Abstract
Background
Long intergenic non-coding RNA 00511 (LINC00511) is highly expressed in diverse cancers and has a correlation with poor clinical outcomes for cancer patients. In view of contradictory data among published data, we aim to evaluate the prognostic role of LINC00511 for cancer patients.
Methods
In the present study, a meta-analysis of related studies has been performed to investigate the prognostic significance of LINC00511 in cancer patients. Relevant studies published before December 22, 2019 were systematically searched online in PubMed, EMBASE, Web of Science, and the Cochrane Library databases. The relationship between LINC00511 expression and cancer patients’ survival, including overall survival (OS), disease-free survival (DFS)/relapse-free survival (RFS) and progression-free survival (PFS), was evaluated using pooled hazard ratios (HRs) with their corresponding 95% confidence intervals (CIs). The association between LINC00511 expression and clinicopathological features was assessed using odd ratios (ORs) and their corresponding 95% CIs.
Results
A total of 14 eligible studies with 1883 patients were enrolled in the present meta-analysis. The results demonstrated that elevated expression of LINC00511 was significantly associated with poor OS (HR = 2.62; 95% CI: 2.00–3.45; p < 0.001), PFS (HR = 1.80; 95% CI: 1.29–2.51; p = 0.001) and DFS/RFS (HR = 2.90; 95% CI: 1.04–8.12; p = 0.04). Additionally, High LINC00511 expression was associated with large tumor size (OR = 3.10; 95% CI: 1.97–4.86; p < 0.00001), lymph node metastasis (OR = 3.11; 95% CI: 2.30–4.21; p < 0.00001), advanced clinical stage (OR = 3.95; 95% CI: 2.68–5.81; p < 0.00001), distant metastasis (OR = 2.39; 95% CI: 1.16–4.93; p = 0.02), and disease recurrence (OR = 4.62; 95% CI: 2.47–8.65; p < 0.00001). Meanwhile, no correlation was found between LINC00511 expression and age, gender, and histological grade. These findings were consolidated by the results of bioinformatics analysis.
Conclusions
Based on our findings, LINC00511 may serve as a novel prognostic biomarker for cancer patients.
Keywords: LINC00511, Prognostic biomarker, Survival, Meta-analysis, TCGA, Cancer
Background
Long non-coding RNAs (lncRNAs) are referred to as the non-protein coding portion of the transcriptome with length of more than 200 nucleotides. They represent, together with small noncoding RNAs (< 200 nucleotides), the biggest part of the transcriptome, since only 1 to 3% of the transcriptome can code for protein synthesis [1–3]. Long noncoding RNAs are involved in several biological processes including cell proliferation, cell differentiation, cell cycle progression, cell apoptosis and metastasis [4–10]. Notably, they were identified as oncogenes or tumor suppressors, prognostic and diagnostic biomarkers and as therapeutic targets [11–13]. For instance, Tang and colleagues reviewed the implication of lncRNAs in colorectal cancer progression and figured out their potential clinical applications as novel diagnostic and prognostic biomarkers and therapeutic targets [14].
Long intergenic non-coding RNA 00511 (LINC00511) is a 2265-bp lncRNA mapped to chromosome 17q24.3 with five exons. Recent studies found that LINC00511 is overexpressed in various type of cancers including breast cancer, ovarian cancer, liver cancer, pancreatic cancer, lung cancer and glioma. LINC00511 is an oncogene which plays a negative regulatory role in cell proliferation, cell cycle progression, apoptosis, invasion, migration, metastasis and chemoresistance [15–23]. Noteworthily, overexpression of LINC00511 was shown to be a predictor for cancer prognosis [16, 18, 20, 22, 24–28]. However, the results of the studies were not consistent. For instance, the studies of Deng et al. [27], Zhao et al. [20] and Wang et al. [18] showed no correlation between the expression level of LINC00511 and tumor size; in contrast the studies of Sun et al. [22], Yu et al. [25] and Zhang et al. [26] demonstrated an association between LINC00511 expression and tumor size. Similarly, Lu et al. [16] detected significant correlation between LINC00511 overexpression and lymph node metastasis in breast cancer, whereas the study conducted by Zhang et al. [26] provided a contradictory result. In view of these contradictory outcomes, we conducted a systematic review and meta-analysis to evaluate the prognostic role of LINC00511 for cancer patients. Next, we validated our results by using The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) datasets.
Methods
Literature search strategies
The present study was performed according to the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) [29]. Relevant studies published before December 22, 2019 were systematically searched online in PubMed, EMBASE, Web of Science and the Cochrane Library databases. The following search terms were used in different combinations: (LINC00511 OR “long intergenic noncoding RNA 00511” OR “long intergenic non-protein coding RNA 511”) AND (cancer OR tumor OR tumour OR carcinoma OR neoplasm OR adenoma OR sarcoma OR malignancy). The reference lists of the full-text articles were also checked to find relevant studies that pointed out the relation between LINC00511 expression and clinical outcomes. No language restriction was applied.
Inclusion and exclusion criteria
The eligibility criteria were as follows: (1) the study subjects were patients with any type of cancer; (2) the studies investigated association between LINC00511 expression levels and clinical outcomes in cancer; (3) patients were separated into LINC00511 high expression level and LINC00511 low expression level groups; (4) the LINC00511 expression levels were measured by quantitative method (e.g., real-time reverse transcription polymerase chain reaction (qRT-PCR)); and (5) sufficient information and data were provided to calculate a hazard ratios (HRs) or odd ratios (ORs) and their corresponding 95% confidence intervals (CIs). Exclusion criteria were as follows: (1) duplicate publications; (2) reviews and meta-analyses; (3) studies without patient samples; (4) studies not relevant to cancer, LINC00511 or prognosis.
Publication quality assessment
The quality of included studies was assessed using the Newcastle-Ottawa Scale (NOS). Three main categories have been considered including selection, comparability and outcome, and the stars rating system has been used. The scores of NOS were ranged from 0 star (lowest score) to 9 stars (highest score). A study with a NOS score higher than 5 was considered as a high-quality study [30, 31].
Data extraction
Two investigators reviewed all eligible studies independently and extracted the following data: the first author’s name, year of publication, country of origin, the tumor type, sample size, cut-off value, detection method, HRs and corresponding 95% CIs, and clinicopathological features. Any controversial issue was resolved by discussion. In case HR value was not provided, Engauge Digitizer version 11.2 (http://markummitchell.github.io/engauge-digitizer/) was used to extract data from Kaplan-Meier curve. The extracted data were then used to estimate HRs and 95% CIs by using the HR calculations spreadsheet provided by Tierney et al. [32].
