Abstract
Background
Gut microbiota play important roles in their co-evolution with mammals. However, little is understood about gut bacterial community of Tibetan sheep compared with other sheep breeds. In this study, we investigated the gut bacterial community in 4 different sheep breeds living in the Qinghai-Tibetan Plateau (QTP) of China using high-throughput sequencing (HTS) technique.
Results
The results suggested that bacterial community abundance and breeds diversity of Tibetan sheep (TS) were significantly lower than that of the other three breeds of sheep [Dorset sheep (DrS), Dorper sheep (DrS) and Small Tail Han sheep (STHS)] (p < 0.05). Principal coordinates analysis (PCoA) and nonmetric multidimensional scaling (NMDS) analysis indicated that microbiome composition of TS was significantly different from that of other three sheep breeds (p < 0.01). Firmicutes was the most predominant microbial phylum in the gut, followed by Bacteroidetes. The gut bacterial community of TS showed higher proportions of phylum Spirochaetes, Proteobacteria and Verrucomicrobia, compared to the other three sheep breeds, but the Deferribacteres was absent in TS. At the genus level, Treponema, Succinivibrio, 5-7 N15 and Prevotella showed significantly higher abundance in TS than in the other three sheep breeds (p < 0.05).
Conclusions
In this study, we first employed HTS to understand the gut microbiomes among different sheep breeds in QTP of China.
Keywords: Tibetan sheep, Gut microbiota, High throughput sequencing, Breeds diversity
Background
The significance of gut microbiome is well known, it is an extremely complicated and diverse population and has been explored extensively [1, 2]. Recently, a new term ‘superorganism’ is applied to describe the strong tie between the gut commensal and its reliable host [3, 4]. The intestinal microbiota is linked to a diverse range of conditions, including gathering energy, promoting intestinal epithelial cell proliferation and enhancing the immune system [5, 6], and can be deservedly regarded as an ‘organ’ playing a significant part in the metabolic process [3, 7, 8]. It has been found that more than 3.3 million commensal genes are resided in humans, which is equivalent to 150 times of human genes [9]. In ruminants, the composition of the gastrointestinal commensals, their effect on host immunity and welfare have been explored for several years. A previous study suggested that it was a great achievement of using gut microbiota by ruminant animals [10]. And it’s true that combining high-throughput ‘omics’ technologies with ruminant’s genomes, the unprecedented well-being of achievement can be harvested [3].
Initially, the studies about gut microorganisms were dependent on the sequence alignment from genomic libraries to screen the functional genes, or through PCR amplification [3]. In Escherichia coli, heterologous gene expression was determined by Sanger Chain Termination Method and obtained the autoradiographic map [11]. However, it could only obtain the genes expressed in E. coli, and certainly leads to a large missing of available genes. Through this method, the first gene was acquired from F. succinogenes and encoding cellulases [12]. In the following years, seven F. succinogenes genes were detected through these traditional genetic methods, which encoded fiber-degrading enzymes [13]. This was certainly an extraordinary progress, but after the sequencing of F. succinogenes S85 genome was completed, it was found that there were 104 open reading frames to participate in the disruption of plant cell wall [14]. Accordingly, the great advantage of genomic sequencing is intuitively clear, more enzymes were detected than that of previous studies in the F. succinogenes genome.
Metagenomics is performed to analyze the genome of microbial communities from an environmental sample, including the genomic sequence-based analysis and functional prediction. It is applied to screen the specific functions and detecting new bio-actives in diverse ecosystems [15]. Moreover, it is a vital step to model and connect the microbial structure and function to that of the host [16, 17]. Some researchers found that the gut microbiota of mammals have a large identical part of their functions, suggesting that the understanding of human researches can provide many common views for ruminants [18]. In addition, relationship between microbial lineages and their specific environment was found [19]. However, those gut microbes are not frequently emerged in other environments [20]. Some studies also showed that gut microbes were extensively shared among various mammals [21], indicating that some views are commonly applied to both humans and domestic animals.
In the present study, to better analyze and clarify the relationship between microbial lineages and the host breeds from a perspective of gut microbiota, V3 and V4 region of the 16 s rDNA was amplified, which was followed by, the most credible techniques, Illumina MiSeq Reagent Kit PE250 sequencing. Apart from the shared characteristics are presented among all sheep, typical microbial population features corresponding to different sheep breeds are also mentioned in our research.
Methods
Description of samples
A total of 40 sheep (male; 1 year old) with a similar feeding pattern were used in this research. Fecal samples including 10 Dorset sheep (DrS), 15 Small Tail Han sheep (STHS), 5 Tibetan sheep (TS) and 10 Dorper sheep (DrS) were obtained from specific farms (Qinghai Province, China). 16S rDNA sequences from 40 individuals were amplified and analyzed. All screened sheep were healthy and no other diseases appeared prior to the sample selection. Table S1 provides the detailed information of each sheep we sampled.
DNA extraction
Following the manufacturer’s instructions of Omega Bio-tek, microbial genomic DNA was extracted from 500 mg of each fecal sample using the fecal DNA kit. Meanwhile, we measured the DNA quality with 1% agarose gel electrophoresis, and examined the concentration through the NanoDrop Spectrophotometer. DNA samples were stored at − 20 °C before use. The experimenters conducting the rDNA extraction of analysis were blinded to the group samples they were testing.
