Abstract
Biallelic inactivation of BRCA1/2 is associated with a pattern of genome-wide mutations known as Signature 3. By analyzing ~1,000 breast cancer samples, we confirmed this association and established that germline nonsense and frameshift variants in PALB2, but not ATM or CHEK2, can also give rise to the same signature. Missense BRCA1/2 variants known to impair homologous recombination (HR) were accurately classified by this signature. Finally, we show that epigenetic silencing of RAD51C and BRCA1 by promoter methylation are strongly associated with Signature 3 and, in our dataset, were highly enriched in basal-like breast cancers in young patients of African descent.
INTRODUCTION
Breast cancer is the most common non-cutaneous malignancy and approximately 10–15% of cases are associated with hereditary syndromes of defective DNA repair1. The most common forms of hereditary breast and ovarian cancer are associated with pathogenic germline variants in two homologous recombination (HR) repair genes, BRCA1 and BRCA22. HR is an evolutionarily-conserved pathway that coordinates high-fidelity repair of double-stranded DNA breaks (DSBs), and functions primarily during the late S and G2 phases of the cell cycle to exploit the intact sister chromatid as a template for error-free repair3. Although germline alterations that abrogate the tumor suppressor functions of BRCA1 and BRCA2 play a role in breast cancer pathogenesis, comprehensive sequencing studies have demonstrated that somatic mutations and epigenetic events in these genes can also drive tumorigenesis4–7. Deficiency of HR leads to cellular dependence on alternate error-prone DNA repair pathways, yielding characteristic genomic alterations, higher overall mutational rates, and unique dependencies used for therapeutic targeting8.
Analyses of tumor-derived genome sequences have revealed that loss of BRCA1 or BRCA2 yields a distinct pattern of base-substitution mutations, termed “Signature 3” 9,10. This pattern, among others, can be discerned using non-negative matrix factorization (NMF), a technique used to identify recurring patterns (i.e. signatures) in the spectra of mutations from a set of tumors and to estimate the contributions of these signatures to the mutational landscape of the individual tumors. The etiology of some signatures was revealed by associating their activities with additional data10–13. While recent efforts have observed a significant association between BRCA1/2 loss and Signature 39,10, several tumors exhibit high levels of Signature 3 without discernible alterations in HR-pathway genes9,10. Furthermore, Signature 3 has not been extensively correlated with other genetic and epigenetic events in HR genes. A comprehensive understanding of the link between HR pathway and Signature 3 is of clinical relevance given the implications for underlying cancer risk and treatment selection.
Current clinical practice in testing for breast cancer susceptibility relies largely on identifying known pathogenic variants in the germline sequence of BRCA1/2 and a limited number of related genes14,15. Such germline testing may uncover germline pathogenic variants that are likely to contribute to HRD in the associated breast tumor. The presence of HRD may expose specific therapeutic vulnerabilities. We, therefore, wanted to assess whether integrating germline and somatic data can improve the detection of HRD. Somatic events, such as loss-of-heterozygousity, epigenetic silencing and other mechanisms that can decrease gene expression, can serve as the “second-hit” in these genes.16. Moreover, integrating somatic data can help interpret poorly-defined germline variants17. Here, we hypothesize that Signature 3 could represent a more reliable readout of HR status than germline sequencing alone and could enhance prediction of breast cancer risk. Moreover, Signature 3 may aid in classifying variants that have yet to be functionally characterized. We, therefore, sought to identify the mechanisms by which Signature 3 could arise in the absence of pathogenic germline alterations in BRCA1/2. Specifically, we analyzed the association between Signature 3 and multi-dimensional events (germline variants, somatic mutations, epigenetic silencing, and allelic imbalance) in HR pathway components aside from BRCA1/2.
A distinct mutational signature is associated with biallelic inactivation of BRCA1/2 and epigenetic silencing of BRCA1
We first characterized the underlying landscape of genomic lesions operating in breast cancer by performing mutational signature analysis of 995 invasive breast carcinoma samples from The Cancer Genome Atlas (TCGA)4. We stratified single-nucleotide variants (SNVs) by base substitutions in 96 possible tri-nucleotide contexts (Figure 1, top row “SNVs”) and applied our method, SignatureAnalyzer12,13, which users a Bayesian derivative of NMF to infer the number of operating signatures as well as the activity of each signature in each tumor (Methods).
Figure 1: Characterization of four distinct mutational signatures in breast cancer.
A Bayesian non-negative matrix factorization approach (Methods) was employed to decompose the overall mutational spectrum across a cohort of 992 breast cancer samples into distinct mutational signatures (“SNVs” – top row). Each color represents one of the six potential base substitutions, with each substitution further stratified by the adjacent 5’ and 3’ flanking nucleotides. The pattern of each signature was matched against a database of known signatures and their etiologies (Signature 3, C>T_CpG, APOBEC, MSI). A signature characterized by C>G transversions and C>T transitions at TC[A/T] motifs (in which the altered cytosine is flanked by a 5’ thymine and a 3’ adenine or thymine) is likely attributable to mutagenesis via endogenous APOBEC machinery (“APOBEC”; COSMIC signatures 2 and 13). A second signature primarily consisting of C>T transitions at CpG dinucleotides has been described among all tumor types and is thought to arise from spontaneous 5-methyl-cytosine deamination (“C>T CpG”; COSMIC signature 1). The third signature resembles previously characterized signatures identified in samples known to have microsatellite instability (MSI) (“MSI”; COSMIC signature 6). A recurrent signature in the cohort closely approximates Signature 3, and is characterized by mutations in all 96 nucleotide contexts, with an increased rate among C:G basepairs as compared to A:T ( “Signature 3”).
We identified seven distinct mutational signatures across the 995 TCGA breast cancers, largely corresponding to previous analyses of this series10 (Supplementary Figure 1 and Methods). Of these, three signatures arose each in a single case, which we excluded from subsequent analyses (Methods), thereby yielding four recurrent signatures across 992 samples (Figure 1; Supplementary Figure 2): (i) Signature 1 (C>T in CpG sites); (ii) an APOBEC related signature (combination of Signature 2 and Signature 13); (iii) a signature associated with microsatellite instability (Signature 6); and (iv) a largely uniform signature similar to the previously reported Signature 3, which was associated with mutations in BRCA1/2 9,10,18.