Bioinformatics analysis
We used GEPIA, a web server for cancer and normal gene expression profiling and interactive analyses, to perform bioinformatics analysis. GEPIA uses the data from both TCGA (The Cancer Genome Atlas) and GTEx (Genotype-Tissue Expression) to perform several analyses. The datasets were computed by the UCSC Xena project based on a standard pipeline [33, 34]. Therefore, we directly use GEPIA to perform our analyses. GEPIA used Kaplan-Meier method and log-rank test for the survival analysis and one-way ANOVA for the gene expression analysis. First, the expression levels of LINC00511 in the different cancer types involved in our meta-analysis was analyzed by matching TCGA and GTEx data. Next, overall survival and disease-free survival curves were retrieved.
Statistical analysis
All statistical analyses were performed with RevMan 5.3 software (Cochrane community, https://community.cochrane.org/help/tools-and-software/revman-5/revman-5-download/) and STATA 15.0. The pooled HRs and their 95% CIs were used to evaluate the association of LINC00511 expression with the cancer patients’ survival. Odd ratios (ORs) analyses were performed to assess the correlation between LINC00511 expression and clinicopathological parameters. The heterogeneity between studies results was examined by Cochran’s Q test and Higgins I-squared (I2) statistic. In the absence of heterogeneity (p ≥ 0.10 and/or I2 < 50%), the fixed-effect model was used; otherwise the random-effect model was applied [35, 36]. Potential publication bias was evaluated using the funnel plot, Egger’s test and Begg’s test. Sensitivity analysis was also performed to assess the impact of individual study on the pooled effect [37]. A p-value less than 0.05 was considered to be statistically significant.
Results
Characteristics of included studies
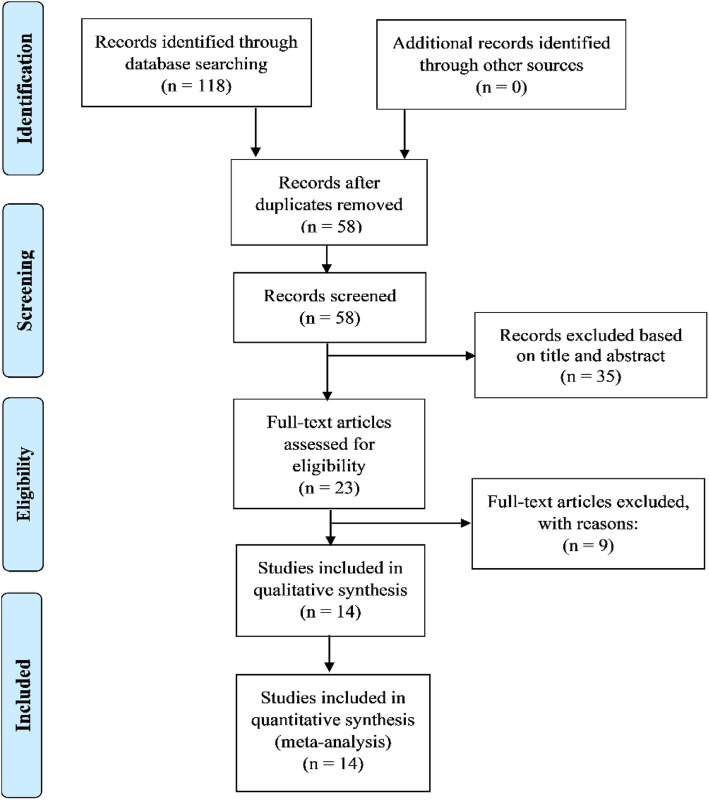
A flowchart of the literature search strategy is shown in Fig. 1. According to the described search strategy, 118 articles were retrieved. Sixty articles were excluded based on duplication criteria. The 58 articles remained, underwent an initial screening. On the basis of title and abstract, 35 articles (non-cancer publications, non-LINC00511 publications, reviews and meta-analyses, retracted article, articles without prognosis and clinical data) were excluded. A total of 23 full-text articles were eventually assessed for eligibility. After reading full-text articles, 9 articles were excluded for reasons: articles without prognosis and clinical data in respect to LINC00511, not available full-text article and non-patient samples. At the end, 14 articles were used for the quantitative synthesis [16–22, 24–28, 38, 39]. The 14 studies involved a total of 1883 patients and all came from China. The sample size of included studies ranged from 36 to 412 patients and the type of cancer included non-small-cell lung cancer, pancreatic cancer, breast cancer, cervical cancer, hepatocellular carcinoma, ovarian cancer, and brain tumors. The articles included in the present study were all written in English and published between 2016 and 2019. The expression level of LINC00511 was mainly determined by qRT-PCR. The cut-off values included mean, median and the P25 value. In our study, the quality of all included studies is high (NOS score > 5). The overall characteristics of included studies are summarized in Table 1.
Fig. 1.
Flowchart of the literature search strategy
Table 1.
Characteristics of the studies included in the meta-analysis
| First author | Year | Country | Tumor type | Sample size (n) |
Gender (Male/Female) (n) |
LINC00511 expression (High/low) (n) |
Detection method | Tumor stage | Cut-off value | Survival analysis | Outcome | Quality score | Data extraction method |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sun C.-C. | 2016 | China | NSCLC | 124 | 58/66 | 93/31 | qRT-PCR | I-IV | P25 value | Univariate/Multivariate | OS/CP | 8 | Direct |
| Zhao X. | 2017 | China | PDAC | 140 | 78/62 | 102/38 | qRT-PCR | I-III | Not available | Univariate/Multivariate | OS/PFS/CP | 7 | Direct |
| Lu G. | 2018 | China | BC | 39 | 0/39 | 23/16 | qRT-PCR | I-IV | Mean | Kaplan-Meier | OS/CP | 7 | Indirect |
| Mao B.-D. | 2019 | China | CC | 84 | 0/84 | 49/35 | qRT-PCR | I1a-II2b | Median (0.996) | Kaplan-Meier | OS/RFS/CP | 7 | Indirect |
| Wang R.-P. | 2019 | China | HCC | 127 | 76/51 | 64/63 | qRT-PCR | I-IV | Median | Univariate/Multivariate | OS/CP | 8 | Direct |
| Zhang J. | 2019 | China | BC | 70 | 0/70 | 24/46 | qRT-PCR | I-IV | Mean | Kaplan-Meier | OS/CP | 8 | Indirect |
| Liu L. | 2019 | China | BC | 98 | 0/98 | 49/49 | qRT-PCR | I-IV | Median | Kaplan-Meier | OS/CP | 7 | Indirect |
| Yu C.-L. | 2019 | China | CC | 92 | 0/92 | 46/46 | qRT-PCR | I-IV | Not available | Univariate/Multivariate | OS/CP | 8 | Direct |
| Deng H. | 2019 | China | ccRCC | 49 | 21/28 | 25/24 | qRT-PCR | I-IV | Median | Kaplan-Meier | OS/CP | 6 | Indirect |
