Abstract
Purpose
To explore the whole-chromosome status, origins, and mechanisms of chromosomal abnormalities in good-quality cleavage embryos using multiple annealing and looping-based amplification cycle (MALBAC) sequencing.
Methods
The embryos studied came from7 patients (maternal aged 26–35) who had healthy birth from the same IVF cycles. These 21 frozen day 3 good-quality embryos were thawed and disaggregated into individual blastomere. Each blastomere was collected and analyzed by MALBAC sequencing.
Results
Conclusive results were obtained from a high percentage of blastomeres (95.3%). A total of 46.6% of blastomeres were diploid, 53.4% were abnormal, and 28.0% had complex aneuploidy. Out of 21 embryos, 3 (14.3%) were normal and 18 (85.7%) were mosaics, showing the occurrence of mitotic errors; aneuploidy was confirmed in all cells of 4 of the 18 embryos, which showed the coexistence of meiotic errors. Conclusive results were obtained from all blastomeres of 15 embryos (71.4%, 15/21), which enabled us to reconstruct the cell lineage on the basis of the chromosomal content of the blastomeres in each division. There were 9 mitotic errors (8.7%, 9/103): nondisjunction accounted for 88.9% (8/9), and endoreplication accounted for 11.1% (1/9).
Conclusions
In good-quality embryos, there was a high rate and diverse array of chromosomal abnormalities. Morphological evaluation does not appear to assist in the reduction in meiotic errors from parental origins. Mitotic errors were common, and nondisjunction was found to be the main mechanism causing malsegregation during the cleavage divisions.
Electronic supplementary material
The online version of this article (10.1007/s10815-020-01803-9) contains supplementary material, which is available to authorized users.
Keywords: Cleavage embryo, Aneuploidy, Preimplantation genetic screening, Multiple annealing and looping-based amplification cycles (MALBAC), Mosaicism
Introduction
Although in vitro fertilization (IVF) technology has rapidly developed, the success rate is still relatively low. The cleavage embryo implantation rate is approximately 36% [1]. The aneuploidy incidence in aborted fetuses is very high (> 50%) [2]. Chromosomal abnormalities in live-born infants primarily originate from meiotic errors. These data suggest that early-stage embryos suffer from a wide range of chromosomal abnormalities and that this might be one of the main causes of the negative outcomes following IVF. For many years, good-quality embryos, based on morphological criteria, including cell number, cell size, and fragmentation, have commonly been selected for transplantation. Understanding the types, incidence, and origin of aneuploidy in good-quality cleavage-stage embryos is of great importance.
The introduction of preimplantation genetic screening (PGS) made it possible to study the chromosomal condition of embryos [3]. Typically, fluorescence in situ hybridization (FISH) as the main method for PGS is performed to identify ploidy states of only 5–12 chromosomes. High-resolution methods for the complete karyotyping of a cell, such as comparative genomic hybridization (CGH; including metaphase CGH and array CGH), single-nucleotide polymorphism (SNP) arrays, and next-generation sequencing (NGS), remedy the limitations of FISH. These methods enable the detection of 24 types of chromosome and chromosome breakage leading to segmental aberrations in one cell, and more abnormalities can be detected than with FISH. Multiple annealing and looping-based amplification cycles (MALBAC) is a newly developed amplification method that offers high genomic coverage with much reduced sequence-dependent bias [4]. MALBAC sequencing was introduced for genome detection of single gametes [5], single blastomeres [6], and several trophectoderm cells [7] and was validated to be an accurate and cost-effective method for whole-genome detection of embryos.
Although many cytogenetic studies using high-resolution methods demonstrated that aneuploidy and mosaicism were common and insinuated chromosome instability in IVF embryos [8–10], the exact whole-chromosome status of good-quality cleavage-stage embryos remains poorly defined. The majority of studies are performed on only single- or dual-biopsied blastomeres or on all blastomeres from discarded embryos. Moreover, the majority of the studied embryos were from women at risk of embryonic aneuploidy (such as those with advanced maternal age, recurrent implantation failure, and recurrent miscarriages) [11]. Only a few studies have obtained complete information on normally developing cleavage-stage embryos from young fertile women [12–14]. Two studies have provided information on every blastomere from good-quality embryos using metaphase CGH, which has a lower resolution than NGS [12, 13]. One study has performed array CGH testing of all blastomeres of high-quality embryos, but conclusive results were not obtained from a significant proportion of the blastomeres in this study [14].
In the present study, we applied MALBAC sequencing to analyze the genomic status of all blastomeres of 21 good-quality cleavage embryos donated by couples who had healthy births from the same IVF cycles. The data in this article can provide insight into the cytogenetic constitution of day 3 good-quality embryos based on morphological characteristics and can define the types and frequencies of chromosomal abnormalities. The results can also shed light on the origins of aneuploidy, the mechanisms of chromosomal malsegregation, and the developmental potential of mosaic embryos.
Material and methods
Embryos
All embryos used in this study were surplus from IVF couples who had delivered babies from the same IVF cycle. The study was approved by the Institutional Review Board of the 3rd Affiliated Hospital of Guangxi Medical University. We obtained signed consent from all of the donors before the treatment. Seven couples donated 21 good-quality frozen cleavage-stage embryos. The mean age of the women was 29 (range 26 to 35) years. General information about the 7 couples is summarized in Table 1. All of the embryos were fertilized by intracytoplasmic sperm injection (ICSI) and exhibited two pronuclei on day 1, normally developed to good-quality cleavage-stage embryos (3–5 blastomeres on day 2, 6–10 equal-sized blastomeres with less than 20% fragmentation on day 3), and were cryopreserved by vitrified cryopreservation [15] (Table 2).