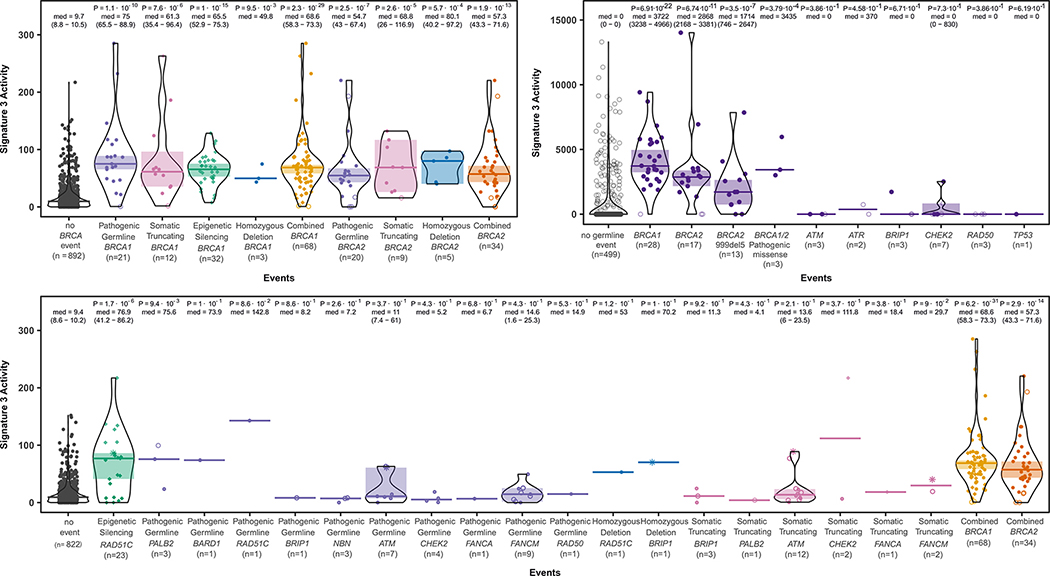
Over 10% (100/992) of the samples harbored clinically-relevant germline or somatic frameshift/nonsense variants in BRCA1/2, well-characterized pathogenic missense BRCA1/2 germline variants19 and/or epigenetic silencing of the BRCA1 promoter16. As previously demonstrated, a variety of BRCA1/2 inactivating variants are associated with Signature 3 (Figure 2; Supplementary Figure 3) 9,10,18. Fifty samples with somatic or germline nonsense/frameshift variants in BRCA1/2 exhibited a ~6-fold increase in levels of Signature 3 in comparison to samples without BRCA1/2 alterations (p <0.001, rank sum test, Figure 3A), consistent with the enrichment of Signature 3 reported in a recent data set9 (Figure 3B). Aside from single nucleotide variants, epigenetic silencing of BRCA1 (n = 32) and homozygous deletions of BRCA1/2 (n = 8) were also associated with Signature 3 (Figure 3A). Moreover, 18 samples harboring the lower-risk truncating BRCA2 variant p.Lys3326Ter20 (c.9976A>T, chr13:g.32972626A>T (hg19);NM_000059.3) did not exhibit elevated levels of Signature 3 (median of 13.4 Signature 3 mutations/sample; p = 0.4; data not shown).
Figure 2: Overall mutation rates, mutational signature contributions, and clinicopathologic features per patient as sorted by descending Signature-3 activity.
Each column represents a tumor sample. Overall indel burden is shown in the top panel with insertion or deletion designated by color. The second panel demonstrates absolute mutation burden per sample, with the estimated contribution of each of the four mutational signatures designated by color (samples are arranged in descending order of Signature 3 activity). The third panel shows sample-level annotations with regard to lesions in known homologous recombination (HR) pathway genes, along with PAM50 intrinsic subtype data, and triple negative status by immunohistochemistry. The bottom panel exhibits genetic lesions by type in select HR genes with mutation type indicated by color. Shown are the first 250 samples by Signature 3 activity; see Supplementary Figure 3 for the rest 742 samples.
Figure 3: Signature 3 activity in tumors with somatic, germline and epigenetic alterations in HR-pathway genes.
(A) Signature 3 activity is elevated in tumors with somatic, germline and epigenetic alterations in BRCA1/2. Tumors were stratified by germline, somatic or epigenetic events in BRCA1/2 and plotted by number of mutations attributable to Signature 3. Pathogenic germline events included those described by ClinVar, in addition to frameshift and nonsense variants. The median signature count among each group was tested against samples lacking any discernible BRCA1/2 alterations (denoted as “no BRCA event”) with two-sided Wilcoxon rank-sum test results as indicated. Open circles represent samples with mono-allelic lesions (i.e. without evidence of biallelic inactivation). Samples with epigenetic silencing are indicated by diamonds. Two samples which harbored a somatic and a germline truncating event in BRCA1/2 and are included in both somatic and germline categories (B) Signature 3 activity in patients with germline alterations as previously denoted9. (C) Signature 3 activity is associated with somatic, germline and epigenetic alterations in HR components beyond BRCA1/2. Signature 3 activity was compared between samples lacking any of the noted alterations (“no event”) versus those with any genetic or epigenetic event in the HR pathway. Samples with an event in BRCA1/2 were noted by an asterisk. Comparison of Signature 3 activity was conducted with two-sided Wilcoxon rank-sum results noted by group
A single functional copy of BRCA1/2 is generally sufficient to maintain normal HR function while biallelic inactivation of BRCA1/2 contributes to tumorigenesis by several mechanisms, including genomic instability that arises from HR deficiency (HRD)21. To investigate whether monoallelic loss of BRCA1/2 is associated with Signature 3, or whether HRD (biallelic loss) is necessary for Signature 3, we analyzed loss of heterozygosity (LOH) at the BRCA1/2 loci (Methods), as these are the most common “second-hit” events22,23. We identified 23 samples with LOH in which the retained BRCA1/2 allele contained a germline frameshift/nonsense variant while the intact allele was lost (Supplementary Figure 4). All samples but one exhibited Signature 3 activity in the top quartile for the series (Figures 2, Supplementary Figure 4; p = 8×10−15; median of 68.9 vs. 9.7 for no BRCA1/2 events). Similarly, all 18 samples with LOH that also harbored a somatic frameshift/nonsense/splice-site BRCA1/2 variant had high Signature 3 activity (Supplementary Figure 4; p = 2.8×10−10).
