Abstract
HSV-1 encephalitis (HSE) is typically sporadic. Inborn errors of TLR3- and DBR1-mediated central nervous system (CNS) cell-intrinsic immunity can account for forebrain and brainstem HSE, respectively. We report five unrelated patients with forebrain HSE, each heterozygous for one of four rare variants of SNORA31, encoding a snoRNA of the H/ACA class that are predicted to direct the isomerization of uridine residues to pseudouridine in snRNA and rRNA. We show that CRISPR/Cas9-introduced biallelic and monoallelic SNORA31 deletions render human pluripotent stem cells (hPSCs)-derived cortical neurons susceptible to HSV-1. Accordingly, SNORA31-mutated patient hPSCs-derived cortical neurons are susceptible to HSV-1, like those from TLR3- or STAT1-deficient patients. Exogenous IFN-β renders SNORA31- and TLR3- but not STAT1-mutated neurons resistant to HSV-1. Finally, transcriptome analysis of the SNORA31-mutated neurons reveal normal responses to TLR3 and IFN-α/β stimulation, but abnormal responses to HSV-1. Human SNORA31 thus controls CNS neuron-intrinsic immunity to HSV-1 by a distinctive mechanism.
Introduction
In the course of primary infection, HSV-1 infects epithelial cells in the oral or nasal cavities, and spreads via the trigeminal nerves to the trigeminal ganglion, where it establishes latency. In about 1 in 10,000 individuals, HSV-1 invades the central nervous system (CNS) via the olfactory bulb, causing forebrain HSE (>95%) or, more rarely, via the trigeminal nerves, causing brainstem HSE (<5%) 1-4. Although rare, HSE is the most common sporadic, as opposed to epidemic, form of viral encephalitis in Western countries and, perhaps, worldwide. HSE typically strikes otherwise healthy individuals. It is almost always fatal if left untreated, and most acyclovir-treated survivors present severe neurological sequelae. The determinism of HSE remained a mystery 4, until our discovery of two genetic etiologies of “syndromic” HSE in children who also had mycobacterial disease (with mutations in NEMO and STAT1 disrupting IFN-α/β, -λ, and IFN-γ immunity) 5,6, and that of six genetic etiologies of “isolated” forebrain HSE (with mutations of TLR3, UNC93B1, TRIF, TRAF3, TBK1, and IRF3, encoding molecules governing the TLR3-dependent IFN-α/β and -λ pathway) 7-14. We also recently discovered the first genetic etiology of brainstem HSE: autosomal recessive (AR) partial DBR1 deficiency impairing RNA lariat metabolism and cell-intrinsic immunity to viruses 15.
Most forebrain HSE-predisposing genotypes display incomplete clinical penetrance for HSE and there are both recessive and dominant forms for two loci (TLR3, TRIF) 8,10,12,13. TLR3 is an endosomal receptor for dsRNA intermediates and by-products generated during viral infection 16,17. TLR3-mediated immunity is critical for mouse and human neuron-intrinsic defense against HSV-1 infection 18,19. IFN-α/β and IFN-λ are antiviral cytokines that activate and signal through the pathway mediated by type I (IFNAR1/IFNAR2) and III IFN receptors (IFNLR), both of which activate STAT1-containing complexes20. We showed that these mutations impaired TLR3-dependent IFN-α/β-mediated cell-intrinsic immunity to HSV-1 in human induced pluripotent stem cell (iPSC)-derived CNS cortical neurons and oligodendrocytes, whereas IFN-λ did not confer antiviral immunity in these conditions 19. In contrast, leukocytes did not require TLR3 to respond to dsRNA or HSV-1 8. These data accounted for the CNS-specific susceptibility of these patients to HSV-1, with a lack of virus dissemination to the bloodstream and other tissues in the course of HSE. However, the genetic basis of HSE remains unknown for most patients (~95%). We tested the hypothesis that some patients develop forebrain HSE due to novel inborn errors of CNS-intrinsic immunity, not necessarily related to the TLR3-IFN-α/β circuit.
Results
Heterozygous SNORA31 mutations in five unrelated HSE patients
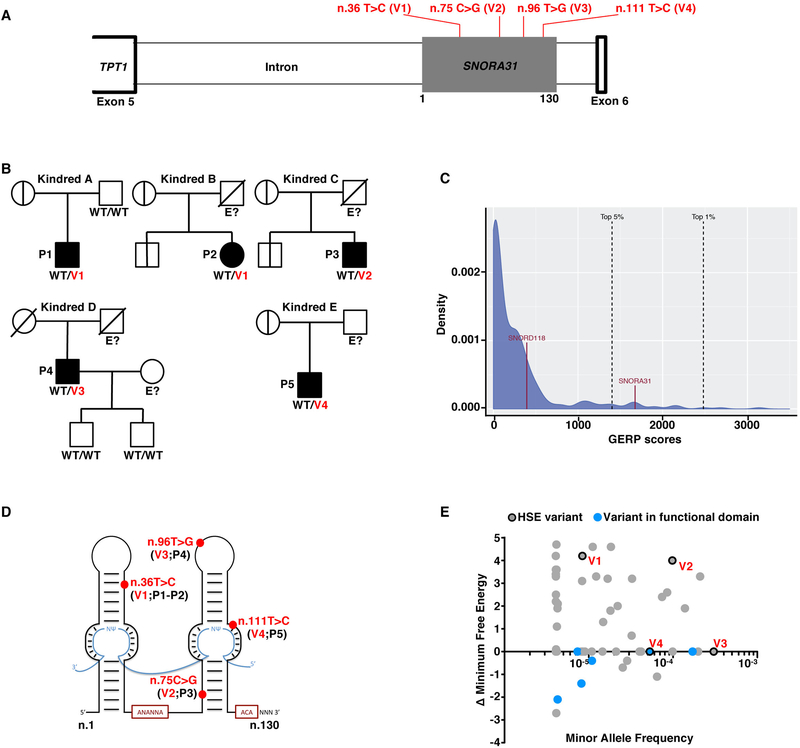
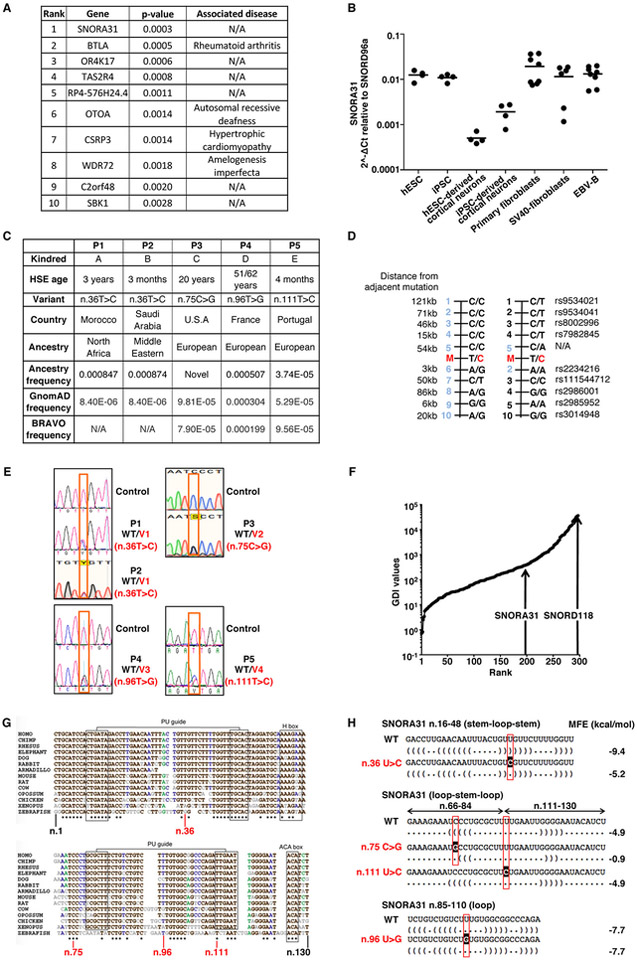
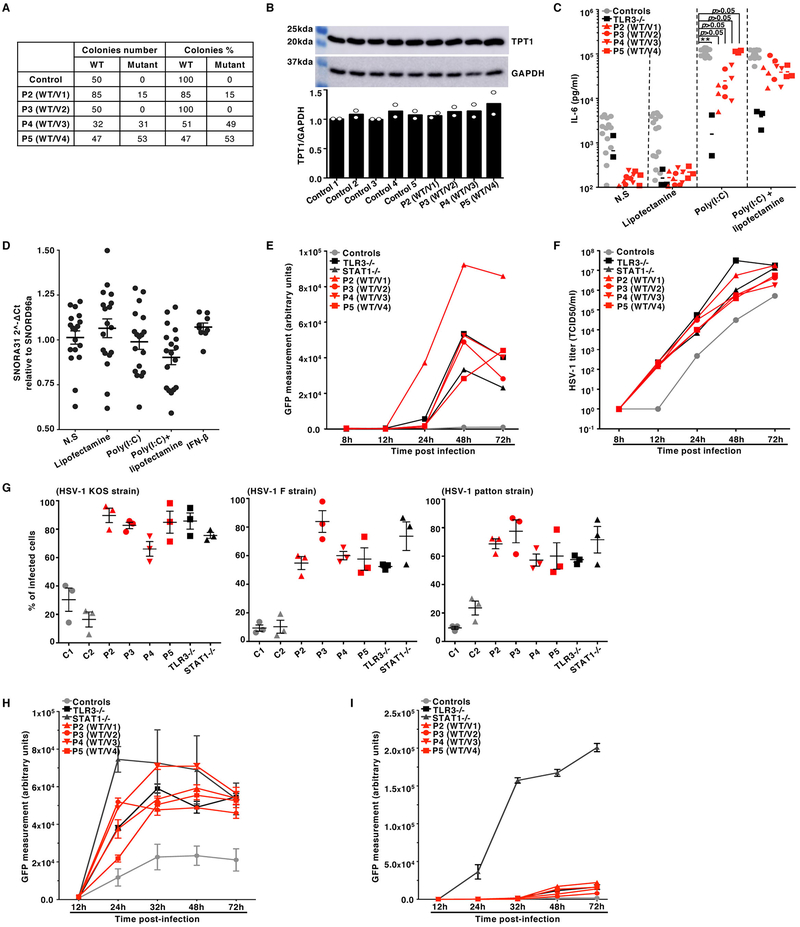
We analyzed the exomes of 205 unrelated HSE patients, testing a hypothesis of genetic homogeneity under an autosomal dominant (AD) model. We searched for genes with an enrichment of very rare heterozygous variants 21 in HSE patients relative to 2,756 individuals from other in-house cohorts of patients with non-viral infectious diseases and 1,511 individuals from the 1,000 genomes (1KG) project database 22. We considered variants with minor allele frequencies (MAF) < 0.001 in the ExAC database 23 that were predicted to be deleterious, as defined by a Combined Annotation Dependent Depletion (CADD) score 24 higher than the gene-specific mutation significance cutoff 25,26. This analysis revealed a small nucleolar RNA (snoRNA)-encoding gene, SNORA31, with the most significant variant enrichment in the HSE cohort (p-value: 0.00029; OR: 11.4; 95% CI: 3.5–32.9) (Extended Data Fig.1A). SnoRNA31 is a 130-nucleotide snoRNA of the H/ACA box class, which is ubiquitously expressed across various human cell types (27 and Extended Data Fig.1B). The only predicted function of snoRNA31 is that of a guide RNA directing the chemical modification of target uridine residues into pseudouridine in ribosomal RNA (rRNA) and small nuclear RNA (snRNA) 28. Five unrelated patients (P), each born to non-consanguineous parents (Suppl. Clinical Information), harbor one of four heterozygous single-nucleotide substitutions: n.36 T>C (v1, in P1, from Morocco, and P2, Saudi Arabia), n.75 C>G (v2, in P3, USA), n.96 T>G (v3, in P4, France), and n.111 T>C (v4, in P5, Portugal) (Fig.1A,B, Extended Data Fig.1C). Variant v1 is recurrent due to a mutational hotspot rather than a founder effect, as the haplotypes encompassing SNORA31 differ in the two patients (Extended Data Fig.1D). Each of the four variants has a MAF below 0.0009 in both the gnomAD and BRAVO databases, and in the corresponding ethnic groups of the patients (Extended Data Fig.1C). All variations were confirmed by Sanger sequencing, and their familial segregation showed incomplete clinical penetrance, as six healthy relatives were heterozygous, including four seropositive for antibodies against HSV-1 (Fig.1B, Extended Data Fig.1E, Suppl. Clinical Information). These findings suggested that heterozygous SNORA31 variants may be HSE-causing.
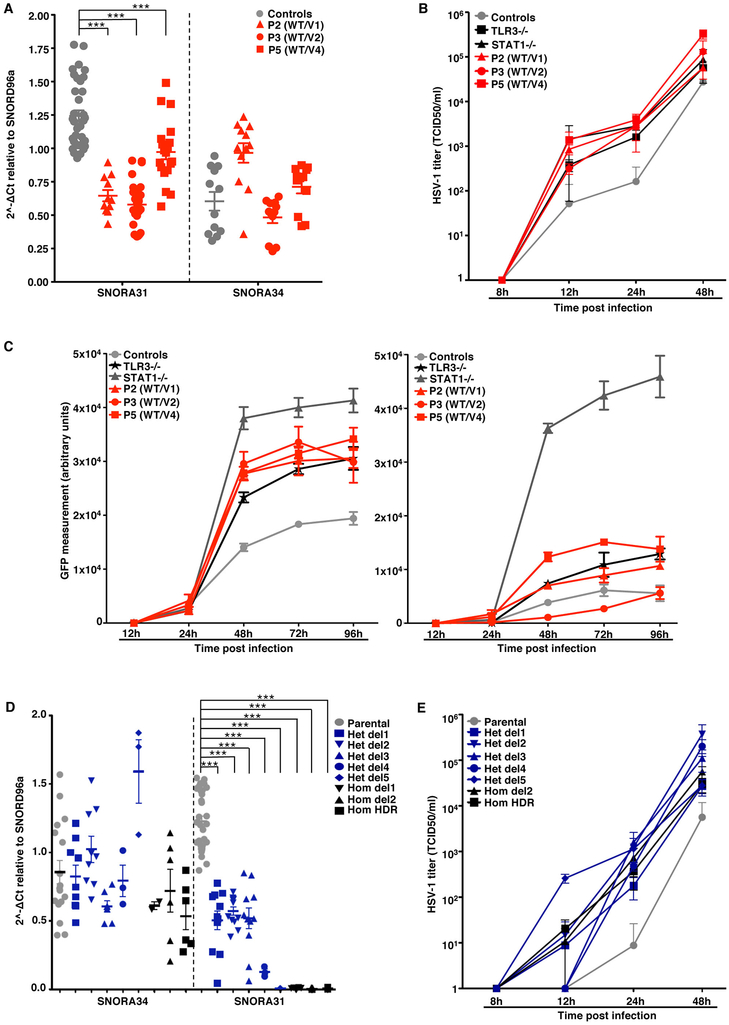
Figure 1. Heterozygous SNORA31 mutations in herpes simplex encephalitis patients from five unrelated kindreds.