Table 1.
Information of donated couples
| Couple | Age of woman | Cause of infertility | No. of donated embryos |
|---|---|---|---|
| Y | 28 | Male factor | 1 |
| L | 28 | Male factor | 5 |
| B | 35 | Male factor | 1 |
| W | 26 | Male factor | 1 |
| LX | 29 | Male factor | 2 |
| D | 30 | Male factor | 3 |
| K | 27 | Male factor | 8 |
Table 2.
MALBAC-sequencing results of 169 analyzed blastomeres from 21 embryos
| Couple | Embryo no. | D2 cell number (grade) | D3 cell number (grade) | MALBAC-sequencing results [no. of cells) |
|---|---|---|---|---|
| Y | 1 | 4(II) | 8 (II) | 46,XY [3] |
| 51,XYY,+Y(×2),+3(×3),+9(×3),+13(×3), +17(×3) [2] | ||||
| 41,X,-Y(×0),-3(×1),-9(×1),-13(×1),-17(×1) [2] | ||||
| 46,XY,+6q(q12→q22.31,~55M,×3) [1] | ||||
| L | 2 | 4(II) | 8 (II) | 46,XX [8] |
| L | 3 | 4(II) | 8 (I) | 46,XX [2] |
| 46,XX,+17p(pter→p11.2,~23M,×3),-20(p11.21→qter,~36M,×1)[1] | ||||
| 46,XX,-17p(pter→p11.2,~23M,×1),+20(p11.1→qter,~32M,×3)[1] | ||||
| 46,XX,-9q(q21.11→qter,~67M,×1) [1] | ||||
| 46,XX,+9q(q21.11→qter,~65M,×3)[1] | ||||
| Complex abnormality[2] | ||||
| L | 4 | 3(II) | 6 (II) | 46,2Y,-X(×0) [1] |
| 46,XX,-16q(q11.2→qter,~43M,×0) [1] | ||||
| 46,XX,+16q(q11.2→qter,~43M,×4) [1] | ||||
| 48,XX, +1(×4) [1] | ||||
| 44,XX,-1(×0) ,+3q(q11.2→q13.33,~24M,×3) [1] | ||||
| 44,XX,-1(×0) ,+13q(q13.1→q21.2,~30M,×3) [1] | ||||
| L | 5 | 4(I) | 8 (I) | 46,XX[5] |
| 46,XX,+5q(q22.3→qter,~66M,×3) [1] | ||||
| 46,XX,-5q(q22.3→qter,~65M,×1) [1] | ||||
| 46,XX,-1(p12→q24.2,~52M,×1) [1] | ||||
| 6 | 4(II) | 8 (II) | 46,XX[2] | |
| 46,XX,-X(p11.3→q13.1,~27M,×1) [1] | ||||
| Complex abnormality[5] | ||||
| B | 7 | 5(II) | 7 (II) | 46,XY[3] |
| 48,XY,+8(×3) +20(×3)[1] | ||||
| 46,XY,-8(×1),+20(×3)[1] | ||||
| Complex abnormality[1] | ||||
| No result[1] | ||||
| W | 8 | 5(II) | 8 (II) | 46,XY[8] |
| LX | 9 | 4(I) | 8 (II) | 46,XX[4] |
| 46,XX,+15(pter→q24.1,~45M,×3) [1] | ||||
| 46,XX,+12(pter→q12,~45M,×3) [1] | ||||
| No result [2] | ||||
| LX | 10 | 4(I) | 8 (II) | 46,XY[7] |
| 46,XY,-13q(q32.1→qter,~20M,×1)[1] | ||||
| D | 11 | 4(II) | 8 (II) | 45,XX,-16(×1)[7] |
| 45,XX,-16(×1),-19(p13.11→q13.2,~24M,×1)[1] | ||||
| D | 12 | 4(II) | 8 (II) | Complex abnormality[7] |
| No result[1] | ||||
| D | 13 | 4(II) | 8 (II) | 46,XX[4] |
| 46,XX,+9q(q21.11→qter,~66M,×3)[1] | ||||
| 46,XX,-9(p13.3→qter,~100M,×1) [1] | ||||
| 45,XX,-17(×1)[1] | ||||
| 47,XX,+17(×3) [1] | ||||
| K | 14 | 3(II) | 8 (II) | Complex abnormality[8] |
| K | 15 | 3(II) | 8 (II) | 46,XX[8] |
| K | 16 | 4(II) | 8 (I) | 46,XX[4] |
| 47,XX,+18(×3)[1] | ||||
| Complex abnormality[1] | ||||
| No result[2] | ||||
| K | 17 | 4(II) | 8 (I) | 46,XY[7] |
| 46,XY,-4q(q12→q22.2,~40M,×1),+14(pter→q23.3,~43M,×3)[1] | ||||
| K | 18 | 4(II) | 10 (II) | 46,XX[6] |
| Complex abnormality[2] | ||||
| No result[2] | ||||
| K | 19 | 4(II) | 8 (II) | 44,XY,-2(×1),-12(×1)[6] |
| Complex abnormality[2] | ||||
| K | 20 | 4(II) | 8 (II) | 45,XY,-16(×1)[4] |
| Complex abnormality[4] | ||||
| K | 21 | 4(II) | 10 (II) | 46,XX[4] |
| Complex abnormality[6] |
Cell isolation and lysis
Twenty-one embryos were thawed, and morphology grade was assigned. The embryos were incubated in 0.6% pronase for 2–3 min to remove the zona pellucida and then gently pipetted to disaggregate the blastomeres. Each blastomere was washed in two 30-μl droplets of sterile phosphate-buffered saline and then placed into 5 μl of lysis buffer (200 mM KOH) and stored at − 20 °C until experimentation.
