Lignocellulases are important industrial enzymes for sustainable production of biofuels and bio-products. Insoluble cellulose has been commonly used to induce the production of lignocellulases in filamentous fungi, which causes a difficult fermentation operation and enzyme loss due to adsorption to cellulose. The disadvantages can be overcome by using soluble inducers, such as the disaccharide cellobiose. Quantitative proteome profiling of the model filamentous fungus Neurospora crassa revealed cellobiose-dependent pathways for cellulase production, including protein processing and export. A protein (CWH43) potentially involved in protein processing was found to be a positive regulator of lignocellulase production. The cellobiose-dependent mechanisms provide new opportunities to improve the production of lignocellulases in filamentous fungi.
KEYWORDS: Neurospora, cellobiose, cellulase, cellulose, glycosylphosphatidylinositols, protein folding, protein translocation, protein transport, proteomics, ribosome synthesis
ABSTRACT
Filamentous fungi are intensively used for producing industrial enzymes, including lignocellulases. Employing insoluble cellulose to induce the production of lignocellulases causes some drawbacks, e.g., a complex fermentation operation, which can be overcome by using soluble inducers such as cellobiose. Here, a triple β-glucosidase mutant of Neurospora crassa, which prevents rapid turnover of cellobiose and thus allows the disaccharide to induce lignocellulases, was applied to profile the proteome responses to cellobiose and cellulose (Avicel). Our results revealed a shared proteomic response to cellobiose and Avicel, whose elements included lignocellulases and cellulolytic product transporters. While the cellulolytic proteins showed a correlated increase in protein and mRNA levels, only a moderate correlation was observed on a proteomic scale between protein and mRNA levels (R2 = 0.31). Ribosome biogenesis and rRNA processing were significantly overrepresented in the protein set with increased protein but unchanged mRNA abundances in response to Avicel. Ribosome biogenesis, as well as protein processing and protein export, was also enriched in the protein set that showed increased abundance in response to cellobiose. NCU05895, a homolog of yeast CWH43, is potentially involved in transferring a glycosylphosphatidylinositol (GPI) anchor to nascent proteins. This protein showed increased abundance but no significant change in mRNA levels. Disruption of CWH43 resulted in a significant decrease in cellulase activities and secreted protein levels in cultures grown on Avicel, suggesting a positive regulatory role for CWH43 in cellulase production. The findings should have an impact on a systems engineering approach for strain improvement for the production of lignocellulases.
IMPORTANCE Lignocellulases are important industrial enzymes for sustainable production of biofuels and bio-products. Insoluble cellulose has been commonly used to induce the production of lignocellulases in filamentous fungi, which causes a difficult fermentation operation and enzyme loss due to adsorption to cellulose. The disadvantages can be overcome by using soluble inducers, such as the disaccharide cellobiose. Quantitative proteome profiling of the model filamentous fungus Neurospora crassa revealed cellobiose-dependent pathways for cellulase production, including protein processing and export. A protein (CWH43) potentially involved in protein processing was found to be a positive regulator of lignocellulase production. The cellobiose-dependent mechanisms provide new opportunities to improve the production of lignocellulases in filamentous fungi.
INTRODUCTION
Plant cell wall-degrading enzymes, including cellulases, are produced mainly by filamentous fungi for the production of biofuels and bio-products (1). Insoluble cellulose has been used to induce cellulases, but its insolubility causes disadvantages such as difficult fermentation operations and enzyme loss (2–4). These drawbacks can be overcome by replacing insoluble cellulose with soluble inducers (5–7). Cellobiose, a soluble major cellulolytic product, can induce the production of cellulases (8, 9). Understanding the underlying mechanisms is valuable for rational optimization of filamentous fungi for improved fermentation performance (10).
It is still under investigation how filamentous fungi sense the presence of cellulose and cellobiose. The cellobiose transporters CDT-1 and CDT-2 in Neurospora crassa were proposed to have a dual function, transporting cellobiose into fungal cells and acting as transceptors to activate downstream cellulolytic gene expression (11, 12). Similar transceptor functions were also proposed for the cellobiose transporters Crt1 and CltB from Trichoderma reesei and Aspergillus nidulans, respectively (13, 14). The cellobionic acid transporter SUT-12, formerly named CBT-1 and CLP-1, was also suggested to be involved in cellulase induction and to act to repress cdt-1 expression in N. crassa (15, 16).
Major cellulase transcription factors are CLR-1 and CLR-2 (17–19). CLR-3 is a repressor of CLR-1 activity in the absence of an inducer (20). Another cellulase transcript factor, CLR-4, is found to bind to the promoter region of CLR-1 (21). Hydrolysis of cellobiose by β-glucosidase produces glucose, which triggers the repression of cellulolytic genes, a mechanism known as carbon catabolite repression (CCR). CCR is partially mediated by the transcription factor CreA/CRE-1 (22). In N. crassa, CRE-1 is negatively regulated by the transcription factors VIB1 and COL26 (23). Recent studies suggest that the expression of cellulase genes is also regulated by the chromatin status, such as histone methylation (24, 25).
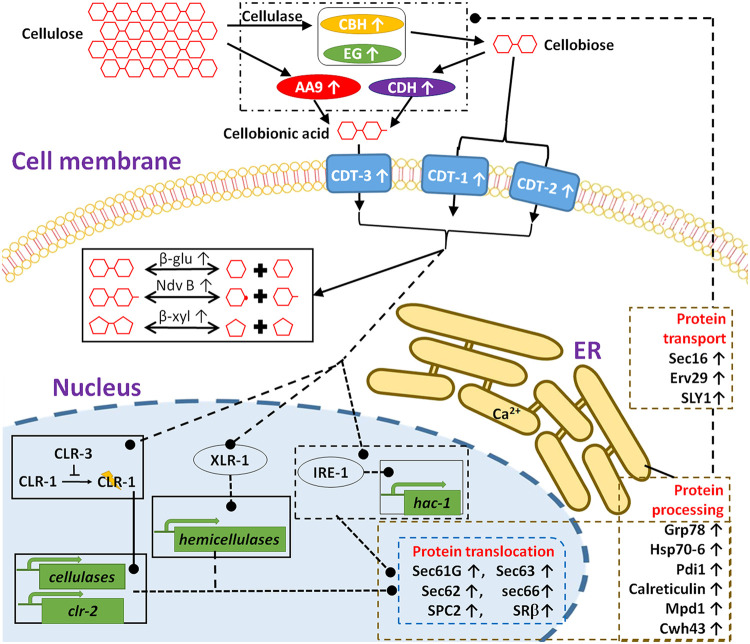
Recent studies suggest a role for transcriptional control as well as for processes downstream of transcription in controlling the production of cellulolytic enzymes (26). Deletion of the light-responsive G-protein signaling components GNB1 and GNG1 increased cellulase activities, while transcript abundance of cellulases decreased (27). The posttranslational pathways, such as protein processing and export, are considered additional limiting factors for the production of cellulolytic enzymes (28, 29). Recent studies suggest that the secretion of cellulases occurs mainly through the conventional endoplasmic reticulum (ER)-Golgi secretory pathway in N. crassa (30). When grown on cellulose, cellulolytic fungi produce relatively high levels of extracellular cellulases for cellulose degradation, which increases the demand for protein synthesis and folding, disulfide bond formation, glycosylation, and sorting in the ER, causing ER stress and activating the unfolded protein response (UPR) (29, 31, 32). In Saccharomyces cerevisiae and filamentous fungi, the UPR mainly depends on an evolutionarily conserved signaling cascade mediated by inositol-requiring enzyme-1 (Ire1) and the transcription factor HAC1 (33).