A) Schematic representation of the genomic structure of human SNORA31. Human SNORA31 is located on chromosome 13, between exons 5 and 6 of the host gene TPT1. The mutations found in five HSE patients are shown in red: n.36T>C (V1) in patient 1 (P1) and P2; n.75C>G (V2) in P3; n.96T>G (V3) in P4; n.111T>C (V4) in P5. B) Family pedigrees with allele segregation in the five families. The patients, in black, are heterozygous for the following mutations (‘V’ in red): n.36T>C (V1) in kindreds A and B; n.75C>G (V2) in kindred C; n.96T>G (V3) in kindred D; n.111T>C (V4) in kindred E. Vertical bars indicate the same SNORA31 genotype as the patient from the corresponding family. “E?” indicates that the individual’s SNORA31 genotype is unknown. C) Conservation score ranking of the known human snoRNA genes, as assessed by the GERP++ method. Density (y-axis) of GERP scores (x-axis) for conserved elements overlapping snoRNAs. D) Schematic representation of the canonical secondary structure of H/ACA class snoRNAs, including snoRNA31. The positions of the patients’ SNORA31 variants are indicated in red. E) Frequency and predicted impact on the secondary structure of snoRNA31, as measured by the calculated change in minimum free energy of mutant sequences relative to wild type, for all variants found in gnomAD. All variants associated with a change in minimum free energy of more than 1 were considered possibly damaging.
Human SNORA31 is highly conserved in the general population
No computational approaches have ever been used to assess the degree of selective constraint operating on snoRNA-encoding genes 29. We initially adapted the gene damage index (GDI), which we previously introduced for protein-coding genes 30, to estimate the extent of structural variation and negative selection on the 327 snoRNA-coding genes, based on the 1KG database. Most snoRNA-coding genes, including SNORA31, displayed very low levels of genetic diversity (Extended Data Fig.1F). Like most known pathogenic genes 30, SNORA31 does not have a high GDI value. We then applied the GERP++ method, based on conservation between the human genome and the genomes of other mammalian species, to look for long continuous conserved elements (CEs) under negative selection 31. We observed that 70% of the snoRNAs intersected with CEs (a percentage close to that for exonic regions, 84.6% of which intersect with CEs). Remarkably, SNORA31 is entirely encompassed by a 764 bp segment under very strong negative selection. It is one of the 5% most strongly conserved snoRNAs (Fig.1C). We used SNORD118, encoding a C/D class snoRNA U8, mutations of which cause autosomal recessive (AR) susceptibility to cerebral microangiopathy leukoencephalopathy with calcifications and cysts (LCC) 32, as a control. This gene has the second highest GDI value of all snoRNAs and a GERP conservation score placing it among the 27% most conserved snoRNAs, consistent with AR inheritance for LCC. Two gene-level approaches therefore suggested that heterozygous variants of SNORA31 predicted to be deleterious might be detrimental to the host.
The patients’ SNORA31 variants are predicted to be deleterious
We then analyzed the HSE-relevant SNORA31 variants with variant-level approaches. The four variants affect nucleotides that are highly conserved in humans. Variant v1 (P1, P2) affects a nucleotide that is highly conserved among 14 vertebrate species (78% conservation), whereas v2 (P3) affects a nucleotide that is strictly conserved (100%). Both are located within the stems of the snoRNA secondary structure (Fig.1D, Extended Data Fig.1G). By contrast, v3 (P4) and v4 (P5) are located in loops, with v3 modifying a strongly conserved nucleotide (64%) in a domain of unknown significance, and v4 altering a strictly conserved (100%) nucleotide within a predicted functional domain. We calculated the change in the minimum free energy of the secondary structure of snoRNA31 for the four variants and for all the other 49 variants found in gnomAD (Fig.1E, Extended Data Fig.1H). Variants v1 and v2 were found to be among the 10 most destabilizing variants, whereas v3 and v4 were not predicted to have any significant effect on RNA structure 33. Overall, the four variants are predicted to be deleterious, further suggesting that these heterozygous SNORA31 variants in five unrelated families are HSE-causing.
Two SNORA31 variants from three patients impair snoRNA31 expression
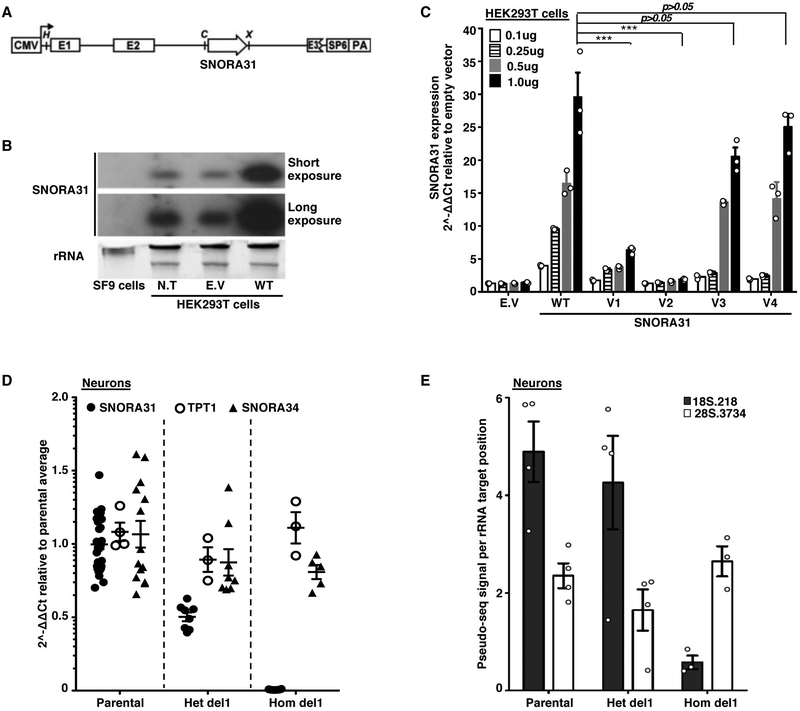
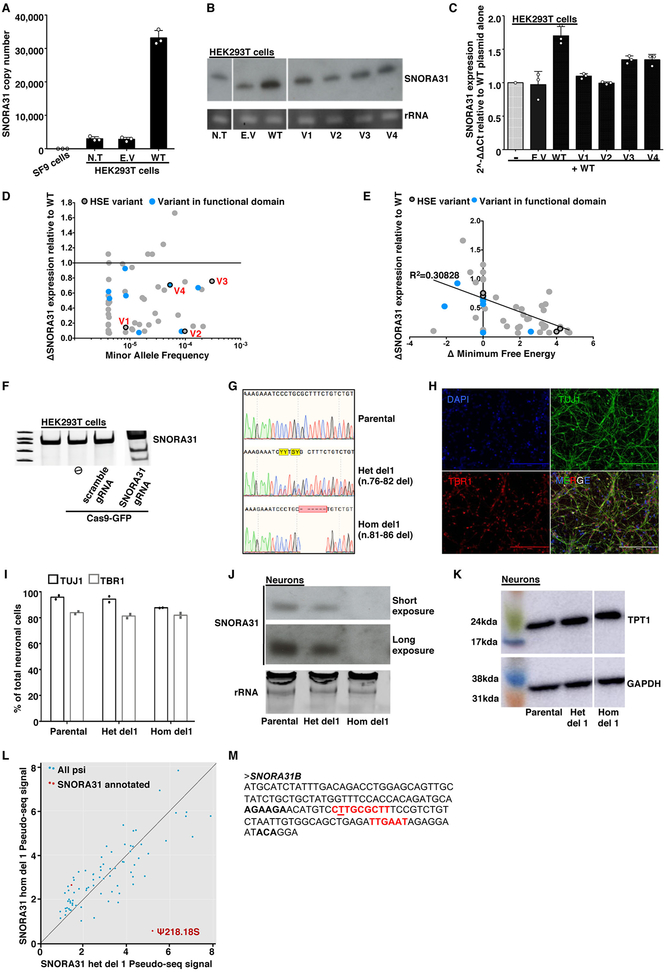
For experimental investigation of the SNORA31 variants, we first overexpressed them in vitro, using a snoRNA expression vector 34 containing wild-type (WT) or mutant SNORA31 sequences (Fig.2A). After transient transfection of HEK293T cells with various amounts of vectors, v1 and v2 displayed much lower levels of expression than the WT SNORA31, as shown by RT-qPCR and northern blotting (Fig.2B,C and Extended Data Fig.2A,B), consistent with in silico predictions (Fig.1E). By contrast, v3 and v4 were produced in normal amounts. When the WT and mutant alleles were co-expressed, no dominant-negative effect for any mutant was observed, as shown by snoRNA31 levels (Extended Data Fig.2C). However, this experiment did not exclude the possibility of heterozygous SNORA31 mutations underlying AD snoRNA31 deficiency through a negative dominance mechanism affecting the function of WT snoRNA31. Our data indicated that two of the four variants (v1, v2) are deleterious by disrupting SNORA31 expression. We then overexpressed the other 49 SNORA31 variants from the gnomAD database. We found an overall correlation between the structural stability of the variant predicted in silico and the corresponding level of expression of the mutant allele in vitro (Extended Data Fig.2D,E). Only 16 of the 49 variants were as disruptive, and their individual MAFs were as low as 4.063E-06 in gnomAD. All known variants disrupting the expression of snoRNA31 (including v1 and v2) have a collective maximum cumulative MAF of 0.00069 in the gnomAD database. These findings are consistent with the estimated prevalence of HSE of 1–2 per 10,0007-14.
Figure 2. Impaired expression of two SNORA31 variants found in three HSE patients and the loss of a ribosomal modification in SNORA31-null hESC-derived CNS neurons.
A) Schematic diagram of the snoRNA expression vector, into which the wild-type (WT) or mutant sequences of SNORA31 were inserted. Upon expression of the beta-globulin transgene, the immature SNORA31 RNA is spliced at the E2-E3 intron and then processed by the cell machinery into a mature snoRNA molecule. B) Expression of WT snoRNA31 in HEK293T cells, with and without transient transfection with the WT SNORA31 plasmid, as assessed by northern blotting. Insect SF9 cells, which do not express a homolog of SNORA31, were used as a negative control in this experiment. N.T: not transfected. E.V: empty vector. The data are representative of n=2 independent experiments. C) RT-qPCR measurement of snoRNA31 expression in HEK293T cells transfected with the indicated amounts of WT or HSE patient-specific SNORA31 mutant plasmids. The data are expressed relative to SNORD96a expression, with normalization against empty vector transfection. Means and standard deviations from n=3 independent experiments are shown. Mean values of SNORA31 expression levels upon the same amount of WT or mutant plasmid transfection were compared in one-way ANOVA (F=47.51, total df=17) followed by Dunnett’s multiple comparison tests, and the results are shown for the highest amount plasmid transfection group. ***, p<0.001. D) RT-qPCR measurement of relative snoRNA31 expression levels, TPT1 mRNA, and an unrelated snoRNA, SNORA34, in isogenic hESC-derived CNS neurons carrying either a heterozygous deletion (het del1) or a homozygous deletion (hom del1) in SNORA31, or the WT allele (parental). For SNORA31 and SNORA34, data are expressed relative to SNORD96a and normalized relative to the mean value for control cells. For TPT1, the data are expressed relative to GUS mRNA, with normalization relative to the mean value for control cells. The mean and standard deviation from n=3 independent experiments are shown. Each point represents one biological replicate from an independendent experiment, with n=2 (for hom del1), n=3 (het del 1) or n=10 (parental) biological replicates per genotype tested for snoRNA31 expression per experiment. E) Quantification of the pseudo-seq signal at two different rRNA sites in isogenic hESC-derived CNS neurons, either WT for SNORA31 or carrying a heterozygous (het del 1) or homozygous (hom del 1) deletion in SNORA31. Means and standard deviations for n=4 libraries each for the parental and het del1 lines and n=3 libraries for the hom del1 line are shown.
SNORA31 deletion does not impair CNS neuron differentiation from stem cells
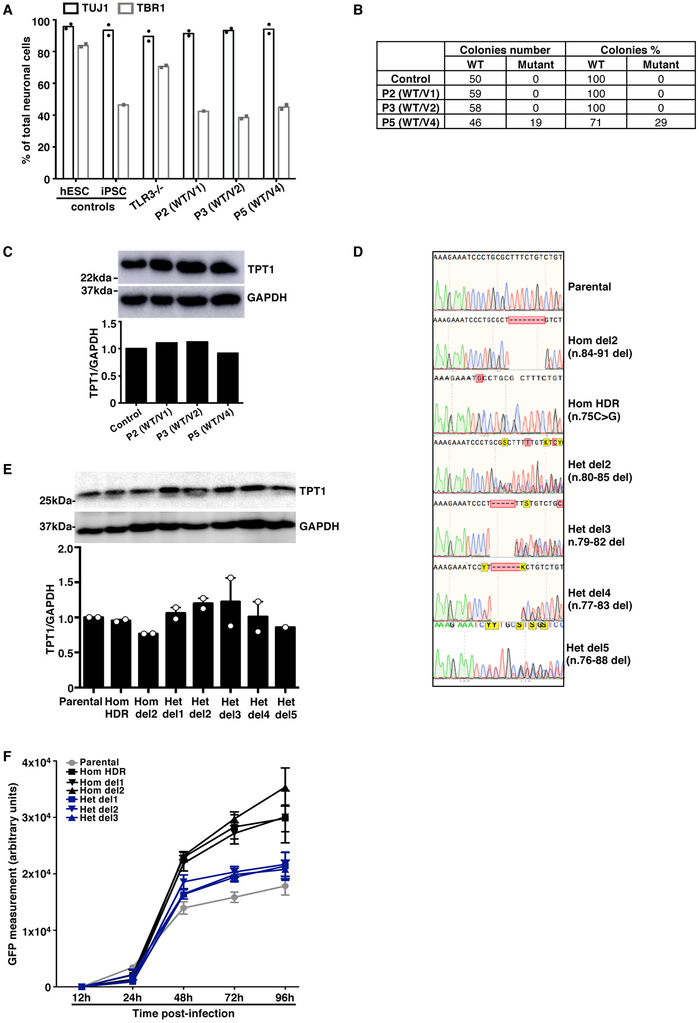
We then investigated the functionality of human SNORA31 in vitro in stem cell-derived CNS cortical neurons, a cell type affected by inborn errors of TLR3 immunity underlying HSE 19. SnoRNA31 is predicted to direct reactions modifying two rRNA species 28, specifically at uridine residues in position 218 of the 18S rRNA and position 3,713 of the 28S rRNA. The precise functions of individual rRNA pseudouridine residues have yet to be elucidated, but the pseudouridylation of rRNAs is thought to be involved in ribosome maturation and function 35, possibly via the stabilization of RNA-RNA interactions 36. We used CRISPR-Cas9 37 to generate isogenic human embryonic stem cell (hESC) lines harboring heterozygous or homozygous deletions in SNORA31 (Extended Data Fig.2F,G). We then generated highly pure populations of CNS neurons positive for TBR1, a marker of cortical neuronal cells (Extended Data Fig.2H,I) 38,39. As SNORA31 is located in the 5th intron of TPT1, we analyzed the expression of both genes. Neurons with a heterozygous deletion (het del1, n.76–82) had 50% lower levels of SNORA31 expression, whereas cells with a homozygous deletion (hom del1, n.81–86) displayed a complete loss of expression (Fig.2D, Extended Data Fig.2J). By contrast, TPT1 mRNA and protein levels were normal in both cell lines (Fig.2D, Extended Data Fig.2K). The expression of another H/ACA class snoRNA, SNORA34, used as a control, was similar to that in the parental line, in all isogenic lines (Fig.2D). Thus, SNORA31 is not required for the maintenance of hESCs or their differentiation into cortical neurons.
