Extended-spectrum β-lactamase (ESBL)- and AmpC β-lactamase (AmpC)-producing E. coli isolates have emerged in recent years as some of the fastest spreading antimicrobial resistance determinants in humans and food-producing animals, becoming a concern for animal and public health. This study provided insight into the prevalence of cefotaxime-resistant E. coli in cattle and sheep in the Basque Country and the associated genetic determinants of antimicrobial resistance. These constituted an important contribution to the limited repository of such data for cattle in the region and for sheep worldwide. Antimicrobial susceptibility testing by phenotypic and molecular methods is key in surveillance programs to enhance early detection of resistance development, monitor resistance trends, and provide guidance to clinicians in selecting the adequate therapy.
KEYWORDS: AmpC β-lactamase-producing E. coli, AmpC, extended-spectrum β-lactamase-producing E. coli, ESBL, minimum inhibitory concentration, MIC, antimicrobial resistance, AMR, beef cattle, dairy cattle, sheep, whole-genome sequencing, WGS
ABSTRACT
In order to estimate herd-level prevalence of extended-spectrum β-lactamase/AmpC β-lactamase (ESBL/AmpC)- and carbapenemase-producing commensal Escherichia coli in ruminants in the Basque Country (northern Spain), a cross-sectional survey was conducted in 2014 to 2016 in 300 herds using selective isolation. ESBL-/AmpC-producing E. coli was isolated in 32.9% of dairy cattle herds, 9.6% of beef cattle herds, and 7.0% of sheep flocks. No carbapenemase-producing E. coli was isolated. Phenotypic antimicrobial susceptibility determined by broth microdilution using EUCAST epidemiological cutoff values identified widespread coresistance to extended-spectrum cephalosporins and other antimicrobials (110/135 isolates), particularly tetracycline, sulfamethoxazole, trimethoprim, and ciprofloxacin. All isolates were susceptible to tigecycline, imipenem, meropenem, and colistin. The genomes of 66 isolates were sequenced using an Illumina NovaSeq 6000 and screened for antimicrobial resistance determinants against ResFinder and PointFinder. The plasmid/chromosomal locations of resistance genes were predicted with PlasFlow, and plasmid replicons were identified using PlasmidFinder. Fifty-two acquired resistance genes and point mutations in another four genes that coded for resistance to 11 antimicrobial classes were identified. Fifty-five genomes carried ESBL-encoding genes, blaCTX-M-14 being the most common, and 11 carried determinants of the AmpC phenotype, mostly the blaCMY-2 gene. Additionally, genes coding for β-lactamases of the CTX-M group 9 were detected as well as the sporadic presence of blaSHV-12, blaCMY-4, and a point mutation in the ampC promoter. Only a bovine isolate coharbored more than one ESBL/AmpC genetic determinant (blaCTX-M-14 and a mutation in the ampC promoter), confirming its ESBL- and AmpC β-lactamase-producing phenotype. Most ESBL/AmpC genes were located in IncI1 plasmids, which also carried a great variety of other antimicrobial resistance genes.
IMPORTANCE Extended-spectrum β-lactamase (ESBL)- and AmpC β-lactamase (AmpC)-producing E. coli isolates have emerged in recent years as some of the fastest spreading antimicrobial resistance determinants in humans and food-producing animals, becoming a concern for animal and public health. This study provided insight into the prevalence of cefotaxime-resistant E. coli in cattle and sheep in the Basque Country and the associated genetic determinants of antimicrobial resistance. These constituted an important contribution to the limited repository of such data for cattle in the region and for sheep worldwide. Antimicrobial susceptibility testing by phenotypic and molecular methods is key in surveillance programs to enhance early detection of resistance development, monitor resistance trends, and provide guidance to clinicians in selecting the adequate therapy.
INTRODUCTION
Antimicrobial resistance (AMR) is considered an emerging problem on a worldwide scale. It has been estimated that unless actions are taken, AMR could cause up to 10 million deaths each year by 2050 (1). The WHO list of critically important antimicrobials for human medicine includes third-generation and higher cephalosporins as well as carbapenems, as these antimicrobials are either the sole or one of the limited therapies available to treat multidrug-resistant (MDR) bacteria in human infections (2). In Escherichia coli, the most important mechanism of resistance to those critically important β-lactam antimicrobials is the production of extended spectrum β-lactamases (ESBLs), followed by the production of AmpC cephalosporinases and carbapenemase enzymes. ESBLs are capable of hydrolyzing penicillins, third-generation cephalosporins, and monobactams (e.g., aztreonam). They are not active against cephamycins (e.g., cefoxitin) or carbapenems but are susceptible to β-lactamase inhibitors like clavulanic acid. AmpC-type β-lactamases, unlike ESBLs, are active against cephamycins and resistant to inhibition by clavulanate. Carbapenemases are carbapenem-hydrolyzing β-lactamases that confer resistance to a broad spectrum of β-lactams, including carbapenems.
ESBL/AmpC-producing E. coli isolates have emerged in recent years, becoming one of the fastest-spreading AMR determinants not only in humans but also in food-producing, companion, and wild animals as well as the environment (3–6). Despite the wide distribution of ESBL/AmpC-producing E. coli isolates in livestock, their contribution as a source of human infection remains controversial (7). Although humans seem to be the main source of community-acquired infections by ESBL/AmpC-producing E. coli, nonhuman sources act as important reservoirs that contribute to further spread of the infection (8–10). Moreover, the prevalence of cefotaxime-resistant bacteria in food-producing animals varies by country and animal species, and some of the animals, like small ruminants, have received less attention than others (4). On the other hand, carbapenemase-producing E. coli isolates (CPEs) are more prevalent in humans than in animals. The use of carbapenems in livestock is banned, and CPEs have only rarely been identified in food-producing animals in Europe (4, 11).
ESBLs are mostly plasmid-mediated enzymes, with CTX-M-1, CTX-M-14, and CTX-M-15 being the most frequently described in E. coli isolated from cattle (12). In fact, the first description of ESBL-producing E. coli in cattle in Spain was a CTX-M-1 cephalosporinase-bearing strain isolated from a cattle with mastitis (13). AmpC enzymes in E. coli from livestock are mainly encoded by blaCMY genes located in plasmids (14) and also by mutations in the promoter region of the chromosomal ampC gene. The latter, normally repressed or only weakly expressed, leads to constitutive hyperexpression of the gene, resulting in β-lactam resistance (15). The most frequently detected carbapenemases in livestock in Europe are OXA-48 and VIM-1, but evidence of the dissemination of NDM, KPC, and IMP carbapenemases has also been reported globally (11).
Phenotypic detection of ESBL, AmpC, and carbapenemase producers among E. coli isolates from food-producing animals is important for epidemiological purposes. However, molecular determination of AMR genetic determinants provides insight into the resistance mechanisms. For this purpose, whole-genome sequencing (WGS) has proven to provide a practical advantage compared to other commonly used molecular methods. The aim of this study was to determine the occurrence of ESBL-, AmpC-, and carbapenemase-producing commensal E. coli in dairy cattle, beef cattle, and sheep without clinical signs of disease in farms in the Basque Country (northern Spain) by using selective isolation methods and to characterize the AMR profiles of the isolates obtained. Phenotypic antimicrobial susceptibility was tested, and a selection of isolates was subjected to WGS in order to assess not only the carriage of the ESBL/AmpC and carbapenemase coding genes but also genetic determinants for resistance to other antimicrobials.
RESULTS
ESBL/AmpC- and carbapenemase-producing E. coli herd prevalence.
E. coli was isolated in cefotaxime-containing medium in samples collected from 15.0% (45/300) of the herds/flocks, with different prevalence distributions according to the production system (Table 1). No E. coli isolates were recovered from the chromogenic medium used to screen for CPEs. Univariate analyses performed to assess factors associated with shedding prevalence of cefotaxime-resistant E. coli identified season, presence of other species in the farm, and herd size as potential confounder variables, but only season passed to the final model (see Table S1 in the supplemental material). Multivariate logistic regression analysis indicated that bovines were more likely to shed cefotaxime-resistant E. coli than sheep (adjusted odds ratio [ORadj], 3.55 [range, 1.57 to 8.04], P = 0.002). When the host was categorized according to the production system, the herd prevalence of cefotaxime-resistant E. coli was significantly higher in dairy cattle than in beef cattle (ORadj, 3.71 [1.60 to 8.58], P = 0.002) and sheep (ORadj, 6.11 [2.55 to 14.60], P < 0.001). Shedding of cefotaxime-resistant E. coli was higher during spring, summer, and autumn than in winter, this difference being largest between autumn and winter (Table S1).
TABLE 1.