| Zhu F.-Y. | 2019 | China | NSCLC | 57 | 28/29 | 29/28 | qRT-PCR | I-IV | Median | Kaplan-Meier | OS/CP | 8 | Direct |
| Du X. | 2019 | China | GBM | 36 | 24/12 | 18/18 | qRT-PCR | Not available | Median | Kaplan-Meier | OS/PFS/CP | 7 | Indirect |
| Wang J. | 2019 | China | OC | 378 | Not available | 189/189 | qRT-PCR | Not available | Median | Univariate | OS | 7 | Direct |
| Wang Z. | 2019 | China | PC | 177 | Not available | 134/43 | RNA sequencing expression data | Not available | Not available | Kaplan-Meier | OS | 7 | Direct |
| Li C. | 2019 | China | GL | 412 | Not available | 206/206 | qRT-PCR | Not available | Not available | Kaplan-Meier | OS | 6 | Indirect |
Abbreviations: NSCLC Non-small-cell lung cancer, PDAC Pancreatic ductal adenocarcinoma, BC Breast cancer, CC Cervical cancer, HCC hepatocellular carcinoma, ccRCC Clear cell renal cell carcinoma, GBM Glioblastoma multiforme, OC Ovarian cancer, PC Pancreatic cancer, GL Glioma, qRT-PCR Quantitative real time polymerase chain, OS Overall survival, PFS Progression-free survival, RFS Relapse-free survival, DFS Disease-free survival, CP Clinicopathological parameters
Association between LINC00511 expression and cancer patients’ survival
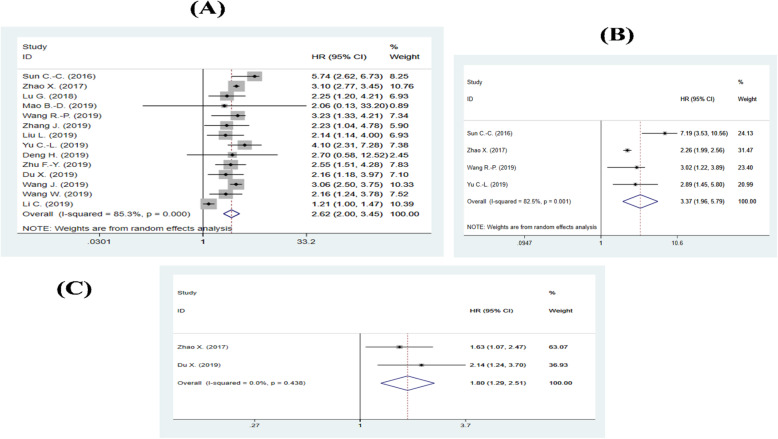
Elevated LINC00511 expression was significantly predictive of poor overall survival (HR = 2.62; 95% CI: 2.00–3.45; p < 0.001). A high heterogeneity was observed among the studies (PH < 0.001; I2 = 85.3%), so the random-effect model was used (Fig. 2a). In addition, high LINC00511 expression has been proven to have an independent prognostic value in four studies by processing the multivariate analysis [18, 20, 22, 25]. As previously reported [40], we conducted the meta-analysis of these studies and found that high LINC00511 expression may serve as an independent predictor for poor overall survival prognosis in cancers (HR = 3.37; 95% CI: 1.96–5.79; p < 0.001; I2 = 82.5%; PH = 0.001; Fig. 2b).
Fig. 2.
Forest plots of combined analyses on the association of survival with LINC00511 expression. a Forest plot of OS analysis, b Forest plot of meta-analysis of the independent predictive value of LINC00511 for OS, and c forest plot of PFS analysis
The relationship between LINC00511 expression and cancer progression was investigated by combining progression-free survival (PFS) studies. The results, from only two studies with available related data, demonstrated that high LINC00511 expression was predicted to be associated significantly with worse PFS (HR = 1.80; 95% CI: 1.29–2.51; p = 0.001; I2 = 0%; PH = 0.44; Fig. 2c). In addition, one study found that high LINC00511 may be associated with worse relapse-free survival (HR = 2.90; 95% CI: 1.04–8.12; p = 0.04).
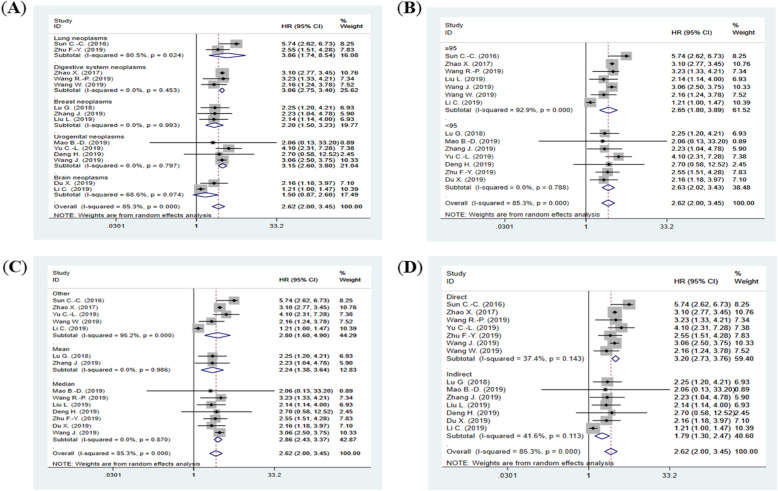
Subgroup meta-analyses based on cancer types (according to NCBI’s medical subject headings (MeSH) [41]) (Fig. 3a), sample size (Fig. 3b), cut-off value (Fig. 3c) and data extraction method (Fig. 3d) were conducted. The results showed that high LINC00511 expression was significantly associated with poor overall survival in lung neoplasms (HR = 3.86; 95% CI: 1.74–8.54; p = 0.001; I2 = 80.5%; PH = 0.02), digestive system neoplasms (HR = 3.06; 95% CI: 2.76–3.40; p < 0.001; I2 = 0%; PH = 0.45), breast neoplasms (HR = 2.20; 95% CI: 1.50–3.23; p < 0.001; I2 = 0%; PH = 0.99) and urothelial neoplasms (HR = 3.15; 95% CI: 2.60–3.80; p < 0.001; I2 = 0%; PH = 0.80). Regarding the brain neoplasms subgroup, despite the relatively high HR, the relationship cannot be considered robust because the p-value is higher than 0.05 (HR = 1.50; 95% CI: 0.87–2.60; p = 0.15; I2 = 68.6%; PH = 0.07). In respect to sample size subgroups, there was significant correlation between high LINC00511 expression and poor overall survival prognosis of cancer patients in both large sample size (n ≥ 95) (HR = 2.65; 95% CI: 1.81–3.89; p < 0.001; I2 = 92.9%; PH < 0.001) and small sample size (n < 95) (HR = 2.63; 95% CI: 2.02–3.43; p < 0.001; I2 = 0%, PH = 0.79). Concerning the cut-off value subgroups analysis, whether the median was used as cut-off value (HR = 2.86; 95% CI: 2.43–3.38; p < 0.001; I2 = 0%; PH = 0.87) or the mean as cut-off value (HR = 2.24; 95% CI: 1.38–3.64; p = 0.001; I2 = 0%, PH = 0.99) or other cut-off values were used (HR = 2.80; 95% CI: 1.60–4.90; p < 0.001; I2 = 95.2%; PH < 0.001), high LINC00511 expression correlated with poor overall survival in cancer patients. Similarly, the subgroup meta-analysis based on the data extraction method showed that whether the data were extracted directly from the literature (HR = 3.21; 95% CI: 2.73–3.76; p < 0.001; I2 = 37.4%; PH = 0.14) or indirectly (HR = 1.80; 95% CI: 1.30–2.47; p < 0.001; I2 = 41.6%; PH = 0.11), high LINC00511 expression was significantly associated with poor overall survival in cancer patients. Of note, the heterogeneity is likely to come from diverse sources.