MALBAC and NGS of single blastomeres
We used MALBAC (20) to amplify the whole genome of a single blastomere, which can generate the micrograms of DNA required for NGS. Then, the amplified genome was sequenced at ~ 0.04× depth on an Illumina HiSeq2500 platform. Therefore, approximately 40 million bases for each blastomere were sequenced, obtaining genomic coverage of 3% on average. The sequencing throughput achieved the standard for variation screening of chromosomal copy number (> 0.01× genome depth) and yielded reproducible copy number variation results with approximately 1 Mb of resolution to detect the variation [6].
Results
MALBAC-sequencing results
Altogether, 21 embryos were thawed, surviving with each blastomere intact. A total of 169 blastomeres were obtained. A total of 164 cells (97.0%, 164/169) were successfully amplified, and conclusive results, which were the results of ploidy determination by MALBAC sequencing, were obtained from 161 cells (95.3%, 161/169). Five cells (3.0%, 5/169) did not yield the expected amount of DNA, and 3 cells (1.8%, 3/169) did not provide analyzable results. Table 2 shows the MALBAC-sequencing results of each blastomere.
Embryonic ploidy
Among the 21 embryos, 9 embryos (42.9%, 9/21) were fertilized by Y sperm and 12 embryos (57.1%, 12/21) were fertilized by X sperm. Of the 21 embryos, 3 embryos (14.3%, 3/21) were normal diploid (embryos 2, 8, and 15), and 18 embryos (85.7%, 18/21) were mosaic; aneuploidy was confirmed in all cells of 4 of the 18 embryos (embryos 11, 14, 19, and 20), which showed the coexistence of meiotic errors. Among the remaining 14 (77.8%, 14/18) mosaic embryos, which consisted of normal blastomeres as well as blastomeres with either whole-chromosome or segmental aneuploids or both, 7 (38.9%, 7/18) were diploid/aneuploid mosaics (≥ 50%) (embryos 5, 9, 10, 13, 16, 17, and 18), 5 (27.8%, 5/18) were diploid/aneuploid mosaics (< 50%) (embryos 1, 3, 6, 7, and 21), and 2 were aneuploidy mosaics (11.1%, 2/18) (embryos 4 and 12) (Table 3).
Table 3.
The incidence rates of diploidy, aneuploidy, and mosaicism in 21 embryos
| Embryo status | n (%) |
|---|---|
| Diploid | 3 (14.3) |
| Aneuploid | 4 (19.0) |
| Mosaic | 14 (66.7) |
| Diploid/aneuploid (≥ 50%) | 7 (33.3) |
| Diploid/aneuploid (< 50%) | 5 (23.8) |
| Aneuploid mosaic | 2 (9.5) |
Blastomere euploidy
Of the 161 analyzed blastomeres, 75 (46.6%, 75/161) were diploid. Forty-six (28.6%, 46/161) had only whole-chromosome abnormalities, out of which 12 (7.5%, 12/161) blastomeres had monosomy and 2 (1.2%, 2/161) blastomeres had trisomy. Nineteen (11.8%, 19/161) blastomeres contained only structural aberrations. Twenty-one (13.0%, 21/161) blastomeres contained structural aberrations coexisting with whole-chromosome abnormalities.
Among the 86 aneuploidy blastomeres, 26 (16.1%, 26/161) had whole or structural aneuploidy of one chromosome, 15 (9.3%, 15/161) had aneuploidy of two chromosomes, and 45 (28.0%, 45/161) had complex aneuploidy.
The incidence of whole-chromosome aneuploidy
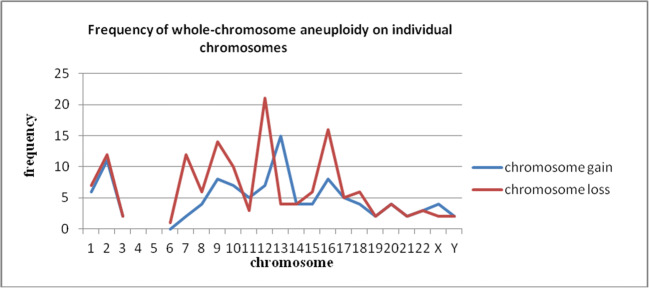
Sixty-seven (41.6%, 67/161) blastomeres were found to have whole-chromosome abnormalities distributed in 13 (61.9%, 13/21) embryos. Whole-chromosome aneuploidy was distributed at all autosomes (except for chromosomes 4 and 5) and sex chromosomes. The abnormal frequencies were not identical on different chromosomes (Fig. 1). The gain and loss of most chromosomes were approximately equal, but the loss of chromosomes 7, 9, 12, and 16 was significantly greater than their gain, and the situation was contradictory for chromosome 13.
Fig. 1.