Proportion of herds/flocks in which E. coli was isolated from cefotaxime-containing medium and distribution of phenotypically inferred phenotypes
| Host | Growth in cefotaxime-containing medium |
Inferred phenotype |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| ESBL |
AmpC |
ESBL+AmpC |
|||||||
| n | % | 95% CI | n | % | n | % | n | % | |
| Beef cattle (n = 104) | 10 | 9.6 | 4.1–15.2 | 9 | 8.7 | 1 | 1.0 | 0 | 0.0 |
| Dairy cattle (n = 82) | 27 | 32.9 | 23.8–42.1 | 20 | 24.4 | 5 | 6.1 | 2b | 2.4 |
| Sheep (n = 114) | 8 | 7.0 | 2.8–11.2 | 7a | 6.1 | 1 | 0.9 | 1a | 0.9 |
| Total (n = 300) | 45 | 15.0 | 11.2–18.9 | 36 | 12.0 | 7 | 2.3 | 3 | 1.0 |
In one sheep flock, two of the three isolates characterized had an ESBL phenotype and another isolate had the ESBL+AmpC phenotype.
Only one was confirmed as ESBL+AmpC by WGS analyses (see the text).
Antimicrobial resistance phenotype as determined by broth microdilution (MICs).
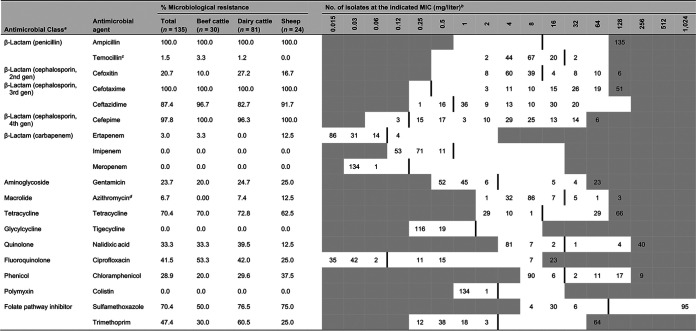
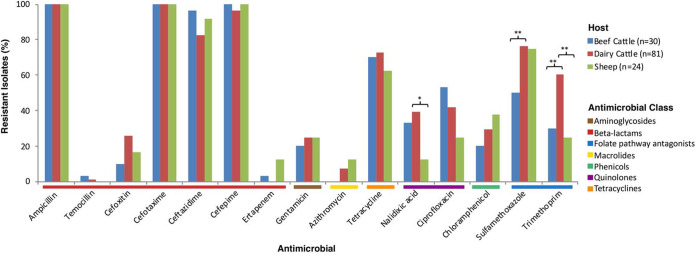
MICs were determined for a total of 135 isolates (3 per positive herd/flock). Antimicrobials tested, distribution of MICs, and interpretation of results are shown in Table 2. All isolates were susceptible to imipenem, meropenem, tigecycline, and colistin. Since isolates had been obtained by selective isolation in medium containing cefotaxime (FOT), they were all resistant to cefotaxime and also to ampicillin. Most isolates (97.8%) were also resistant to the fourth-generation cephalosporin cefepime, while resistance to ceftazidime (TAZ; third-generation cephalosporin) was present in 87.4% of the isolates and resistance to cefoxitin (FOX; second-generation cephalosporin) was present in 20.7% (Fig. 1). Two isolates were resistant to temocillin (MIC = 32 mg/liter). Although no carbapenemase-producing E. coli isolates were found, one isolate from beef cattle and three from sheep were resistant to ertapenem (MIC = 0.12 mg/liter). In addition to this high resistance to β-lactams, coresistance to other antimicrobial classes was also observed in most cases (110/135 isolates). Resistance to other antimicrobials included tetracycline (70.4%), sulfamethoxazole (70.4%), trimethoprim (47.4%), ciprofloxacin (41.5%), nalidixic acid (33.3%), chloramphenicol (28.9%), gentamicin (23.7%), and azithromycin (6.7%; MIC >16 mg/liter). Thus, coresistance to cephalosporins and tetracycline occurred in 70.4% of isolates, coresistance to cephalosporins, tetracycline, and ciprofloxacin occurred in 34.8%, and coresistance to cephalosporins, tetracycline, ciprofloxacin, and sulfamethoxazole-trimethoprim was found in 24.4%. Significant differences among hosts in AMR rates were observed only against nalidixic acid, trimethoprim, and sulfamethoxazole, with dairy cattle presenting a significantly higher prevalence of resistance (Fig. 1). Specifically, proportions of resistant isolates were higher in dairy cattle than in sheep for nalidixic acid (OR = 4.81, P = 0.017) and trimethoprim (OR = 4.59, P = 0.004) and were higher in dairy cattle than in beef cattle for sulfamethoxazole (OR = 3.26, P = 0.009) and trimethoprim (OR = 3.57, P = 0.006).
TABLE 2.
Distribution of MIC values for the 135 E. coli isolates from cefotaxime-containing medium
gen, generation.
White fields denote range of dilutions tested for each antimicrobial agent. MICs above the range are given as the concentration closest to the range, except for that for sulfamethoxazole, which is given as the highest concentration tested. MICs equal to or lower than the lowest concentration tested are given as the lowest concentration tested. Vertical lines indicate European Committee for Antimicrobial Susceptibility Testing (EUCAST) epidemiological cutoff (ECOFF) values.
Since 23 March 2020, the ECOFF for temocillin has been fixed at 16 mg/liter.
FIG 1.
Proportion of isolates microbiologically resistant to different antimicrobials in the different production systems based on phenotypic characterization by broth microdilution. Antimicrobials are grouped according to their corresponding antimicrobial classes, which are color coded. The asterisks denote significant differences (*, P ≤ 0.05; **, P ≤ 0.01).
When MIC data were used for the phenotypic detection of ESBL and AmpC production, most of the 135 tested isolates presented a characteristic ESBL phenotype (107/135, 79.3%), 21 (15.6%) had an AmpC phenotype, and the remaining 7 (5.2%) had an ESBL- and AmpC β-lactamase-producing (ESBL+AmpC) phenotype. The prevalence of inferred phenotypes within each production system is shown in Table 1.
WGS and antimicrobial resistance genotype.
Sixty-six isolates (13 beef cattle, 43 dairy cattle, and 10 sheep) were selected for whole-genome sequencing (WGS) based on their presumptive phenotypic AMR profiles (52 ESBL, 11 AmpC, and 3 ESBL+AmpC). The sequencing facility provided an average of 8.6 million ± 1.6 million reads per sample (range, 5.2 to 12.1 million) corresponding to an average coverage of 258× ± 48× (range, 157× to 364×) in a 5-Mb genome with a mean of quality reads of 36.1. The median N50 of assemblies was 154 kb (interquartile range [IQR], 117 to 185 kb). The median number of contigs recovered per sample was 277 (IQR, 189 to 337), with an average contig length of 442 ± 130 kb (range, 202 to 736 kb) (Table S2).