SNORA31 deletion impairs pseudouridylation of the ribosomal 18S RNA U218
We performed pseudouridine profiling and sequencing, pseudo-seq 40, on rRNA obtained from CNS cortical neurons derived from parental, heterozygous, and homozygous SNORA31-deletion isogenic hESC lines. Reproducible pseudo-seq signals were obtained for 75 of the 93 annotated pseudouridine sites (Extended Data Fig.2L), including the two predicted snoRNA31 targets, U218 in 18S rRNA and U3713 in 28S rRNA. The pseudouridylation signal at the U218 site was almost abolished in homozygous SNORA31-deletion cells (Fig.2E). However, heterozygous deletion had no significant effect on rRNA pseudouridylation. Unexpectedly, the U3713 site of 28S rRNA was unaffected, even in cells with a homozygous deletion. Inspection of an updated human snoRNA database identified a second H/ACA snoRNA gene, SNORA31B, with a guide sequence complementary to U3713 but not U218 (Extended Data Fig.2M), suggesting that SNORA31B might instead drive modification of the U3713 site in CNS neurons. Our results thus directly demonstrate a role for snoRNA31 in driving pseudouridylation in human CNS cortical neurons, at least for residue 218 of 18S rRNA. SNORA31 mutations are therefore plausible candidates for a role in HSE pathogenesis, which may or may not involve the impaired pseudouridylation of U218.
Impaired expression of snoRNA31 in patient fibroblasts heterozygous for v1 or v2
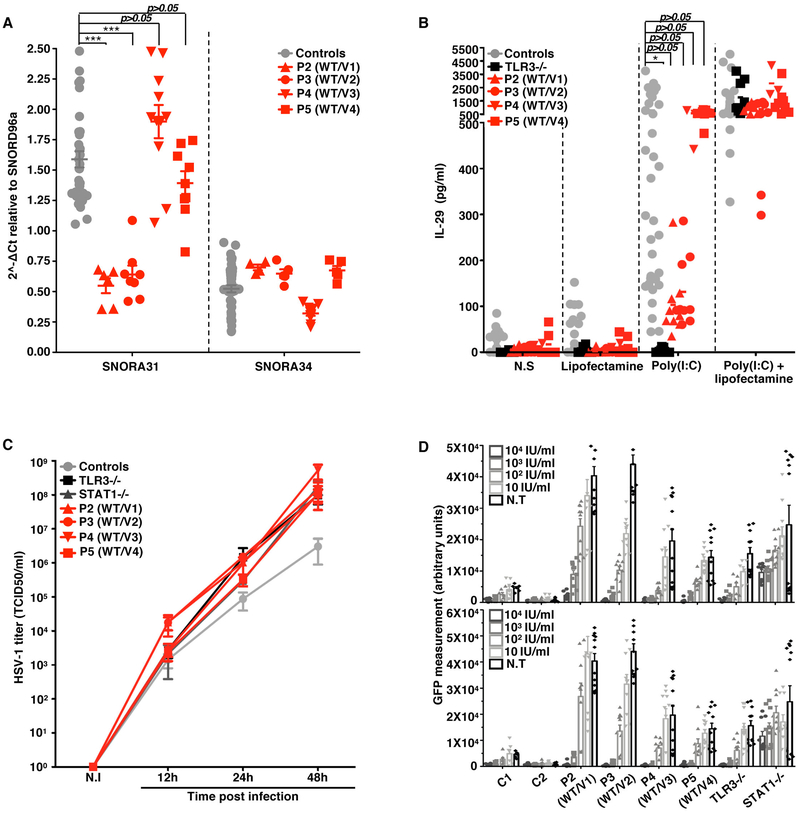
We analyzed the cellular impact of the heterozygous SNORA31 mutations found in patients. Dermal fibroblasts are appropriate surrogate cells for studies of the cellular basis of inborn errors of TLR3 underlying HSE 7-13. We measured the expression of snoRNA31, and of snoRNA34 as a control, in fibroblasts transformed with simian virus 40 (SV40) T antigen (SV40-fibroblasts) from P2, P3, P4, and P5, and healthy control cells. Cells from P2 and P3, heterozygous for variants weakly expressed in vitro (Fig.2C), produced no more than half the normal amount of snoRNA31, but normal amounts of snoRNA34 (Fig.3A). Cells from P4 and P5, heterozygous for variants normally expressed in vitro had normal levels of snoRNA31. Moreover, cDNA sequencing showed that most sequenced SNORA31 clones in P2 and P3 cells (85% and 100%, respectively) did not harbor the corresponding variant, whereas about half the cDNAs from P4 and P5 carried the patient’s variant (Extended Data Fig.3A). Finally, TPT1 protein levels were normal in the four patients’ cells (Extended Data Fig.3B). Thus, the expression of the four variants in heterozygous cells from the patients was consistent with individual data for overexpression in HEK293T cells (Fig.2C). Moreover, the normal expression of the WT allele confirmed that snoRNA31 deficiency was AD, and that heterozygous mutations probably underlie AD snoRNA31 deficiency by haploinsufficiency, at least in the three patients heterozygous for the loss-of-expression alleles v1 and v2 (P1–3).
Figure 3. Cellular phenotypes of the SV40-fibroblasts of patients with SNORA31 mutations.
A) The expression of SNORA31, as measured by RT-qPCR in SV40-fibroblasts from patients (red dots) and four controls (grey dots). The data are expressed relative to SNORD96a expression. Means and standard deviations from three independent experiments are shown. Each point represents one biological replicate from an independent experiment, with n=2 (for P2) or n=3 (all other cell lines) biological replicates per cell line tested per experiment. Mean values were compared between control cells and cells from the patients, in one-way ANOVA (F=26.01, total df=65) followed by Dunnett’s multiple comparison tests. *** 0.0001<p<0.001. B) IL-29 production in SV40-fibroblasts from patients (P2-P5, red dots), a TLR3−/− control (black dots), and four healthy controls (grey dots), as measured by ELISA, in the supernatant of control and patient SV40-fibroblasts stimulated with 25 ng/mL poly(I:C) alone, Lipofectamine alone, or both. N.S: not stimulated. N=3 independent experiments were performed, with n=1 (for P4) or n=3 (all other cell lines) biological replicates per cell line tested per experiment. Means and standard deviations from three independent experiments are shown. Mean values were compared between control cells and cells from the patients, in one-way ANOVA (F=3.254, total df=84) followed by Dunnett’s multiple comparison tests. * 0.01<p<0.05. C) HSV-1 propagation in SV40-fibroblasts from patients (P2-P5), an autosomal recessive (AR) complete TLR3-deficient (TLR3−/−) and an AR complete STAT1-deficient (STAT1−/−) patient as susceptible controls, and two healthy controls. HSV-1 titers were determined at the indicated time points post infection, at a multiplicity of infection (MOI) of 0.001, as quantified by calculating the 50% end point (TCID50) in Vero cells. N.I: not infected. Means and standard deviations from n=3 independent experiments are shown. D) HSV-1 abundance was determined by assessing GFP-capsid expression in patient (P2-P5) and control (C1 and C2) SV40-fibroblasts 24 h post-infection at a MOI of 1. Cells were treated with the indicated doses of IFN-α2b (top) or IFN-β (bottom) for 16 hours before infection (N.T: not pretreated). STAT1−/− cells were used as a control displaying no response to IFN-β in this assay. N=3 independent experiments were performed, with n=3 biological replicates tested per condition per experiment. Means values from n=3 independent experiments are shown with standard deviations.
Impaired cell-intrinsic immunity to HSV-1 in the patients’ fibroblasts
We then tested the responses of the patients’ fibroblasts to TLR3 and IFN-α/β receptor stimulation, and their control of HSV-1. Upon stimulation with the dsRNA mimic and TLR3 agonist polyinosinic-polycytidylic acid (poly(I:C)), SV40-fibroblasts from P2, P3, P4, and P5 produced normal amounts of IL-29 (IFN-λ1) and IL-6, unlike TLR3 pathway-deficient fibroblasts (Fig.3B, Extended Data Fig.3C). Moreover, snoRNA31 expression was unaltered by stimulation with extracellular or cytoplasmic poly(I:C), activating the TLR3 16 and MAVS pathways 41, respectively, or by IFN-β stimulation, which activates the IFNAR1/IFNAR2 pathway (Extended Data Fig.3D). We also found that SV40-fibroblasts from the four patients tested were highly susceptible to HSV-1 infection, as HSV-1 (KOS strain) replicated to levels similar to or higher than those in cells from patients with AR complete TLR3 or STAT1 deficiency, as demonstrated by the titration of infectious HSV-1 by the TCID50 method on Vero cells (Fig.3C), and the levels of GFP expression (Extended Data Fig.3E), which correlated with the titers of the infectious virus (Extended Data Fig.3F), following infection with a GFP-expressing HSV-1. Higher numbers of HSV-1-positive cells were observed in SNORA31-mutated, TLR3-, and STAT1-deficient fibroblasts, than in control cells, upon infection with different strains of HSV-1 (KOS, F, and Patton) (Extended Data Fig.3G). Furthermore, this HSV-1 susceptibility phenotype was rescued by pretreatment with IFN-α2b or -β in a dose-dependent manner in the patients’ cells, as in TLR3-deficient cells but not STAT1-deficient cells (Fig.3D, Extended Data Fig.3H,I). These data further suggested that AD snoRNA31 deficiency underlies HSE by a novel mechanism, different than that of TLR3 and STAT1 deficiencies.
Normal susceptibility of the patients’ iPSC and B cells to HSV-1
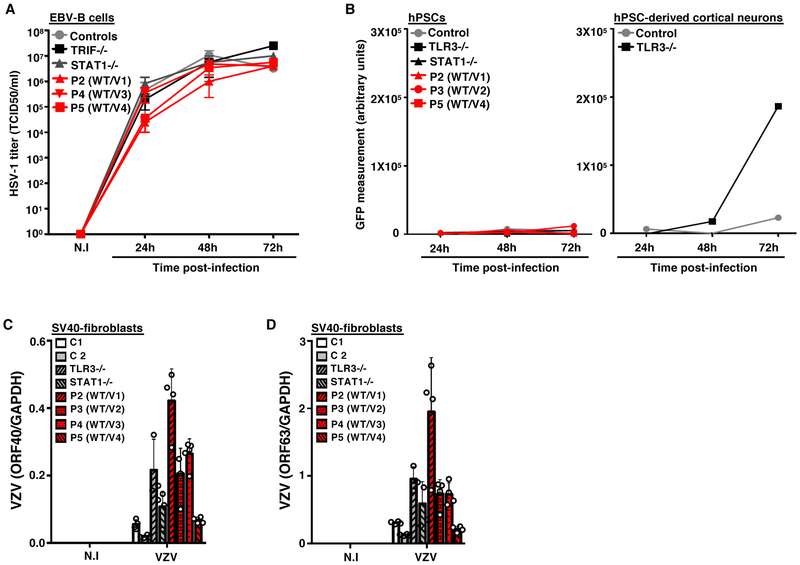
Previous studies of HSE-causing mutations in the TLR3 pathway suggested that impaired TLR3-dependent IFN-α/β-mediated cell-intrinsic immunity to HSV-1 was critical in CNS cortical neurons and oligodendrocytes but redundant in leukocytes 8,19,42. We tested whether snoRNA31 was also essential for antiviral immunity in cell types other than fibroblasts and CNS cells, by studying Epstein Barr virus (EBV)-transformed B (EBV-B) cells and iPSCs. In P2, P4, and P5 EBV-B cells, HSV-1 replication levels were similar to those observed in control cells, an AR TRIF-deficient HSE patient and an AR STAT1-deficient patient (Extended Data Fig.4A). Human iPSCs were generally more resistant to HSV-1 infection than iPSC-derived cortical neurons, and no increase in HSV-1 susceptibility was observed in SNORA31-mutated iPSCs from P3 and P5, or in TLR3- or STAT1-deficient iPSCs, whereas TLR3-deficient iPSC-derived cortical neurons were highly susceptible to HSV-1 infection, as previously reported (Extended Data Fig.4B). Therefore, snoRNA31 haploinsufficiency does not seem to impair anti-HSV-1 immunity in EBV-B cells and iPSCs, suggesting that not all human cell types require the integrity of snoRNA31 expression and function to control HSV-1.
Enhanced susceptibility of the patients’ fibroblasts to other neurotropic viruses
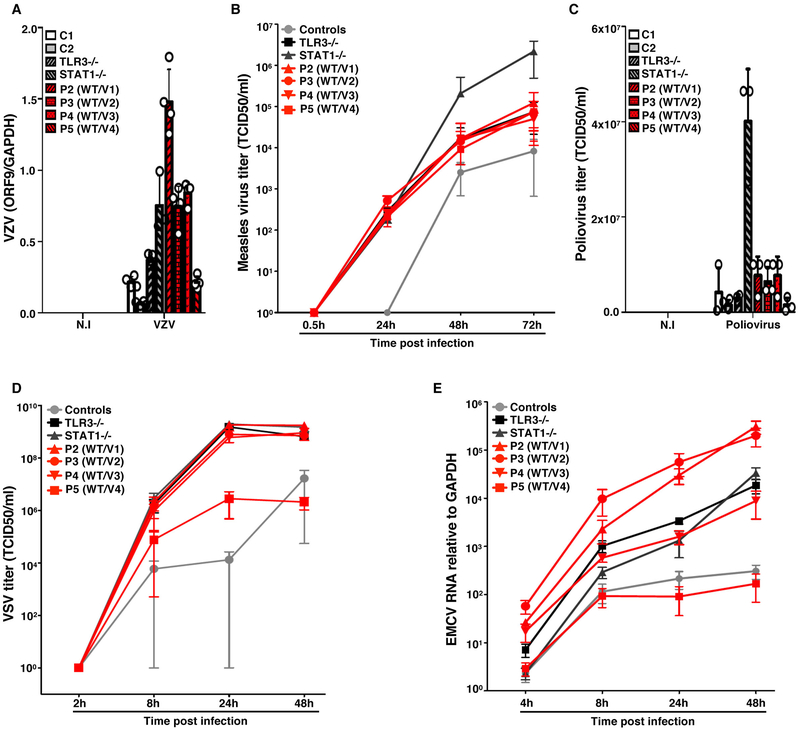
We then used SV40-fibroblasts, a surrogate cell type displaying snoRNA31-dependent anti-HSV-1 immunity, to assess the role of snoRNA31 in cell-intrinsic immunity to other neurotropic viruses, including varicella zoster virus (VZV), measles virus (MeV), poliovirus, vesicular stomatitis virus (VSV) and encephalomyocarditis virus (EMCV). With the exception of poliovirus, these viruses replicated to higher levels in SNORA31-mutated patient SV40-fibroblasts than in healthy control cells, as also observed for TLR3- or STAT1-deficient cells (Fig.4A-E, Extended Data Fig.4C,D). SV40-fibroblasts from P5 displayed levels of viral replication similar to those in cells from the other SNORA31-mutated patients (HSV-1, MeV), intermediate between controls and patients (VSV), or similar to those in cells from controls (VZV, EMCV), suggesting that the P5 SNORA31 mutant (v4) may be the least deleterious of the four variants considered in fibroblasts, or that v4 may have the strongest HSV-1-specific impact on viral growth. SnoRNA31 may be a cell-intrinsic antiviral factor active against a broad range of neurotropic viruses, at least in fibroblasts, although it is unknown whether heterozygosity for deleterious SNORA31 variants underlies a broader viral clinical phenotype than HSE.