In contrast, monoallelic germline inactivation of BRCA1/2 was not associated with Signature 3 (Supplementary Figure 4), consistent with recent reports9. Of 5 samples harboring pathogenic germline variants without loss of the intact allele, 4 showed no increase in Signature 3 activity (p = 0.42). Two samples with a somatic truncating mutation and without evidence of biallelic inactivation also failed to show Signature 3 elevation (Supplementary Figure 4). RNA-seq data for these 5 cases had very low coverage of BRCA1/2 at the variant site (≤2 reads), and could not be used to examine expression of the mutant vs. wildtype alleles. In addition, samples with LOH of BRCA1/2 that retained an allele with a benign missense germline variant did not exhibit strong enrichment of Signature 3 (Supplementary Figure 4; median of 12.9 vs 68.9 where LOH retained a truncating event). These findings suggest that Signature 3 may be a reliable readout of biallelic, and not monoallelic, loss of BRCA1/2 and/or of HRD.
Despite the association between BRCA1/2 inactivation (germline, somatic or epigenetic silencing) and Signature 3 (88 of 100 BRCA1/2 events were in the top quartile of Signature 3 activity), most samples in the top quartile (159 of 247) did not have such events, suggesting that other lesions might contribute to Signature 3 activity. Clinical assays for the assessment of breast cancer risk include gene panels of varying breadth, nearly all of which include a variety of DNA repair genes. As discussed above, numerous germline variants of BRCA1/2 confer high cancer risk, however, deleterious variants in several other genes confer “moderate risk”15 (Supplementary Figure 5). Therefore, we sought to determine whether alterations in these latter genes are associated with Signature 3 (Figure 3C), with a particular focus on germline events in which the intact allele was lost (Supplementary Table 1; Supplementary Figure 6). Below, we consider the association between deleterious variants in these genes and Signature 3.
Signature 3 is associated with lesions in established breast cancer gene PALB2
In search of additional germline variants that could be associated with Signature 3, we examined all 992 cases for any deleterious alterations in other known genes of the HR pathway, including PALB2, a binding partner and nuclear localizer of BRCA2 (Supplementary Figure 6). In recent studies, germline truncating variants in PALB2 have been associated with a relative risk for breast cancer between 3.4 and 6.615,24,25. Three samples among our cohort harbored germline nonsense/frameshift variants in PALB2, and all exhibited elevated Signature 3 activity (Figure 3C). We could not validate this association in a recent report of 560 breast cancers9 due to lack of these germline events. Consistent with our results, however, in a recent pancreatic study26,27, two cases with PALB2 germline truncating variants had high levels of Signature 3.
RAD51C methylation as a key alteration underlying deficient homologous recombination repair in basal-like breast carcinoma
RAD51 is involved in DNA repair by HR, promoting DNA strand invasion and homology search14. This process requires RAD51 in complex with BRCA2 and, to a lesser extent, PALB228, both of which can promote RAD51 loading onto single-stranded DNA. There are several RAD51-related genes, including RAD51B, RAD51C, RAD51D, DMC1, XRCC2 and XRCC3. RAD51C facilitates the accumulation of RAD51 to sites of DNA damage by complexing with several RAD51-like proteins (Figure 4A).
Figure 4: Association of promoter methylation of RAD51C with elevated Signature 3 in basal-like tumors.
(A) Schematic representation of RAD51C architecture and its known interacting proteins that promote HR. (B) Scatter plot of RAD51C expression as compared to promoter methylation levels (methylation values higher than 0.2 were considered for epigenetic silencing). Samples with high Signature 3 activity (top quartile) are denoted by triangles (C) Elevated Signature 3 activity is associated with alterations among multiple components of the HR-pathway and a diversity of alteration types. Samples were ranked by the number of Signature 3 associated mutations and plotted in descending order in bins of 50. The top panels show alterations among various HR-pathway genes, while the bottom panel shows the types of alterations in these genes, demonstrating enrichment of events among the samples with the highest Signature 3 activity. (D) Signature 3 activity was significantly associated with RAD51C promoter methylation among basal-like breast cancers, but not among other intrinsic subtypes.
It is generally thought that germline truncating variants in RAD51C do not confer an increased risk of breast cancer29, although widespread panel-based genetic testing has revealed deleterious RAD51C variants among 0.3% of triple negative breast cancer patients30 and in high-risk breast pedigrees31. In our data, one case displayed biallelic inactivation of RAD51C (a germline variant with LOH that deleted the intact allele), and another case presented somatic homozygous deletion. Both tumors exhibited high levels of Signature 3 (Figures 2 and 3C), suggesting that inactivation of RAD51C can lead to HRD.
As BRCA1 promoter hypermethylation can promote breast cancer development, we interrogated whether such events also occur in other HR genes. Indeed, RAD51C promoter methylation correlated with downregulated gene expression in our series (Figure 4B). Of 23 cases of RAD51C promoter methylation, 19 showed ≥ 2-fold decrease in mRNA expression (Figure 4B). Seventeen of the 23 samples were in the top quartile of Signature 3 activity and all but one did not have any BRCA1/2 event (the one with a BRCA1/2 event had elevated RAD51C expression). Thus, 16 cases of RAD51C epigenetic silencing explain 10% (16/159) of the top quartile without BRCA1/2 events (Figure 4C). Moreover, the number of samples with high Signature 3 that harbor epigenetic events in BRCA1 and RAD51C is comparable to the number of germline events in BRCA1/2, suggesting a similar contribution of germline and epigenetic events to carcinogenesis (Figure 4C).