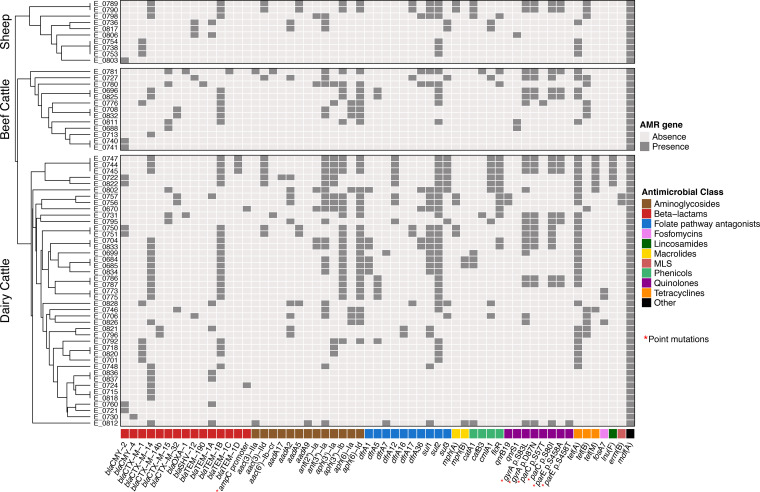
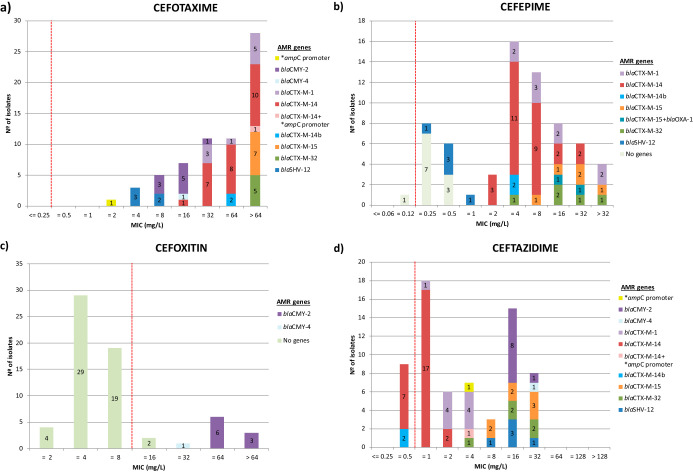
WGS analyses identified 52 acquired AMR genes along with point mutations in another four genes (Fig. 2) that code for resistance to antimicrobials representing 11 different classes. The presence of ESBL-encoding genes was detected in 55 of the 66 genomes. Most of them carried blaCTX-M type genes (50/55, 90.9%), the most abundant being blaCTX-M-14 (n = 27), followed by blaCTX-M-1 (n = 9), blaCTX-M-15 (n = 7), blaCTX-M-32 (n = 5), and blaCTX-M-14b (n = 2). blaSHV genes were only sporadically found (blaSHV-12, n = 5). Ten isolates carried the AmpC-encoding genes blaCMY-2 (n = 9) and blaCMY-4 (n = 1). blaCMY-2 was found in combination with blaTEM-1B in four isolates and with blaTEM-1A in one. Additionally, a point mutation in the ampC promoter (nt 42 C→T) was found in two isolates, in one of them in combination with blaCTX-M-14, this being the only isolate coharboring more than one ESBL/AmpC genetic determinant. In this isolate, the presence of both blaCTX-M-14 and a mutation in the ampC promoter resulted in a much higher MIC value for cefotaxime (MIC > 64 mg/liter) than that of the isolate that carried only the ampC mutation (MIC = 2 mg/liter). However, no difference was observed in ceftazidime and cefoxitin MIC values. The distribution of MIC values and the presence of the different ESBL/AmpC coding genes associated with some of the β-lactams tested are shown in Fig. 3. Other genes coding only for resistance to narrow-spectrum β-lactamases were also detected, such as TEM type genes (41/60, 68.3%), including blaTEM-1B (n = 30), blaTEM-1A (n = 9), blaTEM-1D (n = 3), and blaTEM-1C and blaTEM-190 (one isolate each), as well as blaOXA-1 (n = 2). The majority of ESBL/AmpC genes (61/67) were associated with plasmid-derived contigs; the only chromosomally located ESBL gene was blaCTX-M-15 in a single isolate. Most of the ESBL/AmpC gene-carrying plasmids were identified as IncI1 (Data Set S1). Thus, blaCMY-2, blaCTX-M-1, blaCTX-M-14, and blaSHV-12 were always associated with IncI1 plasmids. On the other hand, blaCTX-M-14 was found not only in IncI1 (n = 3) but also in other rec types (IncB/O/K/Z, n = 9; IncHI2, n = 2), while plasmid-located blaCMY-4 and blaCTX-M-15 genes were associated with IncQ1 and p0111, respectively.
FIG 2.
Heat map showing the distribution of AMR genes detected by WGS in each isolate stratified by production system. Within each production system, samples were grouped based on their antimicrobial resistance pattern according to the result of the hierarchical clustering using the average linkage method (UPGMA) on the Euclidean distance matrix. Genetic determinants of resistance are grouped according to their corresponding antimicrobial classes, which are color coded. Point mutations are indicated by red asterisks.
FIG 3.
Bar plots illustrating the phenotypic and genotypic characterization of resistance to the β-lactams cefotaxime (a), cefepime (b), cefoxitin (c), and ceftazidime (d). Numbers within the stacked bar plots indicate the number of isolates observed with a particular MIC and genotype. ECOFF values are indicated with red dashed lines. Point mutations are indicated by asterisks. “No genes” refers to those isolates lacking any genetic determinant of resistance for the corresponding antimicrobial.
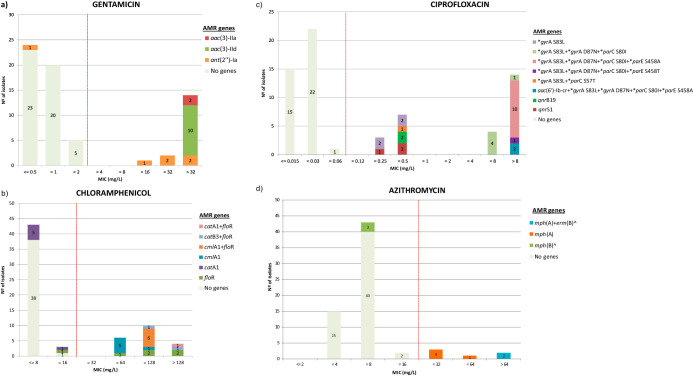
Other genetic determinants of AMR found in the isolates included those associated with resistance to tetracyclines, aminoglycosides, phenicols, quinolones, sulfamethoxazole, trimethoprim, macrolides, lincosamide, and fosfomycin, many of them located in plasmids (Data Set S1). Briefly, all tetracycline-resistant isolates (n = 48) carried a tetracycline efflux gene, tet(A) being the most prevalent (37/48), although tet(B) and tet(M) were also detected, in 14 and 7 isolates, respectively. Thirteen genes associated with resistance to aminoglycosides were detected, including genes encoding aminoglycoside acetyltransferases (aac), nucleotidyltransferases (ant), phosphotransferases (aph), and adenylyltransferases (aad). The distribution of those associated with gentamicin resistance in relation to their MIC values is shown in Fig. 4a. Resistance to phenicols was encoded by four genes, mainly those that activate efflux of phenicols (floR and cmlA) and also genes that mediate the enzymatic inactivation by chloramphenicol acetyltransferases (catA and catB) (Fig. 4b). Resistance to (fluoro)quinolones was associated with mutations in the gyrase (gyrA) gene alone and mostly in combination with different types of mutations in the topoisomerase genes, parC (codons 57 and 80) and parE (codons 355 and 458) (Fig. 4c). In addition, two isolates harbored the aac(6´)-Ib-cr acetyltransferase gene, which confers resistance to ciprofloxacin. Another five isolates carried qnr genes (qnrB19 and qnrS1), which confer resistance to ciprofloxacin (MIC = 0.25 mg/liter, n = 1; MIC = 0.5 mg/liter, n = 4). Resistance to sulfamethoxazole was in all cases (49 isolates from which 35 were also resistant to trimethoprim) mediated by one or different combinations of the three sul genes (sul1, 42.9%; sul2, 81.6%; sul3, 24.5%), whereas resistance to trimethoprim was in all cases encoded by different dfr genes coding for dihydrofolate reductases. However, four isolates with reduced susceptibility to trimethoprim did not present any genes coding for a phenotype of trimethoprim resistance when the ResFinder database was searched. Nevertheless, they all carried a gene that showed 100% homology with dfrA36 (GenBank accession number CP038791), which was also found in another three isolates that also carried the dfrA1 gene. Macrolide resistance-encoding genes were sporadically identified and included mph(A) (n = 6), mph(B) (n = 3), and also erm(B) (n = 2). All six azithromycin isolates with an MIC of >16 mg/liter carried the mph(A) gene, and the two with the highest MIC value (>64 mg/liter) also carried erm(B) (Fig. 4d). The gene lnu(F), which confers resistance to lincomycin, was found in five isolates, and fosA7, which codes for fosfomycin resistance, was detected in three isolates, all obtained from dairy cattle. Finally, mdf(A) was present in all isolates.
FIG 4.
Bar plots illustrating the phenotypic and genotypic characterization of resistance to gentamicin (a), chloramphenicol (b), ciprofloxacin (c), and azithromycin (d). Numbers within the stacked bar plots indicate the number of isolates observed with a particular MIC and genotype. ECOFF values are indicated with red dashed lines. “No genes” refers to those isolates lacking any genetic determinant of resistance for the corresponding antimicrobial. Point mutations associated with ciprofloxacin resistance are indicated by asterisks. Genes marked with a caret symbol are not specific genetic determinants for azithromycin resistance but are associated with resistance to other macrolides.
Among the plasmids found, IncI1, IncQ1, and IncFIC were the ones that carried the greatest variety of AMR genes. Thus, besides several ESBL/AmpC genes, the IncI1 plasmid harbored other AMR coding genes, such as aadA2, ant(3′′)-Ia, cmlA, dfrA16, sul1, sul2, sul3, and tet(A); IncQ1 carried ant(3′′)-Ia, aph(3′′)-Ib, aph(6)-Id, mph(B), dfrA1, sul1, sul2, and tet(A), as well as blaCMY-4; and IncFIC carried aph(6)-Ia, aph(6)-Id, tet(A), and blaTEM-1B.