PCR amplification of 16 s rDNA
The V3-V4 region, a 468 bp within the 16 s rDNA gene, was used to build the illumine sequencing library and amplified with the broadly conserved primers 341F (5′–CCTACGGGNGGCWGCAG-3′) and 805R (5′-GACTACHVGGGTATCTAATCC-3′). Different identifier codes were added at each primer for the further illumina sequencing. Polymerase chain reaction (PCR) was applied in a 50 μl reaction system including 25 μl 2x Phanta Max Master Mix (Vazyme, China), 10 mM each primer, 16 μl each ddH2O and 5 μl DNA template. The PCR program was initial denaturation at 95 °C, with 8 cycles of denaturation at 95 °C for 30s, annealing at 55 °C for 30 s, extension at 72 °C for 45 s, with a final elongation phase at 72 °C for 5 min. The PCR products were analyzed by Quant-It Pico Green kit (Invitrogen, United States) and library was prepared. Barcoded samples were combined equal concentrations according to volume of sequencing. The library concentration was measured using Agilent 2100 Bioanalyzer (Agilent Technologies, United States), and followed by elution with Tris_HCl (pH 8.5). After denaturation, barcoded samples were combined following the volume of sequencing and sequenced on a PE250 v3 instrument using 600 cycles MiSeq Reagent Kit on a MiSeq Platform (Illumina; United States).
Bioinformatics and statistical analysis
In our research, all sequences have been deposited to the National Center for Biotechnology Information (NCBI) database under accession number AR180907. The QIIME (Quantitative Insights Into Microbial Ecology, v1.8.0) was performed to process the raw reads, and then the paired reads were assembled by FLASH v1.2.7 [22, 23]. Subsequently, QIIME was used to filter and analyze the joined sequences. By UPARSE 7.0, operational taxonomic units (OTUs) were obtained with based on a 97% identity threshold. Eventually, the whole OTUs were categorized to distinct taxonomic levels by Ribosomal Database Project (RDP) classifier 2.2 [24]. Based on the OTUs information, R package VennDiagram was performed to complete the venn diagram. In addition, the phylogenetic tree was obtained by MAGA 5.2 after sequences alignment. Alpha diversity was measured by MOTHUR, which was referred to the microbial community diversity. Bray-Curtis distance and unweighted Unifrac was evaluated the similarities of different samples with R package vegan. The Bray-Curtis distance was estimated based on common OTUs among samples to provide equal weight to differences in each taxa [1, 25]. The Unifrac was used to construct the phylogenetic tree for samples. Taxa which are phylogenetically closely related will give less divergent Unifrac values, while the unrelated taxa will generate larger values [26]. OIIME was performed to generate phylogenetic beta diversity. Principal coordinate analysis (PCoA) and hierarchical clustering analysis by R program was conducted based on Bray-Curtis distance and unweighted Unifrac. PERMANOVA and student’s t-test were performed to exam significant differences between various groups.
Results
Description of the sequencing data
Fecal samples were collected from four different sheep breeds (10 Dorset sheep (DrS), 15 Small Tail Han sheep (STHS), 5 Tibetan sheep (TS) and 10 Dorper sheep (DrS)). We retrieved 1,694,264 raw bases from the sequencing platform, as mentioned above. After quality–filtering (also as described in the methods), 1,359,405 total sequences with an average of 433 bp in length were obtained for the following analysis.
Gut microbiota is associated with sheep breeds
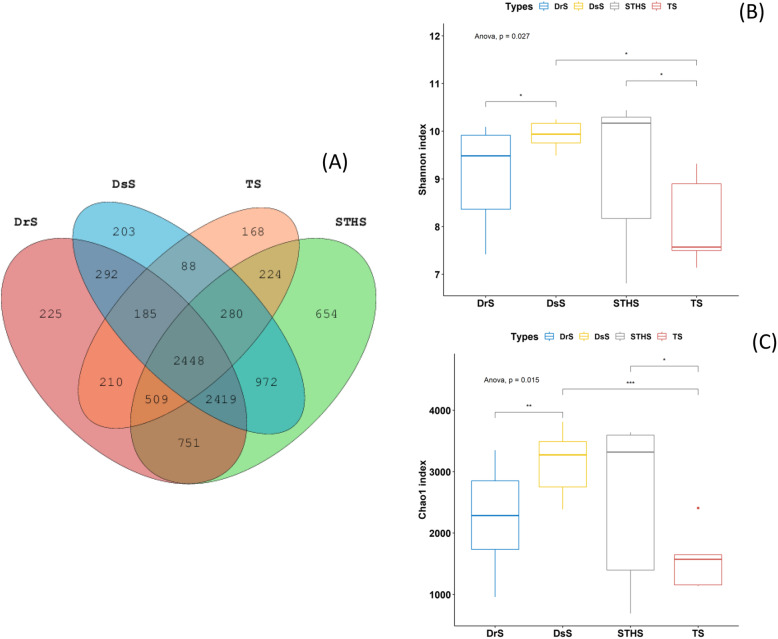
After cleaning the original data, 1,359,405 high-quality available sequences were obtained. According to 97% breeds similarity, 7039, 6887, 4112, and 8257 OTUs were acquired from samples at groups DrS, DsS, TS, and STHS (Table S2), respectively. A total of 26,295 OTUs were detected from all samples, of which 2448 were core OTUs (Fig. 1a). The core OTUs accounts for nearly 9.31% of the entire OTUs. Furthermore, 225,203 OTUs were uniquely detected in DrS and DsS groups, and 168,654 unique OTUs were found in group TS and STHS, respectively. The number of observed OTUs in the TS samples was fewer than that of the other three breeds.