Fig. 3.
Forest plots of the association between LINC00511 expression and overall survival in subgroups based on a cancer types; b sample size; c different cut-off value and d different extraction method
Association between LINC00511 expression and clinicopathological features
The meta-analysis of the correlation between LINC00511 expression and clinicopathological parameters are summarized in Table 2. The pooled odds ratio (OR) values revealed significant association between high LINC00511 expression and large tumor size (OR = 3.10; 95% CI: 1.97–4.86; p < 0.00001; I2 = 50%; PH = 0.03), lymph node metastasis (OR = 3.11; 95% CI: 2.30–4.21; p < 0.00001; I2 = 42%; PH = 0.08), advanced clinical stage (OR = 3.95; 95% CI: 2.68–5.81; p < 0.00001; I2 = 0%; PH = 0.48), distant metastasis (OR = 2.39; 95% CI: 1.16–4.93; p = 0.02; I2 = 21%; PH = 0.28), and disease recurrence (OR = 4.62; 95% CI: 2.47–8.65; p < 0.00001; I2 = 0%; PH = 0.66). Meanwhile, the results showed no statistically significant correlation between LINC00511 expression and age, gender, and histological grade.
Table 2.
Meta-analyses of the correlation between LINC00511 expression and clinicopathological features
| Variables | Number of studies | Number of patients | Model | OR | Heterogeneity | ||
|---|---|---|---|---|---|---|---|
| 95% CI | p-value | I2 (%) | PH | ||||
| Gender (Male vs. Female) | 6 | 498 | Random | 0.98 (0.53–1.85) | 0.96 | 58 | 0.04 |
| Age | |||||||
| ≥45 vs. < 45 | 1 | 84 | – | 0.66 (0.28–1.59) | 0.36 | – | – |
| ≥50 vs. < 50 | 6 | 462 | Fixed | 0.69 (0.47–1.01) | 0.06 | 0 | 0.98 |
| ≥60 vs. < 60 | 1 | 140 | – | 0.72 (0.34–1.52) | 0.39 | – | – |
| ≥65 vs. < 65 | 2 | 173 | Fixed | 0.59 (0.29–1.20) | 0.15 | 0 | 0.89 |
| Histological grade (poor vs. well and moderate) | 8 | 725 | Fixed | 1.34 (0.97–1.85) | 0.07 | 0 | 0.75 |
| Histological tumor type (adenocarcinoma vs. squamous cell carcinoma) | 3 | 300 | Random | 2.80 (0.58–13.59) | 0.20 | 71 | 0.03 |
| Tumor size (large vs. small) | 11 | 916 | Random | 3.10 (1.97–4.86) | < 0.00001 | 50 | 0.03 |
| Lymph node metastasis (positive vs. negative) | 10 | 880 | Fixed | 3.11 (2.30–4.21) | < 0.00001 | 42 | 0.08 |
| Clinical stage (late vs. early) | 8 | 643 | Fixed | 3.95 (2.68–5.81) | < 0.00001 | 0 | 0.48 |
| Distant metastasis (Yes vs. No) | 3 | 215 | Fixed | 2.39 (1.16–4.93) | 0.02 | 21 | 0.28 |
| Recurrence (Yes vs. No) | 3 | 274 | Fixed | 4.62 (2.47–8.65) | < 0.00001 | 0 | 0.66 |
Abbreviations: OR Odd ratio, PHP-value of heterogeneity
Publication bias
Funnel plot analysis, Egger’s test and Begg’s test were performed to evaluate potential publication bias. There was no obvious asymmetry on the funnel plot of OS (Fig. 4) and the tests of publication bias (Begg’s test: p = 0.161; Egger’s test: p = 0.630) indicated that no publication bias existed in studies reported OS.
Fig. 4.
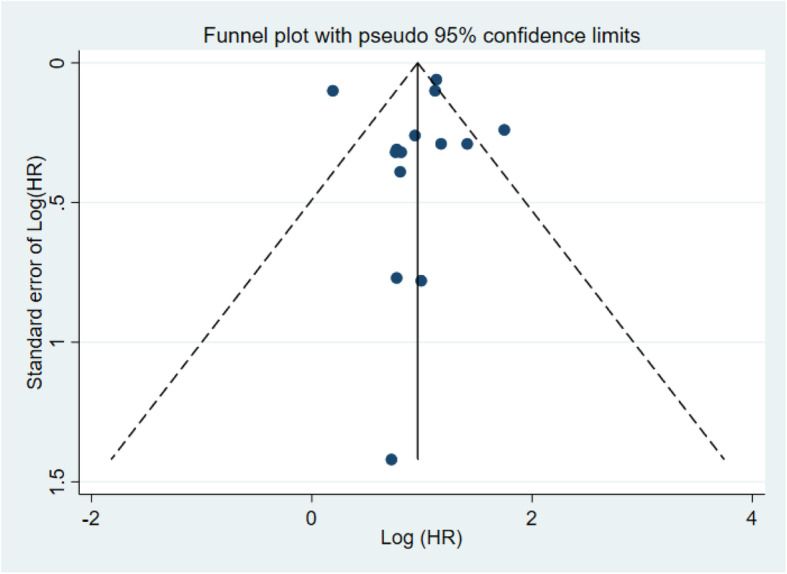
Funnel plot of publication bias based on Overall survival (OS)
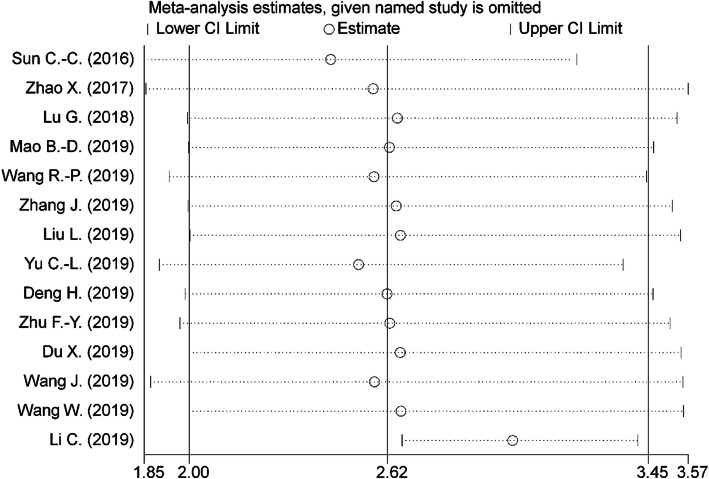
Sensitivity analysis
The sensitivity analysis was performed by omitting each individual study to test the stability of the pooled result of the association between LINC00511 expression and OS. As shown on Fig. 5, when “Li C. (2019)” was omitted, the pooled result oscillated. Subsequently, we evaluated the pooled HR after removing “Li C. (2019)” and found that elevated LINC00511 expression still predicted worse OS (HR = 3.06; 95% CI: 2.80–3.33; p < 0.001; I2 = 14.8%; PH = 0.30). Therefore, the influential study didn’t alter the significance of the pooled result, sustaining the reliability of our pooled result.