In general, a strong agreement between gene presence and phenotypic susceptibility was observed, as supported by kappa scores (Table S3), the only exception being cefepime, which presented the lowest agreement value. This was due to 10 isolates with reduced susceptibility to cefepime (MIC = 0.25 mg/liter, n = 7; MIC = 0.5 mg/liter, n = 3) that did not carry any genes described as conferring resistance to this antimicrobial (Fig. 3b). However, these isolates carried blaCMY-2 (n = 9) and blaCMY-4 (n = 1) genes and displayed an AmpC phenotype.
DISCUSSION
In this cross-sectional survey, herd-level prevalence of ESBL/AmpC- and carbapenemase-producing E. coli was estimated in beef cattle, dairy cattle, and sheep without clinical signs of disease in the Basque Country (northern Spain). A large and representative number of herds was tested, and selective isolation media were used to increase sensitivity. Using cefotaxime-containing medium, presumptive ESBL/AmpC producers were isolated in 15% of the herds, prevalence being significantly higher in dairy cattle (32.9%) than in beef cattle (9.6%) and sheep (7.0%). Although differences in sampling strategies and isolation methods among studies hamper comparisons, prevalence rates of ESBL/AmpC-producing E. coli in food-producing animals has been reported to vary by country and animal species. In Europe, prevalence in individual veal calves under 1 year of age ranged from 7.1% in Denmark to 89.0% in Italy (mean in European Union, 44.5%) in 2017 (4). Herd-level prevalence of ESBL/AmpC-producing E. coli was reported to be 30.0% (3/10) in cattle farms in eastern England (10) and 41.0% (41/100) in a cross-sectional survey carried in dairy cattle in The Netherlands (16). In Germany, cefotaxime-resistant E. coli isolates were found in 70% (42/60) and 85% (44/52) of beef and dairy cattle units, respectively (17). Studies in sheep are scarce, and it is difficult to find herd prevalence data. In Switzerland, ESBL-producing E. coli was isolated in 6.9% of 58 sheep samples (18), similar to what we found in the present study at the herd level.
A higher prevalence of ESBL/AmpC-producing E. coli in dairy cattle than in beef cattle has already been reported (17). This might be associated with the different antimicrobial treatments used in the different management systems. In beef cattle, antimicrobial treatments are mostly implemented in young animals to treat diarrhea and respiratory diseases, while dairy cattle suffer from a wider diversity of pathologies during their longer life span that need to be treated with antimicrobials. In dairy cattle, β-lactams are used to treat mastitis (mainly penicillins but also cephalosporins such as ceftiofur and cefquinome) and also during dry-off to control and prevent intramammary infections following the last milking of the lactation period (mostly penicillins) (19). However, intramammary application might be expected to have less effect than oral administration on the spread of AMR in the intestinal microbiota. In any case, the relationship between antimicrobial use and AMR is a complex process that differs depending on the bacterial species and the AMR involved. Most studies support the association between the use of third- or fourth-generation cephalosporins and the occurrence of ESBL/AmpC-producing E. coli, but the occurrence and persistence of ESBL- and/or AmpC-producing E. coli in the apparent absence of extended-spectrum cephalosporin use has been reported (20). In the Basque Country, management of beef cattle and sheep is semi-intensive and animals graze in farmland or mountain pastures most of the year, while dairy cattle spend most of their time housed in pens. Less-intensive farm management systems have been associated with lower prevalence of infection with cefotaxime-resistant E. coli (17), maybe due to reduced stress and lower infection pressure and probability of recirculation of resistant isolates.
WGS provided insight into ESBL/AmpC resistance genes in ruminants in the Basque Country and identified blaCTX-M-14 as the most common ESBL gene and blaCMY-2 as the most common resistance determinant of the AmpC phenotype. Here, blaCTX-M-14 was significantly more prevalent than other CTX-M type genes like blaCTX-M-1 and blaCTX-M-15, which have been reported to be the most prevalent ones in cattle in other countries (10, 16, 21–23). In The Netherlands, although blaCTX-M-1 still prevails, an increasing trend in prevalence of blaCTX-M-15 and blaCTX-M-14 has been reported in recent years (24). In our study, AmpC-type β-lactamases were mostly associated with the presence of plasmid-borne genes (mostly IncI1−blaCMY-2 but also IncQ1−blaCMY-4), whereas a promoter mutation at position −42 of the chromosomally encoded ampC gene was detected only in two isolates. Opposite results were found in Dutch cattle, where point mutations were more prevalent, while blaCMY-2 predominated in avian hosts (24). In Europe, the three most frequent bla genes in extended-spectrum cephalosporin-resistant E. coli isolates from humans have been reported to be, in descending order, blaCTX-M-15, blaCTX-M-14, and blaCMY-2 (20). In Spain, despite the increasing prevalence of blaCTX-M-15 in human clinical samples, blaCTX-M-14 is still a very prevalent CTX-M type (25–27). On the other hand, E. coli strains harboring blaSHV-12 are mostly isolated from poultry and have been sporadically isolated from cattle (13, 18, 22, 24, 28) but are commonly found in community-acquired E. coli infections in Spain (25, 27). Here, the ESBL gene blaSHV-12 was only sporadically detected in two bovine and three ovine isolates but represented a high proportion of the ovine isolates tested (3/10).
Acquired carbapenemases in E. coli have been rarely identified in food-producing animals (11), and the prevalence of CPEs among livestock seemed to be low (<1%) in European countries (4). Here, no E. coli isolates were recovered from the carbapenem-containing medium used to screen for CPEs, but four isolates displayed a MIC value just above the epidemiological cutoff (ECOFF) for ertapenem while being susceptible to imipenem and meropenem. WGS, however, did not identify any known carbapenemase-encoding gene in any of these isolates, but they were all AmpC producers (blaCMY-2 gene carriers). In fact, AmpC β-lactamase production has been linked to ertapenem resistance due to loss or downregulation of outer membrane porins (29). In the present study, two isolates were phenotypically resistant to temocillin based on the recently set ECOFF for E. coli (MIC > 16 mg/liter) but did not harbor any temocillin resistance-encoding gene. Still, this result was not unexpected, as MICs for temocillin in the range of 16 to 128 mg/liter have been described in CTX-M-producing E. coli (30).
Comparison of WGS and phenotypic resistance profiles showed an overall very good agreement. However, presumptive discrepancies were also noticed in some instances. Thus, nine isolates that carried blaCTX-M-14 were resistant to cefotaxime and cefepime but tested susceptible to ceftazidime. This was, however, not unexpected, since CTX-M enzymes, and specifically CTX-M-14, have been reported to confer higher levels of resistance to cefotaxime than to ceftazidime, whose MICs sometimes remain within the susceptible range (31, 32). Costa Ramos et al. (33) demonstrated that E. coli blaCTX-M-14-bearing isolates switched from ceftazidime-susceptible to ceftazidime-resistant phenotypes under selective pressure by mechanisms yet unknown. On the other hand, 10 isolates resistant to cefepime carried blaCMY-2 (n = 9) or blaCMY-4 (n = 1) genes but no ESBL coding gene. Even though this phenomenon is rare, the potential development of cefepime resistance in CMY-2-producing E. coli isolates has already been reported (34).
Interestingly, four trimethoprim-resistant isolates did not harbor any of the genes coding for a phenotype of trimethoprim resistance included in ResFinder (updated on 5 December 2019). However, they all carried dfrA36, a dihydrofolate reductase gene which has been recently described in E. coli isolated from healthy Swiss fattening calves (35). In addition, another three isolates carried dfrA36 in combination with dfrA1. All seven dfrA36-carrying isolates also harbored floR and sul2, which are integrated along with dfrA36 within the florfenicol/chloramphenicol-sulfonamide resistance ISCR2 element (35). E. coli isolates are typically intrinsically resistant to macrolides (attributable to natural low macrolide permeability and multidrug efflux systems), with azithromycin displaying certain activity against some Gram-negative bacteria (36). Although no ECOFF for azithromycin resistance in E. coli has been established, a MIC of >16 mg/liter has been proposed as the azithromycin resistance breakpoint in some Enterobacteriaceae (37, 38). In the present study, all six isolates with a MIC of >16 mg/liter carried the mph(A) gene, whereas two isolates that solely harbored the mph(B) gene had a low MIC (8 mg/liter). These results confirm the relevant role of mph(A) in macrolide susceptibility previously reported (39, 40). The presence of mph(A) together with another gene [erm(A), erm(B), or ere(A)] has been reported to result in slightly higher MIC values (>32 mg/liter) (39). Here, the two isolates that carried mph(A) in combination with erm(B) had the highest MIC value (>64 mg/liter). The increased MIC value observed in isolates harboring mph(A) together with erm(B) suggests a slight contribution of 23S RNA methylation encoded by the erm gene to an increase in resistance.