Fig. 1.
The community composition and microbial diversity index analysis. a Venn diagram showing overlap in OTUs of differential abundance in DrS, DsS, TS and STHS. b Shannon index. c Chao1 index. Different asterisks represent statistical significance (*p < 0.05, ** p < 0.01, *** p < 0.001)
To confirm the quality of our sequencing data, we examined alpha and beta diversities of bacterial fraction of the sheep microbiota. Several alpha diversity indices diverged significantly between the four sheep breeds (Fig. 1b and c). The Shannon-Wiener index may straightly indicate the heterogeneity of a community according to the OTU counts of breeds and their related abundance [27]. The Shannon-Wiener indicator of groups DsS, STHS, TS and DrS were 9.92, 9.33, 8.09 and 9.06, respectively. Within the groups, Chao1 and Shannon index visually reflected that the abundance and diversity of intestinal microbial population in the TS group were lower than those in groups DsS, STHS and DrS, and the difference among these four groups was significant (P < 0.05; Fig. 1b and c). The Chao1 index of DsS, STHS, TS and DrS breeds was 3160, 2598, 1585 and 2207 respectively. The ACE indices of these four breeds were 3263, 2646, 1627 and 2267 (Table S3), which was consistent with the Chao1 results, suggesting that the OTU richness of TS samples was lower than other three breeds (Fig. 1b and c). Collectively, these data pointed towards a more diverse bacterial population in TS compared to others, and also showed that differences in intestinal microbial composition associates sheep breeds.
Comparison of bacterial microbiome diversity among different sheep breeds
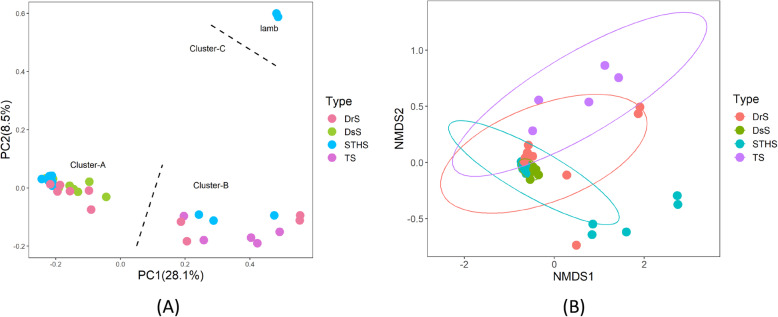
The Bray-Curtis distance matrices were measured according to the OTUs abundance of each sample. Based on the distance matrices, the unweighted Unifrac similarity analysis indicated that the similarities among different sheep breeds were significant. The principal coordinates analysis (PCoA) was performed according to the phylogenetic-tree-based Unifrac metric. As shown in Fig. 2a, samples were sequestered into three clusters. Scattered points in the principal component denoted different breeds and their relationship between each other. There were significant differences among breeds in relation to microbiome composition (PERMANOVA, p < 0.01). TS were mainly aggregated in cluster B, whereas DsS were mostly converged in cluster A. Moreover, DrS and STHS were more scattered and found between cluster A and cluster B. Both principal components accounted for 28.1% (PC1) and 8.5% (PC2) of the explained variance. Interestingly, two lambs in STHS were clustered separately as shown in Fig. 2a, suggesting that there was a general difference in gut microbiome between adult sheep and lambs.
Fig. 2.
Compositional analyses of the gut microbiome of different sheep breeds. a PCoA plot of similarities between the different groups. Principal component (PC) 1 and 2 accounted for 28.1 and 8.5% of the variance, respectively. b NMDS showing the alteration of bacterial population based on Bray-Curtis distance
Nonmetric multidimensional scaling (NMDS) was used to further clarify the difference among the bacterial population of all breeds, which was performed using the Bray-Curtis similarity for all samples at OTU level [1]. As a dominant ordination method that could exhibit the non-linear relationship among samples, NMDS has been widely applied in the study of gut microbiome. As shown in Fig. 2b, there was distinguishing clustering of TS samples, meanwhile samples from DsS were very close to DrS. However, the samples from STHS were more dispersed (Fig. 2b).