Figure 4. Transcription and propagation of various viruses in the SV40-fibroblasts of patients with SNORA31 mutations.
A) Levels of VZV ORF9 transcript, as determined by RT-qPCR, on SV-40 fibroblasts from patients (P2-P5, red bars with black shadings), a TLR3−/− and a STAT1−/− patient (grey bars with black shadings), and healthy controls (C1 and C2, white and grey bars), 48 h after exposure to VZV-infected MeWo cells (VZV) or MeWo cells that were not infected (N.I). The data are expressed relative to GAPDH expression. Means and standard deviations from n=4 independent experiments are shown. N=1 biological replicate was tested per condition per experiment. B-D) Propagation of measles virus (B), poliovirus (C) and VSV (D), in SV40-fibroblasts from patients and two healthy controls at the indicated times post infection with measles virus at a MOI of 0.5, poliovirus at a MOI of 1, or VSV at a MOI of 1, as assessed by the TCID50 virus titration method. Means and standard deviations from n=3 independent experiments are shown. N=1 biological replicate was tested per condition per experiment. E) Expression levels of the 3D region of the EMCV genome, as measured by RT-qPCR in SV40-fibroblasts from patients (P2-P5), a TLR3−/− and a STAT1−/− patient as susceptible controls, and healthy controls, at the indicated times post infection with EMCV at a MOI of 0.1. The data are expressed relative to GAPDH expression. Means and standard deviations from n=3 independent experiments are shown. N=2 biological replicates were tested per condition per experiment.
Enhanced susceptibility to HSV-1 in the patients’ iPSC-derived CNS neurons
We reprogrammed the patients’ primary fibroblasts to obtain iPSCs, which were then differentiated into cortical CNS neurons 38,39 (Extended Data Fig.5A). The relative levels of snoRNA31 expression in P2 and P3 neurons were similar to those in SV40-fibroblasts, at about 50% normal levels (Fig.5A). P5 neurons had significantly lower levels of snoRNA31 expression than healthy controls, suggesting a potential cell type-specific impact of v4 on the stability of its product. Sequencing of individual cDNAs in neurons from P2 and P3 showed that none of these neurons carried v1 or v2, respectively (Extended Data Fig.5B). The lower levels of snoRNA31 in P5 cells were reflected in the composition of cDNAs, only a third of which carried the v4 variant. SnoRNA34 and TPT1 levels were normal in the neurons of all patients (Extended Data Fig.5C). Moreover, unlike healthy control cortical neurons, neurons from all three patients with SNORA31 mutations tested were susceptible to HSV-1 (KOS strain), like TLR3- and STAT1-deficient neurons (Fig.5B). Finally, this phenotype was rescued by pretreatment with IFN-β in SNORA31-mutated and TLR3-deficient neurons, but not in STAT1-deficient neurons (Fig.5C). IPSC-derived cortical neurons from the SNORA31-mutated patients were therefore, susceptible to HSV-1 infection, providing a plausible cellular mechanism of disease.
Figure 5. Cellular phenotypes in CNS neurons derived from patient iPSCs and isogenic hESCs.
A) Expression of SNORA31 and SNORA34, as measured by RT-qPCR in hPSC-derived CNS neurons for patients (P2, P3, P5) and controls. Four healthy control lines were used in the experiments: two control hESC lines and two control iPSC lines. Means and standard deviations from three independent experiments are shown. Each point represents one biological replicate, with n=3 (control lines) n=4 (P2), n=8 (P3) or n=6 (P5) biological replicates per cell line tested for snoRNA31 expression per experiment. Mean values were compared for SNORA31 expression between control cells and cells from the patients, in one-way ANOVA (F=50.84, total df=89)) followed by Dunnett’s multiple comparison tests. ***, p<0.001. B) Quantification of HSV-1 in CNS neurons derived from control and patient hPSCs, at the indicated times post infection at a MOI of 0.001, as quantified by the TCID50 virus titration method. TLR3−/− and STAT1−/− cells, which are highly susceptible to HSV-1, were used as controls. Means and standard deviations from n=3 independent experiments are shown. N=1 biological replicate was tested per condition per experiment. C) HSV-1 abundance was assessed by measuring GFP-capsid expression in iPSC-derived CNS neurons from patients (P2, P3, P5), a TLR3−/− patient and a STAT1−/− patient, and four healthy control lines. Measurements were made at the indicated times post infection with a MOI of 1, without (left) or with (right) IFN-β pretreatment for 16 hours. Means and standard deviations from n=3 independent experiments, with n=3 biological replicates for each set of conditions in each experiment, are shown. D) SNORA31 and SNORA34 expression in CNS neurons derived from gene-edited isogenic hESCs (blue and black dots) and the parental line hESCs (grey dots). Means and standard deviations from n=3 independent experiments are shown. Each point represents one biological replicate, with n=10 (for parental line), n=1 (het del4, het del5), n=2 (hom del1), n=3 (hom del2) or n=4 (het del1, het del2, het del3, hom HDR) biological replicates per genotype tested per experiment. Mean values were compared for SNORA31 expression between SNORA31-mutated and parental cells, in one-way ANOVA (F=106.6, total df=101) followed by Dunnett’s multiple comparison tests. ***, p<0.001. E) Quantification of HSV-1 in CNS neurons derived from isogenic hESCs, either WT (parental), or carrying a homozygous HDR-introduced patient-specific point mutation (hom HDR), a homozygous deletion (hom del2), or various heterozygous mutations (het del1, het del2, het del3, het del4, het del5) in SNORA31. HSV-1 levels were quantified at the indicated times post infection at a MOI of 0.001. HSV-1titers were determined by the TCID50 method. For the hom HDR, hom del2, het del1, het del2 and het del3 lines, means and standard deviations from n=3 independent experiments, with n=1 biological replicate in one experiment and n=2 biological replicates in two experiments for each set of conditions, are shown. For the het del4 and het del5 lines, the mean and standard deviation for n=3 biological replicates from n=4 independent experiments are shown.
Enhanced susceptibility to HSV-1 in hESC-derived CNS neurons harboring CRISPR/Cas9-introduced SNORA31 deletion
We used CRISPR/Cas9 to generate novel isogenic hESC lines carrying alterations of the SNORA31 gene and studying the cortical neurons derived from multiple isogenic hESC lines. In addition to het del1 (n.76–82 del, guide 1) and hom del1 (n.81–86 del, guide 1) (Extended Data Fig.2G), we generated four new lines with heterozygous deletions at the SNORA31 locus: het del2 (n.77–85 del, guide 1), het del3 (n.79–82 del, guide 2), het del4 (n.77–83 del, guide2), het del5 (n.76–88 del, guide2). We also generated one new line with a homozygous deletion at this locus, hom del2 (n.84–91 del, guide 2). Furthermore, we introduced v2 into cells in the homozygous state by homology-directed repair (HDR) (Extended Data Fig.5D). We then differentiated these isogenic lines into cortical neurons and measured snoRNA31 expression and HSV-1 susceptibility. As previously shown for hom del1, SNORA31 expression was abolished in hom del1 and del2 neurons, whereas neurons with any of the other heterozygous deletions in SNORA31 contained half as much snoRNA31 as the parental line (het del1–4) or displayed almost no SNORA31 expression (het del5). Isogenic neurons harboring homozygous v2 also displayed a complete loss of snoRNA31 expression (Fig.5D). Editing of the SNORA31 gene had no effect on snoRNA34 expression or on TPT1 mRNA or protein levels (Fig.2D, Extended Data Fig.2K, S5E). Finally, neurons homozygous for v2 or harboring hom del1–2, het del1–5, all had similar HSV-1 levels, higher than those in parental cells (Fig.5E, Extended Data Fig.5F). Thus, low levels of snoRNA31 expression underlie enhanced susceptibility to HSV-1 in isogenic cortical neurons heterozygous or homozygous for deleterious variants of SNORA31. The expression of two copies of WT SNORA31 is required to ensure CNS neuron-intrinsic immunity to HSV-1.
Normal TLR3 and IFNAR1/IFNAR2 responses in SNORA31-mutated hPSC-derived cortical neurons
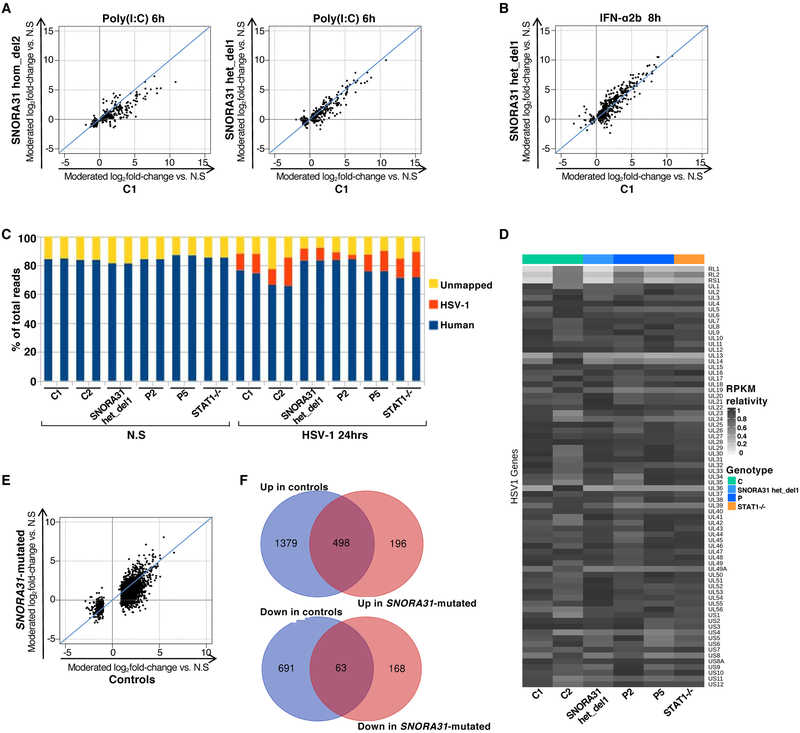
Given the structure of snoRNA31, it might serve as a cell-endogenous source of dsRNA regulating TLR3 activity. Alternatively, as in other human genetic disorders affecting rRNA pseudouridylation 43, snoRNA31 deficiency may impair the translation of certain cellular mRNAs encoding molecules crucial for the integrity of TLR3 or IFNAR1/IFNAR2 pathway signaling. We performed a comprehensive assessment of TLR3 and IFNAR1/IFNAR2 responses, using RNA-Seq to measure gene expression in SNORA31-mutated hPSC-derived cortical neurons from P2, P5, from isogenic hPSC lines with or without CRISPR/Cas9-introduced SNORA31 deletions (hom del2, het del1), and other two control hPSC lines, with and without stimulation with poly(I:C) or IFN-α2b. The stimulation of patient-specific SNORA31-mutated cortical neurons with poly(I:C) or IFN-α2b induced changes in gene expression similar to those observed in healthy control cells (Fig.6A-C). Similar results were obtained with isogenic hPSC-derived neurons, with and without SNORA31 deletions (Fig.6A,B, Extended Data Fig.6A,B). Furthermore, gene ontology enrichment testing revealed no obvious difference. TLR3- or STAT1-deficient cells served as negative controls for poly(I:C) and IFN-α2b responses, respectively (Fig.6A,B). These data suggest that the activation of TLR3- and IFNAR1/IFNAR2-mediated signaling pathways remains intact in SNORA31-mutated cortical neurons. The mechanism of forebrain HSE in patients with SNORA31 mutations is therefore unlikely to involve a defect in the activation of these pathways.
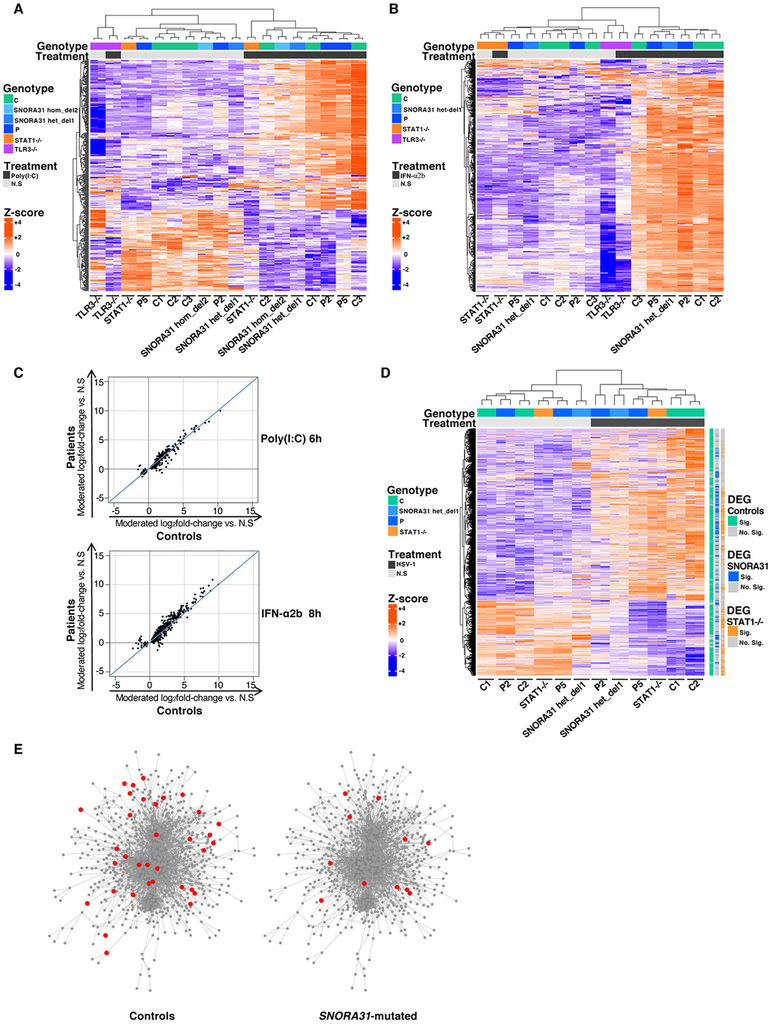
Figure 6. Transcriptome responses to stimulations with poly(I:C), IFN-α2b or HSV-1 in SNORA31-mutated hPSC-derived cortical neurons.