The fact that the majority of ESBL/AmpC genes were plasmid located was not unexpected, and neither was the widespread distribution of IncI1 plasmids, since they are the most common plasmid type in E. coli isolated from animals in Europe (41). In addition, IncI1 plasmids also carried the greatest variety of other AMR genes, including genes that code for resistance to aminoglycosides, chloramphenicol, trimethoprim, and sulfamethoxazole. IncQ1, a mobilizable nonconjugative plasmid, also carried several AMR genes as well as blaCMY-4 (42). This would explain the commonly observed coresistance to extended-spectrum cephalosporins and other antimicrobials.
Genetic determinants associated with ciprofloxacin resistance consisted mostly of mutations in the chromosomally encoded quinolone resistance-determining regions (QRDRs) of the DNA gyrase and DNA topoisomerase IV genes, whereas plasmid-mediated quinolone resistance (PMQR) markers were less common (qnrS1, n = 3; qnrB19, n = 2; aac(6’)-Ib-cr, n = 2). Interestingly, the presence of aac(6’)-Ib-cr was associated with the accumulation of mutations in gyrA (S83N and D87N), parC (S80I), and parE (S458A) genes, as has been described by Poirel et al. (43). Conversely, qnr genes, when present, were the only genetic determinants of fluoroquinolone resistance. A gene that codes for a lincosamide nucleotidyltransferase [lnu(F)] conferring resistance to lincosamides was detected in five dairy cattle isolates. In cattle, lincosamides are used to treat mastitis caused by Gram-positive pathogens (44). Resistance to lincosamides is not routinely tested in E. coli, but considering that lnu(F) has been detected in E. coli, the potential risk for dissemination to other pathogens is worrisome. The presence of fosA7 in the chromosome of three isolates was striking, because fosA7 codes for resistance to fosfomycin, an antibiotic that is not used in cattle in Spain. In humans, fosfomycin is a first-line antimicrobial for the empirical treatment of uncomplicated urinary tract infections, currently being reconsidered as an alternative for the treatment of multidrug-resistant Gram-negative pathogens (45). Many Gram-negative species carry the fosA gene in the chromosome, but it is not frequently found in the E. coli chromosome (46). In E. coli, fosA3 is the most common plasmid-mediated FosA-encoding gene, particularly in East Asia (47). This gene was first detected as a chromosomal gene in Salmonella enterica serovar Heidelberg isolated from chickens (48) and was later also found in E. coli. In S. Heidelberg, the fosA7 gene was demonstrated to confer a high level of resistance to fosfomycin and was found to be potentially transferable by horizontal gene transfer (48). This is a concern that requires surveillance to monitor for the spread of fosfomycin resistance in bacteria.
In conclusion, this study provided insight into the prevalence of cefotaxime-resistant E. coli in ruminants in the Basque Country and the associated genetic determinants of AMR. Results in cattle were similar to those found in other European countries, whereas those in sheep constituted an important contribution to the limited repository of sheep data. Overall, these results showed that ruminants are reservoirs for MDR commensal E. coli. However, all isolates were susceptible to tigecycline, imipenem, meropenem, and colistin, which is reassuring, because some of these compounds are last-line antimicrobial agents for the treatment of human infections. The results of this regional, short-term study highlighted the need to turn this investigation into a long-run surveillance program to monitor trends over time. Antimicrobial susceptibility testing by phenotypic and molecular methods is key in surveillance programs to enhance early detection of resistance development, monitor resistance trends, and provide guidance to clinicians in selecting the adequate therapy, all with the final aim of mitigating resistance spread.
MATERIALS AND METHODS
Sampling design.
A cross-sectional survey was carried out in ruminant herds in the Basque Country, a 7,234-km2 region located in northern Spain. Ruminant production is one of the pillars of the rural economy of the region, with ca. 260,000 sheep and 135,000 cattle (dairy and beef) according to the 2015 census (https://www.eustat.eus/banku/id_4017/indexLista.html). Dairy cattle are managed under an intensive system, whereas semi-extensive production predominates for sheep and beef cattle; animals graze in farmland pastures in spring and part of the summer and in communal mountain pastures from the middle of July until the end of November and are housed in winter. Further details on general husbandry systems for beef cattle, dairy cattle, and sheep in the Basque Country have been reported elsewhere (49, 50).
The census of beef cattle, dairy cattle, and sheep farms was obtained from the Department of Agriculture of the Basque Government. Since this survey was part of a larger study designed to also estimate the prevalence of Salmonella, Listeria monocytogenes (49), thermophilic campylobacters (50), and Shiga toxin-producing Escherichia coli (STEC) (51), the number of herds to sample was calculated separately for each animal category for an expected herd prevalence of 50%, a 95% confidence level, and an accuracy of 10% using Win Episcope 2.0. A sample size of 25 animals per herd was selected after estimating a within-herd prevalence of 10% and a level of confidence of at least 90% in detecting a positive. Thus, a total of 300 herds (104 beef cattle, 82 dairy cattle, and 114 dairy sheep) were sampled once between February 2014 and June 2016. Rectal fecal samples from 25 animals randomly selected per herd were collected with a gloved hand and analyzed in a single 25-g pool (1 g per animal per herd).
Sample collection was carried out by veterinary practitioners as part of the usual screening scheme performed on farms, strictly following Spanish ethical guidelines and animal welfare regulations (Real Decreto 53/2013). The collection of this material, considered routine veterinary practice, did not require the approval of the Ethics Committee for Animal Experimentation. Informed oral consent was obtained from the farmers at the time of sample collection.
ESBL/AmpC- and carbapenemase-producing E. coli selective isolation.
Feces (25 g of pooled rectal fecal samples) were diluted 1:10 in modified tryptic soy broth (mTSB; bioMérieux) supplemented with novobiocine (Biolife) and incubated at 41 ± 1°C for 6 to 7 h. For the selective isolation of cefotaxime-resistant E. coli, samples were then preenriched in MacConkey broth supplemented with cefotaxime at 1 mg/liter (37 ± 1°C, 24 h) and subcultured onto MacConkey agar with cefotaxime (1 mg/liter). For the selective isolation of OXA-48- and other carbapenemase-producing E. coli (CPE), a preenrichment with unsupplemented MacConkey broth was carried out (37 ± 1°C, 24 h), followed by subculturing 50 μl on a biplate selective chromogenic medium (Chromid Carba Smart; bioMérieux). Both plates were incubated at 37 ± 1°C for 24 h. Three colonies per plate were selected based on colony morphology diversity and were further confirmed as E. coli by species-specific real-time PCR targeting the uidA gene (52).
Antimicrobial susceptibility test (AST) determination by broth microdilution.
MICs were determined by broth microdilution by following the recommendations of the Commission Decision 2013/652/EU (https://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:32013D0652&from=EN). Isolates were tested using two Sensititre MIC susceptibility plates (Thermo Fisher Scientific, Waltham, MA): one (EUVSEC1) that contains 14 antimicrobial agents (10 classes) and a second panel (EUVSEC2) with 10 antimicrobial substances for the phenotypic characterization of presumptive ESBL, AmpC, and carbapenemase producers. The second panel includes cefoxitin (FOX) as well as cefotaxime (FOT) and ceftazidime (TAZ) with and without clavulanic acid (CLV) to investigate clavulanate synergy for phenotypic characterization of ESBL and AmpC production, along with imipenem, meropenem, and ertapenem to phenotypically verify the presumptive carbapenemase producers. MIC results were interpreted using epidemiological cutoff values (ECOFF) as developed by the European Committee for Antimicrobial Susceptibility Testing (EUCAST; http://www.eucast.org) to define microbiological resistance to the antimicrobial in question, that is, to discriminate those microorganisms with and without acquired resistance mechanisms (non-wild type and wild type, respectively). For azithromycin resistance (no cutoff assigned by EUCAST), a MIC of ≤16 mg/liter for wild-type isolates was used as a reference, as proposed for Salmonella spp. (37, 38). Here, the terms susceptible and resistant refer to isolates without (wild type) and with (microbiologically resistant) phenotypically expressed resistance mechanisms, respectively.