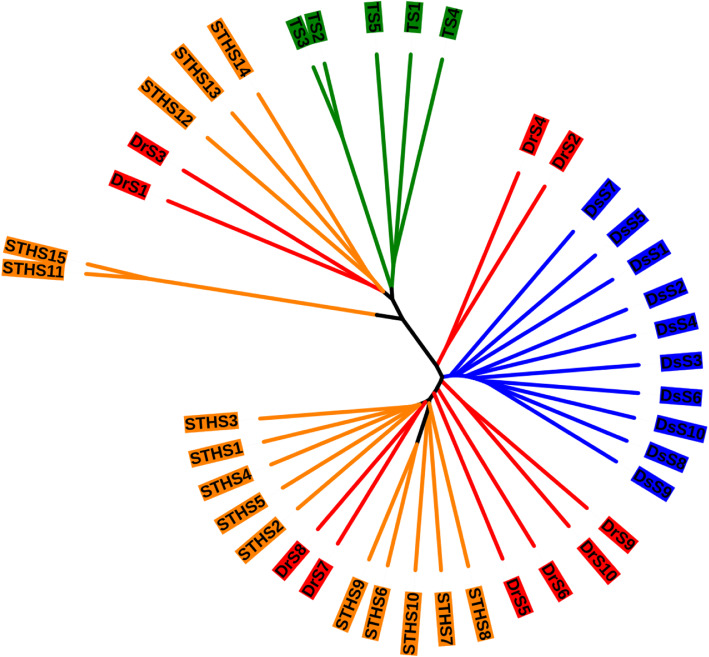
Additionally, we did hierarchical clustering analysis of all samples to exhibit the similarity among samples, which was performed with Unweighted pair-group method with arithmetic means (UPGMA) and the Bray-Curtis similarity. Two primary groups were perceived in this analysis. One cluster contains all TS samples and the other cluster contains all samples from DrS (Fig. 3). Consist with the results above, TS samples were distinctive compared with other breeds. In general, the composition of gut microbiota is greatly influenced by the breed of sheep.
Fig. 3.
Hierarchical clustering of bacterial communities according to Bray-Curtis distance
Gut microbial diversities and community composition among different sheep breeds
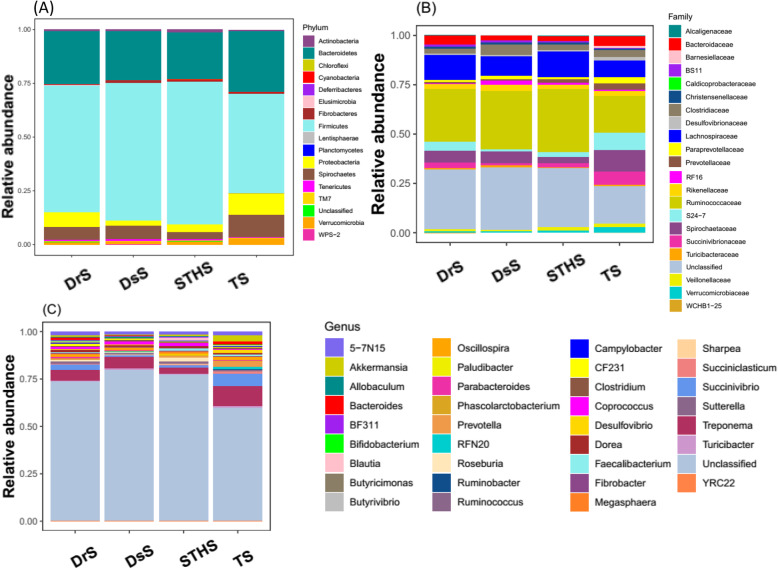
In order to clarify the diversity of gut bacterial composition in different sheep breeds, we estimated the gut microbiota in different taxonomical levels. The overall bacterial composition of different groups at the phylum level is illustrated in Fig. 4a, which shows that Firmicutes was the most predominant phylum in all samples, followed by Bacteroidetes. Higher abundance of phylum Spirochaetes, Proteobacteria and Verrucomicrobia was found in TS than those in other three breeds, but the Deferribacteres was absent in TS (Fig. 4a).
Fig. 4.
Microbial composition of different samples. Each bar represents the average relative abundance of each bacterial taxon within a group. a Taxa assignments at Phylum level. b Taxa assignments at Family level. c Taxa assignments at Genus level
When analyzed on the family level, as shown in Fig. 4b, no significant differences were detected among these four groups. Ruminococcaceae and WCHB1–25 were the most abundant families in DsS, DrS and STHS group, whereas Alcaligenaceae, Desulfovibrionaceae and Barnesiellaceae were almost absent. As for TS group, the most abundant families were Spirochaetaceae, S24–7, Prevotellaceae, Barnesiellaceae and Succinivibrionaceae, while BS11 and WCHB1–25 were almost absent in the TS samples (Fig. 4b).
In contrast to the family level, there are significant differences between TS group and the other three groups on the genus level. The main genera in TS group included Treponema, Succinivibrio, 5-7 N15 and Prevotella (Table 1), while Bifidobacterium, Sharpea and YRC22 were absent (Fig. 4c). Moreover, in DsS, STHS and DrS group, Treponema remained the predominant population, and Coprococcus and Roseburia were relatively less abundant. However, it is worth mentioning that a large number of microbes in TS samples were relatively abundant, when compared to other three groups.
Table 1.