The morphological state of filamentous fungi is another limiting factor for the production of cellulolytic enzymes in submerged or solid-state fermentations (34, 35). A major determinant of cellular morphology is the cell wall, which is regulated by the cell wall integrity (CWI) signaling pathway (36). Disruption of a nonanchored cell wall protein Ncw1 has been shown to promote cellulase production in N. crassa (37).
Taken together, current evidence supports the hypothesis of a regulatory network coordinating transcription and its downstream pathways for lignocellulolytic responses in filamentous fungi (22, 38). To examine how cellobiose mediates lignocellulolytic responses, we applied tandem mass tag (TMT)-based proteome quantification with synchronous precursor selection (SPS)-based MS3 technology for quantitative profiling of N. crassa proteomes in response to cellobiose and cellulose (Avicel). The SPS-MS3 technology significantly improves the sensitivity, resolution, and dynamic range of mass spectrometry (MS) analysis (39). A triple deletion mutant of β-glucosidases (Δ3βG) was employed in this study, as the strain is devoid of rapid turnover of cellobiose (8, 9, 40) and thus allows profiling of the proteomes of this mutant strain in response to the disaccharide (8). The knowledge deduced is important for a systems-level understanding of cellobiose-mediated lignocellulolytic responses for strain optimization to improve the production of lignocellulases and bio-based products.
RESULTS AND DISCUSSION
Quantitative proteome profiling reveals cellobiose-dependent translational and posttranslational pathway for lignocellulolytic responses.
The proteome analysis of this study was performed in duplicates 4 h after the Δ3βG strain was switched from minimal medium to Avicel (AV) or cellobiose (CB)-containing medium. The workflow for this study is summarized in Fig. S1 in the supplemental material. As reported previously, the transcriptional response of the Δ3βG mutant to cellobiose is similar to that of the wild-type N. crassa to Avicel, and the identity and amount of proteins secreted in the Δ3βG strain on cellobiose mimic the wild-type response to Avicel (8, 16, 40). Our proteome profiling of the Δ3βG strain revealed differential abundance of 188, 125, and 581 proteins in the comparisons of CB versus no carbon (NC), AV versus NC, and NC versus sucrose (P < 0.05), respectively (Fig. 1A). A high overlap was observed for the proteins induced by cellobiose and Avicel and the targets of the essential cellulolytic regulators CLR-1 and CLR-2 (Fig. 1B and C) (Table S1). Correlation coefficient analysis between mRNA and protein abundance was further performed with the proteome data of this study and the transcriptome data from a previous study with a similar experimental setup (18). The derived correlation coefficient (R2) value was approximately 0.31 (Fig. 2A), indicating a moderate correlation between changes in protein and mRNA levels in N. crassa under cellulolytic conditions. This is consistent with previous reports on the correlation between protein and mRNA levels in various species (16, 41). The discrepancy between mRNA and protein levels is probably due to several factors, including differences in posttranscriptional, translational, and posttranslational pathways (42). Improved prediction of protein abundance from mRNA abundance was achieved recently by incorporating posttranscriptional processes and protein synthesis (43, 44).
FIG 1.
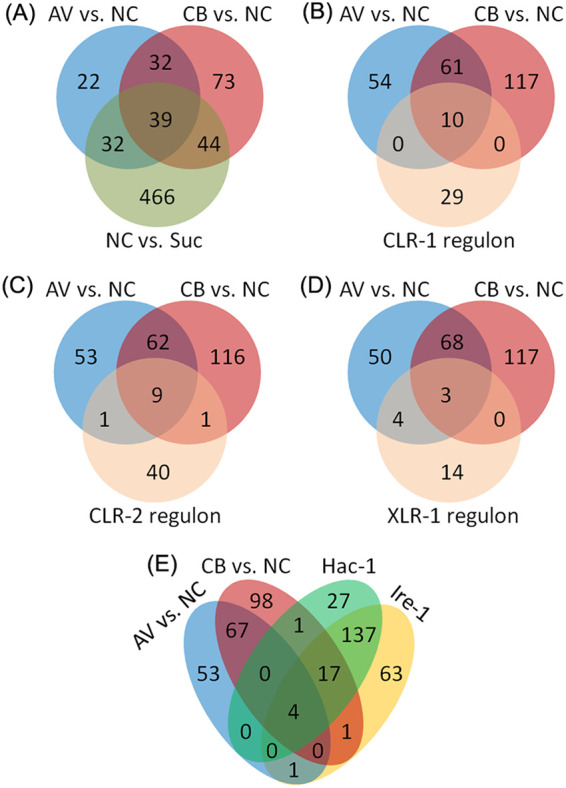
Venn diagram analysis comparing proteins affected by alternate carbon sources. (A) Comparison of proteins that increased in abundance upon exposure to cellobiose (CB) versus no carbon (NC), upon exposure to Avicel (AV) versus NC, and under carbon starvation (NC versus Suc). (B to E) Comparison of proteins that increased in abundance by exposure to cellobiose or Avicel with the targets of the CLR-1 direct regulon (B), the CLR-2 direct regulon (C), the XLR-1 direct regulon (D), or the Ire-1/HAC-1 regulon (E). Quantification of proteins was performed with a P value of <0.05.
FIG 2.
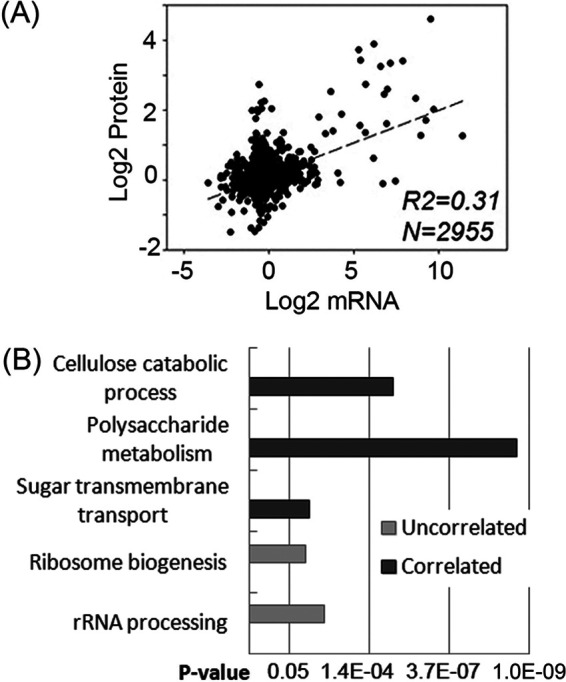
Functional enrichment analysis of the gene/protein sets with correlated or uncorrelated abundance of proteins and mRNAs. (A) Correlation between changes in mRNA levels and protein abundances upon exposure to Avicel (AV) versus no carbon (NC). (B) Functional enrichment analysis of the gene/protein sets whose protein levels correlated with mRNA levels (correlated) and those regulated differentially only at the protein level but not at the mRNA level (uncorrelated).