Interpretation of resistance profiles for phenotypic detection of ESBL and AmpC production was based on EUCAST guideline for the detection of resistance mechanisms and specific resistances of clinical and/or epidemiological importance (53). Briefly, an ESBL phenotype was inferred if isolates were resistant to FOT (>1 mg/liter) or TAZ (>1 mg/liter) but susceptible to FOX (≤8 mg/liter) and showed clavulanate synergy with FOT and/or TAZ (≥8-fold reduction in the MIC of the cephalosporin combined with 4 mg/liter CLV compared with the MIC of the cephalosporin alone). Isolates were considered to have the AmpC phenotype if clavulanate synergy was not shown and they were resistant to FOT (>1 mg/liter) or TAZ (>1 mg/liter) and FOX (>8 mg/liter). An ESBL+AmpC phenotype was inferred if isolates were resistant to FOT (>1 mg/liter) or TAZ (>1 mg/liter), resistant to FOX (>8 mg/liter), and showed clavulanate synergy with FOT and/or TAZ. Meropenem resistance (>0.12 mg/liter) was used to infer a carbapenemase-producing phenotype.
WGS, genome assembly, and analysis.
Bacterial genomic DNA was extracted with a Wizard genomic DNA purification kit (Promega, Madison, WI, USA). Whole-genome sequencing (WGS) was carried out at a commercial facility using an Illumina NovaSeq 6000 system (150-bp pair-end reads). The quality assessment of the raw reads was performed using a FastQC v.0.11.9 quality control tool (Babraham Bioinformatics, Cambridge, United Kingdom) (54). Data were analyzed by using the automated pipeline TORMES v.1.0 (https://github.com/nmquijada/tormes; 55). Briefly, reads were quality filtered using Trimmomatic v.0.38 (56) and de novo assembled into a draft genome using SPAdes v.3.13.0 (57) with the default parameters and in careful mode. QUAST v.5.0.2 (58) was used to evaluate the quality of the assemblies, and contigs below 200 bp in length were discarded. The draft genomes were screened for acquired AMR genes using BLASTn v.2.7.1+ (59) and ABRicate v.0.8.10 (T. Seemann, https://github.com/tseemann/abricate) against ResFinder (60) (last updated on 5 December 2019). Chromosomal point mutations associated with quinolone resistance and β-lactams (ampC promoter) were investigated using the E. coli point mutations database PointFinder (61) (last updated on 4 June 2019). PlasFlow v.1.1 (62) was used to predict plasmid- and chromosome-derived contigs. The presence of plasmid replicons was identified using PlasmidFinder v.2.0.1 (last updated on 4 September 2018) (63). Any hit with coverage below 60% and/or identity below 90% was removed. A dendrogram was generated to illustrate the similarity among isolates based on their AMR pattern. Hierarchical clustering analysis was performed with the unweighted pair group method with arithmetic mean (UPGMA) based on the Euclidean distance matrix, using the function hclust of the R statistical package v.3.6.3 (64).
Statistical analysis.
Herd-level prevalence was expressed as the percentage of herds/flocks that tested positive in each farm system out of all herds/flocks that were examined in the respective farm system, with 95% confidence intervals adjusted for the population size, using the software EpiInfo2. To assess factors associated with shedding prevalence of cefotaxime-resistant E. coli, selected variables were categorized as follows: (i) host species (cattle, sheep), (ii) production system (beef cattle, dairy cattle, and sheep), (iii) sampling season (spring, summer, autumn, winter), (iv) geographical location of the farm (oceanic, continental), (v) presence of other species in the farm, such as cattle, sheep, goats, horses (presence, absence), (vi) herd size stratified according to farm system management (beef cattle, <50, 50 to 100, and >100; dairy cattle, <50, 50 to 150, and >150; sheep, <150, 150 to 300, and >300), and (vii) year of sampling (2014, 2015, 2016). First, univariate logistic regressions were conducted to explore the unadjusted association between herd positivity and variables. Only significant factors (P ≤ 0.20; likelihood-ratio test) were included for further multivariate logistic regression analyses. Test of overall significance (chunk test) was performed to assess any possible effect modifiers that could bias the magnitude of associations, and interactions with a P value of >0.05 were excluded until no significant difference between the full and the reduced models was observed. To identify confounding variables, the measure of association was estimated before and after adjusting for the potential confounder, and variables causing a change of ≥10% in the estimated measure were retained. Adjusted odds ratios (ORadj) were used as the measure of association between positivity and the explanatory variable and were expressed with their confidence interval at 95% (95% CI). To evaluate differences in the distribution of AMR among production systems, simple logistic regressions were performed.
Phenotypic (broth microdilution AST-based) and genotypic (WGS-based) susceptibility results were compared. Resistant WGS genotypes were defined by the presence of one or more resistance genes and/or point mutation for each antimicrobial tested in the AST. The sensitivity, specificity, and positive (PPV) and negative (NPV) predictive values for the genotypic prediction were calculated for each antimicrobial tested for their corresponding phenotypic AST reference. Interrater agreement analyses were performed for each antimicrobial using Cohen’s kappa (κ) method. Interpretation of kappa values to assess the strength of agreement between techniques was based on the one proposed by Altman (65), which is as follows: κ ≤ 0.20, poor; κ = 0.21 to 0.40, fair; κ = 0.41 to 0.60, moderate; κ = 0.61 to 0.80, good; and κ = 0.81 to 1.00, very good. Analyses were conducted using statistical software Stata/IC version 13.1 (StataCorp LP, College Station, TX, USA).
Data availability.
Sequencing data of the 66 genomes analyzed in this study have been deposited at the NCBI Sequence Read Archive (SRA) database under accession numbers SRR11810138 to SRR11810203, associated with BioProject accession number PRJNA633740.
Supplementary Material
ACKNOWLEDGMENTS
We thank José Luis Lavín (NEIKER) for helpful advice on bioinformatic analyses and the veterinary staff from the Diputaciones Forales de Gipuzkoa, Bizkaia, and Araba and the farmers for their collaboration. We also thank Montserrat Agüero-García (Laboratorio Central de Veterinaria, Madrid, Spain) for kindly providing control strains to validate the selective isolation media.
This work was funded by the Basque Government. M.T. is the recipient of a predoctoral fellowship from the Basque Government (Departamento de Desarrollo Económico e Infraestructuras, Ekonomiaren Garapen eta Lehiakortasun Saila, Eusko Jaurlaritza).
We declare no conflicts of interest.
A.H. conceived the study and coordinated the project. M.T. and B.O. performed laboratory analyses. M.O. carried out statistical and bioinformatic data analyses. A.H., M.T., and M.O. interpreted the data and wrote the manuscript. All authors read and approved the final manuscript.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.O’Neill J. 2014. Antimicrobial resistance: tackling a crisis for the health and wealth of nations. The review on antimicrobial resistance chaired by Jim O’Neill https://amr-review.org/sites/default/files/AMR%20Review%20Paper%20-%20Tackling%20a%20crisis%20for%20the%20health%20and%20wealth%20of%20nations_1.pdf.