The relative abundance of top 10 genera bacteria in four sheep breeds, expressed as an average percentage of the total (n = 40)
| Genera | Group | Class | Phylum | |||
|---|---|---|---|---|---|---|
| DsS | STHS | TS | DrS | |||
| Treponema | 6.11 | 3.09 | 10.63 | 5.90 | Spirochaetes | Spirochaetes |
| Succinivibrio | 0.82 | 1.15 | 6.52 | 2.96 | Gammaproteobacteria | Proteobacteria |
| 5-7N15 | 1.36 | 1.35 | 1.91 | 1.84 | Bacteroidia | Bacteroidetes |
| Akkermansia | 0.70 | 1.06 | 2.95 | 0.87 | Verrucomicrobiae | Verrucomicrobia |
| Coprococcus | 1.66 | 1.36 | 0.79 | 1.60 | Clostridia | Firmicutes |
| Oscillospira | 1.05 | 1.19 | 1.49 | 1.22 | Clostridia | Firmicutes |
| Ruminococcus | 0.87 | 1.33 | 0.86 | 1.25 | Clostridia | Firmicutes |
| Prevotella | 0.29 | 0.83 | 2.82 | 0.35 | Bacteroidia | Bacteroidetes |
| Dorea | 0.79 | 1.12 | 0.79 | 1.20 | Clostridia | Firmicutes |
| Bacteroides | 0.14 | 0.22 | 1.80 | 1.58 | Bacteroidia | Bacteroidetes |
| Other | 86.21 | 87.3 | 69.44 | 81.23 | ||
Discussion
This study was aimed at acquiring insight into the gut bacterial composition of four sheep breeds living in the QTP of China using next-generation sequencing technique. In our study, the gut bacterial community of four different sheep breeds was estimated by PCR-retrieved microbial 16S rDNA gene libraries. In our research, intestinal microflora of four sheep breeds has been examined by bacterial diversity and abundance. These results indicated that there are significant differences of the gut microbiota between TS and the other three sheep breeds (DrS, DsS and STHS), besides, the bacterial diversity and composition of TS are relatively lower. However, the bacterial abundance in TS are higher than those in the other three sheep breeds. The microbial diversity of TS altered significantly compared with the other three breeds, which is similar with the earlier reports in high-altitude mammals [28, 29]. PCoA clustering analysis revealed that the microbial structure is distinct between the TS and the other three breeds (Fig. 2a). Besides, hierarchical clustering analysis showed that TS samples clearly cluster together, indicating that the intestinal microbial population of TS are highly conserved for the comparison between interbreeds. On different taxonomical levels, the abundance of gut bacterial composition is also distinct among different sheep breeds. This phenomenon is probably due to the fact that Tibetan sheep have adapted to the high-altitude environment, while the other three breeds, as introduced later, are convergent to the commensal composition of Tibetan sheep. In addition, our study showed that the gut bacterial composition of lambs is quite different from that in adult sheep. We assumed that the gut microbial composition in lambs will develop towards adult sheep under their living environment.
This study suggested that Firmicutes and Bacteroidetes are the most abundant phyla in the gut microbiomes of all samples, which accounted for 55.83 and 24.39% of the total microbial abundance, respectively, which was consistent with the prior studies in herbivores [30]. The functions of Firmicutes and Bacteroidetes are closely associated with carbohydrate, protein, and fiber metabolism [31]. The crop straws mainly contained cellulose, hemicellulose and lignin, and the Firmicutes contained a lot of fiber-decomposing bacteria, including Butyvibrio, Ruminococcus, Oscillibacter and Eubacterium, which may explain why the Firmicutes are dominant in the rumen bacterial community of ruminants [30]. Bacteroidetes are the major degraders for decomposing non-fibrous plant components in sheep gut, and the Prevotella have the highest composition in this bacteria group. It has been suggested that Prevotella can account for 60–70% of the overall microbial communities in rumen, and it includes highly active hemicellulose decomposing microbes, which was important for the decomposition of non-fibrous polysaccharides or proteins in crops [32, 33].
In this study, the relative abundance of Firmicutes and Bacteroidetes were not consistent among the different sheep breeds, Bacteroidetes had higher abundance in TS than the other three breeds. In order to investigate the reasons of this result, we estimated the genera level of taxonomy and found that within the phylum Bacteroidetes, three genera had significantly higher abundance in TS than the other three sheep breeds, including 5-7 N15, Prevotella and Bacteroides. These three genera belong to the class Bacteroidia (Table 1). Previous study has shown that the Prevotella played an important influence on the fermentation process of feed in the rumen of sika deer [34]. Other studies also suggested that Prevotella was predominant in ruminants [33, 35, 36]. Prevotella was used to degrade lignocellulosic feedstock with xylanase and carboxymethylcellulase [37]. Besides, some genera of the class Bacteroidia had high active hemicellulose decomposition which provides hosts with the abilities to digest and extract nutrition from fibrous plants [38]. In our research, we hypothesized that this phenomenon is probably due to the fact that TS have unique microbiota structure which was adapted to the high-altitude environment, while the other three breeds TS share greater similarity.
A special bacterial phylum called Proteobacteria was identified in the four breeds, which was observed in various ruminants, such as sheep and cattle. But a relative low proportion (0.8%) was found in yaks [39–41]. In this study, Proteobacteria had significantly higher abundance in TS than the other three breeds (p < 0.01). It has been reported that phylum Proteobacteria had the highest richness in the bovine rumen [42], and played a crucial role in the biofilm formation, fermentation and the soluble carbohydrates digestion [43]. We inferred that the reason for the higher abundance of Proteobacteria in TS was related to the different breeds of sheep. TS have adapted to survive in this harsh plateau environment, where Tibetans raise these animals for food and sustenance [28, 44]. Several studies revealed that the gut microbes of ruminants help them survival at high altitude, which involve in the energy metabolism pathway [28, 29, 45].