Of note, 52% (12/23) of samples with RAD51C silencing exhibited the basal-like expression pattern and all of these basal-like cases were among the 17 RAD51C-silenced samples in the top Signature 3 quartile (70%; p = 0.003, Fisher’s Exact Test). Examining an independent series32,33 with methylation and expression data showed a similar correlation between promoter methylation and expression of RAD51C (Supplementary Figure 7). Consistent with our findings, cases with RAD51C promoter methylation and low expression levels were highly enriched with basal-like tumors (Figure 4B; Supplementary Figure 7). Basal-like expression is characteristic of breast cancers with BRCA1 defects34, seen in 81% (17/21) of cases with germline BRCA1 deleterious variants and in 81% (25/32) of cases with BRCA1 promoter hypermethylation35,36; the last association was also observed in the independent series (Supplementary Figure 7). In contrast, only 10% (2/20) of tumors with BRCA2 germline deleterious variants were basal-like, suggesting that inactivation of RAD51C may be biologically closer to cases with BRCA1 defects rather than to those with BRCA2 inactivation. RAD51C promoter methylation was only associated with increased level of Signature 3 in basal-like tumors (Figure 4D), consistent with the strong reduced expression in this subtype, suggesting that RAD51C promoter methylation plays a role in HRD mainly in the context of basal-like tumors.
No association of Signature 3 with mutations in DNA damage signaling pathway genes
The role of BRCA1/2 in PARP inhibitor susceptibility is established. However, the therapeutic significance of deleterious variants in HR components upstream of BRCA1/2 (Supplementary Figure 5), such as ATM, NBN and CHEK2, that are also implicated in DNA damage signaling and double-strand break detection and increase the risk of breast cancer15 (relative risk [RR] 2–3), is less clear. In our dataset, we identified 17 germline pathogenic variants in six established or candidate breast cancer susceptibility genes (7 in ATM, 4 in CHEK2, 3 in NBN and one in BARD1, BRIP1 and RAD50; see Supplementary Note for discussion). Overall, germline pathogenic variants in ATM and CHEK2 were not associated with a high level of Signature 3 (Supplementary Table 1; Figure 3C) and this result was replicated in the previously published dataset9 (Figure 3B). The single case with BARD1 truncating variant and LOH (losing the wild-type allele), had a high level of Signature 3 (Figure 3C and Supplementary Figure 6).
Promoter methylation events of BRCA1 and RAD51C are enriched among young black patients
We next sought to assess whether self-reported racial differences underlie various Signature 3 etiologies. The largest racial groups within our series consisted of 699 white patients and 142 black/African American patients (the remaining 181 included 57 Asian patients). We evaluated differences in the frequencies of genetic (germline and somatic) and epigenetic events that contribute to Signature 3 between these racial groups, focusing on the cases in the top quartile of Signature 3 activity (159 white and 41 black/African American).
Among the 41 black patients in the top quartile, 36% (15/41) showed promoter methylation in either BRCA1 or RAD51C, 9.8% (4/41) harbored germline or somatic deleterious mutations in BRCA1/2 and the remaining 22 had no discernible lesion (Figure 5A). In contrast, among the 159 white patients in the top quartile, 13%, (22/159) exhibited an epigenetic event, while 32% (56/159) had germline events or somatic deleterious events and the 81 remaining cases had no event (Figure 5A, Freeman-Halton 3×2 exact test p = 3×10−4 for whites compared to blacks). Reduced mRNA levels of RAD51C and BRCA1 in cases with promoter methylation were similar among white and black patients, suggesting similar function effects.
Figure 5: Analysis of genetic and epigenetic events in the 250 samples with highest Signature 3 levels, broken by largest racial subgroups in our cohort (white and African American).
(A) Distribution of various types of alterations among patients stratified by racial groups show an enrichment of promoter methylation in African American patients. The inset shows the distribution of various types of alterations in individual genes. The enrichment of promoter methylation among African American patients is most pronounced in women aged 40–49, both when looking at all breast cancer subtypes (B) and only basal-like cancers (C).
Among premenopausal women, African Americans are at higher risk of developing basal-like breast cancers than their Non-Hispanic White counterparts37. Given the association between HR-deficiency and basal-like biology, and the observed differences between racial groups in the prevalence of Signature 3-associated genetic and epigenetic events, we further investigated differences by age subgroups (Figure 5B) and within the basal-like subtype (Figure 5C). The most pronounced differences arose among women aged 40–49 years. Of black patients in this age group, 80% (8/10) exhibited promoter methylation (6 BRCA1 and 2 RAD51C) in contrast to only 18% (7/38) of white patients. Conversely, genetic events were enriched among white women (36%; 14/ 38), while only one black woman in this age group had a genetic alteration (1/10). This suggests that the mechanism of HR deficiency differs between white patients, in whom genetic mutations predominate, and black patients in whom promoter methylation is more frequent (p = 0.0009). We further examined if this difference also arises in basal-like patients aged 40–49 and found a similar disparity (Figure 5C, p = 0.0065).
Signature 3 activity correlates with the pathogenic potential of rare germline variants
We next sought to evaluate whether Signature 3 could be used to characterize rare missense SNVs in BRCA1/2. Understanding the pathogenicity of rare germline variants of BRCA1/2 has become the focus of multiple international consortia38,39 and has led to the assignment of consensus annotations to rare variants into five classes: benign, likely benign, variants of uncertain significance (VUS), likely pathogenic and pathogenic40. Over the entire series of 992 cases, 262 (26.3%) harbored at least one rare (prevalence <0.1%) missense SNV in BRCA1/2.
As discussed above, the biallelic inactivation of BRCA1/2, particularly via an inherited pathogenic variant and LOH is considered a hallmark of HRD. As shown above, nearly all samples with pathogenic BRCA1/2 germline variants and loss of the intact allele exhibited high levels of Signature 3, while those harboring pathogenic variants without LOH had no elevation of Signature 3. Conversely, samples with LOH that otherwise retained an allele harboring a silent germline variant were not associated with a significant increase in Signature 3 (Supplementary Figure 4), thereby suggesting that the presence of LOH alone cannot accurately identify samples with HRD. Signature 3 in conjunction with LOH, however, could represent a robust readout of HRD and of the pathogenic potential of indeterminate variants. As a result, we sought to apply a “bi-allelic inactivation” model with high Signature 3 levels to potentially discriminate between pathogenic and benign variants. We estimated the likely pathogenicity of a variant by employing two methodologies based on the bi-allelic inactivation model for HRD (Methods).