A, B) Heatmaps of RNA-Seq-quantified gene expression (z-score scaled log2 read counts per million, cpm) in hPSC-derived CNS cortical neurons from healthy controls (C1: a H9 hESC control line, C2 and C3: two iPSC control lines), SNORA31-mutated patients (P2 and P5), isogenic SNORA31-mutated hESC lines (SNORA31 hom del2, het del 1), a TLR3−/− and a STAT1−/− patient, not stimulated (N.S), stimulated with 25 μg/ml poly(I:C) for 6 hours (A), or stimulated with 100 IU/ml IFN-α2b for 8 hours (B). Each RNA-Seq library was sequenced twice. Each heatmap includes genes differentially expressed (FDR<=0.05) in response to the indicated stimulus relative to N.S samples in the healthy control or SNORA31-mutated groups. C) Scatter plots of average log2 fold-changes in RNA-Seq-quantified gene expression following stimulation with 25 μg/ml poly(I:C) for 6 hours (upper panel) or 100 IU/ml IFN-α2b for 8 hours (lower panel), in hPSC-derived cortical neurons from two SNORA31-mutated patients (P2 and P5) versus three healthy controls (C1, C2 and C3). Each point represents a single gene. D) Heatmaps of RNA-Seq-quantified gene expression (z-score scaled log2 read counts per million, cpm) in hPSC-derived cortical neurons from healthy controls (C1 and C2), SNORA31-mutated patients (P2 and P5), an isogenic SNORA31-mutated hESC line (SNORA31 het del 1), and a STAT1−/− patient, not stimulated (N.S), or infected with HSV-1 for 24 hours at a MOI of 1. The heatmap includes genes differentially expressed (FDR<=0.05, >2-fold difference) in response to HSV-1 relative to N.S samples in the healthy control or SNORA31-mutated groups. Each gene is annotated as significantly (Sig.) or not significantly (No. Sig.) differentially expressed (DEG = differentially expressed gene) in control, SNORA31-mutated or STAT1−/− neurons by the colored bars to the right of the heatmap. E) Upregulation of host immune response-related genes in healthy control and SNORA31-mutated hPSC-derived cortical neurons, after 24 hours of infection with HSV-1 at a MOI of 1. Each node represents one gene, and each edge represents the protein-protein interaction. The genes significantly upregulated by HSV-1 infection in healthy control or SNORA31-mutated hPSC-derived cortical neurons are highlighted in red.
Impaired response to HSV-1 in SNORA31-mutated hPSC-derived cortical neurons
snoRNA31 may interfere with one or more key effectors induced by the TLR3 or IFNAR1/IFNAR2 pathways, or other key molecules crucial for other known or unknown antiviral immune pathways. We performed transcriptome analysis by RNA-Seq in SNORA31-mutated hPSC-derived cortical neurons from P2, P5, from isogenic hPSC lines with and without a CRISPR/Cas9-introduced SNORA31 deletion (het del1), another control hPSC line, and one STAT1-deficient patient, with and without infection with HSV-1. After 24 hours of HSV-1 infection, a similar redistribution of RNA-Seq reads from host mRNA to HSV-1 mRNA was observed in cells from all hPSC lines (Extended Data Fig.6C). None of the 73 HSV-1 transcripts detected displayed significantly different relative expression levels in SNORA31-mutated neurons and control neurons (Extended Data Fig.6D). HSV-1 infection led to the up- or downregulation of 1,877 and 754 host transcripts, respectively, in control neurons. Among these transcripts, 1,379 and 691 were significantly up- and down-regulated, respectively, in control but not in SNORA31-mutated neurons (Fig.6D, Extended Data Fig.6E,F). Gene ontology enrichment testing of the up-regulated, but not the down-regulated, transcripts showed a significant enrichment in genes with functions related to immune responses, in control but not SNORA31-mutated neurons (Fig.6E, Table S1). Different gene expression regulation patterns were observed for STAT1−/− and SNORA31-mutated neurons, although there was some similarity for a subset of transcripts (Fig.6D, Table S1). Thus, transcriptome responses to HSV-1 infection are altered in SNORA31-mutated cells. This may have contributed to the greater susceptibility of SNORA31-mutated hPSC-derived cortical neurons.
Discussion
We report five unrelated patients from different ethnic groups, living in different countries, who developed forebrain HSE during primary infection with HSV-1 at various ages, due to heterozygous mutations of SNORA31. The role of snoRNA genes in human physiology and disease remains largely unknown 44. It may not be coincidental that both box H/ACA snoRNA31 and box C/D snoRNA 32,45,46 deficiencies result in conditions affecting the CNS. We establish causality between SNORA31 mutations and HSE here by showing that (i) the HSE cohort is enriched in very rare SNORA31 variants, (ii) SNORA31 and the residues mutated in our patients are highly conserved in humans, (iii) WT snoRNA31 is functional in cortical neurons, (iv) two of the four mutant alleles are loss-of-expression, and all other loss-of-expression variants in the general population have a very low collective MAF, (v) both fibroblasts and iPSC-derived neurons from patients are susceptible to HSV-1, (vi) fibroblasts from patients are also susceptible to other neurotropic viruses, including VZV, MeV, VSV, and EMCV, (vii) snoRNA31 is a cell-intrinsic antiviral factor in cortical neurons, acting via a mechanism not dependent on the TLR3 and IFNAR1/IFNAR2 response pathways, (viii) SNORA31-mutated hPSC-derived cortical neurons display impaired transcriptome responses to HSV-1 infection, (ix) isogenic hESC-derived cortical neurons heterozygous for SNORA31 null mutations are also highly susceptible to HSV-1.
The trait is AD with incomplete clinical penetrance, consistent with the typically sporadic nature of HSE 1,2,4. Various environmental (e.g. virus load and isolate) and host (e.g. age at infection, modifier genes) factors may explain the incomplete clinical penetrance. The mechanism of dominance appears to involve haploinsufficiency, rather than negative dominance. AD snoRNA31 deficiency modifies the human transcriptome response and the control of HSV-1 infection in cortical neurons in vitro, thereby potentially accounting for the failure to protect the forebrain from HSV-1 in vivo. We speculate that the modified transcriptome response due to snoRNA31 deficiency may affect the expression of one or more effectors induced by TLR3 or IFNAR1/IFANR2, produced within the cortical neurons or secreted by them into the extracellular medium, thereby impairing anti-HSV-1 immunity in these cells. Alternatively, snoRNA31 may interfere with HSV-1 propagation directly by interacting with viral transcripts. Future studies will address the molecular mechanism by which snoRNA31 contributes to the control of HSV-1 in CNS neurons. The role of snoRNA31 in anti-HSV-1 immunity in other CNS-resident cell types also remains to be assessed. In conclusion, we report a new genetic etiology and immunological mechanism of HSE, involving a disruption of CNS cortical neuron intrinsic immunity to HSV-1 with maintenance of the TLR3 and IFNAR1/IFNAR2 response pathways.
Materials and Methods
Patients
All patients were living in and were followed up in their countries of origin. Informed consent was obtained in the home country of each patient, in accordance with local regulations and the requirements for institutional review board (IRB) approval from The Rockefeller University and INSERM. Experiments were conducted in the United States and France, in accordance with local regulations and with the approval of the IRB of The Rockefeller University and INSERM, respectively.
Whole-exome sequencing and identification of the mutations
DNA was extracted from cells and sheared with a Covaris S2 Ultrasonicator (Covaris). An adapter-ligated library was prepared with the TruSeq DNA Sample Prep Kit (Illumina). Exome capture was performed with the SureSelect Human All Exon 71 Mb kit. Paired-end sequencing was performed on an Illumina HiSeq 2000 (Illumina), generating 100-base reads. The sequences were aligned with the human genome reference sequence (hg19 build), with BWA. Downstream processing was carried out with the Genome Analysis Toolkit (GATK), SAM tools and Picard Tools (http://picard.sourceforge.net). Variant calls were made with GATK UnifiedGenotyper (see also Reporting Summary NMED-A92100A_RS). All variant calls with a variant quality (QUAL) ≤60, depth of coverage (DP) <10, or a mapping quality (MQ) ≤40 were filtered out. Our cohort of HSE patients contained 205 subjects from diverse ethnic groups, but there were none from Asia, and none of the patients had known HSE-causing mutations of TLR3 pathway-related genes. The control cohort consisted of the 1,511 individuals from the 1000G database (http://www.internationalgenome.org/) who were not of Asian origin, together with 2,756 in-house non-Asian patients without viral diseases (total=4,267 individuals). For enrichment analysis, we considered high-quality rare heterozygous variants (MAF<0.001 in the Exac database (http://exac.broadinstitute.org/)) annotated as missense, indel-frameshift, start-lost, nonsense, stop-lost, stop-gained, synonymous splice-site (within the first or last two bases of the exon), or essential splice-site mutations, and predicted to be damaging, with a CADD score >MSC 25. The proportion of individuals with mutant alleles in each cohort was compared by logistic regression analysis, with the likelihood ratio test, as previously described 47. We accounted for cohort ethnic heterogeneity, by systematically including the first three principal components of the principal component analysis in the logistic regression model 47.
SNORA31 overexpression in HEK293T cells by plasmid transfection
The snoRNA expression vector pCMV-globin was a gift from Tamar Kiss (Toulouse, France). The human SNORA31 gene was inserted into the second intron of the human β-globin gene, which had been placed under the control of the cytomegalovirus (CMV) promoter. The genomic sequence of SNORA31 was first amplified by PCR with primers harboring Nco1 and Xho1 sites. The PCR product was ligated into a pGEM-T easy vector (Promega). The amplified SNORA31 cassette was then cut from the pGEM vector with Nco1 (blunting with the Klenow fragment of DNA polymerase I), and Xho1. The cassette containing SNORA31 was finally ligated into the pCMV-β-globin vector after the linearization of this vector with Cla1 (blunting with the Klenow fragment) and digestion with Xho1, to yield the pCMV-SNORA31-β-globin vector. Site-directed mutagenesis was performed to create mutations with the QuikChange Site-Directed Mutagenesis Kit (Agilent Technologies), with sequencing to check for correct mutagenesis. HEK293T cells (ATCC, USA) were transfected in the presence of Lipofectamine 2000 reagent (Invitrogen). We used 500 ng to 1 μg of plasmid for the transfection of 5×105 cells.
Fibroblast cell lines from controls and patients
Primary human fibroblasts were obtained from skin biopsy specimens from patients, and were cultured in DMEM (Gibco BRL, Invitrogen) supplemented with 10% fetal calf serum (FCS) (Gibco BRL, Invitrogen). For the creation of immortalized SV40-transformed fibroblast cell lines (SV40-fibroblasts), we used 4 μg of a plasmid containing T antigen DNA to transfect about 5 million cells by electroporation. The cells were then placed in two fresh 75 cm2 flasks containing 12 ml of DMEM medium (Gibco BRL, Invitrogen) supplemented with 10% FCS (Gibco BRL, Invitrogen). SV40-fibroblast clones appeared after about 15 days. They were then grown and passaged for experimental usage. Four healthy control SV40-fibroblast cell lines were also used in this study. They were generated in the Laboratory of Human Genetics of Infectious Diseases by the same method. Patient-specific cell lines derived through this study are available from the authors upon request.
RT-qPCR
RNA was extracted with the miRNeasy Mini Kit (Qiagen, #217004) and treated with DNase (Qiagen, #79254) before elution according to the manufacturer’s protocol. Reverse transcription was performed with the miScript II RT Kit (Qiagen, #218161) and HiFlex buffer. For small non-coding RNAs, qPCR was performed with the QuantiTect SYBR Green PCR Kit (Qiagen, #204143). The following primers were used: SNORD96a, miScript Primer Assay (Qiagen, #MS00033733). SNORA31 forward primer, 5’-AAAGAAAGAAATCCCTGCG-3’, SNORA34 forward primer, 5’-CTGACTGAAGACCAGCAGTTGTA-3’. For normalization of the data, the expression of five different snoRNA genes (SNORD61, SNORD68, SNORD72, SNORD95 and SNORD96a) previously shown to be suitable controls and suggested as such in the protocols of the miScript PCR system (www.qiagen.com), was assessed under multiple conditions in different cell types. SNORD96a was chosen as the most reliable control for the cell types and stimulations used in our experiments (data not shown). For protein-coding genes, qPCR was performed with the QuantiTect SYBR Green PCR Kit for TPT1 and 18S. The following primers were used: TPT1 forward primer, 5’-TCCAGATGGCATGGTTGCTCT-3’, reverse primer, 3’-AACAATGCCTCCACTC CAAA-5’; 18S primers (Sigma, #KSPQ12012G), forward primer, 5’-CAGAAGGATGTAAAGGATG G-3’, reverse primer, 5’-TATTTCTTCTTGGACACACC-3’. All qPCRs were analyzed in an Applied Biosystems 7500 Fast Real-Time PCR System. Results are expressed according to the ∆Ct method, as described by the manufacturer.
TOPO cloning and sequencing of cDNAs from the patients’ cells
The SNORA31 sequence was amplified by RT-PCR. The following primers were used: forward primer: 5’-CTGCATCCACTGATAGACCTTGA-3’, reverse primer: 10x miScript universal primer or 5’-AATTCAATCTGGGCCGCCA-3’ for nested PCR. PCR products were inserted into the pGEM-T Easy vector (Promega, #A1360) and amplified in competent E. coli. The sequence of SNORA31 for each colony was determined with M13 primers: forward primer: 5’-GTTGTAAAACGACGGCCAGT-3’, reverse primer: 5’-TCACACAGGAAACAGCTATGA-3’.
Western blotting
Total cell extracts were prepared from SV40-fibroblasts or human pluripotent stem cell (hPSC)-derived CNS neuronal cells from healthy controls, patients, and gene-edited lines. Equal amounts of protein from each sample were separated by SDS-PAGE and blotted onto polyvinylidene difluoride membrane (Bio-Rad, Hercules, CA). These PVDF membranes were then probed with a polyclonal goat anti-human TPT1 antibody (ABCAM, #ab37506). Antibody binding was detected by enhanced chemiluminescence (ECL; Amersham-Pharmacia-Biotech). Membranes were stripped and reprobed with an antibody against GAPDH (Santa-Cruz, USA), to control for protein loading.