A large body of evidence has shown that gut microbial community composition is affected by animal breed and several other factors. For example, a study on Yak and Tibetan sheep demonstrated that the difference in bacterial compositions of the gut was mainly attributed to host breed [46]. Similarly, a report about the gut microbiota of Bos taurus and Bubalus bubalis demonstrated that rumen microbiota community varied with both breeds and feeding patterns [47]. Consistent with these studies, our results suggested that sheep breeds may critically determine their gut bacterial community. Other factors influencing gut microbiota structure includes geographical environment, which especially affects the richness of Prevotellaceae, Butyrivibrio, and Campylobacter [48]. Meanwhile, feeding regimens not only affect gut microbial composition, but also influence metabolic homeostasis in sheep [49, 50]. Another study also demonstrated that feeding with rosemary leaves could alter the abundance of rumen microbial community into one that was dominant with bacterial species involved in protein degradation and methane production [51].
Conclusion
To conclude, our 16S rDNA analyses reflected the gut microbiome of different sheep breeds and highlighted the difference between TS and the other three sheep breeds (DsS, STHS and DrS). This study also showed a lot of high abundance species, which may play important roles in the host. The fluctuation of gut bacteria composition indicated that gut microbes could be changed along with the differences of sheep breeds. Different breeds caused a shift on the microbial community structure and decreased the bacterial species diversity in the gut of TS. Furthermore, the investigation of these distinct microbial structures may provide a better understanding for the light on mechanisms of these comparatively abundant, but enigmatic microbial symbionts of ruminants.
Supplementary information
Additional file 1: Table S1. The information of sheep in this study. Table S2. Sequence data of samples. Table S3. The estimators of sequence diversity and richness.
Acknowledgments
Not applicable.
Abbreviations
- DrS
Dorset sheep
- STHS
Small Tail Han sheep
- TS
Tibetan sheep
- DrS
Dorper sheep
- NCBI
National Center for Biotechnology Information
- QIIME
Quantitative Insights Into Microbial Ecology
- OTU
Operational taxonomic units
- RDP
Ribosomal Database Project
- PCoA
Principal coordinate analysis
- PICRUSt
Phylogenetic Investigation of Communities by Reconstruction of Unobserved States
- NMDS
Nonmetric multidimensional scaling
- UPGMA
Unweighted pair-group method with arithmetic means
- QTP
Qinghai-Tibetan Plateau
Authors’ contributions
WTM and DKC conceived and designed experiments; XTY and JJC performed all experiments. XTY, CXZ and YXQ collected and analyzed the data. XTY and JJC drafted the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by Qinghai province Major R&D and Transformation Project (2018-NK-125) and National Natural Science Foundation of China (31902282). The funding bodies provided all the help in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
All data generated or analyzed during this study are included in this published article, and also available from the corresponding author on reasonable request.
Ethics approval and consent to participate
All experiments were approved by the Ministry of Health in China for the care and use of laboratory animals and supervised by the Research Ethics Committee of Northwest A&F University. Informed consent was obtained from the animal owners in advance.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jianjun Chang and Xiaoting Yao contributed equally to this work.
Contributor Information
Jianjun Chang, Email: 383322290@qq.com.
Xiaoting Yao, Email: yaoxiaoting@nwafu.edu.cn.
Chenxiang Zuo, Email: zcx1997@nwafu.edu.cn.
Yuxu Qi, Email: qyx@nwafu.edu.cn.
Dekun Chen, Email: cdk@nwafu.edu.cn.
Wentao Ma, Email: mawentao@nwafu.edu.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12917-020-02477-2.
References
- 1.Ma X, Wang Q, Li H, Xu C, Cui N, Zhao X. 16S rRNA genes Illumina sequencing revealed differential cecal microbiome in specific pathogen free chickens infected with different subgroup of avian leukosis viruses. Vet Microbiol. 2017;207:195–204. doi: 10.1016/j.vetmic.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 2.Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977;31:107–133. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- 3.Morgavi DP, Kelly WJ, Janssen PH, Attwood GT. Rumen microbial (meta) genomics and its application to ruminant production. Animal. 2013;7(Suppl 1):184–201. doi: 10.1017/S1751731112000419. [DOI] [PubMed] [Google Scholar]
- 4.Lederberg J. Infectious history. Science. 2000;288(5464):287–293. doi: 10.1126/science.288.5464.287. [DOI] [PubMed] [Google Scholar]