Frequency of whole-chromosome aneuploidy on individual chromosomes
The incidence of structural aberration
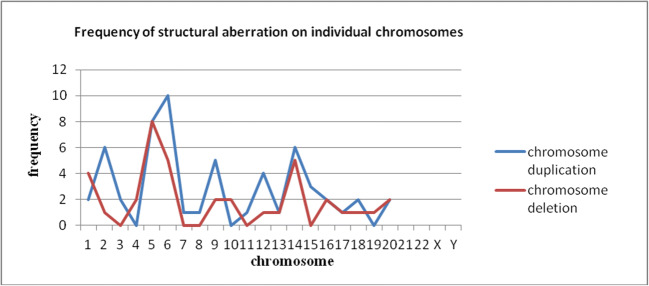
Forty (24.8%, 40/161) blastomeres were found to have structural aberrations, which were distributed in 16 (76.2%, 16/21) embryos. Out of a total of 92 unbalanced segments detected, 43 (46.7%, 43/92) were deletions and 49 (53.3%, 49/92) were duplications. Fifteen (16.3%, 15/92) were p segments, 44 (47.8%, 44/92) were q segments and 33 (35.9%, 33/92) were segments spanning the centromeric region. Structural aberrations were distributed on all chromosomes except chromosomes 21, 22, and Y. Figure 2 shows the frequencies on different chromosomes.
Fig. 2.
Frequency of structural aberration on individual chromosomes
Cell lineage analysis
Among the 21 embryos, conclusive results from all blastomeres were obtained from 15 embryos. All embryos were evaluated daily after fertilization. Based on the information regarding their size and cell numbers, the number of cleavages they underwent can be estimated. Along with the chromosome content of each blastomere, the developmental history of each embryo can be reflected and the cell lineages of their day-by-day chromosomal status from zygote until day 3 can be reconstructed. The simplest possible mechanisms (either nondisjunction, anaphase lagging, chromosome breakage or endoreplication) were always chosen to explain the final outcome [16]. The supplemental figure shows the cell lineages of the 15 embryos.
The 15 embryos underwent 103 mitotic divisions during the 3 culturing days. There were 9 (8.7%, 9/103) mitotic errors that led to whole-chromosome imbalances. The frequency of nondisjunctions was 7.8% (8/103), which was more than that of endoreduplication (1.0%, 1/103), and no anaphase lag was found. Chromosomal breakages leading to structural aberrations occurred in 17 (16.5%, 17/103) divisions.
Discussion
Understanding the whole genetic status of good-quality embryos based on morphological criteria that are selected for transplantation provides insight into the cause of the low utilization rate of IVF embryos and the origin and mechanisms of chromosomal abnormalities. To the best of our knowledge, this is the first study of the chromosomes of whole blastomeres in normally developing and good-quality embryos from young women using MALBAC sequencing. The strength of this study is that we obtained conclusive results from over 95.0% of the blastomeres studied, and conclusive results were obtained from all blastomeres in more than 70.0% of the embryos analyzed. Thus, our data present a more complete picture of the genomic status in good-quality embryos.
In our study, 46.6% of the blastomeres were chromosomally normal, which was in line with the rates reported by previous studies [17, 18]. Of the remaining blastomeres, 16.1% had whole or structural aneuploidy of one chromosome, 9.3% had aneuploidy of two chromosomes, and 28.0% had complex aneuploidy associated with a disturbance of cell division. Comparable data were obtained using CGH. Wells et al. [13] found that 56% of blastomeres were normal, 25% had aneuploidy of one chromosome, and 14% had aneuploidy of two or more chromosomes. In a study performing an SNP array [17], it was reported that 48% of blastomeres were euploid, 32% had one or two aneuploidies, and 20% carried complex aneuploidy in high-quality cleavage-stage embryos. The findings of Mertzanidou et al. [14] after array CGH analysis were comparable; they reported 55.7% diploid cells, 2.7% cells with two aneuploidies, and 11.4% cells with complex aneuploidies in good-quality cleavage-stage embryos.
For chromosomal mosaicism in cleavage-stage embryos, a systematic review on the chromosomal constitution of each cell from 815 cleavage embryos found that only 22% were diploid, 73% were mosaic, 59% were diploid-aneuploid mosaic, and 5% contained other abnormalities [19]. However, almost all of the studies in this review mainly used FISH to analyze spare embryos after IVF rather than embryos that were transferred or cryopreserved. Only a few studies have paid attention to the 24 chromosomes of whole blastomeres from high-quality embryos. Mertzanidou et al. [14] analyzed the chromosomal status of 66.7% (70/105) of blastomeres from 14 high-quality embryos using array CGH and showed 28.6% diploid embryos and 71.4% mosaic embryos. Chow et al. [18] obtained results from 92.8% (90/97) of blastomeres from 12 good-quality embryos using array CGH and found that 16.7% were diploid and 58.3% were diploid-aneuploid mosaic.
In the present study, we investigated all 24 chromosomes of whole blastomeres from 21 donated good-quality embryos and obtained conclusive results from 95.3% of blastomeres. Our results indicated that only 14.3% (3/21) of embryos were normal diploid, and 85.7% (18/21) of embryos had mosaicism. In the mosaic embryos, 12 diploid-aneuploidy embryos were observed, accounting for 57.1% of the total embryos. Thus, the rate of diploid-aneuploidy embryos is consistent with expectations based on prior data, but the proportions of mosaicism are higher than those in previous studies. This finding may be associated with the detection of more chromosomes and more blastomeres in our study. The high rate of chromosomal mosaicism found in our study indicates that mitotic errors are common in good-quality cleavage embryos.