Fig. 5.
Sensitivity analysis of pooled HR for overall survival
Datasets analyses by GEPIA
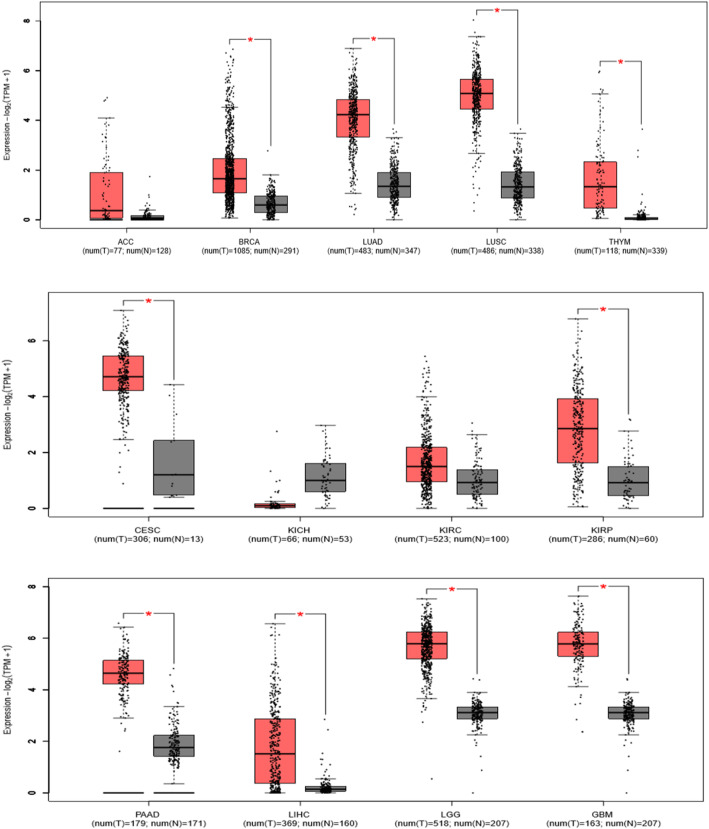
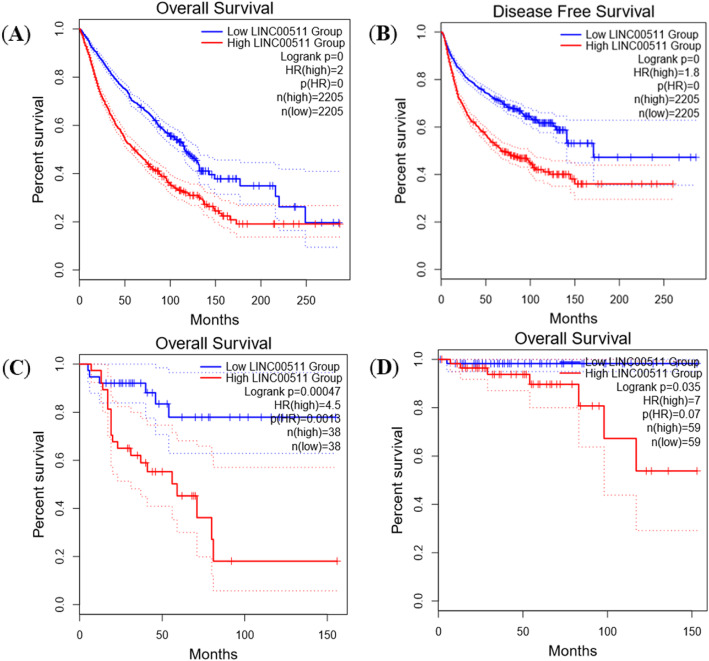
To strengthen the results of our meta-analysis, we used GEPIA web server which contain a larger amount of sample size and performed bioinformatics analysis. The expression level of LINC00511 was analyzed in eleven different type of cancers (cancer types included in the meta-analysis). As shown on the Fig. 6, LINC00511 was highly expressed in almost all the related cancers, including lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), pancreatic adenocarcinoma (PAAD), breast invasive carcinoma (BRCA), cervical squamous cell carcinoma (CESC), liver hepatocellular carcinoma (LIHC), kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), kidney chromophobe (KICH), glioblastoma multiforme (GBM), and brain lower grade glioma (LGG) (log2FC value > 1 and p-value < 0.01). The Fig. 7 shows the Kaplan-Meier curves of the overall survival (Fig. 7a) and disease-free survival (Fig. 7b) analyses. A total of 4410 patients were divided into LINC00511 low expression and high expression groups based on the median LINC00511 expression. The patients in high expression group showed a worst overall survival (HR = 2.00; log-rank p-value < 0.001) and disease-free survival (HR = 1.8; log-rank p-value < 0.001) than those in the low expression group. Furthermore, we explored the possibility for any association of the LINC00511 expression to overall cancer survival in other cancer types that are not involved in the meta-analysis. It was found that LINC00511 overexpression was significantly associated with worse OS in adrenocortical carcinoma (ACC) (HR = 4.5; log-rank p-value = 0.00047; Fig. 7c), and thymoma (THYM) (HR = 7.00; log-rank p-value = 0.035; Fig. 7d). These results confirmed that LINC00511 is overexpressed in different types of cancers and this is correlated with poor survival. Of note, the GEPIA web server was accessed on December 28, 2019.
Fig. 6.
Validation of the expression levels of LINC00511 in TCGA normal and GTEx datasets: “*” indicates log2FC value > 1 and p-value < 0.01. TCGA, The Cancer Genome Atlas; GTEx, Genotype-Tissue Expression; ACC, adrenocortical carcinoma; THYM, thymoma; LGG, brain lower grade glioma; GBM, glioblastoma multiforme; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; PAAD, pancreatic adenocarcinoma; BRCA, breast invasive carcinoma; CESC, cervical squamous cell carcinoma; LIHC, liver hepatocellular carcinoma; KIRC, Kidney renal clear cell carcinoma; KIRP, Kidney renal papillary cell carcinoma; KICH, Kidney Chromophobe; TPM, transcripts per million; T, tumor; N, normal
Fig. 7.
