program Accessed August 1, 2019. https://www.cdc.gov/hai/epicenters/index.html
- 3.Lindenauer PK, Lagu T, Shieh MS, Pekow PS, Rothberg MB. Association of diagnostic coding with trends in hospitalizations and mortality of patients with pneumonia, 2003-2009. JAMA. 2012;307(13):1405-1413. doi: 10.1001/jama.2012.384 [DOI] [PubMed] [Google Scholar]
- 4.Aronsky D, Haug PJ, Lagor C, Dean NC. Accuracy of administrative data for identifying patients with pneumonia. Am J Med Qual. 2005;20(6):319-328. doi: 10.1177/1062860605280358 [DOI] [PubMed] [Google Scholar]
- 5.Rothberg MB, Pekow PS, Priya A, et al. Using highly detailed administrative data to predict pneumonia mortality. PLoS One. 2014;9(1):e87382. doi: 10.1371/journal.pone.0087382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.The US Census Bureau Urban and rural. Accessed June 24, 2020. https://www.census.gov/programs-surveys/geography/guidance/geo-areas/urban-rural.html
- 7.Gupta NM, Lindenauer PK, Yu PC, et al. Association between alcohol use disorders and outcomes of patients hospitalized with community-acquired pneumonia. JAMA Netw Open. 2019;2(6):e195172. doi: 10.1001/jamanetworkopen.2019.5172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.The US Census Bureau Statistical groupings of states and counties. Accessed June 24, 2020. https://www2.census.gov/geo/pdfs/reference/GARM/Ch6GARM.pdf
- 9.Guevara RE, Butler JC, Marston BJ, Plouffe JF, File TM Jr, Breiman RF. Accuracy of ICD-9-CM codes in detecting community-acquired pneumococcal pneumonia for incidence and vaccine efficacy studies. Am J Epidemiol. 1999;149(3):282-289. doi: 10.1093/oxfordjournals.aje.a009804 [DOI] [PubMed] [Google Scholar]
- 10.Schweizer ML, Eber MR, Laxminarayan R, et al. Validity of ICD-9-CM coding for identifying incident methicillin-resistant Staphylococcus aureus (MRSA) infections: is MRSA infection coded as a chronic disease? Infect Control Hosp Epidemiol. 2011;32(2):148-154. doi: 10.1086/657936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burnham JP, Kwon JH, Babcock HM, Olsen MA, Kollef MH. ICD-9-CM coding for multidrug resistant infection correlates poorly with microbiologically confirmed multidrug resistant infection. Infect Control Hosp Epidemiol. 2017;38(11):1381-1383. doi: 10.1017/ice.2017.192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramirez JA, Wiemken TL, Peyrani P, et al. ; University of Louisville Pneumonia Study Group . Adults hospitalized with pneumonia in the United States: incidence, epidemiology, and mortality. Clin Infect Dis. 2017;65(11):1806-1812. doi: 10.1093/cid/cix647 [DOI] [PubMed] [Google Scholar]
- 13.Smith SB, Ruhnke GW, Weiss CH, Waterer GW, Wunderink RG. Trends in pathogens among patients hospitalized for pneumonia from 1993 to 2011. JAMA Intern Med. 2014;174(11):1837-1839. doi: 10.1001/jamainternmed.2014.4344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kobayashi D, Shindo Y, Ito R, et al. Validation of the prediction rules identifying drug-resistant pathogens in community-onset pneumonia. Infect Drug Resist. 2018;11:1703-1713. doi: 10.2147/IDR.S165669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Webb BJ, Dascomb K, Stenehjem E, Dean N. Predicting risk of drug-resistant organisms in pneumonia: moving beyond the HCAP model. Respir Med. 2015;109(1):1-10. doi: 10.1016/j.rmed.2014.10.017 [DOI] [PubMed] [Google Scholar]
- 16.McDonald LC. Trends in antimicrobial resistance in health care-associated pathogens and effect on treatment. Clin Infect Dis. 2006;42(suppl 2):S65-S71. doi: 10.1086/499404 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eFigure. Venn Diagram of Etiology and Laboratory Result
eTable. Tests Required for Study Inclusion