Of the 262 cases discussed above, we observed 143 distinct missense germline SNVs in BRCA1/2, 74 were present in >1 sample, of which 8 also exhibited LOH in at least two tumors by losing the intact allele (Supplementary Table 2). Of these 8, one is considered pathogenic and 7 are thought to be benign19. The pathogenic variant, BRCA1, p.Cys61Gly (Supplementary Table 2 for HGSV nomenclature), was present in four samples, all of which exhibited high Signature 3 activity, and biallelic inactivation (3 with LOH, 1 with an additional somatic truncating variant, p-value = 0.004, Supplementary Table 2). p.Cys61Gly in BRCA1 is a founder variant among Poles and disrupts the RING-domain interaction between BRCA1 and BARD141,42. Here, it was identified in a largely unbiased approach by seeking rare germline missense SNVs with concomitant LOH, without familial, pathological or other clinically-relevant data. The remaining 7 germline variants, previously described as benign, were not associated with an increase in Signature 3 activity.
We hypothesized that LOH status in tandem with Signature 3 might further inform the clinical implications of poorly-characterized BRCA1/2 germline variants. To test this approach, we used the set of germline missense SNVs in our data for which a functional implication had been classified by the ENIGMA consortium (n=142): 5 as pathogenic variants (class 5 in ClinVar) and the remainder 137 as benign (class 1)39 (Figure 6). Therefore, in our dataset the fraction of pathogenic missense variants was 3.5% (CI95% [1%, 8%]). However, among samples with germline variants in which allelic imbalance favored the alternate allele, 22% harbored pathogenic variants (4/18; CI95% [6%, 47%]; p <0.001 by Hypergeometric test). Overlaying the yet more stringent requirement that a variant exhibit both allelic imbalance and high Signature 3 activity (top quartile) yielded a set in which 66% of samples harbored pathogenic alleles (4/6, CI95% [22%, 99%]; p < 4.1×10−6; Figure 6); one of the two apparently-benign variants also harbored RAD51C hypermethylation, which can underlie the high Signature 3 activity. By considering both LOH and Signature 3 activity, the likelihood of pathogenicity for a given allele may be improved considerably. Conversely, among 16 samples with an excess of the alternate allele, but with low levels of Signature 3, all BRCA1/2 variants were benign, suggesting that this method is not only sensitive, but also highly specific. Moreover, in an independent dataset of whole genomes9, we observed that two pathogenic variants (class 5) and one likely pathogenic (class 4) variant (Figure 3B) had high levels of Signature 3 concomitant with an LOH event that favored retention of the mutated allele 9, thereby demonstrating the sensitivity of this approach, independent of method or platform.
Figure 6: A framework for enhancing the classification of BRCA1/2 germline missense variants using signature 3 and biallelic inactivation.
Beginning with all rare BRCA1/2 missense germline variants (n=137; 3.5% of randomly selected variants in TCGA breast cancer dataset are pathogenic), the group was dichotomized by presence of LoH at the BRCA1/2 locus. Those without LOH (n = 82) included only one sample with a pathogenic allele (i.e. 1.2% probability of pathogenicity), although this sample also harbored an additional variant with the potential for biallelic inactivation (indicated by red frame). Conversely, among those with LOH (n= 60; 6.6% of variants are pathogenic), samples were further sorted by allelic imbalance (22.2% of variants are pathogenic), elevated Signature 3 activity (66.6% probability of pathogenicity, indicated by green frame) and the absence of lesions among other HR-pathway components
Considering that either LOH or a second deleterious mutation that results in biallelic inactivation, our approach captured the known pathogenic BRCA1/2 mutations among the series and accurately classified BRCA1/2 VUSs. Overall, 12 unclassified missense variants fulfilled the criteria (Supplementary Note). Downstream, only 3 potentially pathogenic rare missense mutations had supportive evidence (Supplementary Table 3). Thus, in a dataset of ~1000 breast cancers, only 8 rare missense SNVs (of which 5 were previously known) are likely to have contributed to the development of HR-deficient tumors. A similar approach was applied to somatic events and suggested that 4 somatic missense mutations (all in BRCA1) may disrupt critical functions within the HR pathway (Supplementary Note).
Signature 3 is a robust and independent biomarker of HR deficiency
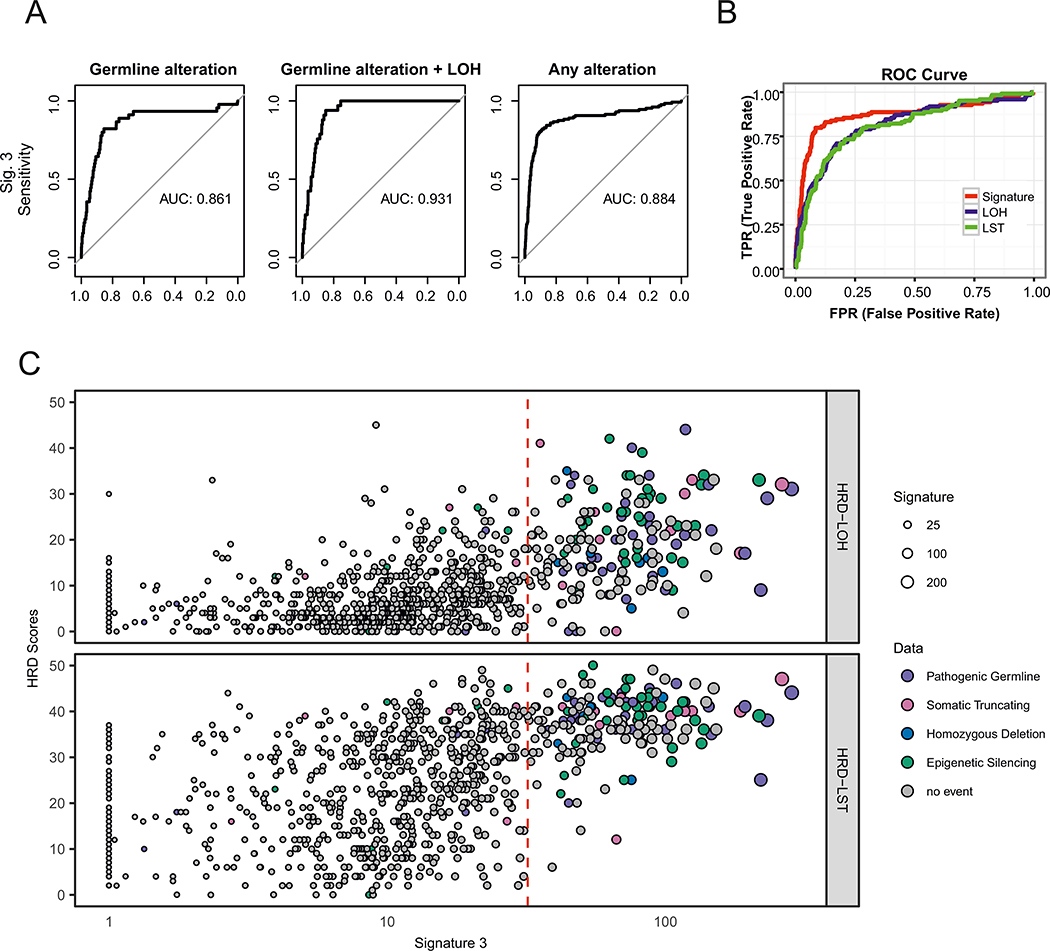