In our study, among the 18 mosaic embryos, 4 embryos (embryos 11, 14, 19, and 20) had consistent aneuploidy of one or more chromosomes in all blastomeres, which can be assumed to also suffer from meiotic errors of paternal or maternal origins. Since aneuploidy occurs relatively infrequently in spermatozoa [20, 21], the meiotic errors that occur during oogenesis may play a more important role. Maternal meiotic errors are well known to be positively correlated with advanced maternal age [22, 23]. Our studied embryos were donated from young patients (aged 26–35 years). Previous studies measured whole-chromosome aneuploidy in polar bodies and/or oocytes and showed aneuploidy rates of 3–20% for young women [5, 24]. The aneuploidy rate derived from meiotic errors found in our study is in agreement with that of previous studies about aneuploidy rates of gametes. The data are evidence that good-quality embryos still harbor meiotic errors from paternal or maternal origins. It may be that morphological evaluation does not appear to assist the reduction in embryos with meiotic errors, but this remains to be conclusively demonstrated.
The embryos used for this study were surplus good-quality embryos from the same IVF cycles that resulted in healthy births. In our reproductive center, the implantation rate in the frozen-thawed embryo cycle for young women is 36.1%, which is much higher than the percentage of normal diploid embryos in our study. Our observation suggests that some of the mosaic embryos could have implantation. Many studies have provided evidence that diploid-aneuploid mosaic embryos might undergo self-correction during their development [25–28]. Early cleavage embryos exhibit a high rate of mosaicism due to incomplete activation of the embryonic genome [29]. Once the embryo successfully activates its genome around the 4- to 8-cell stage, mechanisms by which diploid cells proliferate and allocate to the inner cell mass and by which aneuploid cells decrease through cell death or aneuploidy rescue have been suggested for the selection of diploid cells versus aneuploid cells during development. This brings up the question of which mosaic embryos will result in live birth. Bialanska et al. [30] suggested that the developmental potential of the pre-embryo might be impaired if the majority of its cells are abnormal. Therefore, it is reasonable to assume that embryos with less than 50% chromosomally abnormal blastomeres could be implanted. Previous research suggested that mosaic embryos affected by complex abnormalities might suffer reduced viability [31–33], as the presence of complex abnormalities in cells caused by a disturbance of replication and/or cell division might disrupt intercellular interactions, leading to failures in cellular reorganization and differentiation, which are necessary for embryo development. In our study, mosaic embryos 5, 9, 10, 13, and 17 had < 50% abnormal blastomeres, excluding complex abnormalities. Thus, the percentage of studied embryos with implantation potential, i.e., diploid and diploid-aneuploidy mosaics with < 50% abnormal blastomeres, was 38.1% (8/21), which was consistent with the implantation rate of frozen-thawed embryos. In contrast, > 60% of day 3 good-quality embryos consisting of chromosomal abnormalities due to meiotic and/or mitotic embryos always fail to develop.
In our hands, over 40% of the analyzed cells were found to have whole-chromosome abnormalities. The aneuploidy was distributed at all autosomes (except for chromosomes 4 and 5) and sex chromosomes, although the abnormal frequencies were not identical. The gains and losses of most chromosomes were approximately equal, which indicated that these chromosomes were involved in mitotic errors. However, the losses of chromosomes 7, 9, 12, and 16 were significantly greater than their gains and the situation was contradictory for chromosome 13. The loss or gain of these chromosomes was caused by the aneuploidy in almost all cells of some embryos. Thus, this finding might be the result of involvement in meiotic errors or in mitotic errors of the zygote. As previously reported, some types of aneuploidy displayed dramatic fluctuations in frequency as preimplantation development progressed [34, 35]. Aneuploidies involving larger chromosomes (such as chromosomes 1, 3, 4, 6, and 8) are not commonly seen in spontaneously aborted fetuses or in live births [2]. It can be reasonably deduced that some whole-chromosome aneuploidy would diminish as cleavage embryos developed.
There are three main mechanisms by which a gain and/or loss of whole chromosomes can occur in mitosis: anaphase lagging, nondisjunction, and endoreplication [36]. Studies observed a slight overrepresentation of monosomy from PGS data that detected one or two cells of an embryo and suggested that anaphase lagging accounted for the majority of mitotic errors during the cleavage stage, while nondisjunction occurred to a lesser extent [37]. Our data might appear controversial. In our study, there was an excess (18:2) showing single monosomy over single trisomy, but this excess was attributed to the repeat count of monosomy due to meiotic errors. As we obtained conclusive results from over 95% of the blastomeres from 21 embryos, in which intact genome information was obtained for each blastomere from 15 embryos, we reconstructed the cell lineages of the 15 embryos and deduced the cell divisions. Among the 15 embryos, we found 9 mitotic errors leading to whole-chromosome imbalance, out of which 8 were nondisjunction errors and 1 was an endoreplication error. One nondisjunction error was found at the zygote stage. In the other 6 embryos that lost the message of one or two cells, if a gain of a chromosome was observed in one blastomere, a loss of the corresponding chromosome would be detected in the sister blastomere. Even in the two sister blastomeres with complex abnormalities, the corresponding gain or loss of the chromosomes would also be detected. We can reasonably deduce that nondisjunction is the main mechanism for mitotic errors in day 3 good-quality embryos.