To examine if specific functional categories are associated with the identified correlated and uncorrelated protein/gene lists (Data Sets S10, S11, and S12), functional enrichment analysis was performed, which revealed that in the list with increased abundance of both proteins (fold changes of ≥1.3, P < 0.05) and mRNAs (fold changes of ≥2.0, P < 0.05) (Data Set S10), the significantly enriched functional groups were cellulose catabolism, polysaccharide metabolism, and sugar transmembrane transport (Fig. 2B) (Table S1). However, in the list that showed increased protein but not mRNA levels (P ≥ 0.05) (Data Set S11), the enriched functional groups were ribosome biogenesis and rRNA processing (Fig. 2B). The results corroborate the idea that cellulose degradation and utilization is controlled primarily at the transcriptional level (see Fig. 4) (22), but that the processes after transcription also play a role in controlling lignocellulolytic responses. This is consistent with the observation that the processes of ribosome biogenesis, protein processing, and protein export were also enriched in the identified proteome showing increased abundance in response to 0.2% cellobiose (Table 1). Uncorrelated levels of proteins and mRNAs have also been observed in previous studies of filamentous fungi under cellulolytic conditions (27). In the Trichoderma reesei deletion mutants of the G-protein signaling components gnb1 and gng1, increased cellulase activities were observed while transcript abundance of major cellulases was significantly decreased, suggesting the importance of additional mechanisms downstream of transcription in lignocellulase production in filamentous fungi.
FIG 4.
Schematic model illustrating cellobiose-elicited action of ER stress and lignocellulase synthesis. Cellobiose, the major cellulolytic product by endo-glucanases (EG) and exo-glucanases (CBH), is transported by the cellodextrin transporters/transceptors CDT-1 and CDT-2. Cellobionic acid, derived from catalytic reactions of CDH and LPMO, is transported into the cell by CBT-1. Signals, derived from cellobiose, a modified version of cellobiose, and/or cellobiose interaction with CDT-1 and/or CDT-2 (40) or other intracellular proteins results in the activation of downstream events, including transcriptional activation and gene expression, modulation of intracellular metabolism, and translation and secretion of lignocellulases. In particular, cellobiose signaling induces the activation of the Ire-1/HAC-1-mediated UPR pathway (29), particularly proteins involved in translocation, folding, and protein glycosylation. Upright arrows indicate increases in protein abundance in response to cellobiose.
TABLE 1.
Gene ontology terms enriched in the set of proteins with increased abundance in response to cellobiosea
| GO term | Count | % | P value | Fold enrichment | FDR |
|---|---|---|---|---|---|
| Protein processing in endoplasmic reticulum | 13 | 6.91 | 1.04E−05 | 4.59 | 0.01 |
| Protein export | 6 | 3.19 | 1.92E−04 | 9.88 | 0.19 |
| Ribosome biogenesis in eukaryotes | 7 | 3.72 | 3.40E−02 | 2.79 | 28.45 |
| Cell wall organization | 4 | 2.13 | 5.13E−03 | 10.77 | 5.89 |
| Cellulose catabolic process | 4 | 2.13 | 5.13E−03 | 10.77 | 5.89 |
| Conidium formation | 3 | 1.60 | 3.08E−02 | 10.51 | 30.86 |
| Sporulation resulting in formation of a cellular spore | 3 | 1.60 | 4.35E−02 | 8.75 | 40.85 |
| Endoplasmic reticulum membrane | 8 | 4.26 | 9.83E−03 | 3.29 | 9.46 |
| Endoplasmic reticulum | 5 | 2.66 | 3.44E−02 | 4.00 | 29.69 |
| Extracellular region | 7 | 3.72 | 1.60E−02 | 3.38 | 15.00 |
| Small-subunit processome | 4 | 2.13 | 3.96E−02 | 5.20 | 33.38 |
GO, gene ontology; FDR, false-discovery rate.
Cellobiose-dependent protein processing and export for lignocellulolytic responses.
When grown on cellulose, filamentous fungi produce relatively high levels of extracellular cellulases for cellulose degradation and utilization, which causes ER stress and activates the UPR response (29, 32). It is still not clear how the ER stress response is controlled under cellulolytic conditions. Previous studies suggest that the major cellulolytic product, cellobiose or a modified version of cellobiose, functions as an inducer of cellulolytic gene expression (8, 40). It is possible that cellobiose, or a modified version of cellobiose, acts as not only the inducer of cellulolytic gene expression but also a messenger to mediate the feedback cross talk between the levels of extracellular cellulases and the intracellular processes involved in cellulase production. This is consistent with the finding that exposure to 0.2% cellobiose increased the abundance of 23 identified targets of the Ire-1/HAC-1 regulon (29) (Fig. 1E, Table 2). The identified ER stress-responsive proteins are involved in different stages of protein export, including translocation, folding, glycosylation, degradation, and transport of proteins. During early adaptation to cellulose, cellobiose concentrations were relatively low in the cultures of wild-type and β-glucosidase deletion strains (8, 9, 40), which may explain the observation that exposure to Avicel increased only four of the targets of the Ire-1/HAC-1 regulon (Fig. 1E, Table 2).
TABLE 2.
Overlapped elements of the Ire-1/Hac-1 pathway and the proteomes upregulated by cellobiose and/or Avicela
| NCU no. | Locus | Annotation or domain | Target of the Ire-1/Hac-1 pathway | Upregulation condition |
|---|---|---|---|---|
| Protein translocation | ||||
| NCU04127 | sec61g | Probable protein transport protein Sec61 subunit gamma Sss1 | yes | CB vs. NC |
| NCU06333 | sec62 | Translocation protein SEC62 | yes | CB vs. NC |
| NCU00169 | sec63 | Translocation protein SEC63 | yes | CB vs. NC |
| NCU02681 | sec66 | Translocation protein SEC66 | yes | CB vs. NC |
| NCU08217 | srβ | SRP receptor β subunit | yes | CB vs. NC |
| NCU00965 | spc2 | Signal peptidase complex subunit Spc2 | yes | CB vs. NC, AV vs. NC |
| NCU04519 | Microsomal signal peptidase 18 kDa subunit | yes | CB vs. NC, AV vs. NC | |
| Protein folding | ||||
| NCU03982 | grp-78 | Glucose regulated protein 78 | yes | CB vs. NC |
| NCU09265 | Calreticulin | yes | CB vs. NC | |
| NCU09485 | HSP70-6 chaperone dnaK | yes | CB vs. NC | |
| NCU09223 | pdi-1 | Protein disulfide isomerase | yes | CB vs. NC |
| NCU00813 | mpd-1 | Disulfide isomerase | yes | CB vs. NC, AV vs. NC |
| Protein glycosylation | ||||
| NCU05895 | cwh43 | Putative ceramide-conjugation protein | no | CB vs. NC, AV vs. NC |
| NCU03503 | alg-2 | α-1,3/1,6-Mannosyltransferase | yes | CB vs. NC, AV vs. NC |
| NCU06057 | gim-1 | GPI mannosyltransferase 1 | yes | CB vs. NC |
| NCU00924 | gpit-2 | Glycosylphosphatidylinositol transamidase-2 | No | AV vs. NC |
| NCU02349 | UDP-glucose:glycoprotein glucosyltransferase | yes | CB vs. NC | |
| Proteolytic degradation | ||||
| NCU11102 | scj1 | DnaJ-related protein | yes | CB vs. NC |
| NCU10789 | Ubiquitin-protein ligase Sel1/Ubx2 | yes | CB vs. NC | |
| NCU04193 | Uncharacterized protein | yes | CB vs. NC | |
| NCU07672 | Uncharacterized protein | no | CB vs. NC | |
| Protein transport | ||||
| NCU03819 | sec16 | COP II coat assembly protein sec-16 | yes | CB vs. NC |
| NCU03319 | erv29 | COP II-coated vesicle protein SurF4/Erv29 | yes | CB vs. NC |
| NCU04252 | sly1 | SNARE docking complex subunit | yes | CB vs. NC |
| Unknown function in protein processing/secretion | ||||
| NCU02153 | Uncharacterized protein | yes | CB vs. NC | |
| NCU10546 | 4-Coumarate-CoA ligase | yes | CB vs. NC |
CB, cellobiose; AV, Avicel; NC, no carbon.