Northern blotting
Total RNA was extracted in Qiazol (Qiagen, #79306). Acrylamide gels were prepared with the SequaGel UreaGel System (National Diagnostics), according to the manufacturer’s protocol. Equal amounts of RNA sample and RNA loading dye (BioLabs, #B0363A) were mixed and heated at 70°C for 5 minutes to denature the RNA. After migration in the gel, the RNA was transferred to a nylon membrane (GE Healthcare, #RPN303B) and fixed with an EDC solution for cross-linking. The gel was stained with ethidium bromide for a few minutes to visualize the ribosomal RNA bands. The immobilized RNA membrane was then wetted with SSC solution (Invitrogen, #15557–044) placed in a hybridization tube with hybridization solution (Molecular Research Center, #HS114F), and heated for 3 hours at 42°C. The 32P-labeled double-stranded full-length SNORA31 probe, labeled with the Prime-It II Random Primer Labeling Kit (Agilent Technologies, #300385), was heated at 100°C for 10 minutes, added to the hybridization solution and the hybridization tube was then incubated overnight at 42°C. The membrane was washed several times with SSC solutions of increasing stringency, and the radioactive signal was detected by placing the membrane against photographic film.
GDI calculation for snoRNA genes
For each snoRNA gene, we summed the CADD scores of each variant multiplied by the corresponding number of alleles in the 1000 Genomes database, to generate the crude GDI score. . As previously described 25, genes with very low GDI values, below the lower limit of the 95% confidence interval, are probably subject to purifying selection. Conversely, genes with very high GDI values, above the upper limit of this confidence interval, are probably under positive selection.
In silico analysis of SNORA31 variants
In silico analysis of variants identified in patients with HSE or in public databases was carried out with RNAfold (version 2.1.9), from the ViennaRNA suite. The change in minimum free energy was assessed by first determining the basal minimum free energy of four different domains of SNORA31: 5’ lower stem n.1–15/n.49–72, 5’ top stem-loop n.16–48, 3’ lower stem n.73–84/n.111–130 and 3’ top stem-loop n.85–110. The minimum free energy for the mutant sequences was determined and the difference in energy between the mutant and wild-type sequences was calculated.
Patient-specific iPSC reprogramming and characterization
Induced pluripotent stem cells (iPSCs) were obtained by reprogramming the patients’ primary fibroblasts by infection with the non-integrating CytoTune Sendai viral vector kit (Life Technologies, USA). Reprogrammed cells were karyotyped to ensure that the genome was intact. Patient-specific SNORA31 mutations were confirmed by the Sanger sequencing of genomic DNA extracted from the iPSC lines. Patient-specific cell lines derived through this study are available from the authors upon request.
HESC or iPSC culture and CNS neuron differentiation
Human embryonic stem cells (hESC) or iPSC cultures were maintained in Essential 8 medium (Life Technologies, A1517001). Two healthy control hESC lines (H9, RUES) and two healthy control iPSC lines (J1, J2) were used in this study. The differentiation of hESCs or iPSCs (Together referred as hPSC) into CNS neurons was promoted as previously described 38,39. In short, for the first 10 days, the serum-free differentiation medium used consisted of Essential 6 (Life Technologies, A1516401) supplemented with 500 nM LDN189193 (Stemgent, #04–0074), 10 μM SB431542 (STEMCELL Technologies, #72232) and 5 μM XAV939 (Tocris, #3748/10). On day 10, the medium was replaced with N2 (STEMCELL Technologies, #07156), supplemented with 1:50 B-27 (Life Technologies, #12587–100), with daily replacement for eight days. The cells were then detached with Accutase (Innovative Cell Technologies, AT-104) and replated in N2 supplemented with 10 μM Y-27632 (R&D Systems, #1254) and 6 μM CHIR 99021(Tocris, #4423). Finally, after the amplification of neural progenitor cells for one or two passages, the cells were detached with Accutase, counted, and plated in wells of an appropriate size, in N2 supplemented with 10 μM Y-27632, B-27 (1:50) and 10 μM DAPT (R&D Systems, #2634/50). Neurons were left to mature for four weeks before the experiments.
Gene editing
Gene-editing experiments were performed as previously described 37. Briefly, guide RNA sequences were generated with the CRISPR design tool (http://crispr.mit.edu/). Guide 1, AAAGACAGACAGAAAGCGCA, was selected on the basis of the overlap between the corresponding PAM sequence and P3’s n.75 C/G mutation. Guide 2, CAAAGACAGACAGAAAGCGC, was selected on the basis of its proximity to the previously chosen guide RNA and its completely different set of potential off-target sequences. Forward and reverse oligonucleotides for each guide RNA were then inserted into the MLM3636 vector (a gift from Keith Joung, Addgene, #43860). The activity of each guide RNA was assessed in HEK293T cells. Cells were transfected at 50–70% confluence in a 9 cm2 dish with 1 μg of Cas9-GFP vector (a gift from Kiran Musunuru, Addgene, #44719) and 0.5 μg of guide RNA plasmid. After 48 hours, genomic DNA was extracted for the Surveyor assay (IDT, 706025). PCR products were separated by electrophoresis in 4–20% Bio-Rad precast TBE Mini-Protean gels (Bio Rad, #4565014). For experiments in hESCs, we electroporated 2 million cells with 20 μg Cas9-GFP plasmid and 5 μg guide RNA plasmid mixed in electroporation buffer (BTX, #45–0805). Green cells were sorted by FACS 48 hours post-electroporation. We replated 50,000 cells of moderate GFP intensity and cultured them for 72 hours. The cells were then detached with Accutase, counted and plated at clonal density in 96-well plates. Ten days later, the colonies were passaged and amplified. Genomic DNA was then extracted from each clone and a genomic region of about 450 bp surrounding SNORA31 was subjected to Sanger sequencing. The forward primer was 5’-TTGTTGGTAGAGCAGGGTGTG-3’, and the reverse primer was 5’-ACACTGCAAAAGTTACATTACCAT-3’. The sequence of the 100 bp oligomer used for the HDR experiment to introduce P3’s n.75 C>G mutation into control hESCs was: 5’-CAATTTACTGTTGTTCTTTTGGTTTGCACTAGGATGCAAAAGAAAGAAATGCCTGCGCTTTCTGTCTGTCTTTGTGGCGGCCCAGATTGAATTGGGGAAT-3’. The gene-edited cell lines derived through this study are available from the authors upon request.
Immunostaining
Cells were fixed by incubation in 4% paraformaldehyde at room temperature for 15 minutes and washed with PBS. They were permeabilized at room temperature and incubated with PBS supplemented with 0.3% Triton X100, 0.5% BSA, and 1% goat serum for 30 minutes for antigen blocking. The cells were then washed and stained by overnight incubation with primary antibodies at 4°C. The primary antibodies used were: anti-TBR1 rabbit IgG (AbCam, ab31940), anti-TUJ1 mouse IgG2a (Covance, MMS-435P), and anti-Nestin mouse IgG1 (Neuromics, MO15012). The signals were visualized by staining the cells with Alexa Fluor-tagged secondary antibodies (Invitrogen) and DAPI, and imaging under an EVOS FL fluorescence microscope (Thermo Fisher Scientific). For quantification purposes, we used a minimum of three images of CNS neurons, counting the DAPI/Nestin, DAPI/TUJ1, or TUJ1/TBR1 double-positive cells on these images by eye. For each image, we counted a minimum of 100 cells.
Stimulations and ELISA
We used a synthetic analog of dsRNA, polyinosine-polycytidylic acid (poly(I:C)), as a nonspecific agonist of TLR3 and MDA5/RIG-I. SV40-fibroblast cells were activated in 48-well plates, at a density of 50,000 cells/well, by incubation for 24 hours with poly(I:C) at a concentration of 25 μg/mL. In parallel, cells were stimulated with 25 μg/mL poly(I:C), in the presence of Lipofectamine 2000, to activate MDA5/RIG-I signaling. The protocol for measuring IFN-λ1 production by ELISA was developed in the laboratory. Briefly, plates were coated by overnight incubation at 4°C with 1 μg/mL anti-human IFN-λ1 mAb (R&D Systems, Minneapolis, MN), and the concentration of IFN-λ1 in the supernatant was determined by incubation with a biotinylated secondary antibody (R&D Systems, Minneapolis, MN) at a concentration of 400 μg/mL. The production of IL-6 was assessed with an IL-6 ELISA kit (Invitrogen, USA).
Quantification of viral replication
For infections with HSV-1 strain KOS encoding GFP-capsids (a gift from Dr. Prashant Desai 48), 2×104 SV40-transformed fibroblasts or 7.5×104 hPSC-derived neurons were added to the wells of 96-well plates and infected with HSV-1 at various multiplicities of infection (MOI) in DMEM supplemented with 2% FCS (for fibroblasts) or neuron culture medium (for neurons). Cells were incubated for two hours then washed and incubated in 200 μL of culture medium. GFP emissions from the wells were quantified at various time points, in a fluorescence plate reader (Victor™X4 2030 Multilabel Reader). The output is expressed as the difference between infected and non-infected wells. For wild-type HSV-1 (KOS strain, ATCC) infection, 0.5×105 SV40-fibroblasts or 1.75×105 neurons per well were added to 48-well plates and infected at various MOI in DMEM supplemented with 2% FCS (for fibroblasts) or neuron culture medium (for neurons). After 2 hours, the cells were washed and transferred to 500 μL of fresh medium. Cells and supernatants were collected at various time points and frozen. HSV-1 titers were determined by calculating the TCID50/ml, as described by Reed and Muench 49, after the inoculation of 96-well plates with Vero cells. For HSV-1 (KOS, Patton and F strain) infection, 1×104 SV40-fibroblasts were used to seed coverslips in the wells of 6-well plates and were infected with HSV-1 at a MOI of 3 in DMEM supplemented with 2% FCS and imaged 24 hours post infection. Images were captured with a CoolSnap HQ2 camera (Photometrics). The MetaMorph software package was used for image acquisition and processing (Molecular Devices). Fluorescence images were captured with an exposure time of 500 ms. Individual cells were considered to be positive for fluorescence if emissions were 1.5 times above background. At least two hundred cells were counted per replicate. The HSV-1 strain KOS encoding GFP-capsids is described above. HSV-1 strain F encoding RFP-capsids has been described elsewhere 50. The HSV-1 strain Patton recombinant encoding the mCherry/UL25 fusion was isolated by plaque purification following the infection of cells transiently transfected with a construct encoding the fusion.
For measles virus (MeV) and vesicular stomatitis virus (VSV) infections, 5×104 SV40-fibroblasts per well were added to 48-well plates and infected with MeV at a MOI of 1 or VSV at a MOI of 1, in DMEM supplemented with 2% FCS. After 2 hours, the cells were washed and transferred to 500 μL of fresh medium. Cells and supernatants were collected at various time points and frozen. Virus titers were determined by calculating the TCID50/ml, as described by Reed and Muench, after the inoculation of 96-well plates with Vero cells.
For encephalomyocarditis virus (EMCV) infection, 1×105 SV40-fibroblasts per well were added to 24-well plates and infected with EMCV at a MOI of 0.1, in DMEM supplemented with 2% FCS. After 2 hours, the cells were washed and transferred to 500 μL of fresh medium. Cells were collected at various time points and frozen. DNA was extracted from fibroblasts and EMCV genome copy number was measured by qPCR as previously described 51.
For VZV infection, SV40-fibroblasts were used to seed 24-well tissue culture plates, at a density of 7.5×104 cells per well. The cells were infected with VZV- infected MeWo cells (ROka strain) (fibroblasts: VZV-MeWo ratio, 1:1) for 48 h, and total RNA was isolated. RNA was extracted from fibroblast whole-cell lysate and purified with the NucleoSpin 96 RNA Core kit (Macherey-Nagel, # 740466.4) before elution according to the kit manufacturer’s instructions. Before cDNA synthesis, VZV-infected fibroblasts underwent DNAse treatment and removal (Turbo-DNA-free Kit, Thermo Fischer Scientific # AM1907). The isolated RNA was then used for cDNA synthesis with the QuantiTect Reverse Transcription Kit (Qiagen, # 205314). The synthesized cDNA was subsequently used for real-time quantitative PCR with SYBR green (Agilent, #600882). For the analysis of ORF63, ORF40, ORF9, and GAPDH, we used the following primer sequences: ORF63 forward: GCGCCGGCATGATATACC and ORF63 reverse: GACACGAGCCAAACCATTGTA; ORF40 forward ACTTGGTAACCGCCCTTGTG and ORF40 reverse: CGGGCTACATCATCCATTCC; ORF9 forward: GGGAGCAGGCGCAA TTG and ORF9 reverse: TTTGGTGCAGTGCTGAAGGA; GAPDH forward TCT TTT GCG TCG CCA GCC GAG and GAPDH reverse ACC AGG CGC CCA ATA CGA CCA.
For poliovirus infection, SV40-fibroblasts were used to seed 24-well tissue culture plates, at a density of 7.5×104 cells per well. The cells were infected with poliovirus 1 (LSa strain) at a MOI of 1 and incubated for 24 h. Yield was determined by end-point titration on HeLa cells. Briefly, serial dilutions of culture supernatants were added to the HeLa cells and incubated for 3 days. Wells were evaluated for the presence of infection, and TCID50/ml was calculated by the Reed-Muench method.
Pseudo-seq library preparation and analysis
Pseudo-seq libraries were prepared as previously described 40, but with the following modifications. RNA was fragmented by heating at 60°C for 20 min. CMC modification was reversed by overnight incubation at 50°C. We used 50–70 nt RNA fragments to make libraries. An RT primer with 10 random nucleotides at the 5’ end was used to collapse PCR duplicates (/5Phos/NNNNNNNNNNGATCGTCGGACTGTAGAACTCTGAACCTGTCGGTGGTCGCCGTATCATT/iSp18/CACTCA/iSp18/GCCTTGGCACCCGAGAATTCCA). Circularization was achieved by heating at 60°C for 6 hours. The data were analyzed as previously described 40, but with the following modifications. PCR duplicates were removed with fastx_collapser. The first 10 bases of the collapsed reads, corresponding to the 10 Ns in the RT primer, were trimmed with cutadapt 52. Reads were mapped to GRCh37 with tophat2 53. The pseudo-seq signal was calculated as previously described 54. The Pseud-seq data have been deposited in the Gene Expression Omnibus database, under accession number GSE102078.
Total RNA sequencing and analysis
Total RNA was extracted from hPSC-derived cortical neurons from (1) two SNORA31-mutated hPSC lines from P2 and P5; (2) three isogenic hPSC lines with (two lines: hom del2 and het del 1) or without (one line) CRISPR/Cas9-introduced SNORA31 deletions; (3) one TLR3-deficient patient line; (4) one STAT1-deficient patient line; and (5) two other control hPSC lines. Neurons were left untreated, or were stimulated with poly(I:C) or IFN-α2b, or infected with HSV-1. RNA was extracted with the miRNeasy Mini Kit (Zimo), and treated with DNAse (Qiagen) to remove residual genomic DNA. RNA-Seq libraries were prepared with the Illumina RiboZero TruSeq Stranded Total RNA Library Prep Kit (Illumina) and sequenced on the Illumina NextSeq platform in the 150 nt, paired-end configuration. Each library was sequenced twice.