- 5.Kelly D, Conway S, Aminov R. Commensal gut bacteria: mechanisms of immune modulation. Trends Immunol. 2005;26(6):326–333. doi: 10.1016/j.it.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 6.Hooper LV. Bacterial contributions to mammalian gut development. Trends Microbiol. 2004;12(3):129–134. doi: 10.1016/j.tim.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 7.Xu JGJ. Honor thy symbionts. Proc Natl Acad Sci U S A. 2003;100(18):10452–10459. doi: 10.1073/pnas.1734063100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Egert M, de Graaf AA, Smidt H, de Vos WM, Venema K. Beyond diversity: functional microbiomics of the human colon. Trends Microbiol. 2006;14(2):86–91. doi: 10.1016/j.tim.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 9.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Russell JB, Muck RE, Weimer PJ. Quantitative analysis of cellulose degradation and growth of cellulolytic bacteria in the rumen. FEMS Microbiol Ecol. 2009;67(2):183–197. doi: 10.1111/j.1574-6941.2008.00633.x. [DOI] [PubMed] [Google Scholar]
- 11.Sanger FNS, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cavicchioli RWK. Molecular cloning, expression, and characterization of endoglucanase genes from Fibrobacter succinogenes AR1. Appl Environ Microbiol. 1991;57(2):359–365. doi: 10.1128/aem.57.2.359-365.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forsberg CW, Forano E, Chesson A. Microbial adherence to the plant cell wall and enzymatic hydrolysis. In: Cronje PB, editor. Ruminant physiology digestion metabolism, growth and reproduction. 2000. pp. 79–98. [Google Scholar]
- 14.Jun HS, Qi M, Ha JK, Forsberg CW. Fibrobacter succinogenes, a dominant fibrolytic ruminal bacterium: transition to the post genomic era*. Asian Aust J Anim Sci. 2007;20(5):802–810. [Google Scholar]
- 15.Handelsman JRM, Brady SF, Clardy J, Goodman RM. Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem Biol. 1998;5(10):R245–R249. doi: 10.1016/s1074-5521(98)90108-9. [DOI] [PubMed] [Google Scholar]
- 16.Raes JBP. Molecular eco-systems biology: towards an understanding of community function. Nat Rev Microbiol. 2008;6(9):693–699. doi: 10.1038/nrmicro1935. [DOI] [PubMed] [Google Scholar]
- 17.Karlsson FH, Nookaew I, Petranovic D, Nielsen J. Prospects for systems biology and modeling of the gut microbiome. Trends Biotechnol. 2011;29(6):251–258. doi: 10.1016/j.tibtech.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 18.Muegge BD, Kuczynski J, Knights D, Clemente JC, Gonzalez A, Fontana L, Henrissat B, Knight R, Gordon JI. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science. 2011;332(6032):970–974. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.von Mering CHP, Raes J, Tringe SG, Doerks T, Jensen LJ, Ward N, Bork P. Quantitative phylogenetic assessment of microbial communities in diverse environments. Science. 2007;315(5815):1126–1130. doi: 10.1126/science.1133420. [DOI] [PubMed] [Google Scholar]
- 20.Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat Rev Microbiol. 2008;6(10):776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, et al. Evolution of mammals and their gut microbes. Science. 2008;320(5883):1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Magoc T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27(21):2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, Brown CT, Porras-Alfaro A, Kuske CR, Tiedje JM. Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014;42(Database issue):D633–D642. doi: 10.1093/nar/gkt1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mills DK, Entry JA, Voss JD, Gillevet PM, Mathee K. An assessment of the hypervariable domains of the 16S rRNA genes for their value in determining microbial community diversity: the paradox of traditional ecological indices. FEMS Microbiol Ecol. 2006;57(3):496–503. doi: 10.1111/j.1574-6941.2006.00135.x. [DOI] [PubMed] [Google Scholar]
- 26.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71(12):8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lopez-Gonzalez JA, Suarez-Estrella F, Vargas-Garcia MC, Lopez MJ, Jurado MM, Moreno J. Dynamics of bacterial microbiota during lignocellulosic waste composting: studies upon its structure, functionality and biodiversity. Bioresour Technol. 2015;175:406–416. doi: 10.1016/j.biortech.2014.10.123. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Z, Xu D, Wang L, Hao J, Wang J, Zhou X, Wang W, Qiu Q, Huang X, Zhou J, et al. Convergent evolution of rumen microbiomes in high-altitude mammals. Curr Biol. 2016;26(14):1873–1879. doi: 10.1016/j.cub.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 29.Ge RL, Cai Q, Shen YY, San A, Ma L, Zhang Y, Yi X, Chen Y, Yang L, Huang Y, et al. Draft genome sequence of the Tibetan antelope. Nat Commun. 2013;4:1858. doi: 10.1038/ncomms2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hook SESM, Northwood KS, Dijkstra J, France J, Wright AD, McBride BW. Impact of subacute ruminal acidosis (SARA) adaptation and recovery on the density and diversity of bacteria in the rumen of dairy cows. FEMS Microbiol Ecol. 2011;78(2):275–284. doi: 10.1111/j.1574-6941.2011.01154.x. [DOI] [PubMed] [Google Scholar]
- 31.Huo W, Zhu W, Mao S. Impact of subacute ruminal acidosis on the diversity of liquid and solid-associated bacteria in the rumen of goats. World J Microbiol Biotechnol. 2014;30(2):669–680. doi: 10.1007/s11274-013-1489-8. [DOI] [PubMed] [Google Scholar]
- 32.Hess MSA, Egan R, Kim TW, Chokhawala H, Schroth G, Luo S, Clark DS, Chen F, Zhang T, Mackie RI, Pennacchio LA, Tringe SG, Visel A, Woyke T, Wang Z. EM. R. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science. 2011;331(6016):463–467. doi: 10.1126/science.1200387. [DOI] [PubMed] [Google Scholar]