The first step in biogenesis of secretory proteins is signal sequence-directed translocation of a nascent polypeptide into the ER (45). Exposure to cellobiose increased the abundance of Sec61 (NCU04127), Sec62 (NCU06333), Sec63 (NCU00169), Sec66/Sec71 (NCU02681), and SPC2 (NCU00965) (Table 2, see Fig. 4). In S. cerevisiae, efficient posttranslational translocation requires the assembly of Sec61 with the Sec63 complex, consisting of Sec62, Sec63, Sec71, and Sec72 (46–48). A component of the signal sequence receptor (SSR), SRβ (NCU08217), also increased in abundance in CB-treated cultures (Table 2). The SSR facilitates delivery of the ribosome-nascent chain complex (RNC) to the protein translocation channel. As proteins are transferred into the ER, the signal peptidase complex (SPC) cleaves the signal peptide sequence (49). Two potential signal peptidases, SPC2 and NCU04519, also increased in abundance in response to cellobiose (Table 2).
Some proteins that increased in abundance are potentially involved in the later stages of protein secretion, including Sec16 (NCU03819), the coat protein complex II (COP II)-coated vesicle cargo adapter protein Erv29 (NCU03319) (28), and a SNARE docking complex subunit (NCU04252) (Table 2, see Fig. 4). Protein transport from the ER to the Golgi apparatus has been shown to be dependent on COP II (50). Cargo adaptor proteins, such as Erv29/SurF4, are essential for maintaining the architecture of endoplasmic reticulum-Golgi intermediate compartment (ERGIC) and Golgi by controlling COP I recruitment in mammalian cells (51). In particular, Erv29/SurF4 was identified as a primary mediator of the secretion of protein convertase subtilisin/kexin type 9 (52). In N. crassa, ERV-29 functions as a cargo adaptor that mediates efficient trafficking of cellobiohydrolase II (NCU9680) under Avicel conditions (28). The Sec16 protein defines ER exit sites and is required for secretory cargo export (53). Moderate overexpression of Sec16 was shown to improve the secretion of T. reesei endoglucanase I and Rhizopus oryzae glucan-1,4-α-glucosidase in S. cerevisiae, suggesting a common role of Sec16 in protein secretion in fungi.
Once in the ER, a series of folding and maturation events accompany secretory protein biogenesis, including disulfide bond formation, primary glycosylation, and folding and quality control (45). Exposure to cellobiose increased the abundance of the homologs of the protein disulfide isomerases Pdi1 (NCU09223) and Mpd11 (NCU00813) and the ER chaperones Grp78 (NCU03982), calreticulin (NCU09265), and Hsp70-6 (NCU09485) (Table 2, see Fig. 4). Transcription levels of pdi-1 in the T. reesei hyper-secretion RUT-C30 strain were also found to be significantly elevated in response to increased production of cellulases (54), suggesting a conserved role of Pdi-1 in protein secretion in filamentous fungi during lignocellulolytic responses (32). Two major chaperone systems in the ER, the calnexin/calreticulin system and the BiP system, recognize unfolded proteins and promote their folding by cycles of substrate binding and release (55).
In addition to disulfide bond formation and chaperone folding machinery, our study also demonstrated increased abundance of potential glycosylation enzymes in response to cellobiose, including two mannosyltransferases (Alg2, NCU03503; Gim1, NCU06057), a UDP-glucose glycoprotein glucosyltransferase (NCU02349), and the putative ceramide conjugation protein CWH43 (Table 2). Gim1/Yke2 is a subunit of the heterohexameric Gim/prefoldin protein complex, which acts as a molecular chaperone present in both eukaryotes and archaea for maintaining native protein conformation and preventing nonspecific aggregation (46). Alg2 is a bifunctional enzyme required for the transfer of the α-1,3- and the α-1,6-mannose-linked residue on the cytosolic side of the ER (47). ER-localized mannosidases play an important role in the glycosylation of eukaryotic secretory and membrane-bound proteins (48), which affects protein folding or serves as ligands for other proteins that modulate the folding (56).
CWH43, a putative ceramide conjugation protein, is positively involved in cellulase production.
cwh43 encodes a transmembrane protein that localizes in the ER (57). In budding yeast, CWH43 incorporates the sphingolipid ceramide into a lipid moiety of GPI-anchored proteins in the ER (58–60). GPI anchoring is an evolutionarily conserved posttranslational modification that anchors proteins in the outer leaflet of the plasma membrane. GPI-anchored proteins can function as coreceptors to modulate signaling pathways through interactions with membrane receptors. It is possible that upregulation of CWH43 by cellobiose or Avicel may modulate cellobiose-mediated signaling events under cellulolytic conditions. To examine if CWH43 plays a role in lignocellulolytic responses, a cwh43 deletion strain was compared with a wild-type strain with respect to hyphal growth, cellulase activity, and protein secretion. Growth on minimal medium supplemented with sucrose resulted in a comparable growth rate to the wild-type control (Fig. 3A). However, growth on the medium supplemented with Avicel resulted in significantly reduced levels of endoglucanase activities, as well as secreted proteins (Fig. 3B and C), suggesting a positive role of CWH43 in cellulase production. GPI-anchored proteins, such as the anchored cell wall protein Acw10 and the cell wall glucanosyltransferase Mwg1, also showed increased abundance in response to cellobiose and/or Avicel (Data Set S3). To examine if the observed effects of CWH43 are related to Acw10 and Mwg1, the deletion mutants were examined for the two proteins (Fig. 3). The derived results suggest that disruption of Acw10 and Mwg1 had no significant effects on hyphal growth, protein secretion, or endoglucanase activities (Fig. 3). It is still under investigation how CWH43 modulates cellulase production. CWH43 in N. crassa is 43% identical to the homolog in yeast (Fig. S2). As observed in yeast (57, 61), GPI-anchored proteins in N. crassa are also required for cell wall integrity and, furthermore, cell growth and morphology (62). Therefore, we cannot rule out the possibility that the observed effects of CWH43 on cellulase production is related to cell wall integrity, which has been shown to affect cellulase secretion in N. crassa in previous studies (63, 64).
FIG 3.