Kaplan–Meier plots depicting the prognostic potential of LINC00511 for cancer patients’ survival. a Overall survival plot. b Disease-free survival. c Overall survival plot for adrenocortical carcinoma (ACC). d Overall survival plot for thymoma (THYM)
Discussion
LINC00511, a newly identified lncRNA, has been reported to be upregulated and to have an oncogenic function in diverse cancers including lung cancer, breast cancer, pancreatic cancer, cervical cancer, liver cancer, ovarian cancer and glioma. Its underlying mechanisms of action include promotion of proliferation, tumorigenesis, cell cycle progression, invasion, migration, metastasis and chemoresistance and inhibition of apoptosis [16–22, 24]. More importantly, upregulated LINC00511 has been associated with prognosis, suggesting that it can represent a biomarker for prognosis in cancer patients. However, there is still discrepancy regarding the relationship between LINC00511 expression and clinical outcomes. In the present study, a first-time, comprehensive meta-analysis on the prognostic value of LINC00511 in diverse human cancers is presented. Collectively, 14 eligible studies were systematically included.
The results obtained from this meta-analysis have shown that upregulated LINC00511 is strongly predictive of poor overall survival of cancer patients. Moreover, the combination of multivariate analysis of four relevant studies showed that LINC00511 may serve as an independent predictor of poor overall survival for cancer patients. The subgroup analysis revealed that elevated LINC00511 expression correlated with poor overall survival independently to the tumor type, sample size, cut-off value and the data extraction method. Nevertheless, some subgroup analysis results should be taken with caution, due to the small number of studies included. Subsequently, these findings were corroborated by the results of TCGA analysis. Additionally, it was found that patients with elevated LINC00511 expression were more prone to worse clinicopathological features including larger tumor size, advanced clinical tumor stage, lymph node metastasis, distant metastasis and disease recurrence. Besides, high LINC00511 expression was found to be associated with poor disease-free survival and progression-free survival.
Taken together, the above findings lead to the suggestion that LINC00511 could serve as a potential biomarker and functional regulator in human cancers. Molecular mechanisms investigations highlighted that LINC00511 exerts its oncogenic function mainly by modulating microRNAs functions [42–44]. For instance, via its competing endogenous RNA activity on hsa-miR-29b-3p, LINC00511 induced the expression of VEGFA leading functionally to pancreatic ductal adenocarcinoma progression [20]. Similarly, Lu et al. [16] found that LINC00511 targeted the miR-185-3p/E2F1/Nanog axis in order to promote breast cancer tumorigenesis and stemness. In a recent study, LINC00511 was also found to foster the process of gastric cancer by targeting miR-625-5p/NFIX axis [43]. These mechanistic studies revealed that LINC00511 could act at transcriptional and post-transcriptional level. Moreover, LINC00511 was found to interact with a variety of signaling pathways including JAK2/STAT3 [45], Wnt/β-catenin [46], and PTEN/AKT/FOXO1 [47], in the pathogenesis of cancers.
LINC00511 has been demonstrated to be a poor predictor for both cancer recurrence and progression. These analogous outcomes imply that there might be similar LINC0511-dependent mechanisms underlying these two events. In particular, LINC00511 has been shown to induce radio-resistance in breast cancer resulting in recurrence and progression by regulating STXBP4 expression via miR-185 [28]. Similarly, the silencing of LINC00511 in cervical cancer cells enhanced cancer drug paclitaxel’s sensitivity, suppressed cell viability, cell proliferation, migration and invasion, and promoted apoptosis, thereby preventing progression and recurrence [24]. To achieve those functions, LINC00511 modulated the expression of related proteins, namely Bcl-2, Bax, cleaved caspase-3, metalloproteinases 2 and 9, multidrug resistance protein 1 (MRP1) and P-glycoprotein. The resistance to paclitaxel caused by LINC00511 was also found in breast cancer and is mediated via regulating miR-29c/CDK6 axis [23]. Du et al. found, in glioblastoma multiforme (GBM), that high LINC00511 expression was correlated with recurrence, and ectopic LINC00511 enhanced GBM cells proliferation, EMT, migration and invasion by sponging miR-524-5p to indirectly regulate YB1/ZEB1 [39].
A number of limitations should be addressed when considering the findings of the present study. Firstly, all the studies included in the meta-analysis have been conducted in China, narrowing the representativeness of the results. Secondly, there is a rather limited number of studies available on major cancers, such as lung neoplasms and brain neoplasms. Therefore, the results on subgroup analysis based on cancer types should be taken with cautions. Thus, more clinical studies would be necessary to better assess the relationship between LINC00511 expression and cancer patients’ clinical outcomes.
Conclusions
In summary, this study demonstrates that the overexpression of LINC00511 can strongly predict a poor survival in cancer patients and that it is associated to large tumor size, lymph node metastasis, advanced clinical stage, distant metastasis and disease recurrence. LINC00511 can thus serve as prognostic biomarker for cancer patients. Nevertheless, further large-scale and high-quality studies from different ethnic background are needed to confirm the present analysis.
Acknowledgments
Not applicable.
Abbreviations
- ACC
Adrenocortical carcinoma
- BC
Breast cancer
- BRCA
Breast invasive carcinoma
- CC
Cervical cancer
- ccRCC
Clear cell renal cell carcinoma
- CESC
Cervical squamous cell carcinoma
- CP
Clinicopathological parameters
- DFS
Disease-free survival
- GBM
Glioblastoma multiforme
- GL
Glioma
- GTEx
Genotype-Tissue Expression
- HCC
Hepatocellular carcinoma
- KICH
Kidney chromophobe
- KIRC
Kidney renal clear cell carcinoma
- KIRP
Kidney renal papillary cell carcinoma
- LGG
Brain lower grade glioma
- LINC00511
Long intergenic non-coding RNA 00511
- LIHC
Liver hepatocellular carcinoma
- LUAD
Lung adenocarcinoma
- LUSC
Lung squamous cell carcinoma
- N
Normal
- NOS
Newcastle-Ottawa Scale
- NSCLC
Non-small-cell lung cancer
- OC
Ovarian cancer
- OS
Overall survival
- PAAD
Pancreatic adenocarcinoma
- PC
Pancreatic cancer
- PDAC
Pancreatic ductal adenocarcinoma
- PFS
Progression-free survival
- qRT-PCR
Quantitative real time polymerase chain
- RFS
Relapse-free survival
- T
Tumor
- TCGA
The Cancer Genome Atlas
- THYM
Thymoma
- TPM
Transcripts per million
Authors’ contributions
Conceptualization, YLA and YZ; Formal analysis, YLA, MA and YN; Investigation, YLA, MA, YN, GX, JC and ZY; Methodology, YLA; Resources, QZ, ZY and YZ; Supervision, YK and YZ; Writing – original draft, YLA, MA and QZ; Writing – review & editing, FY, ZY and YZ. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (No. 31660246, 31960200,81960462,31960145,81460421,81760455, 81560037,91660135) and Yunnan Province, Kunming Medical University joint Foundation for Applied Basic Research (No. 2017FE468(− 003), 2018FE468(− 001), 2017FE468(− 132)). The funding bodies played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
All data are included in this article.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yannick Luther Agbana, Email: bioluther@163.com.