Finally, since Signature 3 could be associated with the degree of HRD, we sought to establish a clinically-applicable threshold for binary phenotypic classification. Such a task is difficult since there is no standard to unequivocally establish if HRD is present. However, germline events in BRCA1/2 and PALB2 are known contributors to HRD. To standardize a measure to estimate the predictive power of Signature 3 for identifying samples with events in HR pathway genes, we calculated the Area Under the ROC Curve (AUC) using all cases with deleterious alterations in BRCA1/2, PALB2 and RAD51C as a ground truth positive set. The AUC for detecting deleterious alterations was 0.86 and increased to 0.93 when considering, as the ground truth set, only germline cases with a second hit (Figure 7A). A similar AUC for classifying HRD events was recapitulated upon using whole-genomes or only 8 MB of sequence, and applying a normalized version of Signature 3. In addition, Signature 3 levels were not correlated with purity in our TCGA series (Supplementary Note, Supplementary Figures 8–11). Hence, Signature 3 activity may serve as a robust classifier of HR events across different platforms.
Figure 7: Prediction accuracy of Signature 3 relative to established rearrangement-based HRD scores.
(A) The three panels exhibit receiver operating characteristic (ROC) curves for predicted alterations among BRCA1, BRCA2, RAD51C, and PALB2 using Signature 3 levels. The panels show analyses based on (i) all pathogenic germline mutations (ii) biallelic pathogenic germline mutations and (iii) pathogenic germline, loss of function somatic, or epigenetic alterations. (B) ROC curves comparing Signature 3 performance to LOH and LST scores for identifying samples with the HR-associated lesions denoted above. (C) Scatter plot exhibiting a comparison of Signature 3 levels versus HRD-LOH (top panel) and HRD-LST scores (bottom panel).
Assays for HRD are used to identify samples with underlying events beyond germline lesions in BRCA1/2, e.g. somatic events in HR genes or epigenetic silencing. The calculated AUC using all events is 0.89 (Figure 7A); slightly higher than that calculated based on BRCA1/2 germline events, but lower compared to the estimated germline+LOH AUC. These results improve upon alternate approaches that are based on LOH43 and large scale transition44 scores (referred to as HRD-LOH and HRD-LST, respectively) (Figure 7B), largely because Signature 3 is more sensitive in identifying BRCA2 and RAD51C events (Supplementary Figure 12). We found that a threshold of 31 Signature-3-associated mutations (chosen to maximize Youden’s Index45) detects 82% of the events in our dataset that can give rise to HRD and may identify another 11% of samples with a yet unknown defect in HR (Supplementary Figure 13). Of samples above this 31-mutation threshold, we found a putatively-etiologic lesion in roughly one-half. Most, however, also harbored high HRD-LOH and LST scores (Figure 7C), suggesting elevated degrees of genomic instability. To classify bi-allelic inactivation of BRCA1/2 events the optimal threshold is 37 Signature-3-associated mutations (Supplementary Figure 14; Supplementary Note). Since clinical applications often assay a subset of genes, we tested how many genes would yield an accurate classification of high Signature 3 patients (as defined by exome sequencing) and found that with 1600 genes we can achieve an AUC of ~0.82 (Supplementary Figure 15; Supplementary Note).
DISCUSSION
Our analysis represents an extended characterization of Signature 39,10,18 and its relationship to the functional underpinnings of the HR repair machinery. Using a multi-dimensional approach, we demonstrate that this signature is reliably detected among samples with deleterious germline and somatic mutations in BRCA1 and BRCA2. Consistent with recent results9, we demonstrate and quantify the association of biallelic inactivation of BRCA1/2 with Signature 3 and show lack of association in cases with mono-allelic inactivation.
We further used Signature 3 to identify a large subset of tumors (16%) with no discernible canonical HR defect, yet with a mutational landscape similar to that of samples with HRD. Focusing on this subset, we found that a small number of them harbor rare truncating variants in other components of the HR machinery.
Most importantly, we demonstrated that epigenetic silencing and somatic mutations in RAD51C have similar potential to abrogate HR function and yield the same characteristic mutational signature. The RAD51C promoter methylation events are comparable in frequency to BRCA2 germline events and higher than somatic truncating BRCA1 events in these tumors. RAD51C silencing events are even more frequent in basal-like breast cancers. To emphasize the magnitude of this finding, while PALB2 is now considered to be an important determinant for hereditary breast cancer15, we found only 2 samples harboring germline PALB2 lesions among the top quartile of Signature 3 activity, in contrast to 18 with somatically-acquired RAD51C dysfunction, thus making RAD51C methylation an important determinant of basal-like breast tumors. Preclinical models lacking RAD51C have been reported to exhibit sensitivity to PARP inhibitors46,47, raising potential translational implications for this finding. Therefore, RAD51C methylation could aid in patient stratification for agents that target HRD, as is currently being employed among ovarian cancer patients48.