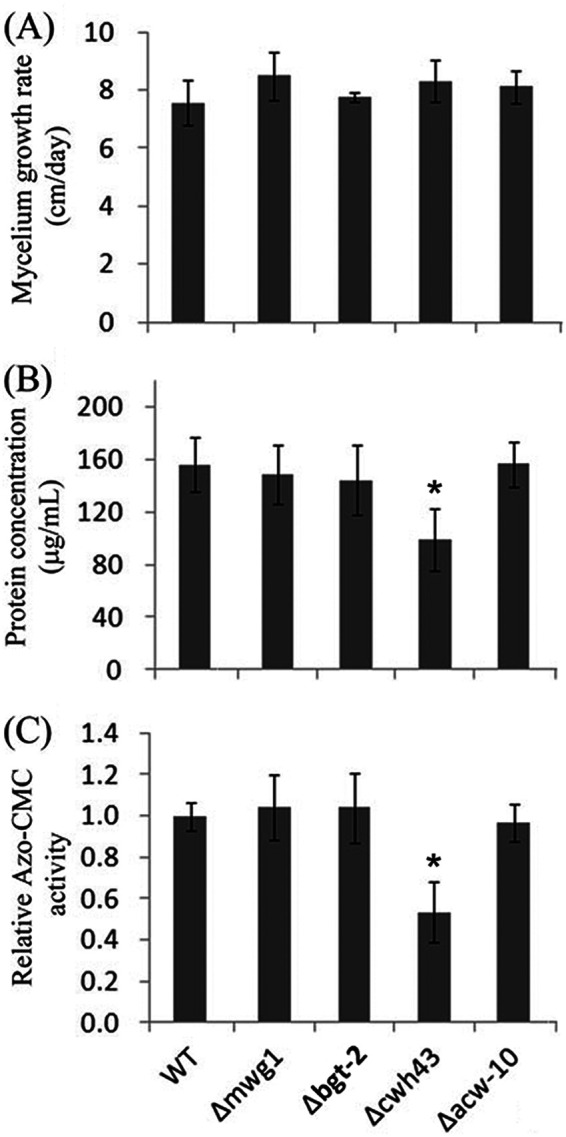
Phenotypic analysis of the deletion mutants of the selected proteins that increased in abundance in the Δ3βG strain upon exposure to cellobiose (CB versus NC). (A) Growth rates of wild type (WT) and mutant strains. A suspension of 103 conidia from the strains indicated was inoculated into race tubes containing VMM and incubated for 3 days. Mycelium length was measured every 24 h. (B and C) Total secreted protein levels (B) and endoglucanase activities (C) in the supernatant of 96-h cultures in VMM with 2% Avicel after a shift from a 16-h VMM culture. Shown are the mean values of at least three replicates. Error bars show the standard deviations between these replicates. The significance of differences between the mutants and the WT was based on t test analysis. Asterisks indicate significant differences (*, P < 0.001). Azo-CMC, azo-carboxymethyl cellulose.
In summary, we applied a TMT-based quantification approach with SPS-MS3 analysis to profile and compare the proteome responses of the N. crassa mutant strain Δ3βG to cellobiose and Avicel. A detailed analysis suggests a cellobiose-dependent response network that integrates transcription and its downstream pathways, such as protein processing and export, for lignocellulase expression and secretion (Fig. 4). The cellobiose-dependent pathways presented here provide a systems-level understanding of how the major cellulolytic product, cellobiose, functions as an inducer for the production of lignocellulases. Given the importance of filamentous fungi for biotechnological enzyme production, the findings should have an impact on systems-engineering approaches to strain improvement for the production of lignocellulases.
MATERIALS AND METHODS
Cell cultures.
An N. crassa strain bearing a deletion of a gene encoding an intracellular β-glucosidase plus two genes encoding extracellular β-glucosidases (Δ3βG) (12) was inoculated onto slants of Vogel’s minimal medium (MM) with 1.5% agar and 2% sucrose and grown for 3 days at 30°C in the dark followed by 7 days at 25°C in light. Conidia were inoculated into 100 ml of liquid Vogel’s minimal medium (VMM) (65) with 2% (wt/vol) sucrose at 106 conidia/ml and grown at 25°C in constant light with shaking (200 rpm) for 16 h. Mycelia were harvested by filtrating through 3 layers of autoclaved Miracloth (Calbiochem, La Jolla, CA) and washed 2 to 3 times with VMM without a carbon source, followed by growth in 100 ml of fresh VMM with either 0.5% sucrose, 0.2% cellobiose, 0.5% Avicel PH-101 (Sigma-Aldrich, MO), or no carbon source added, as described previously (8).
Protein digestion and TMT labeling.
The Δ3βG strain was grown in VMM with 2% sucrose for 16 h before being washed and resuspended in fresh VMM containing sucrose, cellobiose, or Avicel or medium lacking any carbon source. Mycelia were harvested at 4 h after medium shifts by filtering through Miracloth and immediately frozen in liquid nitrogen. Frozen mycelia (∼100 mg) were ground into fine powder with a precooled mortar and pestle. The powder was suspended in 0.5 ml of ice-cold lysis buffer (250 mM sucrose, 20 mM HEPES, 0.1 mM EGTA, 1 mM EDTA, pH 7.4, adjusted with 1 N KOH, 4°C) with freshly added 1× Roche Complete Protease Inhibitor Cocktail. Cell lysate was centrifuged for 20 min at 20,000 × g at 4°C to remove cell debris. Protein concentration was determined using the Bradford assay (Bio-Rad Laboratories). Proteins were reduced by adding dithiothreitol (DTT) to a final concentration of 10 mM and incubating for 30 min at 60°C, and alkylated with iodoacetamide (final concentration 20 mM) for 30 min in the dark at room temperature. Excess iodoacetamide was quenched by the addition of DTT to a final concentration of 20 mM. Acetone (5 volumes, precooled at –20°C) was added to the protein solution, and the solution was incubated at –20°C for 3 h, before being centrifuged at 8,000 × g for 10 min. Protein pellets were washed with precooled 90% acetone and air-dried.
An aliquot of 25 to 100 μg of acetone-precipitated protein was suspended in 100 mM triethylammonium bicarbonate (TEAB), crushed into fine particles, and incubated at 37°C overnight after trypsin was added at a protein/trypsin ratio of 40:1 (wt/wt). Digested peptides were labeled using TMT 6-plex isobaric mass tagging reagents (Thermo Scientific) according to the manufacturer’s instructions. The peptide solution was centrifuged for 10 min at 20,000 × g to remove any insoluble material. Peptides were concentrated and desalted on a peptide trap column (Michrom) and eluted from the trap with 1 to 2 trap volumes of buffer B (80/20/0.1 acetonitrile/H2O/TFA). The peptide solution was acidified with 10% trifluoroacetic acid (TFA), centrifuged, and dried to about 5 μl by vacuum centrifugation.
LC-MS analysis.
An appropriate volume of 5% acetonitrile (vol/vol) solution with 0.1% (vol/vol) trifluoroacetic acid was added to the dried peptide samples for liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis. Samples were fractionated on an EASY-Spray column (50 cm × 75μm ID, PepMap RSLC C18, 2 μm) using an EASY-nLC 1200 system (Thermo Scientific) coupled to an Orbitrap Fusion Tribird mass spectrometer. Mobile phases A and B were water with 0.1% formic acid (phase A) and acetonitrile with 0.1% formic acid (phase B). A 210-min gradient was used with a flow rate of 200 nl min−1.