- 2.WHO. 2019. Critically important antimicrobials for human medicine, 6th revision WHO, Geneva, Switzerland. [Google Scholar]
- 3.Dierikx CM, van Duijkeren E, Schoormans AHW, van Essen-Zandbergen A, Veldman K, Kant A, Huijsdens XW, van der Zwaluw K, Wagenaar JA, Mevius DJ. 2012. Occurrence and characteristics of extended-spectrum-β-lactamase- and AmpC-producing clinical isolates derived from companion animals and horses. J Antimicrob Chemother 67:1368–1374. doi: 10.1093/jac/dks049. [DOI] [PubMed] [Google Scholar]
- 4.European Food Safety Authority, European Centre for Disease Prevention and Control. 2019. The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2017. EFSA J 17:5598. doi: 10.2903/j.efsa.2019.5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gekenidis M-T, Qi W, Hummerjohann J, Zbinden R, Walsh F, Drissner D. 2018. Antibiotic-resistant indicator bacteria in irrigation water: high prevalence of extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli. PLoS One 13:e0207857. doi: 10.1371/journal.pone.0207857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guenther S, Ewers C, Wieler LH, Stefani S. 2011. Extended-spectrum beta-lactamases producing E. coli in wildlife, yet another form of environmental pollution? Front Microbiol 2:246. doi: 10.3389/fmicb.2011.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collis RM, Burgess SA, Biggs PJ, Midwinter AC, French NP, Toombs-Ruane L, Cookson AL. 2019. Extended-spectrum beta-lactamase-producing Enterobacteriaceae in dairy farm environments: a New Zealand perspective. Foodborne Pathog Dis 16:5–22. doi: 10.1089/fpd.2018.2524. [DOI] [PubMed] [Google Scholar]
- 8.Dorado-García A, Smid JH, van Pelt W, Bonten MJM, Fluit AC, van den Bunt G, Wagenaar JA, Hordijk J, Dierikx CM, Veldman KT, de Koeijer A, Dohmen W, Schmitt H, Liakopoulos A, Pacholewicz E, Lam T, Velthuis AG, Heuvelink A, Gonggrijp MA, van Duijkeren E, van Hoek A, de Roda Husman AM, Blaak H, Havelaar AH, Mevius DJ, Heederik D. 2018. Molecular relatedness of ESBL/AmpC-producing Escherichia coli from humans, animals, food and the environment: a pooled analysis. J Antimicrob Chemother 73:339–347. doi: 10.1093/jac/dkx397. [DOI] [PubMed] [Google Scholar]
- 9.Mughini-Gras L, Dorado-García A, van Duijkeren E, van den Bunt G, Dierikx CM, Bonten MJM, Bootsma MCJ, Schmitt H, Hald T, Evers EG, de Koeijer A, van Pelt W, Franz E, Mevius DJ, Heederik D. 2019. Attributable sources of community-acquired carriage of Escherichia coli containing β-lactam antibiotic resistance genes: a population-based modelling study. Lancet Planet Health 3:e357–e369. doi: 10.1016/S2542-5196(19)30130-5. [DOI] [PubMed] [Google Scholar]
- 10.Ludden C, Raven K, Jamrozy D, Gouliouris T, Blane B, Coll F, de Goffau M, Naydenova P, Horner C, Hernandez-Garcia J, Wood P, Hadjirin N, Radakovic M, Brown N, Holmes M, Parkhill J, Peacock S. 2019. One Health genomic surveillance of Escherichia coli demonstrates distinct lineages and mobile genetic elements in isolates from humans versus livestock. mBio 10:e02693-18. doi: 10.1128/mBio.02693-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kock R, Daniels-Haardt I, Becker K, Mellmann A, Friedrich AW, Mevius D, Schwarz S, Jurke A. 2018. Carbapenem-resistant Enterobacteriaceae in wildlife, food-producing, and companion animals: a systematic review. Clin Microbiol Infect 24:1241–1250. doi: 10.1016/j.cmi.2018.04.004. [DOI] [PubMed] [Google Scholar]
- 12.Dantas Palmeira J, Ferreira H. 2020. Extended-spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae in cattle production—a threat around the world. Heliyon 6:e3206. doi: 10.1016/j.heliyon.2020.e03206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Briñas L, Moreno MA, Teshager T, Saenz Y, Porrero MC, Dominguez L, Torres C. 2005. Monitoring and characterization of extended-spectrum β-lactamases in Escherichia coli strains from healthy and sick animals in Spain in 2003. Antimicrob Agents Chemother 49:1262–1264. doi: 10.1128/AAC.49.3.1262-1264.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poirel L, Madec J-Y, Lupo A, Schink A-K, Kieffer N, Nordmann P, Schwarz S. 2018. Antimicrobial resistance in Escherichia coli, p 289–316. In Schwarz S, Cavaco LM, Shen J (ed), Antimicrobial resistance in bacteria from livestock and companion animals. ASM Press, Washington, DC. [Google Scholar]
- 15.Jørgensen RL, Nielsen JB, Friis-Møller A, Fjeldsøe-Nielsen H, Schønning K. 2010. Prevalence and molecular characterization of clinical isolates of Escherichia coli expressing an AmpC phenotype. J Antimicrob Chemother 65:460–464. doi: 10.1093/jac/dkp484. [DOI] [PubMed] [Google Scholar]
- 16.Gonggrijp MA, Santman-Berends I, Heuvelink AE, Buter GJ, van Schaik G, Hage JJ, Lam T. 2016. Prevalence and risk factors for extended-spectrum β-lactamase- and AmpC-producing Escherichia coli in dairy farms. J Dairy Sci 99:9001–9013. doi: 10.3168/jds.2016-11134. [DOI] [PubMed] [Google Scholar]
- 17.Hille K, Ruddat I, Schmid A, Hering J, Hartmann M, von Munchhausen C, Schneider B, Messelhausser U, Friese A, Mansfeld R, Kasbohrer A, Hormansdorfer S, Roesler U, Kreienbrock L. 2017. Cefotaxime-resistant E. coli in dairy and beef cattle farms—joint analyses of two cross-sectional investigations in Germany. Prev Vet Med 142:39–45. doi: 10.1016/j.prevetmed.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 18.Geser N, Stephan R, Hachler H. 2012. Occurrence and characteristics of extended-spectrum β-lactamase (ESBL) producing Enterobacteriaceae in food producing animals, minced meat and raw milk. BMC Vet Res 8:21. doi: 10.1186/1746-6148-8-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simjee S, McDermott P, Trott D, Chuanchuen R. 2018. Present and future surveillance of antimicrobial resistance in animals: principles and practices. Microbiol Spectr 6:ARBA-0028-2017. doi: 10.1128/microbiolspec.ARBA-0028-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seiffert SN, Hilty M, Perreten V, Endimiani A. 2013. Extended-spectrum cephalosporin-resistant gram-negative organisms in livestock: an emerging problem for human health? Drug Resist Updat 16:22–45. doi: 10.1016/j.drup.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 21.Schmid A, Hormansdorfer S, Messelhausser U, Kasbohrer A, Sauter-Louis C, Mansfeld R. 2013. Prevalence of extended-spectrum β-lactamase-producing Escherichia coli on Bavarian dairy and beef cattle farms. Appl Environ Microbiol 79:3027–3032. doi: 10.1128/AEM.00204-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Michael GB, Kaspar H, Siqueira AK, de Freitas Costa E, Corbellini LG, Kadlec K, Schwarz S. 2017. Extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli isolates collected from diseased food-producing animals in the GERM-Vet monitoring program 2008–2014. Vet Microbiol 200:142–150. doi: 10.1016/j.vetmic.2016.08.023. [DOI] [PubMed] [Google Scholar]
- 23.Day MJ, Rodríguez I, van Essen-Zandbergen A, Dierikx C, Kadlec K, Schink AK, Wu G, Chattaway MA, DoNascimento V, Wain J, Helmuth R, Guerra B, Schwarz S, Threlfall J, Woodward MJ, Coldham N, Mevius D, Woodford N. 2016. Diversity of STs, plasmids and ESBL genes among Escherichia coli from humans, animals and food in Germany, the Netherlands and the UK. J Antimicrob Chemother 71:1178–1182. doi: 10.1093/jac/dkv485. [DOI] [PubMed] [Google Scholar]
- 24.Ceccarelli D, Kant A, Van Essen-Zandbergen A, Dierikx C, Hordijk J, Wit B, Mevius DJ, Veldman KT. 2019. Diversity of plasmids and genes encoding resistance to extended spectrum cephalosporins in commensal Escherichia coli from Dutch livestock in 2007–2017. Front Microbiol 10:76. doi: 10.3389/fmicb.2019.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Díaz MA, Spanish Group for Nosocomial Infections (GEIH), Hernández-Bello JR, Rodríguez-Baño J, Martínez-Martínez L, Calvo J, Blanco J, Pascual A. 2010. Diversity of Escherichia coli strains producing extended-spectrum β-lactamases in Spain: second nationwide study. J Clin Microbiol 48:2840–2845. doi: 10.1128/JCM.02147-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cantón R, González-Alba JM, Galán JC. 2012. CTX-M enzymes: origin and diffusion. Front Microbiol 3:110. doi: 10.3389/fmicb.2012.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fernández-Reyes M, Vicente D, Gomariz M, Esnal O, Landa J, Oñate E, Pérez-Trallero E. 2014. High rate of fecal carriage of extended-spectrum-β-lactamase-producing Escherichia coli in healthy children in Gipuzkoa, northern Spain. Antimicrob Agents Chemother 58:1822–1824. doi: 10.1128/AAC.01503-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smet A, Martel A, Persoons D, Dewulf J, Heyndrickx M, Herman L, Haesebrouck F, Butaye P. 2010. Broad-spectrum β-lactamases among Enterobacteriaceae of animal origin: molecular aspects, mobility and impact on public health. FEMS Microbiol Rev 34:295–316. doi: 10.1111/j.1574-6976.2009.00198.x. [DOI] [PubMed] [Google Scholar]