In contrast to BRCA1/2, RAD51C and PALB2, protein-truncating germline variants in ATM, CHEK2 and NBN, that were shown to confer increased risk of developing breast cancer15 (including established pathogenic variants, Supplementary Table 1), were not associated with Signature 3. This finding suggests that these events may drive cancer risk via mechanisms independent of the BRCA1/2 double-strand break repair pathway. Indeed, in two pancreatic cancer studies with 17 combined cases of ATM mutations, 16 of which were biallelic, only one showed a signature consistent with HRD26,27. In contrast, alterations among HR components that are in the same protein complex with BRCA1/2 (e. g. RAD51C, PALB2 and BARD1) are associated with Signature 3, accounting for 8% of samples in the top quartile of Signature 3 activity and 13% of cases without BRCA1/2 events (Figure 4C). These findings may further extend the rational use of novel therapeutics.
“BRCAness” refers to HRD in a tumor that lacks a germline BRCA1 or BRCA2 deleterious variant16. As we have shown, Signature 3 can serve as a novel readout of HRD, and as such contribute to BRCAness, likely reflecting other defects in the BRCA1/2 part of the pathway. It will be important to test the association between different measures of BRCAness including ones based on expression, mutational signatures and other molecular measures, and responses to treatment in clinical trials. Our measure of Signature 3 could represent a putative biomarker for HR status. Current predictors of PARP-inhibitor or cisplatin sensitivity have had notable clinical success in a limited number of patients49–51, indicating the need for additional biomarkers such as HRD49 or Signature 3 to identify other patients who could also benefit from these treatments.
Developing a companion diagnostic based on this work could inform appropriate allocation of HRD-based therapies. Genomic-based scores, especially when based on clonal mutations, reflect the aggregated effect of HRD throughout the development of the cancer, but not necessarily its current HRD status52 Although not stated, ovarian cancers with BRCA1 reversion52 likely retained high levels of Signature 3 inherited from their ancestor cells, and were unlikely to have ongoing HRD. We expect a similar phenomenon with promoter methylation, whereby platinum (or PARP-inhibition) resistance might arise due to demethylation of the relevant promoter (e.g. BRCA1 or RAD51C).
Our findings have significant implications for the analysis of germline variants. When a pathogenic BRCA1/2 germline variant is found in the setting of a breast or ovarian cancer, it is inferred to be the putative driver of carcinogenesis53. However, among 28 carriers of pathogenic germline BRCA1/2 variants in our series whose tumor LOH status was discernible, we identified 4 tumors that exhibited neither loss of the intact BRCA1/2 allele nor elevated Signature 3 activity. In these, the tumors either arose independently of BRCA1/2 inactivation, or BRCA1/2 haploinsufficiency triggered events that, while cancer-initiating, were insufficient to independently generate HRD. BRCA1 haploinsufficiency in breast epithelium can lead to defective repair of stalled replication forks54. Inactivation of BRCA2 among other HR factors may cause similar stalling. As these forks can be repaired by non-HR dependent mechanisms55, BRCA1/2 haploinsufficiency could lead to non-HRD tumor phenotypes. Consequently, integrating Signature 3 with BRCA1/2 germline testing, may affect treatment selection.
Moreover, the classification of missense germline variants remains a challenging prospect given the breadth of uncommon variants and the large sample size often needed to reliably determine pathogenicity. Tumor signatures could be a powerful adjunct to existing methods in determining therapeutic approaches56. Importantly, our findings must be interpreted in the context of the TCGA study design. Namely, the use of samples from the TCGA may have limited generalizability to underrepresented minorities from whom few samples were collected. Similarly, accessibility to TCGA-participating centers and unblinded enrollment in this study may have introduced unforeseen biases into the study cohort. The clinical utility of Signature 3 will require prospective validation given the scarcity of pathogenic missense variants in this cohort (n = 5).
While clinical genetics has long relied on the use of population-level data and family-based segregation analysis to identify high-risk patients, this approach is of somewhat limited utility among those who are part of small pedigrees or who otherwise have limited access to familial information, and in the context of the discovery of increasing numbers of very rare, possibly unique variants. In the near future, with the increasing amount of tumor sequencing data becoming available57, mutation signatures extracted from these clinical sequencing projects can be used to contribute to variant classification. It is possible that some VUSs could have a Signature 3 profile that is sufficiently robust to support classification as likely benign (class 2) or likely pathogenic (class 4), even in the absence of other contributing data. In addition, the contribution of deleterious alterations in other genes, such as PALB2 and RAD51C, to HR defects and hence Signature 3, could lead to the design of therapeutic trials based on the findings described herein.