The introduction of high-resolution methods has enabled the detection of segmental chromosomal abnormalities. Prior data demonstrated that chromosomal structural aberrations were common in cleavage embryos. However, it should be noted that the frequency of structural aberrations is significantly different among studies, and the difference may be associated with the resolution of detection technology. Daphnis et al. [38], using CGH, reported that 28% of embryos carried chromosomal breakages with a resolution of approximately 40 Mb. Using SNP arrays with much higher resolution, Vanneste et al. [9] reported that 31–70% of embryos carried structural abnormalities. Mertzanidon et al. [14] performed array CGH and found that 29% of the blastomeres carried structural aberrations. In our study, we performed MALBAC sequencing, comparable with array CGH and array SNP, and found that 24.8% of cells had structural aberrations, which were distributed in 76.2% of good-quality embryos. A small excess of chromosome duplications (53.3%) compared with deletions (46.7%) and some types of chromosomal segments due to chromosomal breakages resulted in an unstable karyotype for acentric or dicentric chromosomes. All chromosomes (except 21, 22, and Y) participated in partial chromosomal breakages, although to different extents. The incidence of abnormalities increased for chromosomes 5, 6, and 14 compared with other chromosomes. The segmental aneuploidy observed in our study existed in a mosaic form, indicating that the aneuploidy originated during mitotic division and that rearrangement errors are a main factor for mitotic division. However, relatively little is known about their susceptibility, ultimate fate, and biological significance.
Of course, our study had limitations. First, the embryos in this study were frozen embryos. During the thawing process of the embryos in our study, each blastomere survived and was intact with a clear nucleus, and we analyzed the embryos shortly after thawing. Therefore, the influence of freezing-thawing on chromosomes may be small. Second, the IVF cycles all used ICSI for male factors. Although spermatozoa from men with severe oligoasthenoteratospermia have increased aneuploidy rates, the estimated aneuploidy rate is less than 5% [20, 21].
In conclusion, we have shown the most complete chromosome status of day 3 good-quality embryos from young women based on morphological criteria. A high rate and diverse array of chromosomal abnormalities were found. Good-quality embryos still harbor meiotic errors from paternal or maternal origins, and morphological evaluation does not appear to assist in the reduction in meiotic errors. During the first mitotic divisions in human preimplantation development, the checkpoint seems nonexistent, leading to chromosomal malsegregation and subsequent mosaicism [39–41]. The occurrence of mosaicism in > 50% of abnormal blastomeres or the occurrence of complex abnormalities plays an important role in embryo development retardation. Nondisjunction was found to be the main mechanism causing malsegregation. The common existence of mosaicism undermines the reliability of cleavage-stage PGS.
Electronic supplementary material
(DOCX 708 kb)
Funding information
Supported by the Natural Science Foundation of Guangxi Province (grant number 2017GXNSFAA198149, 2017GXNSFAA198163) and the Major Science and Technology of Nanning (grant no. 20153011, 20153124, 20163138)
Compliance with ethical standards
The study was approved by the Institutional Review Board of the 3rd Affiliated Hospital of Guangxi Medical University. We obtained signed consent from all of the donors before the treatment.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ying Qiu, Email: nnqying@sina.com.
Changlong Xu, Email: xuchanglong2011@hotmail.com.
References
- 1.Glujovsky D, Farquhar C, Quinteiro Retamar AM, Alvarez Sedo CR, Blake D. Cleavage stage versus blastocyst stage embryo transfer in assisted reproductive technology. Cochrane Database Syst Rev. 2016;30:CD002118. doi: 10.1002/14651858.CD002118.pub5. [DOI] [PubMed] [Google Scholar]
- 2.Sahoo T, Dzidic NA-O, Strecker MA-O, Commander S, Travis MK, Doherty C, et al. Comprehensive genetic analysis of pregnancy loss by chromosomal microarrays: outcomes, benefits, and challenges. Genet Med. 2017;19:83–89. doi: 10.1038/gim.2016.69. [DOI] [PubMed] [Google Scholar]
- 3.Baart EB, Martini E, van den Berg I, Macklon NS, RJH G, Fauser BCJM, Van Opstal D, et al. Preimplantation genetic screening reveals a high incidence of aneuploidy and mosaicism in embryos from young women undergoing IVF. Hum Reprod. 2006;21:223–233. doi: 10.1093/humrep/dei291. [DOI] [PubMed] [Google Scholar]
- 4.Lu S, Zong C, Fan W, Yang M, Li J, Chapman AR, Zhu P, et al. Probing meiotic recombination and aneuploidy of single sperm cells by whole-genome sequencing. Science. 2012;338:1627–1630. doi: 10.1126/science.1229112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hou Y, Fan W, Yan L, Li R, Lian Y, Huang J, Li J, Xu L, Tang F, Xie XS, Qiao J. Genome analyses of single human oocytes. Cell. 2013;155:1492–1506. doi: 10.1016/j.cell.2013.11.040. [DOI] [PubMed] [Google Scholar]