The RNA-seq FASTQ files were first inspected by fastqc to ensure that the raw data were of high quality. The sequencing reads of each FASTQ file were then aligned to the GENCODE human reference genome GRCh37.p13 55 with STAR aligner v2.6 56 and the alignment quality of each BAM file was evaluated with RSeQC 57. Read quantification was performed to generate the gene-level feature counts from the read alignment, with featureCounts v1.6.0 58 and based on GENCODE GRCh37.p13 gene annotation 55. The gene-level feature counts were then normalized and log2-transformed by DESeq2 59, to obtain the gene expression value for all genes and all samples. The differential gene expression analyses were conducted by contrasting the poly(I:C)-stimulated samples, the IFN-α2b-stimulated samples, and the HSV-1-infected samples, with the non-stimulated samples. In each gene expression analysis, we applied TMM normalization and gene-wise generalized linear model regression by edgeR 60, and the significant differentially expressed genes were selected according to the following criteria: FDR<=0.05 and ∣log2(FoldChange)∣>=1. The differential gene expression was plotted as a heatmap by ComplexHeatmap 61, and the clustering of genes and samples was based on complete linkage and the Euclidean distances of gene expression values. Gene set enrichment was assessed with the DAVID enrichment tool 62.
The immune response gene network was constructed by extracting the human physical protein-protein interactions for which both genes were annotated by Gene Ontology as GO:0006955: Immune Response from the BioGRID Database (build 3.5.175) 63. We then retained the largest connected subnetwork, using PROFEAT 64,65 for visualization. Each node represents one gene, and each edge represents the protein-protein interaction collected from BioGRID. The genes significantly upregulated by HSV-1 infection in healthy control or SNORA31-mutated hPSC-derived cortical neurons are highlighted in red in the immune response gene network, which was visualized with Cytoscape 66.
The RNA-Seq data are available at the NCBI SRA repository under accession number PRJNA580002.
Statistical analysis
Mean values were compared between control cells and patient cells, in one way ANOVA followed by Dunnett’s multiple comparison tests using GraphPad PRISM Version 5.0f, or in two-tailed Student’s t-tests when indicated. Statistical significance is indicated as follows: NS (not significant, p>0.05), * (p<0.05), ** (p<0.01) and *** (p<0.001) in the figures and figure legends.
Data availability statement
For population genetics analyses of SNORA31, we used available data from the gnomAD public database (http://gnomad.broadinstitute.org/about). The Pseudo-seq data reported in this manuscript are available under accession number GSE102078. The RNA-Seq data reported in this paper are available at the NCBI SRA repository under accession number PRJNA580002. Other raw experimental data associated with the figures presented in the manuscript are available from the authors upon request.
Code availability statement
There is no restriction to access to the custom code for the cell lines used in this study. Information is available from the authors upon request.
Extended Data
Extended Data Fig. 1. Heterozygous SNORA31 mutations in HSE patients from five unrelated kindreds.
A) Ranking of the top 10 mutated genes for which enrichment was detected in the HSE cohort (205 patients) versus 4,267 controls. The selection filters for the variants were: MAF ExAC<0.001 and CADD>MSC. This represents 1,274,230 variants in 20,691 genes. For each gene, the number of affected individuals among HSE cases and controls was determined. Individuals were considered affected if they carried at least one mutant allele passing the filters. The genes were ranked by p-values adjusted for ethnic heterogeneity, which was obtained by logistic regression comparing cases and controls adjusted for the three principal components and using the likelihood ratio test (see Methods for more details). B) Expression of snoRNA31 across the different cell types tested. N=4 (for hESC, iPSC, hESC-derived cortical neurons, iPSC-derived cortical neurons), n=6 (SV40-fibroblasts) or n=8 (for primary fibroblasts, EBV-B) biological replicates were tested per cell type, in a single experiment. Each point represents one biological replicate. Means from the biological replicates are indicated with centre lines. C) Patient origin, age at the time of the HSE event, and the MAF of the SNORA31 variant in the GnomAD and BRAVO databases. The ancestry frequencies of the n.36T>C variant were determined from WES data for 500 Moroccan individuals and 1,100 Saudi Arabian individuals. D) Genomic haplotypes of the n.36 T>C regions in kindreds A and B. The n.36 T>C mutation occurred independently in kindreds A and B. Genotypes for the mutation (M, in red) and 10 informative SNPs (black) are represented. Different haplotypes were found in kindreds A and B. E) Histogram representation of the patient-specific SNORA1 mutations, confirmed by Sanger sequencing on genomic DNA from leukocytes and/or fibroblasts from the 5 patients and a healthy control wild type (WT) for SNORA31. F) Plot of GDI values for all reported snoRNA genes (from Ensembl) against their rank, from the least to the most mutated, in the general population (1000 Genomes database). The ranks of SNORA31 and SNORD118 are indicated. G) Phylogenic conservation of the genomic sequence of SNORA31 across 14 different vertebrate species. The data were taken from the snoRNABase (www.snorna.biotoul.fr). Invariant residues are indicated by asterisks below the sequence. Residues conserved in at least 78% of the species are indicated in black. Residues conserved in at least 64% of the species are indicated in blue. Residues conserved in at least 57% of the species are indicated in green. All other residues are indicated in gray. The positions of the variants found in the HSE patients are indicated in red below the sequence. H) Minimum free energy (MFE) of a SNORA31 stem (n.66-84; n.111-130) or loop (n.16-48 and n.85-110), for WT or mutated sequences. The MFE was calculated with the RNAfold program (www.rna.tbi.univie.ac.at). The dot-bracket sequence of SNORA31 is indicated under each nucleotide. The location of each mutation is indicated in red and highlighted.
Extended Data Fig. 2. Expression of WT and mutant SNORA31 in HEK293T cells and in isogenic hESC-derived neurons.
A) Quantification, by qPCR, of SNORA31 copy numbers in 293T cells transfected with an empty plasmid or a plasmid containing the WT sequence of SNORA31. Copy numbers were calculated from a standard curve. Insect SF9 cells were used as a negative control in this experiment as they contain no SNORA31 homolog. N.T: not transfected. E.V: empty vector. Means and standard deviations from n=3 independent experiments are shown. B) Northern blot of SNORA31 in HEK293T cells either not transfected or transfected with an empty vector or a vector containing WT or mutant SNORA31. Cropped images from the same blot are shown. The data presented are representative of n=2 independent experiments. C) RT-qPCR quantification of the fold-change in SNORA31 expression in HEK293T cells. Cells were transfected with a vector containing WT SNORA31 alone or cotransfected with the same amount of empty vector and a vector containing the WT or one of the HSE SNORA31 mutant sequences. The data are expressed relative to SNORD96a expression and normalized relative to the expression of SNORA31 in HEK293T cells transfected with WT SNORA31 vector alone. Means and standard deviations from n=3 independent experiments are shown. D) Plots of the relative expression levels of gnomAD SNORA31 mutant alleles (normalized relative to WT SNORA31 expression level) against minor allele frequency in gnomAD. RT-qPCR quantification of gnomAD SNORA31 mutant allele expression in HEK293T cells transfected with vectors containing each of the mutant alleles. The data are expressed relative to the expression of endogenous SNORD96a and normalized relative to that of SNORA31 in cells transfected with a plasmid carrying the WT sequence. The expression levels of each gnomAD variant were assessed in n=2 independent experiment, and the mean value for each variant is shown as a dot. E) Plot of the relative levels of expression of gnomAD SNORA31 mutant alleles (normalized relative to WT SNORA31 expression levels) against their calculated change in minimum free energy. The expression levels of each gnomAD variant were assessed in n=2 independent experiment, and the mean value for each variant is shown as a dot. F) Surveyor assay for full-length SNORA31 PCR in HEK293T cells transfected with Cas9-GFP vector alone (⦸) or together with a scrambled guide RNA (gRNA) vector, or a SNORA31 gRNA vector. The data shown is representative of data from n=2 independent experiments. G) Histogram representation of the CRISPR-Cas9-introduced homozygous or heterozygous SNORA1 mutations (het del1 n.76-82, hom del1 n.81-86), confirmed by Sanger sequencing on genomic DNA from the gene-edited hESC lines. Sequencing results from the parental line is also shown. H) Demonstration of the CNS cortical identity (TBR1-positive) of neurons (TUJ1-positive) differentiated from the hESC control line H9. The images shown are representative of data from n=6 independent experiments. I) Quantification of the proportion of cortical neurons among total neurons based on the immunostaining of parental and gene-edited hESCs harboring either a heterozygous (het del1) or a homozygous (hom del1) deletion in SNORA31. Results from technical duplicates from a single experiment are shown, representative of n=3 independent neuron differentiations for each line. J) Northern blot of SNORA31 expression in isogenic hESC-derived CNS neurons. SNORA31 expression in parental cells is compared to that in cells carrying heterozygous del 1 (het del1) or homozygous del 1 (hom del1) in the genomic sequence of SNORA31. The data presented are representative of n=2 independent experiments. K) Levels of TPT1 protein, encoded by the host gene of SNORA31, as assessed on western blots for isogenic CNS neurons. Cropped images from the same blot are shown. GAPDH was used as a loading control. The data are representative of n=2 independent experiments. L) Pseudouridylation data obtained by pseudo-seq. Each point represents a pseudouridylation site. Mean values from n=4 libraries each for the parental and het del1 lines and n=3 libraries for the hom del1 line are shown. M) Genomic sequence of SNORA31B. The guide sequences for this putative snoRNA gene are highlighted in red.
Extended Data Fig. 3. Cellular phenotypes of the SV40-fibroblasts of patients with SNORA31 mutations.
A) Sequencing results for SNORA31 cDNA from the SV40-fibroblasts of four patients (P2-P5), after cloning in E. coli. The number of individual WT or mutant colonies detected by sequencing is indicated on the left of the table, whereas the corresponding percentage is indicated on the right. B) Levels of TPT1 protein (upper panel), encoded by the host gene of SNORA31, in the patient and control SV40-fibroblasts, as measured by western blotting. GAPDH was used as a loading control (middle panel). The data presented are representative of n=2 independent experiments. Semi-quantification of TPT1 band density relative to GPADH was performed and the data are shown in the lower panel. C) IL-6 production in SV40-fibroblasts from patients (P2-P5), a TLR3−/− control, and five healthy controls, as measured by ELISA, in the supernatant of control and patient SV40-fibroblasts stimulated with 25 ng/mL poly(I:C) alone, Lipofectamine alone, or both. TLR3−/− cells were used as a negative control for the poly(I:C) response. N.S: not stimulated. N=3 independent experiments were performed, with n=1 biological replicate per cell line tested per experiment. Each point represents one biological replicate. The means of the three independent experiments are shown. The levels of IL-6 production upon poly(I:C) stimulation are compared between controls’ and patients’ cells, in one way ANOVA (F=1.403, total df=29) followed by Dunnett’s multiple comparison tests. ** 0.001<p<0.01. D) Relative expression levels for SNORA31, as determined by RT-qPCR after stimulation with poly(I:C) or IFN-β in control SV40-fibroblasts. The data are expressed relative to SNORD96a expression. N=3 independent experiments were performed, with n=3 (for IFN-β stimulation) or n=6 (all other conditions) biological replicates tested per experiment. Each point represents one biological replicate. Means and standard deviations from three independent experiments are shown. E, F) HSV-1 (strain KOS) abundance, as determined by measurements of GFP-capsid expression in SV40-fibroblasts at the indicated time points post infection with a MOI of 0.01 (E) followed by measurements of the corresponding titers, as determined by calculating the 50% end point (TCID50) in Vero cells (F), for four SNORA31-deficient patients (P2-P5), a TLR3−/− patient and a STAT1−/− patient as susceptible controls, and two healthy controls. Mean values from n=2 independent experiments are shown. N=1 (for TCID50 assay) or n=3 (for measurement of GFP-capsid expression) biological replicates were tested per condition per experiment. G) Percentage of patient and control SV40-fibroblasts producing capsids 24 hours post infection with HSV-1 strain KOS (left), F (middle) or Patton (right) at a MOI of 3 (based on titers measured on Vero cells). Means and standard deviations of n=3 independent experiments are shown. N=1 biological replicate was tested per condition per experiment. H, I) HSV-1 (strain KOS) abundance, as determined by measurements of GFP-capsid expression in patient (P2-P5) and control SV40-fibroblasts at the indicated time points following infection at a MOI of 0.1, without (H) and with (I) IFN-β pretreatment for 16 hours. STAT1−/− cells were used as a control with no IFN-β response. Means and standard deviations from n=3 independent experiments are shown. N=3 biological duplicates were tested per condition per experiment.
Extended Data Fig. 4. HSV-1 propagation in immortalized B cells and iPSCs from patients with SNORA31 mutations.
A) Quantification of HSV-1 (strain KOS) in EBV-immortalized B cells from patients (P2, P4, P5), a TRIF−/− patient and a STAT1−/− patient, and healthy controls, at the indicated time points post infection with a MOI of 0.1. HSV-1 titers were determined by the TCID50 virus titration method. Means and standard deviations from n=3 independent experiments are shown. B) HSV-1 (strain KOS) abundance, as determined by measurements of GFP-capsid expression in iPSCs (left) at the indicated time points post infection with a MOI of 0.1, for three SNORA31-mutated patients (P2, P3, P5), a TLR3−/− patient and a STAT1−/− patient, and hESCs from a healthy control (H9 line). HPSC-derived cortical neurons from a TLR3−/− patient and a healthy control were assessed in the same assay (right). Mean values from n=2 independent experiments are shown. C, D) Expression levels of the VZV ORF40 (C) and ORF63 (D) transcripts, as measured by RT-qPCR on SV40-fibroblasts from patients (P2-P5), a TLR3−/− patient and a STAT1−/− patient, and healthy controls (C1 and C2), 48 h post exposure to VZV-infected MeWo cells (VZV) or MeWo cells that were not infected (N.I.). The data are expressed relative to GAPDH expression. Means and standard deviations from n=3 independent experiments are shown.
Extended Data Fig. 5. Characterization of CNS neurons derived from patient iPSCs and isogenic hESCs.