Supplementary Material
References
- 1.Prakash R, Zhang Y, Feng W & Jasin M Homologous recombination and human health: the roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb Perspect Biol 7, a016600 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antoniou A et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case Series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet 72, 1117–30 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ceccaldi R, Rondinelli B & D’Andrea AD Repair Pathway Choices and Consequences at the Double-Strand Break. Trends Cell Biol 26, 52–64 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas N Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burstein MD et al. Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clin Cancer Res 21, 1688–98 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ellis MJ & Perou CM The genomic landscape of breast cancer as a therapeutic roadmap. Cancer Discov 3, 27–34 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ciriello G et al. Comprehensive Molecular Portraits of Invasive Lobular Breast Cancer. Cell 163, 506–19 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ceccaldi R et al. Homologous-recombination-deficient tumours are dependent on Poltheta-mediated repair. Nature 518, 258–62 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nik-Zainal S et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 534, 47–54 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alexandrov LB et al. Signatures of mutational processes in human cancer. Nature (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lawrence MS et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim J et al. Somatic ERCC2 mutations are associated with a distinct genomic signature in urothelial tumors. Nat Genet 48, 600–6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kasar S et al. Whole-genome sequencing reveals activation-induced cytidine deaminase signatures during indolent chronic lymphocytic leukaemia evolution. Nat Commun 6, 8866 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nielsen FC, van Overeem Hansen T & Sorensen CS Hereditary breast and ovarian cancer: new genes in confined pathways. Nat Rev Cancer 16, 599–612 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Easton DF et al. Gene-panel sequencing and the prediction of breast-cancer risk. N Engl J Med 372, 2243–57 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lord CJ & Ashworth A BRCAness revisited. Nat Rev Cancer 16, 110–20 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Eccles DM et al. BRCA1 and BRCA2 genetic testing-pitfalls and recommendations for managing variants of uncertain clinical significance. Ann Oncol 26, 2057–65 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nik-Zainal S et al. Mutational processes molding the genomes of 21 breast cancers. Cell 149, 979–93 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Landrum MJ et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res 42, D980–5 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meeks HD et al. BRCA2 Polymorphic Stop Codon K3326X and the Risk of Breast, Prostate, and Ovarian Cancers. J Natl Cancer Inst 108(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scully R & Livingston DM In search of the tumour-suppressor functions of BRCA1 and BRCA2. Nature 408, 429–32 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merajver SD et al. Germline BRCA1 mutations and loss of the wild-type allele in tumors from families with early onset breast and ovarian cancer. Clin Cancer Res 1, 539–44 (1995). [PubMed] [Google Scholar]
- 23.Cornelis RS et al. High allele loss rates at 17q12-q21 in breast and ovarian tumors from BRCAl-linked families. The Breast Cancer Linkage Consortium. Genes Chromosomes Cancer 13, 203–10 (1995). [DOI] [PubMed] [Google Scholar]
- 24.Thompson ER et al. Panel Testing for Familial Breast Cancer: Calibrating the Tension Between Research and Clinical Care. J Clin Oncol 34, 1455–9 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Southey MC et al. PALB2, CHEK2 and ATM rare variants and cancer risk: data from COGS. J Med Genet (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Waddell N et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 518, 495–501 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Connor AA et al. Association of Distinct Mutational Signatures With Correlates of Increased Immune Activity in Pancreatic Ductal Adenocarcinoma. JAMA Oncol (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Livingston DM Cancer. Complicated supercomplexes. Science 324, 602–3 (2009). [DOI] [PubMed] [Google Scholar]
- 29.Loveday C et al. Germline RAD51C mutations confer susceptibility to ovarian cancer. Nat Genet 44, 475–6; author reply 476 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Couch FJ et al. Inherited mutations in 17 breast cancer susceptibility genes among a large triple-negative breast cancer cohort unselected for family history of breast cancer. J Clin Oncol 33, 304–11 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blanco A et al. RAD51C germline mutations found in Spanish site-specific breast cancer and breast-ovarian cancer families. Breast Cancer Res Treat 147, 133–43 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Curtis C et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486, 346–352 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pereira B et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nature Communications 7(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Foulkes WD et al. Germline BRCA1 mutations and a basal epithelial phenotype in breast cancer. J Natl Cancer Inst 95, 1482–5 (2003). [DOI] [PubMed] [Google Scholar]
- 35.Esteller M et al. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J Natl Cancer Inst 92, 564–9 (2000). [DOI] [PubMed] [Google Scholar]
- 36.Foulkes WD, Smith IE & Reis-Filho JS Triple-negative breast cancer. N Engl J Med 363, 1938–48 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Dietze EC, Sistrunk C, Miranda-Carboni G, O’Regan R & Seewaldt VL Triple-negative breast cancer in African-American women: disparities versus biology. Nature Reviews Cancer 15, 248–254 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rehm HL et al. ClinGen--the Clinical Genome Resource. N Engl J Med 372, 2235–42 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spurdle AB et al. ENIGMA--evidence-based network for the interpretation of germline mutant alleles: an international initiative to evaluate risk and clinical significance associated with sequence variation in BRCA1 and BRCA2 genes. Hum Mutat 33, 2–7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Richards S et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genetics in Medicine 17, 405–424 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hashizume R et al. The RING heterodimer BRCA1-BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation. J Biol Chem 276, 14537–40 (2001). [DOI] [PubMed] [Google Scholar]
- 42.Gorski B et al. Founder mutations in the BRCA1 gene in Polish families with breast-ovarian cancer. Am J Hum Genet 66, 1963–8 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abkevich V et al. Patterns of genomic loss of heterozygosity predict homologous recombination repair defects in epithelial ovarian cancer. Br J Cancer 107, 1776–82 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Popova T et al. Ploidy and large-scale genomic instability consistently identify basal-like breast carcinomas with BRCA1/2 inactivation. Cancer Res 72, 5454–62 (2012). [DOI] [PubMed] [Google Scholar]
- 45.Youden WJ Index for rating diagnostic tests. Cancer 3, 32–5 (1950). [DOI] [PubMed] [Google Scholar]
- 46.Min A et al. RAD51C-deficient cancer cells are highly sensitive to the PARP inhibitor olaparib. Mol Cancer Ther 12, 865–77 (2013). [DOI] [PubMed] [Google Scholar]
- 47.Somyajit K, Subramanya S & Nagaraju G RAD51C: a novel cancer susceptibility gene is linked to Fanconi anemia and breast cancer. Carcinogenesis 31, 2031–2038 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Evans T & Matulonis U PARP inhibitors in ovarian cancer: evidence, experience and clinical potential. Ther Adv Med Oncol 9, 253–267 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chan SL & Mok T PARP inhibition in BRCA-mutated breast and ovarian cancers. Lancet 376, 211–3 (2010). [DOI] [PubMed] [Google Scholar]
- 50.Fong PC et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med 361, 123–34 (2009). [DOI] [PubMed] [Google Scholar]
- 51.Kaufman B et al. Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J Clin Oncol 33, 244–50 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Patch AM et al. Whole-genome characterization of chemoresistant ovarian cancer. Nature 521, 489–494 (2015). [DOI] [PubMed] [Google Scholar]
- 53.Clamp A & Jayson G PARP inhibitors in BRCA mutation-associated ovarian cancer. Lancet Oncol 16, 10–2 (2015). [DOI] [PubMed] [Google Scholar]
- 54.Pathania S et al. BRCA1 haploinsufficiency for replication stress suppression in primary cells. Nat Commun 5, 5496 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yeeles JT, Poli J, Marians KJ & Pasero P Rescuing stalled or damaged replication forks. Cold Spring Harb Perspect Biol 5, a012815 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lindor NM et al. A review of a multifactorial probability-based model for classification of BRCA1 and BRCA2 variants of uncertain significance (VUS). Hum Mutat 33, 8–21 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hyman DM et al. Precision medicine at Memorial Sloan Kettering Cancer Center: clinical next-generation sequencing enabling next-generation targeted therapy trials. Drug Discov Today 20, 1422–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.