For MS analysis, a data-dependent method of synchronous precursor selection (SPS)-MS3 was used (Data Set S1). Relative quantification of protein abundance between two different media, i.e., no carbon versus sucrose (NC versus Suc), cellobiose versus no carbon (CB versus NC), and Avicel versus no carbon (AV versus NC) conditions, was performed for each replicate.
Data analysis.
Proteome Discoverer 1.4 (Thermo Scientific) was used to search MS/MS spectra against the N. crassa strain OR74A database from NCBI and the resulting peptide hits were filtered for maximum 1% false-discovery rate (FDR). The TMT sixplex quantification method was used to calculate the reporter ratios with mass tolerance ±10 ppm. Only peptide spectra containing all reporter ions were designated “quantifiable spectra.” Spectra of the non-TMT, standard peptides were analyzed manually. For data sets from the MS3 method which contained both MS2 and MS3 spectra, CID (MS2) spectra were used for identification and HCD (MS3) spectra were used for quantification.
The reporter ion intensities in the spectrum of each TMT-labeled peptide represent the total quantification signal for that peptide. The statistical significance of results was evaluated with Student’s t test and a P value of less than 0.05 was considered as statistically significant. Gene ontology enrichment was obtained from DAVID Bioinformatics Resources 6.8 (66) and the FungiFun2 tool (67). A Venn diagram was constructed using the web-based tool (http://bioinformatics.psb.ugent.be/webtools/Venn).
Data availability.
The mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD011598.
Supplementary Material
ACKNOWLEGMENTS
This work was supported by grants from “The Twelfth Five-Year-Plan” in National Science and Technology for the Rural Development under the Ministry of Science and Technology of PRC (approval no. 2015BAD15B0503) (to S.C.), the National Natural Science Foundation of China (approval no. 31770207) (to S.C.), and the Intergovernmental International Cooperation on Science and Technology Innovation under the Ministry of Science and Technology of PRC (approval no. 2018YFE0112400) (to S.C.). N.L.G. is supported by a Laboratory Directed Research and Development Program of the Lawrence Berkeley National Laboratory under U.S. Department of Energy contract no. DE-AC02-05CH11231.
S. Chen, N. L. Glass, and Z. Hao designed the research. D. Liu, Y. Liu, L. Wei, Y. Wang, and B. Xiong performed the research. D. Zhang, X. Chen, Q. Liu, and L. Zhang analyzed the data. S. Chen, Z. Hao, and D. Liu wrote the paper. H. Fang, J. Liesche, and Y. Wei provided critical insights.
We thank Chris R. Somerville (University of California Berkeley) for valuable advice on this work, Phillip Benz (Technical University of Munich) for critical reading of the manuscript, and Juan Wang, Meijuan Ren, and Jidong Feng (the Life Science Core Facility, Northwest A&F University) for technical support.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Payne CM, Knott BC, Mayes HB, Hansson H, Himmel ME, Sandgren M, Ståhlberg J, Beckham GT. 2015. Fungal cellulases. Chem Rev 115:1308–1448. doi: 10.1021/cr500351c. [DOI] [PubMed] [Google Scholar]
- 2.Peitersen N, Medeiros J, Mandels M. 1977. Adsorption of Trichoderma cellulase on cellulose. Biotechnol Bioeng 19:1091–1094. doi: 10.1002/bit.260190710. [DOI] [PubMed] [Google Scholar]
- 3.Sukumaran RK, Singhania RR, Pandey A. 2005. Microbial cellulases—production, applications and challenges. J Sci Ind Res 64:832–844. [Google Scholar]
- 4.Gabelle JC, Jourdier E, Licht RB, Ben Chaabane F, Henaut I, Morchain J, Augier F. 2012. Impact of rheology on the mass transfer coefficient during the growth phase of Trichoderma reesei in stirred bioreactors. Chem Eng Sci 75:408–417. doi: 10.1016/j.ces.2012.03.053. [DOI] [Google Scholar]
- 5.Li Y, Liu C, Bai F, Zhao X. 2016. Overproduction of cellulase by Trichoderma reesei RUT C30 through batch-feeding of synthesized low-cost sugar mixture. Bioresour Technol 216:503–510. doi: 10.1016/j.biortech.2016.05.108. [DOI] [PubMed] [Google Scholar]
- 6.Li C, Lin F, Zhou L, Qin L, Li B, Zhou Z, Jin M, Chen Z. 2017. Cellulase hyper-production by Trichoderma reesei mutant SEU-7 on lactose. Biotechnol Biofuels 10:228. doi: 10.1186/s13068-017-0915-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vaheri MP, Vaheri ME, Kauppinen VS. 1979. Formation and release of cellulolytic enzymes during growth of Trichoderma reesei on cellobiose and glycerol. Eur J Appl Microbiol Biotechnol 8:73–80. doi: 10.1007/BF00510268. [DOI] [Google Scholar]
- 8.Znameroski EA, Coradetti ST, Roche CM, Tsai JC, Iavarone AT, Cate JH, Glass NL. 2012. Induction of lignocellulose-degrading enzymes in Neurospora crassa by cellodextrins. Proc Natl Acad Sci U S A 109:6012–6017. doi: 10.1073/pnas.1118440109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu WH, Hildebrand A, Kasuga T, Xiong XC, Fan ZL. 2013. Direct cellobiose production from cellulose using sextuple beta-glucosidase gene deletion Neurospora crassa mutants. Enzyme Microb Technol 52:184–189. doi: 10.1016/j.enzmictec.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 10.Chen S, Xiong B, Wei L, Wang Y, Yang Y, Liu Y, Zhang D, Guo S, Fang QLH, Wei Y. 2018. The model filamentous fungus Neurospora crassa: progress toward a systems understanding of plant cell wall deconstruction, p 107–134. In Fang X, Qu Y (ed), Fungal cellulolytic enzymes: microbial production and application. Springer Nature, New York, NY. [Google Scholar]
- 11.Galazka JM, Tian CG, Beeson WT, Martinez B, Glass NL, Cate J. 2010. Cellodextrin transport in yeast for improved biofuel production. Science 330:84–86. doi: 10.1126/science.1192838. [DOI] [PubMed] [Google Scholar]