- 6.Huang J, Yan L, Fan W, Zhao N, Zhang Y, Tang F, Xie XS, Qiao J. Validation of multiple annealing and looping-based amplification cycle sequencing for 24-chromosome aneuploidy screening of cleavage-stage embryos. Fertil Steril. 2014;102:1685–1691. doi: 10.1016/j.fertnstert.2014.08.015. [DOI] [PubMed] [Google Scholar]
- 7.Huang J, Yan L, Lu S, Zhao N, Xie XS, Qiao J. Validation of a next-generation sequencing-based protocol for 24-chromosome aneuploidy screening of blastocysts. Fertil Steril. 2016;105:1532–1536. doi: 10.1016/j.fertnstert.2016.01.040. [DOI] [PubMed] [Google Scholar]
- 8.Babariya D, Fragouli E, Alfarawati S, Spath K, Wells D. The incidence and origin of segmental aneuploidy in human oocytes and preimplantation embryos. Hum Reprod. 2017;32:2549–2560. doi: 10.1093/humrep/dex324. [DOI] [PubMed] [Google Scholar]
- 9.Vanneste E, Voet T, Le Caignec C, Ampe M, Konings P, Melotte C, Debrock S, et al. Chromosome instability is common in human cleavage-stage embryos. Nat Med. 2009;15:577–583. doi: 10.1038/nm.1924. [DOI] [PubMed] [Google Scholar]
- 10.Weissman A, Shoham G, Shoham Z, Fishel S, Leong M, Yaron Y. Chromosomal mosaicism detected during preimplantation genetic screening: results of a worldwide Web-based survey. Fertil Steril. 2017;107:1092–1097. doi: 10.1016/j.fertnstert.2017.02.119. [DOI] [PubMed] [Google Scholar]
- 11.Harper J. Sermon K Fau - Geraedts J, Geraedts J Fau - Vesela K, Vesela K Fau - Harton G, Harton G Fau - Thornhill A, Thornhill A Fau - Pehlivan T et al. What next for preimplantation genetic screening? Hum Reprod. 2008;23:478–480. doi: 10.1093/humrep/dem424. [DOI] [PubMed] [Google Scholar]
- 12.Voullaire L, Slater H, Williamson R, Wilton L. Chromosome analysis of blastomeres from human embryos by using comparative genomic hybridization. Hum Genet. 2000;106:210–217. doi: 10.1007/s004390051030. [DOI] [PubMed] [Google Scholar]
- 13.Wells D, Delhanty JD. Comprehensive chromosomal analysis of human preimplantation embryos using whole genome amplification and single cell comparative genomic hybridization. Mol Hum Reprod. 2000;6:1055–1062. doi: 10.1093/molehr/6.11.1055. [DOI] [PubMed] [Google Scholar]
- 14.Mertzanidou A, Wilton L, Cheng J, Spits C, Vanneste E, Moreau Y, Vermeesch JR, et al. Microarray analysis reveals abnormal chromosomal complements in over 70% of 14 normally developing human embryos. Hum Reprod. 2013;28:256–264. doi: 10.1093/humrep/des362. [DOI] [PubMed] [Google Scholar]
- 15.Zhang L, Yilmaz A, Chian R-C, Son W-Y, Zhang XY, Kong D, Dahan M, et al. Reliable preimplantation genetic diagnosis in thawed human embryos vitrified at cleavage stages without biopsy. J Assist Reprod Genet. 2011;28:597–602. doi: 10.1007/s10815-011-9556-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mertzanidou A, Spits C, Nguyen HT, Van de Velde H, Sermon K. Evolution of aneuploidy up to day 4 of human preimplantation development. Hum Reprod. 2013;28:1716–1724. doi: 10.1093/humrep/det079. [DOI] [PubMed] [Google Scholar]
- 17.Johnson DS, Gemelos G, Baner J, Ryan A, Cinnioglu C, Banjevic M, Ross R, et al. Preclinical validation of a microarray method for full molecular karyotyping of blastomeres in a 24-h protocol. Hum Reprod. 2010;25:1066–1075. doi: 10.1093/humrep/dep452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chow JF, Yeung WS, Lau EY, Lee VC, Ng EH, Ho P-C. Array comparative genomic hybridization analyses of all blastomeres of a cohort of embryos from young IVF patients revealed significant contribution of mitotic errors to embryo mosaicism at the cleavage stage. Reprod Biol Endocrinol. 2014;12:105. doi: 10.1186/1477-7827-12-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Echten-Arends J, Mastenbroek S, Sikkema-Raddatz B, Korevaar JC, Heineman MJ, van der Veen F, Repping S, et al. Chromosomal mosaicism in human preimplantation embryos: a systematic review. Hum Reprod Update. 2011;17:620–627. doi: 10.1093/humupd/dmr014. [DOI] [PubMed] [Google Scholar]
- 20.Donate A, Estop AM, Giraldo J, Templado C. Paternal age and numerical chromosome abnormalities in human spermatozoa. Cytogenet Genome Res. 2016;148:241–248. doi: 10.1159/000446724. [DOI] [PubMed] [Google Scholar]
- 21.Ramasamy R, Scovell JM, Kovac JR, Cook PJ, Lamb DJ, Lipshultz LI. Fluorescence in situ hybridization detects increased sperm aneuploidy in men with recurrent pregnancy loss. Fertil Steril. 2015;103:906–9.e1. doi: 10.1016/j.fertnstert.2015.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Christopikou D, Tsorva E, Economou K, Shelley P, Davies S, Mastrominas M, Handyside AH, et al. Polar body analysis by array comparative genomic hybridization accurately predicts aneuploidies of maternal meiotic origin in cleavage stage embryos of women of advanced maternal age. Hum Reprod. 2013;28:1426–1434. doi: 10.1093/humrep/det053. [DOI] [PubMed] [Google Scholar]
