Fig. 2.
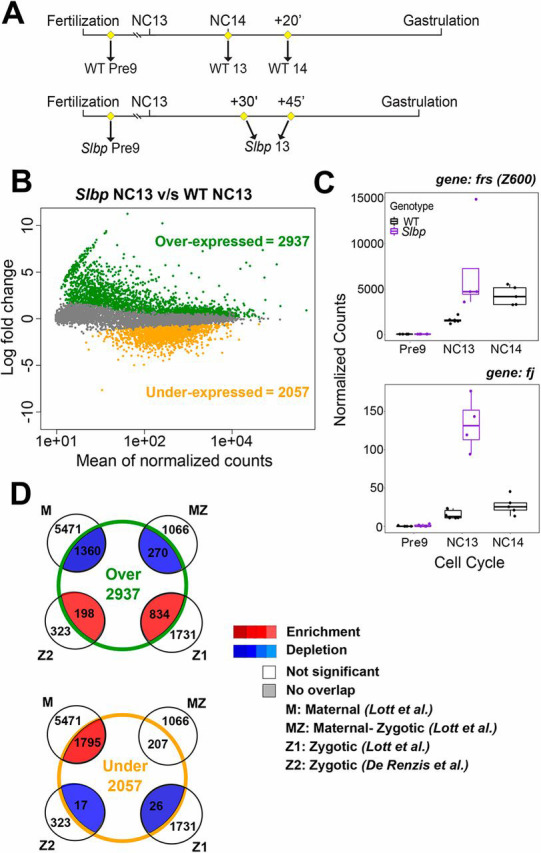
Depletion of maternal histones results in an early ZGA. (A) Schematic of embryo collection for Slbp RNA-seq. Pre-NC9 embryos were collected for both WT (n=5) and Slbp (n=5) to control for maternal loading. Slbp embryos were collected 30 or 45 min into NC13 (n=4) to ensure embryo health and phenotype and compared with NC-matched (mitosis of NC13) (n=6) or time-matched (20 min into NC14) (n=5) WT embryos. (B) When compared with WT NC13, 2937 genes are overexpressed and 2057 genes are underexpressed in Slbp embryos. (C) Traces from two overexpressed genes, frs and fj. Box plots: the box represents the interquartile range (ends of the box representing the upper and lower quartile), the central line is the median and the whiskers extend to the upper- and lowermost values. (D) Data from B compared with previous datasets. When compared with NC-matched controls, overexpressed transcripts are enriched for zygotically expressed genes and de-enriched maternal transcripts. Conversely, underexpressed genes are enriched for maternal transcripts and de-enriched for new zygotic transcripts. This pattern is consistent with premature ZGA. Comparisons with time-matched controls yield similar results (Fig. S3).