- 12.Znameroski EA, Glass NL. 2013. Using a model filamentous fungus to unravel mechanisms of lignocellulose deconstruction. Biotechnol Biofuels 6:6. doi: 10.1186/1754-6834-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.dos Reis TF, de Lima PBA, Parachin NS, Mingossi FB, de Castro Oliveira JV, Ries LNA, Goldman GH. 2016. Identification and characterization of putative xylose and cellobiose transporters in Aspergillus nidulans. Biotechnol Biofuels 9:204. doi: 10.1186/s13068-016-0611-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang W, Kou Y, Xu J, Cao Y, Zhao G, Shao J, Wang H, Wang Z, Bao X, Chen G, Liu W. 2013. Two major facilitator superfamily sugar transporters from Trichoderma reesei and their roles in induction of cellulase biosynthesis. J Biol Chem 288:32861–32872. doi: 10.1074/jbc.M113.505826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cai P, Wang B, Ji J, Jiang Y, Wan L, Tian C, Ma Y. 2015. The putative cellodextrin transporter-like protein CLP1 is involved in cellulase induction in Neurospora crassa. J Biol Chem 290:788–796. doi: 10.1074/jbc.M114.609875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiong Y, Coradetti ST, Li X, Gritsenko MA, Clauss T, Petyuk V, Camp D, Smith R, Cate JHD, Yang F, Glass NL. 2014. The proteome and phosphoproteome of Neurospora crassa in response to cellulose, sucrose and carbon starvation. Fungal Genet Biol 72:21–33. doi: 10.1016/j.fgb.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coradetti ST, Craig JP, Xiong Y, Shock T, Tian C, Glass NL. 2012. Conserved and essential transcription factors for cellulase gene expression in ascomycete fungi. Proc Natl Acad Sci U S A 109:7397–7402. doi: 10.1073/pnas.1200785109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coradetti ST, Xiong Y, Glass NL. 2013. Analysis of a conserved cellulase transcriptional regulator reveals inducer-independent production of cellulolytic enzymes in Neurospora crassa. Microbiologyopen 2:595–609. doi: 10.1002/mbo3.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Craig JP, Coradetti ST, Starr TL, Glass NL. 2015. Direct target network of the Neurospora crassa plant cell wall deconstruction regulators CLR-1, CLR-2, and XLR-1. mBio 6:e01452-15. doi: 10.1128/mBio.01452-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huberman LB, Coradetti ST, Glass NL. 2017. Network of nutrient-sensing pathways and a conserved kinase cascade integrate osmolarity and carbon sensing in Neurospora crassa. Proc Natl Acad Sci U S A 114:E8665–E8674. doi: 10.1073/pnas.1707713114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Q, Li J, Gao R, Li J, Ma G, Tian C. 2019. CLR-4, a novel conserved transcription factor for cellulase gene expression in ascomycete fungi. Mol Microbiol 111:373–394. doi: 10.1111/mmi.14160. [DOI] [PubMed] [Google Scholar]
- 22.Huberman LB, Liu J, Qin L, Glass NL. 2016. Regulation of the lignocellulolytic response in filamentous fungi. Fungal Biol Rev 30:101–111. doi: 10.1016/j.fbr.2016.06.001. [DOI] [Google Scholar]
- 23.Xiong Y, Sun J, Glass NL. 2014. VIB1, a link between glucose signaling and carbon catabolite repression, is essential for plant cell wall degradation by Neurospora crassa. PLoS Genet 10:e1004500. doi: 10.1371/journal.pgen.1004500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, Hu Y, Zhao K, Pan Y, Qu Y, Zhao J, Qin Y. 2019. The indispensable role of histone methyltransferase PoDot1 in extracellular glycoside hydrolase biosynthesis of Penicillium oxalicum. Front Microbiol 10:2566. doi: 10.3389/fmicb.2019.02566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, Hu Y, Zhu Z, Zhao K, Liu G, Wang L, Qu Y, Zhao J, Qin Y. 2019. Normal transcription of cellulolytic enzyme genes relies on the balance between the methylation of H3K36 and H3K4 in Penicillium oxalicum. Biotechnol Biofuels 12:198. doi: 10.1186/s13068-019-1539-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stappler E, Dattenböck C, Tisch D, Schmoll M. 2017. Analysis of light-and carbon-specific transcriptomes implicates a class of G-protein-coupled receptors in cellulose sensing. mSphere 2:e00089-17. doi: 10.1128/mSphere.00089-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tisch D, Kubicek CP, Schmoll M. 2011. The phosducin-like protein PhLP1 impacts regulation of glycoside hydrolases and light response in Trichoderma reesei. BMC Genomics 12:613. doi: 10.1186/1471-2164-12-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Starr TL, Gonçalves AP, Meshgin N, Glass NL. 2018. The major cellulases CBH-1 and CBH-2 of Neurospora crassa rely on distinct ER cargo adaptors for efficient ER-exit. Mol Microbiol 107:229–248. doi: 10.1111/mmi.13879. [DOI] [PubMed] [Google Scholar]
- 29.Fan F, Ma G, Li J, Liu Q, Benz JP, Tian C, Ma Y. 2015. Genome-wide analysis of the endoplasmic reticulum stress response during lignocellulase production in Neurospora crassa. Biotechnol Biofuels 8:66. doi: 10.1186/s13068-015-0248-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li C, Pang A-P, Yang H, Lv R, Zhou Z, Wu F-G, Lin F. 2019. Tracking localization and secretion of cellulase spatiotemporally and directly in living Trichoderma reesei. Biotechnol Biofuels 12:200. doi: 10.1186/s13068-019-1538-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanaka M, Shintani T, Gomi K. 2015. Unfolded protein response is required for Aspergillus oryzae growth under conditions inducing secretory hydrolytic enzyme production. Fungal Genet Biol 85:1–6. doi: 10.1016/j.fgb.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 32.Qin L, Wu VW, Glass NL. 2017. Deciphering the regulatory network between the SREBP pathway and protein secretion in Neurospora crassa. mBio 8:e00233-17. doi: 10.1128/mBio.00233-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hollien J. 2013. Evolution of the unfolded protein response. Biochim Biophys Acta 1833:2458–2463. doi: 10.1016/j.bbamcr.2013.01.016. [DOI] [PubMed] [Google Scholar]
- 34.Cairns TC, Zheng X, Zheng P, Sun J, Meyer V. 2019. Moulding the mould: understanding and reprogramming filamentous fungal growth and morphogenesis for next generation cell factories. Biotechnol Biofuels 12:77. doi: 10.1186/s13068-019-1400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Veiter L, Rajamanickam V, Herwig C. 2018. The filamentous fungal pellet—relationship between morphology and productivity. Appl Microbiol Biotechnol 102:2997–3006. doi: 10.1007/s00253-018-8818-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verdín J, Sánchez-León E, Rico-Ramírez AM, Martínez-Núñez L, Fajardo-Somera RA, Riquelme M. 2019. Off the wall: the rhyme and reason of Neurospora crassa hyphal morphogenesis. Cell Surf 5:100020. doi: 10.1016/j.tcsw.2019.100020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin L, Chen Y, Li J, Wang S, Sun W, Tian C. 2017. Disruption of non-anchored cell wall protein NCW-1 promotes cellulase production by increasing cellobiose uptake in Neurospora crassa. Biotechnol Lett 39:545–551. doi: 10.1007/s10529-016-2274-1. [DOI] [PubMed] [Google Scholar]
- 38.Benocci T, Aguilar-Pontes MV, Zhou M, Seiboth B, de Vries RP. 2017. Regulators of plant biomass degradation in ascomycetous fungi. Biotechnol Biofuels 10:152. doi: 10.1186/s13068-017-0841-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ting L, Rad R, Gygi SP, Haas W. 2011. MS3 eliminates ratio distortion in isobaric multiplexed quantitative proteomics. Nat Methods 8:937–940. doi: 10.1038/nmeth.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Znameroski EA, Li X, Tsai JC, Galazka JM, Glass NL, Cate J. 2014. Evidence for transceptor function of cellodextrin transporters in Neurospora crassa. J Biol Chem 289:2610–2619. doi: 10.1074/jbc.M113.533273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumar D, Bansal G, Narang A, Basak T, Abbas T, Dash D. 2016. Integrating transcriptome and proteome profiling: strategies and applications. Proteomics 16:2533–2544. doi: 10.1002/pmic.201600140. [DOI] [PubMed] [Google Scholar]