Manzama-Esso Abi, Email: mariethereseabi@126.com.
Yueli Ni, Email: 2290078545@qq.com.
Guohang Xiong, Email: xiongguoh90@163.com.
Jing Chen, Email: 17835651766@163.com.
Fang Yun, Email: yunfang52@163.com.
Zihan Yi, Email: yiwei500@126.com.
Qiao Zhang, Email: zhangqiao200824@126.com.
Zhe Yang, Email: zyangpku@163.com.
Yingmin Kuang, Email: yingmin1512@aliyun.com.
Yuechun Zhu, Email: zhuyuechun20091119@163.com.
References
- 1.Frith MC, Bailey TL, Kasukawa T, Mignone F, Kummerfeld SK, Madera M, Sunkara S, Furuno M, Bult CJ, Quackenbush J, et al. Discrimination of non-protein-coding transcripts from protein-coding mRNA. RNA Biol. 2006;3:40–48. doi: 10.4161/rna.3.1.2789. [DOI] [PubMed] [Google Scholar]
- 2.Richtig G, Ehall B, Richtig E, Aigelsreiter A, Gutschner T, Pichler M. Function and clinical implications of long non-coding RNAs in melanoma. Int J Mol Sci. 2017;18 Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5412301/. Cited 2019 Jun 10. [DOI] [PMC free article] [PubMed]
- 3.An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74 Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3439153/. Cited 2019 Jun 10. [DOI] [PMC free article] [PubMed]
- 4.Hu X, Sood AK, Dang CV, Zhang L. The role of long noncoding RNAs in cancer: the dark matter matters. Curr Opin Genet Dev. 2018;48:8–15. doi: 10.1016/j.gde.2017.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen L, Yang H, Xiao Y, Tang X, Li Y, Han Q, Fu J, Yang Y, Zhu Y. Lentiviral-mediated overexpression of long non-coding RNA GAS5 reduces invasion by mediating MMP2 expression and activity in human melanoma cells. Int J Oncol. 2016;48:1509–1518. doi: 10.3892/ijo.2016.3377. [DOI] [PubMed] [Google Scholar]
- 6.Chen L, Yang H, Xiao Y, Tang X, Li Y, Han Q, Fu J, Yang Y, Zhu Y. LncRNA GAS5 is a critical regulator of metastasis phenotype of melanoma cells and inhibits tumor growth in vivo. OncoTargets Ther. 2016;9:4075–4087. doi: 10.2147/OTT.S98203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen L, Yang H, Yi Z, Jiang L, Li Y, Han Q, Yang Y, Zhang Q, Yang Z, Kuang Y, et al. LncRNA GAS5 regulates redox balance and dysregulates the cell cycle and apoptosis in malignant melanoma cells. J Cancer Res Clin Oncol. 2019;145:637–652. doi: 10.1007/s00432-018-2820-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roberts TC, Morris KV, Wood MJA. The role of long non-coding RNAs in neurodevelopment, brain function and neurological disease. Philos Trans R Soc Lond Ser B Biol Sci. 2014;369(1652):20130507. doi: 10.1098/rstb.2013.0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi C, Zhang L, Qin C. Long non-coding RNAs in brain development, synaptic biology, and Alzheimer’s disease. Brain Res Bull. 2017;132:160–169. doi: 10.1016/j.brainresbull.2017.03.010. [DOI] [PubMed] [Google Scholar]
- 10.Li Y, Egranov SD, Yang L, Lin C. Molecular mechanisms of long noncoding RNAs-mediated cancer metastasis. Genes Chromosomes Cancer. 2019;58:200–207. doi: 10.1002/gcc.22691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pecero ML, Salvador-Bofill J, Molina-Pinelo S. Long non-coding RNAs as monitoring tools and therapeutic targets in breast cancer. Cell Oncol Dordr. 2019;42:1–12. doi: 10.1007/s13402-018-0412-6. [DOI] [PubMed] [Google Scholar]
- 12.Li M, Wang Y, Cheng L, Niu W, Zhao G, Raju JK, Huo J, Wu B, Yin B, Song Y, et al. Long non-coding RNAs in renal cell carcinoma: a systematic review and clinical implications. Oncotarget. 2017;8:48424–48435. doi: 10.18632/oncotarget.17053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lorenzi L, Avila Cobos F, Decock A, Everaert C, Helsmoortel H, Lefever S, Verboom K, Volders P-J, Speleman F, Vandesompele J, et al. Long noncoding RNA expression profiling in cancer: challenges and opportunities. Genes Chromosomes Cancer. 2019;58:191–199. doi: 10.1002/gcc.22709. [DOI] [PubMed] [Google Scholar]
- 14.Tang X, Qiao X, Chen C, Liu Y, Zhu J, Liu J. Regulation mechanism of long noncoding RNAs in colon cancer development and progression. Yonsei Med J. 2019;60:319–325. doi: 10.3349/ymj.2019.60.4.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang F, Lyu S, Dong S, Liu Y, Zhang X, Wang O. Expression profile analysis of long noncoding RNA in HER-2-enriched subtype breast cancer by next-generation sequencing and bioinformatics. OncoTargets Ther. 2016;9:761–772. doi: 10.2147/OTT.S97664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu G, Li Y, Ma Y, Lu J, Chen Y, Jiang Q, Qin Q, Zhao L, Huang Q, Luo Z, et al. Long noncoding RNA LINC00511 contributes to breast cancer tumourigenesis and stemness by inducing the miR-185-3p/E2F1/Nanog axis. J Exp Clin Cancer Res CR. 2018;37:289. doi: 10.1186/s13046-018-0945-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J, Tian Y, Zheng H, Ding Y, Wang X. An integrated analysis reveals the oncogenic function of lncRNA LINC00511 in human ovarian cancer. Cancer Med. 2019;8:3026–3035. doi: 10.1002/cam4.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang R-P, Jiang J, Jiang T, Wang Y, Chen L-X. Increased long noncoding RNA LINC00511 is correlated with poor prognosis and contributes to cell proliferation and metastasis by modulating miR-424 in hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2019;23:3291–3301. doi: 10.26355/eurrev_201904_17691. [DOI] [PubMed] [Google Scholar]
- 19.Wang W, Lou W, Ding B, Yang B, Lu H, Kong Q, Fan W. A novel mRNA-miRNA-lncRNA competing endogenous RNA triple sub-network associated with prognosis of pancreatic cancer. Aging. 2019;11:2610–2627. doi: 10.18632/aging.101933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao X, Liu Y, Li Z, Zheng S, Wang Z, Li W, Bi Z, Li L, Jiang Y, Luo Y, et al. Linc00511 acts as a competing endogenous RNA to regulate VEGFA expression through sponging hsa-miR-29b-3p in pancreatic ductal adenocarcinoma. J Cell Mol Med. 2018;22:655–667. doi: 10.1111/jcmm.13351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li C, Liu H, Yang J, Yang J, Yang L, Wang Y, Yan Z, Sun Y, Sun X, Jiao B. Long noncoding RNA LINC00511 induced by SP1 accelerates the glioma progression through targeting miR-124-3p/CCND2 axis. J Cell Mol Med. 2019;23:4386–4394. doi: 10.1111/jcmm.14331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun C-C, Li S-J, Li G, Hua R-X, Zhou X-H, Li D-J. Long Intergenic noncoding RNA 00511 acts as an oncogene in non-small-cell lung cancer by binding to EZH2 and suppressing p57. Mol Ther Nucleic Acids. 2016;5:e385. doi: 10.1038/mtna.2016.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang H, Zhao B, Wang X, Zhang F, Yu W. LINC00511 knockdown enhances paclitaxel cytotoxicity in breast cancer via regulating miR-29c/CDK6 axis. Life Sci. 2019;228:135–144. doi: 10.1016/j.lfs.2019.04.063. [DOI] [PubMed] [Google Scholar]