- 23.Ubaldi FM, Cimadomo D, Capalbo A, Vaiarelli A, Buffo L, Trabucco E, Ferrero S, Albani E, Rienzi L, Levi Setti PE. Preimplantation genetic diagnosis for aneuploidy testing in women older than 44 years: a multicenter experience. Fertil Steril. 2017;107:1173–1180. doi: 10.1016/j.fertnstert.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 24.Obradors A, Rius M, Daina G, Ramos L, Benet J, Navarro J. Whole-chromosome aneuploidy analysis in human oocytes: focus on comparative genomic hybridization. Cytogenet Genome Res. 2011;133:119–126. doi: 10.1159/000324233. [DOI] [PubMed] [Google Scholar]
- 25.Barbash-Hazan S, Frumkin T, Malcov M, Yaron Y, Cohen T, Azem F, Amit A, et al. Preimplantation aneuploid embryos undergo self-correction in correlation with their developmental potential. Fertil Steril. 2009;92:890–896. doi: 10.1016/j.fertnstert.2008.07.1761. [DOI] [PubMed] [Google Scholar]
- 26.Bazrgar M, Gourabi H, Valojerdi MR, Yazdi PE, Baharvand H. Self-correction of chromosomal abnormalities in human preimplantation embryos and embryonic stem cells. Stem Cells Dev. 2013;22:2449–2456. doi: 10.1089/scd.2013.0053. [DOI] [PubMed] [Google Scholar]
- 27.Bolton H, Graham SJL, Van der Aa N, Kumar P, Theunis K, Fernandez Gallardo E, et al. Mouse model of chromosome mosaicism reveals lineage-specific depletion of aneuploid cells and normal developmental potential. Nat Commun. 2016;7:11165. doi: 10.1038/ncomms11165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lagalla C, Tarozzi N, Sciajno R, Wells D, Di Santo M, Nadalini M, et al. Embryos with morphokinetic abnormalities may develop into euploid blastocysts. Reprod BioMed Online. 2017;34:137–146. doi: 10.1016/j.rbmo.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 29.Vassena R, Boué S, González-Roca E, Aran B, Auer H, Veiga A, Izpisua Belmonte JC, et al. Waves of early transcriptional activation and pluripotency program initiation during human preimplantation development. Development. 2011;138:3699–3709. doi: 10.1242/dev.064741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bielanska M, Tan S, Ao A. Chromosomal mosaicism throughout human preimplantation development in vitro: incidence, type, and relevance to embryo outcome. Hum Reprod. 2002;17:413–419. doi: 10.1093/humrep/17.2.413. [DOI] [PubMed] [Google Scholar]
- 31.Delhanty JD, Harper JC, Ao A, Handyside AH, Winston RM. Multicolour FISH detects frequent chromosomal mosaicism and chaotic division in normal preimplantation embryos from fertile patients. Hum Genet. 1997;99:755–760. doi: 10.1007/s004390050443. [DOI] [PubMed] [Google Scholar]
- 32.Gutiérrez-Mateo C, Colls P, Sánchez-García J, Escudero T, Prates R, Ketterson K, Wells D, et al. Validation of microarray comparative genomic hybridization for comprehensive chromosome analysis of embryos. Fertil Steril. 2011;95:953–958. doi: 10.1016/j.fertnstert.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 33.McCoy RC, Demko ZP, Ryan A, Banjevic M, Hill M, Sigurjonsson S, et al. Evidence of selection against complex mitotic-origin aneuploidy during preimplantation development. PLoS Genet. 2015;11:e1005601. doi: 10.1371/journal.pgen.1005601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fragouli E, Alfarawati S, Daphnis DD, Goodall NN, Mania A, Griffiths T, Gordon A, et al. Cytogenetic analysis of human blastocysts with the use of FISH, CGH and aCGH: scientific data and technical evaluation. Hum Reprod. 2011;26:480–490. doi: 10.1093/humrep/deq344. [DOI] [PubMed] [Google Scholar]
- 35.Fragouli E, Alfarawati S, Spath K, Babariya D, Tarozzi N, Borini A, Wells D. Analysis of implantation and ongoing pregnancy rates following the transfer of mosaic diploid-aneuploid blastocysts. Hum Genet. 2017;136:805–819. doi: 10.1007/s00439-017-1797-4. [DOI] [PubMed] [Google Scholar]
- 36.Taylor TH, Gitlin SA, Patrick JL, Crain JL, Wilson JM, Griffin DK. The origin, mechanisms, incidence and clinical consequences of chromosomal mosaicism in humans. Hum Reprod Update. 2014;20:571–581. doi: 10.1093/humupd/dmu016. [DOI] [PubMed] [Google Scholar]
- 37.Fragouli E, Alfarawati S, Spath K, Jaroudi S, Sarasa J, Enciso M, Wells D, et al. The origin and impact of embryonic aneuploidy. Hum Genet. 2013;132:1001–1013. doi: 10.1007/s00439-013-1309-0. [DOI] [PubMed] [Google Scholar]
- 38.Daphnis DD, Fragouli E, Economou K, Jerkovic S, Craft IL, Delhanty JDA, Harper JC, et al. Analysis of the evolution of chromosome abnormalities in human embryos from day 3 to 5 using CGH and FISH. Mol Hum Reprod. 2008;14:117–125. doi: 10.1093/molehr/gam087. [DOI] [PubMed] [Google Scholar]
- 39.Harrison RH, Kuo HC, Scriven PN, Handyside AH, Ogilvie CM. Lack of cell cycle checkpoints in human cleavage stage embryos revealed by a clonal pattern of chromosomal mosaicism analysed by sequential multicolour FISH. Zygote. 2008;8:217–224. doi: 10.1017/s0967199400001015. [DOI] [PubMed] [Google Scholar]
- 40.Kiessling AA, Bletsa R, Desmarais B, Mara C, Kallianidis K, Loutradis D. Evidence that human blastomere cleavage is under unique cell cycle control. J Assist Reprod Genet. 2009;26:187–195. doi: 10.1007/s10815-009-9306-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee A, Kiessling AA. Early human embryos are naturally aneuploid-can that be corrected? J Assist Reprod Genet. 2016;34:15–21. doi: 10.1007/s10815-016-0845-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 708 kb)