- 42.Wethmar K, Smink JJ, Leutz A. 2010. Upstream open reading frames: molecular switches in (patho) physiology. Bioessays 32:885–893. doi: 10.1002/bies.201000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fortelny N, Overall CM, Pavlidis P, Freue G. 2017. Can we predict protein from mRNA levels? Nature 547:E19–E20. doi: 10.1038/nature22293. [DOI] [PubMed] [Google Scholar]
- 44.Magnusson R, Rundquist O, Kim MJ, Hellberg S, Na CH, Benson M, Gomez-Cabrero D, Kockum I, Tegnér Jesper Piehl F, Jagodic M, Mellergård J, Altafini C, Ernerudh J, Jenmalm MC, Nestor CE, Kim M-S, Gustafsson M. 2019. On the prediction of protein abundance from RNA. bioRxiv 599373. doi: 10.1101/599373. [DOI]
- 45.Barlowe CK, Miller EA. 2013. Secretory protein biogenesis and traffic in the early secretory pathway. Genetics 193:383–410. doi: 10.1534/genetics.112.142810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hansen WJ, Cowan NJ, Welch WJ. 1999. Prefoldin–nascent chain complexes in the folding of cytoskeletal proteins. J Cell Biol 145:265–277. doi: 10.1083/jcb.145.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kämpf M, Absmanner B, Schwarz M, Lehle L. 2009. Biochemical characterization and membrane topology of Alg2 from Saccharomyces cerevisiae as a bifunctional α1,3- and 1,6-mannosyltransferase involved in lipid-linked oligosaccharide biosynthesis. J Biol Chem 284:11900–11912. doi: 10.1074/jbc.M806416200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tannous A, Pisoni GB, Hebert DN, Molinari M. 2015. N-linked sugar-regulated protein folding and quality control in the ER. Semin Cell Dev Biol 41:79–89. doi: 10.1016/j.semcdb.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paetzel M, Karla A, Strynadka NCJ, Dalbey RE. 2002. Signal peptidases. Chem Rev 102:4549–4580. doi: 10.1021/cr010166y. [DOI] [PubMed] [Google Scholar]
- 50.Barlowe C, Orci L, Yeung T, Hosobuchi M, Hamamoto S, Salama N, Rexach MF, Ravazzola M, Amherdt M, Schekman R. 1994. COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell 77:895–907. doi: 10.1016/0092-8674(94)90138-4. [DOI] [PubMed] [Google Scholar]
- 51.Mitrovic S, Ben-Tekaya H, Koegler E, Gruenberg J, Hauri H-P. 2008. The cargo receptors Surf4, endoplasmic reticulum-Golgi intermediate compartment (ERGIC)-53, and p25 are required to maintain the architecture of ERGIC and Golgi. Mol Biol Cell 19:1976–1990. doi: 10.1091/mbc.e07-10-0989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Emmer BT, Hesketh GG, Kotnik E, Tang VT, Lascuna PJ, Xiang J, Gingras A-C, Chen X-W, Ginsburg D. 2018. The cargo receptor SURF4 promotes the efficient cellular secretion of PCSK9. Elife 7:e38839. doi: 10.7554/eLife.38839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watson P, Townley AK, Koka P, Palmer KJ, Stephens DJ. 2006. Sec16 defines endoplasmic reticulum exit sites and is required for secretory cargo export in mammalian cells. Traffic 7:1678–1687. doi: 10.1111/j.1600-0854.2006.00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pakula TM, Salonen K, Uusitalo J, Penttilä M. 2005. The effect of specific growth rate on protein synthesis and secretion in the filamentous fungus Trichoderma reesei. Microbiology 151:135–143. doi: 10.1099/mic.0.27458-0. [DOI] [PubMed] [Google Scholar]
- 55.Tremmel D, Duarte M, Videira A, Tropschug M. 2007. FKBP22 is part of chaperone/folding catalyst complexes in the endoplasmic reticulum of Neurospora crassa. FEBS Lett 581:2036–2040. doi: 10.1016/j.febslet.2007.04.042. [DOI] [PubMed] [Google Scholar]
- 56.Helenius A, Aebi M. 2004. Roles of N-linked glycans in the endoplasmic reticulum. Annu Rev Biochem 73:1019–1049. doi: 10.1146/annurev.biochem.73.011303.073752. [DOI] [PubMed] [Google Scholar]
- 57.Nakazawa N, Xu X, Arakawa O, Yanagida M. 2019. Coordinated roles of the putative ceramide-conjugation protein, Cwh43, and a Mn2+-transporting, P-type ATPase, Pmr1, in fission yeast. G3 (Bethesda) 9:2667–2676. doi: 10.1534/g3.119.400281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martin-Yken H, Dagkessamanskaia A, De Groot P, Ram A, Klis F, François J. 2001. Saccharomyces cerevisiae YCRO17c/CWH43 encodes a putative sensor/transporter protein upstream of the BCK2 branch of the PKC1-dependent cell wall integrity pathway. Yeast 18:827–840. doi: 10.1002/yea.731. [DOI] [PubMed] [Google Scholar]
- 59.Ghugtyal V, Vionnet C, Roubaty C, Conzelmann A. 2007. CWH43 is required for the introduction of ceramides into GPI anchors in Saccharomyces cerevisiae. Mol Microbiol 65:1493–1502. doi: 10.1111/j.1365-2958.2007.05883.x. [DOI] [PubMed] [Google Scholar]
- 60.Umemura M, Fujita M, Yoko-O T, Fukamizu A, Jigami Y. 2007. Saccharomyces cerevisiae CWH43 is involved in the remodeling of the lipid moiety of GPI anchors to ceramides. Mol Biol Cell 18:4304–4316. doi: 10.1091/mbc.e07-05-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu X-X, Komatsuzaki A, Chiba Y, Gao X-D, Yoko-o T. 2018. PER1, GUP1 and CWH43 of methylotrophic yeast Ogataea minuta are involved in cell wall integrity. Yeast 35:225–236. doi: 10.1002/yea.3285. [DOI] [PubMed] [Google Scholar]
- 62.Bowman SM, Piwowar A, Al Dabbous M, Vierula J, Free SJ. 2006. Mutational analysis of the glycosylphosphatidylinositol (GPI) anchor pathway demonstrates that GPI-anchored proteins are required for cell wall biogenesis and normal hyphal growth in Neurospora crassa. Eukaryot Cell 5:587–600. doi: 10.1128/EC.5.3.587-600.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lin L, Sun Z, Li J, Chen Y, Liu Q, Sun W, Tian C. 2018. Disruption of gul-1 decreased the culture viscosity and improved protein secretion in the filamentous fungus Neurospora crassa. Microb Cell Fact 17:96. doi: 10.1186/s12934-018-0944-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Herold I, Yarden O. 2017. Regulation of Neurospora crassa cell wall remodeling via the cot-1 pathway is mediated by gul-1. Curr Genet 63:145–159. doi: 10.1007/s00294-016-0625-z. [DOI] [PubMed] [Google Scholar]
- 65.Vogel HJ. 1956. A convenient growth medium for Neurospora (Medium N). Microb Genet Bull 13:42–43. [Google Scholar]
- 66.Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 67.Priebe S, Kreisel C, Horn F, Guthke R, Linde J. 2015. FungiFun2: a comprehensive online resource for systematic analysis of gene lists from fungal species. Bioinformatics 31:445–446. doi: 10.1093/bioinformatics/btu627. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD011598.