- 24.Mao B-D, Xu P, Xu P, Zhong Y, Ding W-W, Meng Q-Z. LINC00511 knockdown prevents cervical cancer cell proliferation and reduces resistance to paclitaxel. J Biosci. 2019;44(2):44. doi: 10.1007/s12038-019-9851-0. [DOI] [PubMed] [Google Scholar]
- 25.Yu C-L, Xu X-L, Yuan F. LINC00511 is associated with the malignant status and promotes cell proliferation and motility in cervical cancer. Biosci Rep. 2019;39(9):BSR20190903. doi: 10.1042/BSR20190903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J, Sui S, Wu H, Zhang J, Zhang X, Xu S, Pang D. The transcriptional landscape of lncRNAs reveals the oncogenic function of LINC00511 in ER-negative breast cancer. Cell Death Dis. 2019;10:599. doi: 10.1038/s41419-019-1835-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deng H, Huang C, Wang Y, Jiang H, Peng S, Zhao X. LINC00511 promotes the malignant phenotype of clear cell renal cell carcinoma by sponging microRNA-625 and thereby increasing cyclin D1 expression. Aging. 2019;11(16):5975–5991. doi: 10.18632/aging.102156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu L, Zhu Y, Liu A-M, Feng Y, Chen Y. Long noncoding RNA LINC00511 involves in breast cancer recurrence and radioresistance by regulating STXBP4 expression via miR-185. Eur Rev Med Pharmacol Sci. 2019;23:7457–7468. doi: 10.26355/eurrev_201909_18855. [DOI] [PubMed] [Google Scholar]
- 29.Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6 Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2707599/. Cited 2019 Jun 20. [PMC free article] [PubMed]
- 30.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25:603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 31.Ottawa Hospital Research Institute. Available from: http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp. Cited 2019 May 31.
- 32.Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47(W1):W556–W560. doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Egger M, Smith GD, Phillips AN. Meta-analysis: principles and procedures. BMJ. 1997;315:1533–1537. doi: 10.1136/bmj.315.7121.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee YH. An overview of meta-analysis for clinicians. Korean J Intern Med. 2018;33:277–283. doi: 10.3904/kjim.2016.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Copas J, Shi JQ. Meta-analysis, funnel plots and sensitivity analysis. Biostat Oxf Engl. 2000;1:247–262. doi: 10.1093/biostatistics/1.3.247. [DOI] [PubMed] [Google Scholar]
- 38.Zhu F-Y, Zhang S-R, Wang L-H, Wu W-D, Zhao H. LINC00511 promotes the progression of non-small cell lung cancer through downregulating LATS2 and KLF2 by binding to EZH2 and LSD1. Eur Rev Med Pharmacol Sci. 2019;23:8377–8390. doi: 10.26355/eurrev_201910_19149. [DOI] [PubMed] [Google Scholar]
- 39.Du X, Tu Y, Liu S, Zhao P, Bao Z, Li C, Li J, Pan M, Ji J. LINC00511 contributes to glioblastoma tumorigenesis and epithelial-mesenchymal transition via LINC00511/miR-524-5p/YB1/ZEB1 positive feedback loop. J Cell Mol Med. Available from: https://onlinelibrary.wiley.com/doi/abs/10.1111/jcmm.14829. Cited 2019 Dec 21. [DOI] [PMC free article] [PubMed]
- 40.Liu F-T, Dong Q, Gao H, Zhu Z-M. The prognostic significance of UCA1 for predicting clinical outcome in patients with digestive system malignancies. Oncotarget. 2017;8:40620–40632. doi: 10.18632/oncotarget.16534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.NCBI Resource Coordinators Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2018;46:D8–13. doi: 10.1093/nar/gkx1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang K, Zhu G, Bao S, Chen S. Long non-coding RNA LINC00511 mediates the effects of ESR1 on proliferation and invasion of ovarian cancer through miR-424-5p and miR-370-5p. Cancer Manag Res. 2019; Available from: https://www.dovepress.com/long-non-coding-rna-linc00511-mediates-the-effects-of-esr1-on-prolifer-peer-reviewed-fulltext-article-CMAR. Cited 2019 Dec 29. [DOI] [PMC free article] [PubMed]
- 43.Chen Z, Wu H, Zhang Z, Li G, Liu B. LINC00511 accelerated the process of gastric cancer by targeting miR-625-5p/NFIX axis. Cancer Cell Int. 2019;19:351. doi: 10.1186/s12935-019-1070-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ding J, Yang C, Yang S. LINC00511 interacts with miR-765 and modulates tongue squamous cell carcinoma progression by targeting LAMC2. J Oral Pathol Med Off Publ Int Assoc Oral Pathol Am Acad Oral Pathol. 2018;47:468–476. doi: 10.1111/jop.12677. [DOI] [PubMed] [Google Scholar]
- 45.Chen Y, Bao C, Zhang X, Lin X, Fu Y. Knockdown of LINC00511 promotes radiosensitivity of thyroid carcinoma cells via suppressing JAK2/STAT3 signaling pathway. Cancer Biol Ther. 2019;20:1249–1257. doi: 10.1080/15384047.2019.1617569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li J, Li Y, Meng F, Fu L, Kong C. Knockdown of long non-coding RNA linc00511 suppresses proliferation and promotes apoptosis of bladder cancer cells via suppressing Wnt/β-catenin signaling pathway. Biosci Rep. 2018;38(4):BSR20171701. doi: 10.1042/BSR20171701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang L, Xie X, Ding F, Mei J, Bi R. Silencing LINC00511 inhibits cell proliferation, migration and EMT via PTEN/AKT/FOXO1 signaling pathway in lung cancer. Biochem Cell Biol. 2019. 10.1139/bcb-2018-0364. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are included in this article.