- 29.Mammeri H, Nordmann P, Berkani A, Eb F. 2008. Contribution of extended-spectrum AmpC (ESAC) β-lactamases to carbapenem resistance in Escherichia coli. FEMS Microbiol Lett 282:238–240. doi: 10.1111/j.1574-6968.2008.01126.x. [DOI] [PubMed] [Google Scholar]
- 30.Cavaco LM, Hansen F, Mushtaq S, Hill RLR, Woodford N, Le Hello S, Hendriksen RS, Hammerum AM, Hasman H. 2019. Evaluation of temocillin for phenotypic carbapenemase screening of Escherichia coli and Salmonella enterica isolates in relation to the presence of genes encoding ESBLs and carbapenemase production. J Antimicrob Chemother 74:639–644. doi: 10.1093/jac/dky493. [DOI] [PubMed] [Google Scholar]
- 31.Bonnet R. 2004. Growing group of extended-spectrum beta-lactamases: the CTX-M enzymes. Antimicrob Agents Chemother 48:1–14. doi: 10.1128/aac.48.1.1-14.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williamson DA, Roberts SA, Smith M, Heffernan H, Tiong A, Pope C, Freeman JT. 2012. High rates of susceptibility to ceftazidime among globally prevalent CTX-M-producing Escherichia coli: potential clinical implications of the revised CLSI interpretive criteria. Eur J Clin Microbiol Infect Dis 31:821–824. doi: 10.1007/s10096-011-1380-1. [DOI] [PubMed] [Google Scholar]
- 33.Costa Ramos JM, Stein C, Pfeifer Y, Brandt C, Pletz MW, Makarewicz O. 2015. Mutagenesis of the CTX-M-type ESBL—is MIC-guided treatment according to the new EUCAST recommendations a safe approach? J Antimicrob Chemother 70:2528–2535. doi: 10.1093/jac/dkv153. [DOI] [PubMed] [Google Scholar]
- 34.Dona V, Scheidegger M, Pires J, Furrer H, Atkinson A, Babouee Flury B. 2019. Gradual in vitro evolution of cefepime resistance in an ST131 Escherichia coli strain expressing a plasmid-encoded CMY-2 β-lactamase. Front Microbiol 10:1311. doi: 10.3389/fmicb.2019.01311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wuthrich D, Brilhante M, Hausherr A, Becker J, Meylan M, Perreten V. 2019. A novel trimethoprim resistance gene, dfrA36, characterized from Escherichia coli from calves. mSphere 4:e00255-19. doi: 10.1128/mSphere.00255-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gomes C, Martínez-Puchol S, Palma N, Horna G, Ruiz-Roldán L, Pons MJ, Ruiz J. 2017. Macrolide resistance mechanisms in Enterobacteriaceae: focus on azithromycin. Crit Rev Microbiol 43:1–30. doi: 10.3109/1040841X.2015.1136261. [DOI] [PubMed] [Google Scholar]
- 37.Clinical and Laboratory Standards Institute. 2015. Performance standards for antimicrobial susceptibility testing, 25th informational supplement. Document M100-S25. CLSI, Wayne, PA. [Google Scholar]
- 38.Sjölund-Karlsson M, Joyce K, Blickenstaff K, Ball T, Haro J, Medalla FM, Fedorka-Cray P, Zhao S, Crump JA, Whichard JM. 2011. Antimicrobial susceptibility to azithromycin among Salmonella enterica isolates from the United States. Antimicrob Agents Chemother 55:3985–3989. doi: 10.1128/AAC.00590-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gomes C, Ruiz-Roldán L, Mateu J, Ochoa TJ, Ruiz J. 2019. Azithromycin resistance levels and mechanisms in Escherichia coli. Sci Rep 9:6089. doi: 10.1038/s41598-019-42423-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nguyen MCP, Woerther PL, Bouvet M, Andremont A, Leclercq R, Canu A. 2009. Escherichia coli as reservoir for macrolide resistance genes. Emerg Infect Dis 15:1648–1650. doi: 10.3201/eid1510.090696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rozwandowicz M, Brouwer MSM, Fischer J, Wagenaar JA, Gonzalez-Zorn B, Guerra B, Mevius DJ, Hordijk J. 2018. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J Antimicrob Chemother 73:1121–1137. doi: 10.1093/jac/dkx488. [DOI] [PubMed] [Google Scholar]
- 42.Kotsakis SD, Tzouvelekis LS, Lebessi E, Doudoulakakis A, Bouli T, Tzelepi E, Miriagou V. 2015. Characterization of a mobilizable IncQ plasmid encoding cephalosporinase CMY-4 in Escherichia coli. Antimicrob Agents Chemother 59:2964–2966. doi: 10.1128/AAC.05017-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poirel L, Cattoir V, Nordmann P. 2012. Plasmid-mediated quinolone resistance; interactions between human, animal, and environmental ecologies. Front Microbiol 3:24. doi: 10.3389/fmicb.2012.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Constable P, Pyörälä S, Smith G. 2008. Guidelines for antimicrobial use in cattle, p 143–160. In Guardabassi L, Jensen LB, Kruse H (ed), Guide to antimicrobial use in animals. Blackwell Publishing Ltd, Oxford, United Kingdom. [Google Scholar]
- 45.Meletis G. 2016. Carbapenem resistance: overview of the problem and future perspectives. Ther Adv Infect Dis 3:15–21. doi: 10.1177/2049936115621709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ito R, Mustapha MM, Tomich AD, Callaghan JD, McElheny CL, Mettus RT, Shanks RMQ, Sluis-Cremer N, Doi Y. 2017. Widespread fosfomycin resistance in Gram-negative bacteria attributable to the chromosomal fosA gene. mBio 8:e00749-17. doi: 10.1128/mBio.00749-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang XM, Dong Z, Schwarz S, Zhu Y, Hua X, Zhang Y, Liu S, Zhang WJ. 2017. Plasmids of diverse Inc groups disseminate the fosfomycin resistance gene fosA3 among Escherichia coli isolates from pigs, chickens, and dairy cows in Northeast China. Antimicrob Agents Chemother 61:e00859-17. doi: 10.1128/AAC.00859-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rehman MA, Yin X, Persaud-Lachhman MG, Diarra MS. 2017. First detection of a fosfomycin resistance gene, fosA7, in Salmonella enterica serovar Heidelberg isolated from broiler chickens. Antimicrob Agents Chemother 61:e00410-17. doi: 10.1128/AAC.00410-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hurtado A, Ocejo M, Oporto B. 2017. Salmonella spp. and Listeria monocytogenes shedding in domestic ruminants and characterization of potentially pathogenic strains. Vet Microbiol 210:71–76. doi: 10.1016/j.vetmic.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 50.Ocejo M, Oporto B, Hurtado A. 2019. Occurrence of Campylobacter jejuni and Campylobacter coli in cattle and sheep in northern Spain and changes in antimicrobial resistance in two studies 10-years apart. Pathogens 8:98. doi: 10.3390/pathogens8030098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oporto B, Ocejo M, Alkorta M, Marimón JM, Montes M, Hurtado A. 2019. Zoonotic approach to Shiga toxin-producing Escherichia coli: integrated analysis of virulence and antimicrobial resistance in ruminants and humans. Epidemiol Infect 147:e164. doi: 10.1017/S0950268819000566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Frahm E, Obst U. 2003. Application of the fluorogenic probe technique (TaqMan PCR) to the detection of Enterococcus spp. and Escherichia coli in water samples. J Microbiol Methods 52:123–131. doi: 10.1016/S0167-7012(02)00150-1. [DOI] [PubMed] [Google Scholar]
- 53.EUCAST. July 2017. EUCAST guidelines for detection of resistance mechanisms and specific resistances of clinical and/or epidemiological importance, version 2.0. https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Resistance_mechanisms/EUCAST_detection_of_resistance_mechanisms_170711.pdf.
- 54.Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 55.Quijada NM, Rodríguez-Lázaro D, Eiros JM, Hernández M. 2019. TORMES: an automated pipeline for whole bacterial genome analysis. Bioinformatics 35:4207–4212. doi: 10.1093/bioinformatics/btz220. [DOI] [PubMed] [Google Scholar]
- 56.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Z, Schwartz S, Wagner L, Miller W. 2000. A greedy algorithm for aligning DNA sequences. J Comput Biol 7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 60.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zankari E, Allesøe R, Joensen KG, Cavaco LM, Lund O, Aarestrup FM. 2017. PointFinder: a novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J Antimicrob Chemother 72:2764–2768. doi: 10.1093/jac/dkx217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Krawczyk PS, Lipinski L, Dziembowski A. 2018. PlasFlow: predicting plasmid sequences in metagenomic data using genome signatures. Nucleic Acids Res 46:e35. doi: 10.1093/nar/gkx1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Carattoli A, Hasman H. 2020. PlasmidFinder and in silico pMLST: identification and typing of plasmid replicons in whole-genome sequencing (WGS). Methods Mol Biol 2075:285–294. doi: 10.1007/978-1-4939-9877-7_20. [DOI] [PubMed] [Google Scholar]
- 64.R Development Core Team. 2016. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 65.Altman DG. 1991. Practical statistics for medical research. Chapman and Hall, London, England. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data of the 66 genomes analyzed in this study have been deposited at the NCBI Sequence Read Archive (SRA) database under accession numbers SRR11810138 to SRR11810203, associated with BioProject accession number PRJNA633740.