- 33.Kim M, Morrison M, Yu Z. Status of the phylogenetic diversity census of ruminal microbiomes. FEMS Microbiol Ecol. 2011;76(1):49–63. doi: 10.1111/j.1574-6941.2010.01029.x. [DOI] [PubMed] [Google Scholar]
- 34.Li Z, Wright AD, Liu H, Bao K, Zhang T, Wang K, Cui X, Yang F, Zhang Z, Li G. Bacterial community composition and fermentation patterns in the rumen of sika deer (Cervus nippon) fed three different diets. Microb Ecol. 2015;69(2):307–318. doi: 10.1007/s00248-014-0497-z. [DOI] [PubMed] [Google Scholar]
- 35.Ishaq SL, Wright AD. High-throughput DNA sequencing of the ruminal bacteria from moose (Alces alces) in Vermont, Alaska, and Norway. Microb Ecol. 2014;68(2):185–195. doi: 10.1007/s00248-014-0399-0. [DOI] [PubMed] [Google Scholar]
- 36.Gruninger RJ, Sensen CW, McAllister TA, Forster RJ. Diversity of rumen bacteria in Canadian cervids. PLoS One. 2014;9(2):e89682. doi: 10.1371/journal.pone.0089682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nyonyo T, Shinkai T, Mitsumori M. Improved culturability of cellulolytic rumen bacteria and phylogenetic diversity of culturable cellulolytic and xylanolytic bacteria newly isolated from the bovine rumen. FEMS Microbiol Ecol. 2014;88(3):528–537. doi: 10.1111/1574-6941.12318. [DOI] [PubMed] [Google Scholar]
- 38.Schnorr SL, Candela M, Rampelli S, Centanni M, Consolandi C, Basaglia G, Turroni S, Biagi E, Peano C, Severgnini M, et al. Gut microbiome of the Hadza hunter-gatherers. Nat Commun. 2014;5:3654. doi: 10.1038/ncomms4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chaucheyras-Durand F, Ossa F. REVIEW: the rumen microbiome: composition, abundance, diversity, and new investigative tools. Prof Anim Sci. 2014;30(1):1–12. [Google Scholar]
- 40.Cunha IS, Barreto CC, Costa OYA, Bomfim MA, Castro AP, Kruger RH, Quirino BF. Bacteria and Archaea community structure in the rumen microbiome of goats (Capra hircus) from the semiarid region of Brazil. Anaerobe. 2011;17(3):118–124. doi: 10.1016/j.anaerobe.2011.04.018. [DOI] [PubMed] [Google Scholar]
- 41.Tajima K, Arai S, Ogata K, Nagamine T, Matsui H, Nakamura M, Aminov RI, Benno Y. Rumen bacterial community transition during adaptation to high-grain diet. Anaerobe. 2000;6(5):273–284. [Google Scholar]
- 42.Jami E, Israel A, Kotser A, Mizrahi I. Exploring the bovine rumen bacterial community from birth to adulthood. ISME J. 2013;7(6):1069–1079. doi: 10.1038/ismej.2013.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pitta DW, Pinchak WE, Indugu N, Vecchiarelli B, Sinha R, Fulford JD. Metagenomic analysis of the rumen microbiome of steers with wheat-induced frothy bloat. Front Microbiol. 2016;7:689. doi: 10.3389/fmicb.2016.00689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Long RJ, Ding LM, Shang ZH, Guo XH. The yak grazing system on the Qinghai-Tibetan plateau and its status. Rangel J. 2008;30:241–246. [Google Scholar]
- 45.Qiu Q, Zhang G, Ma T, Qian W, Wang J, Ye Z, Cao C, Hu Q, Kim J, Larkin DM, et al. The yak genome and adaptation to life at high altitude. Nat Genet. 2012;44(8):946–949. doi: 10.1038/ng.2343. [DOI] [PubMed] [Google Scholar]
- 46.Xue D, Chen H, Zhao X, Xu S, Hu L, Xu T, Jiang L, Zhan W. Rumen prokaryotic communities of ruminants under different feeding paradigms on the Qinghai-Tibetan plateau. Syst Appl Microbiol. 2017;40(4):227–236. doi: 10.1016/j.syapm.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 47.Bohra V, Dafale NA, Purohit HJ. Understanding the alteration in rumen microbiome and CAZymes profile with diet and host through comparative metagenomic approach. Arch Microbiol. 2019;201(10):1385–1397. doi: 10.1007/s00203-019-01706-z. [DOI] [PubMed] [Google Scholar]
- 48.Sbardellati DL, Fischer A, Cox MS, Li W, Kalscheur KF, Suen G. The bovine epimural microbiota displays compositional and structural heterogeneity across different ruminal locations. J Dairy Sci. 2020;103(4):3636–3647. doi: 10.3168/jds.2019-17649. [DOI] [PubMed] [Google Scholar]
- 49.Wang B, Luo Y, Su R, Yao D, Hou Y, Liu C, Du R, Jin Y. Impact of feeding regimens on the composition of gut microbiota and metabolite profiles of plasma and feces from Mongolian sheep. J Microbiol. 2020;58(6):472–482. doi: 10.1007/s12275-020-9501-0. [DOI] [PubMed] [Google Scholar]
- 50.Cui X, Wang Z, Yan T, Chang S, Wang H, Hou F. Rumen bacterial diversity of Tibetan sheep (Ovis aries) associated with different forage types on the Qinghai-Tibetan Plateau. Can J Microbiol. 2019;65(12):859–869. doi: 10.1139/cjm-2019-0154. [DOI] [PubMed] [Google Scholar]
- 51.Cobellis G, Yu Z, Forte C, Acuti G, Trabalza-Marinucci M. Dietary supplementation of Rosmarinus officinalis L. leaves in sheep affects the abundance of rumen methanogens and other microbial populations. J Anim Sci Biotechnol. 2016;7:27. doi: 10.1186/s40104-016-0086-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. The information of sheep in this study. Table S2. Sequence data of samples. Table S3. The estimators of sequence diversity and richness.
Data Availability Statement
All data generated or analyzed during this study are included in this published article, and also available from the corresponding author on reasonable request.