A) Quantification of the proportion of neurons among total neuronal cells, based on immunostaining, for control hESC- and control iPSC-derived CNS neurons (H9 and J2) and for CNS neurons derived from the patients’ iPSCs. Results from technical duplicates from one experiment are shown, representative of n=3 neuron differentiations per line (H9, J1, J2 control lines, and iPSC lines from P2, P3 and P5). B) Sequencing results for the SNORA31 cDNA obtained from the patients’ iPSC-derived neurons, after cloning in E. coli. The number of individual WT or mutant colonies detected by sequencing is indicated on the left side of the table, whereas the corresponding percentage is indicated on the right. C) Levels of TPT1 protein, encoded by the host gene of SNORA31, in control (H9 hESC) and patient iPSC-derived neurons, as assessed by western blotting. GAPDH was used as a loading control. Results of semi-quantification of TPT1 expression levels relative to GAPDH are shown in the lower panel. The data presented are representative of n=2 independent experiments. D) Histogram representation of the CRISPR-Cas9-introduced homozygous or heterozygous SNORA1 mutations (hom del2 n.84-91, hom HDR n.75C>G, het del2 n.80-85, het del3 n.79-82, het del4 n.77-83, het del5 n.76-88), confirmed by Sanger sequencing on genomic DNA from the gene-edited hESC lines. Sequencing results from the parental line is also shown. E) Levels of TPT1 protein, encoded by the host gene of SNORA31, as measured by western blotting (upper panel), in CNS neurons derived from isogenic hESCs carrying a homozygous HDR-introduced patient-specific point mutation (hom HDR), a homozygous deletion (hom del1 or hom del2), or various heterozygous mutations (het del1, het del2, het del3, het del4, het del5) in SNORA31. GAPDH was used as a loading control (lower panel). The data presented are representative of n=3 independent experiments, with n=1 biological replicate tested per condition per experiment. Semi-quantification of TPT1 band density relative to GPADH was performed and the data are shown in the lower panel. F) Quantification of HSV-1 in isogenic hPSC-derived CNS neurons, at various time points after HSV-1 infection at a MOI of 1, as assessed by measuring GFP-capsid expression. Four healthy control lines were used in these experiments: two control hESC lines and two control iPSC lines. Means and standard deviations from n=3 independent experiments are shown.
Extended Data Fig. 6. Transcriptome responses to stimulation with poly(I:C), IFN-α2b or HSV-1 in SNORA31-mutated hPSC-derived cortical neurons.
A, B) Scatter plots of fold-changes in RNA-Seq-quantified gene expression following stimulation with 25 μg/ml poly(I:C) for 6 hours (A) or 100 IU/ml IFN-α2b for 8 hours (B), in hPSC-derived cortical neurons from one healthy control H9 hESC line (C1) versus one isogenic SNORA31-mutated line (hom del2 or het del1). Moderated fold-changes are expressed relative to the corresponding mock-treated samples. Each point represents a single gene, and each plot includes genes identified as differentially expressed (FDR<=0.05) in response to the indicated stimulus relative to non-stimulated (N.S) samples in the isogenic control or SNORA31-mutated line. C) Transcriptome composition analysis of RNA-Seq-quantified gene expression, in hPSC-derived cortical neurons from healthy controls (C1 and C2), an isogenic SNORA31-mutated line, SNORA31-mutated patients (P2 and P5), and a STAT1−/− patient, with or without 24 hours of infection with HSV-1 at a MOI of 1. D) Heatmap of RNA-Seq-quantified HSV-1 transcript expression (RPKM relativity) in hPSC-derived cortical neurons from healthy controls (C1 and C2), SNORA31-mutated patients (P2 and P5), an isogenic SNORA31-mutated hESC line (het del1), and a STAT1−/− patient, following infection with HSV-1 for 24 hours at a MOI of 1. E) Scatter plot of averaged log2 fold-changes in gene expression by 24 hours of HSV-1 infection, in two healthy controls (C1, C2) versus hPSC-derived cortical neurons from three SNORA31-mutated lines (patient-specific lines from P2 and P5, and an isogenic SNORA31-mutated hESC line, SNORA31 het del1). Moderated fold-changes are expressed relative to the corresponding mock-treated samples. Each point represents a single gene, and the plot includes genes identified as differentially expressed (FDR<=0.05, >2-fold difference) in response to the indicated stimulus relative to N.S samples in the healthy control or SNORA31-mutated groups. F) Number of human transcripts up- or downregulated by HSV-1 infection in either or both of control (C1 and C2) and SNORA31-mutated (P2 and P5, and isogenic SNORA31 het del1 line) hPSC-derived cortical neurons.
Supplementary Material
Acknowledgments
We warmly thank the patients and their families for participating in this study. We thank Dr. Fowzan Sami Alkuraya from the Saudi Human Genome Project for providing us with the minor allele frequency of the reported variant. We thank Jason Tchieu and Gabriele Ciceri for providing advice concerning the protocol for iPSC differentiation into CNS neurons. We thank Kevin Bohannon from Feinberg School of Medicine at Northwestern University for providing HSV-1 strain Patton encoding the mCherry-UL25 fusion. We thank the members of both branches of the Laboratory of Human Genetics of Infectious Diseases for helpful discussions; Tatiana Kochetkov for technical assistance; Benedetta Bigio, Vimel Ratinna, Yoann Seeleuthner, Bertrand Boisson and Aurélie Cobat for bioinformatic assistance; Dominick Papandrea, Cécile Patissier, and Yelena Nemirovskaya for administrative assistance. This work was conducted in the two branches of the Laboratory of Human Genetics of Infectious Diseases, and was funded in part by the National Center for Advancing Translational Sciences (NCATS), National Institutes of Health (NIH), Clinical and Translational Science Award (CTSA) program, grant numbers UL1TR000043 and UL1TR001866, NIH grant numbers R01AI088364 (to J.L.C. & S.Y.Z.), R01NS072381 (to J.L.C. & S.Y.Z.) and R01GM101316 (to W.G.), grants from the Integrative Biology of Emerging Infectious Diseases Laboratory of Excellence (ANR-10-LABX-62-IBEID to L.A.) and the French National Research Agency (ANR) under the “Investments for the future” program (ANR-10-IAHU-01 to L.A.), the ANR grant IEIHSEER (ANR-14-CE14-0008-01 to S.Y.Z.), the Lundbeck Foundation grant R268–2016-3927 (to S.R.P.), the Rockefeller University, Institut National de la Santé et de la Recherche Médicale (INSERM), Paris Descartes University, and the St Giles Foundation. The New York Stem Cell Foundation supported FGL and DP. FGL also was supported by a Merck Postdoctoral Fellowship at The Rockefeller University. Funding for this work was also provided in part by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Footnotes
Declaration of Interests
The authors have no competing interests to declare.
References
- 1.Abel L, et al. Age-Dependent Mendelian Predisposition to Herpes Simplex Virus Type 1 Encephalitis in Childhood. J Pediatr 157, 623–629 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Whitley RJ Herpes simplex encephalitis: adolescents and adults. Antiviral Res 71, 141–148 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Jubelt B, Mihai C, Li TM & Veerapaneni P Rhombencephalitis / brainstem encephalitis. Current neurology and neuroscience reports 11, 543–552 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Whitley RJ Herpes Simplex Virus Infections of the Central Nervous System. Continuum 21, 1704–1713 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Audry M, et al. NEMO is a key component of NF-kappaB- and IRF-3-dependent TLR3-mediated immunity to herpes simplex virus. J Allergy Clin Immunol 128, 610–617 e611–614 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dupuis S, et al. Impaired response to interferon-alpha/beta and lethal viral disease in human STAT1 deficiency. Nat Genet 33, 388–391. (2003). [DOI] [PubMed] [Google Scholar]
- 7.Casrouge A, et al. Herpes simplex virus encephalitis in human UNC-93B deficiency. Science 314, 308–312 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Guo Y, et al. Herpes simplex virus encephalitis in a patient with complete TLR3 deficiency: TLR3 is otherwise redundant in protective immunity. J Exp Med 208, 2083–2098 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herman M, et al. Heterozygous TBK1 mutations impair TLR3 immunity and underlie herpes simplex encephalitis of childhood. J Exp Med 209, 1567–1582 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lim HK, et al. TLR3 deficiency in herpes simplex encephalitis: high allelic heterogeneity and recurrence risk. Neurology 83, 1888–1897 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perez de Diego R, et al. Human TRAF3 Adaptor Molecule Deficiency Leads to Impaired Toll-like Receptor 3 Response and Susceptibility to Herpes Simplex Encephalitis. Immunity 33, 400–411 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sancho-Shimizu V, et al. Herpes simplex encephalitis in children with autosomal recessive and dominant TRIF deficiency. J Clin Invest 121, 4889–4902 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang SY, et al. TLR3 deficiency in patients with herpes simplex encephalitis. Science 317, 1522–1527 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Andersen LL, et al. Functional IRF3 deficiency in a patient with herpes simplex encephalitis. J Exp Med 212, 1371–1379 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang SY, et al. Inborn Errors of RNA Lariat Metabolism in Humans with Brainstem Viral Infection. Cell 172, 952–965 e918 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alexopoulou L, Holt AC, Medzhitov R & Flavell RA Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 413, 732–738 (2001). [DOI] [PubMed] [Google Scholar]
- 17.Jacobs BL & Langland JO When two strands are better than one: the mediators and modulators of the cellular responses to double-stranded RNA. Virology 219, 339–349 (1996). [DOI] [PubMed] [Google Scholar]
- 18.Sato R, et al. Combating herpesvirus encephalitis by potentiating a TLR3-mTORC2 axis. Nat Immunol 19, 1071–1082 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Lafaille FG, et al. Impaired intrinsic immunity to HSV-1 in human iPSC-derived TLR3-deficient CNS cells. Nature 494, 769–773 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang S-Y TLR3 immunity to infection in mice and humans. Current Opinion in Immunology 25, 15 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Serkan B Autosomal recessive cardiomyopathy presenting as acute myocarditis. Journal of the American College of Cardiology 69, 13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Consortium TGP A global reference for human genetic variation. Nature 526, 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin K A general framework for estimating the relative pathogenicity of human genetic variants. Nature Genetics 46, 8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Itan Y, et al. The mutation significance cutoff: gene-level thresholds for variant predictions. Nat Methods 13, 109–110 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang P, et al. PopViz: a webserver for visualizing minor allele frequencies and damage prediction scores of human genetic variations. Bioinformatics 34, 4307–4309 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jorjani H, et al. An updated human snoRNAome. Nucleic Acids Res 44, 5068–5082 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiss AM Human Box H/ACA Pseudouridylation Guide RNA Machinery. Molecular and Cellular Biology 24, 11 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quach H, et al. Signatures of purifying and local positive selection in human miRNAs. Am J Hum Genet 84, 316–327 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Itan Y, et al. The human gene damage index as a gene-level approach to prioritizing exome variants. Proc Natl Acad Sci U S A 112, 13615–13620 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davydov EV, et al. Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS computational biology 6, e1001025 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jenkinson EM, et al. Mutations in SNORD118 cause the cerebral microangiopathy leukoencephalopathy with calcifications and cysts. Nat Genet 48, 1185–1192 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marie-Line B Elements essential for accumulation and function of small nucleolar RNAs directing site-specific pseudouridylation of ribosomal RNAs. The EMBO Journal 18, 13 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tamas J.B.E.a.K. A small nucleolar guide RNA functions both in 2’-O-ribose methylation and pseudouridylation of the U5 spliceosomal RNA. The EMBO Journal 20, 11 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ni J, Tien AL & Fournier MJ Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell 89, 565–573 (1997). [DOI] [PubMed] [Google Scholar]
- 36.Ganot P, Bortolin ML & Kiss T Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell 89, 799–809 (1997). [DOI] [PubMed] [Google Scholar]
- 37.Paquet D, et al. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature 533, 125–129 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Qi Y, et al. Combined small-molecule inhibition accelerates the derivation of functional cortical neurons from human pluripotent stem cells. Nat Biotechnol 35, 154–163 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chambers SM, et al. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol 27, 275–280 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carlile TM, et al. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 515, 143–146 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kato H, et al. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med 205, 1601–1610 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang SY, Jouanguy E, Zhang Q, Abel L, Puel A, Casanova JL Human inborn errors of immunity to infection affecting cells other than leukocytes: from the immune system to the whole organism. Curr Opin Immunol 59, 88–100 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Penzo M & Montanaro L Turning Uridines around: Role of rRNA Pseudouridylation in Ribosome Biogenesis and Ribosomal Function. Biomolecules 8(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McMahon M, Contreras A & Ruggero D Small RNAs with big implications: new insights into H/ACA snoRNA function and their role in human disease. Wiley Interdiscip Rev RNA 6, 173–189 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Duker AL, et al. Paternally inherited microdeletion at 15q11.2 confirms a significant role for the SNORD116 C/D box snoRNA cluster in Prader-Willi syndrome. European journal of human genetics : EJHG 18, 1196–1201 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Polex-Wolf J, et al. Hypothalamic loss of Snord116 recapitulates the hyperphagia of Prader-Willi syndrome. J Clin Invest 128, 960–969 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References
- 47.Belkaya S, et al. Autosomal Recessive Cardiomyopathy Presenting as Acute Myocarditis. Journal of the American College of Cardiology 69, 1653–1665 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Desai P & Person S Incorporation of the green fluorescent protein into the herpes simplex virus type 1 capsid. J Virol 72, 7563–7568 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reed LJ & Muench H A simple method of estimating fifty percent endpoints. The American Journal of Hygiene 27, 493–497(1938). [Google Scholar]
- 50.Huffmaster NJ, Sollars PJ, Richards AL, Pickard GE & Smith GA Dynamic ubiquitination drives herpesvirus neuroinvasion. Proc Natl Acad Sci U S A 112, 12818–12823 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Z, Liu Y, Lin W & Cui S A real-time PCR to detect and analyze virulent EMCV loads in sows and piglets. Mol Biol Rep 39, 10013–10017 (2012). [DOI] [PubMed] [Google Scholar]
- 52.Marcel M Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal 17, 3 (2011). [Google Scholar]
- 53.D K TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biology 14, R36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stanley. Loss-of-function mutations in the RNA biogenesis factor NAF1 predispose to pulmonary fibrosis-emphysema. Sci. Transl. Med 8, 351ra107 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Frankish A, et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res 47, D766–D773 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang L, Wang S & Li W RSeQC: quality control of RNA-seq experiments. Bioinformatics 28, 2184–2185 (2012). [DOI] [PubMed] [Google Scholar]
- 58.Liao Y, Smyth GK & Shi W featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 59.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Robinson MD, McCarthy DJ & Smyth GK edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gu Z, Eils R & Schlesner M Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849 (2016). [DOI] [PubMed] [Google Scholar]
- 62.Huang da W, Sherman BT & Lempicki RA Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37, 1–13 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oughtred R, et al. The BioGRID interaction database: 2019 update. Nucleic Acids Res 47, D529–D541 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang P, et al. PROFEAT Update: A Protein Features Web Server with Added Facility to Compute Network Descriptors for Studying Omics-Derived Networks. J Mol Biol 429, 416–425 (2017). [DOI] [PubMed] [Google Scholar]
- 65.Zhang P, et al. A protein network descriptor server and its use in studying protein, disease, metabolic and drug targeted networks. Brief Bioinform 18, 1057–1070 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smoot ME, Ono K, Ruscheinski J, Wang PL & Ideker T Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27, 431–432 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
For population genetics analyses of SNORA31, we used available data from the gnomAD public database (http://gnomad.broadinstitute.org/about). The Pseudo-seq data reported in this manuscript are available under accession number GSE102078. The RNA-Seq data reported in this paper are available at the NCBI SRA repository under accession number PRJNA580002. Other raw experimental data associated with the figures presented in the manuscript are available from the authors upon request.