Abstract
Breast cancer is one of the most common malignancies and the leading cause of cancer-associated death among women. Anterior gradient 3 (AGR3) is a cancer-associated gene and is similar to its homologous oncogene AGR2. However, whether AGR3 participates in breast cancer progression remains unclear. The present study aimed to investigate the function of AGR3 in ER-positive breast cancer. In the present study, reverse transcription-quantitative PCR was used to detect AGR3 mRNA expression in breast cancer tissues and cell lines; linear correlation analysis was used to investigate the correlation between AGR3 and estrogen receptor 1 (ESR1) expression in breast cancer via GEO dataset analysis; western blotting was used to assess the levels of AGR3, ER and GAPDH; small interfering (si)RNA transfection was used to knock down AGR3 and ESR1 expression; and finally the Cell Counting Kit-8 assay was used to evaluate cell viability. In the present study, AGR3 expression was markedly increased in estrogen receptor (ER)-positive breast cancer tissues and cell lines compared with that in ER-negative breast cancer. AGR3 expression was upregulated in estrogen-treated T47D cells, whereas 4-hydroxytamoxifen, an inhibitor of estrogen-ER activity in breast cancer cells, downregulated AGR3 expression in T47D cells. Functional assays demonstrated that knockdown of AGR3 using siRNAs inhibited T47D cell proliferation compared with that of the negative control group. Additionally, AGR3 expression was decreased after knocking down ESR1. The present results suggested that AGR3 may serve an important role in estrogen-mediated cell proliferation in breast cancer and that AGR3 knockdown may be a potential therapeutic strategy for ER-positive breast cancer.
Keywords: anterior gradient 3, estrogen, estrogen receptor-positive breast cancer, proliferation
Introduction
Breast cancer is the most frequently diagnosed cancer and the leading cause of cancer-associated death among women worldwide (1). In 2018, the International Agency for Research on Cancer estimated a disease incidence rate of 24.2% and mortality rate of 15.0% in women across 185 countries (1). Breast cancer is divided into four subtypes according to the 2015 St. Gallen consensus (2): Luminal A-like, luminal B-like, human epidermal growth factor receptor 2 (HER2)-positive and triple-negative (TN) subtypes, which are based on estrogen receptor (ER), progesterone receptor (PR), Ki-67 and HER2 immunohistochemical status (2). ERα is a nuclear transcription factor encoded by the estrogen receptor 1 (ESR1) gene and activated by estrogen (3). ERα has different effects in normal breast epithelial cells and breast cancer cells, and it serves a predictive role in the response to endocrine therapies (4). Estrogen promotes cell proliferation and breast cancer development in an ER-dependent manner (5); in turn, ERα promotes breast cancer tumorigenesis and progression (6,7). Therefore, estrogens serve an essential role in regulating breast cancer cell proliferation, and estrogen-activated ERα is a crucial factor for breast cancer development and therapy.
Anterior gradient protein 3 (AGR3) is a member of the protein disulfide isomerase (PDI) gene family, which consists of 21 members (http://www.genenames.org/cgi-bin/genefamilies/set/692), and AGR3 also has two AGR subfamilies, AGR1 and AGR2 (8,9). AGR2 has been widely investigated in breast cancer and is known to participate in numerous aspects of its development and therapy, including cell proliferation and migration (10,11). Although AGR3 and AGR2 are highly similar homologous genes, the function of AGR3 in cancer may not be the same as that of the metastasis-associated AGR2 (12). AGR3 is upregulated in serous borderline ovarian tumor compared with serous ovarian carcinoma, and high levels of AGR3 predict a longer survival time in patients with serous ovarian carcinoma (13). In prostate cancer cells, AGR3 is upregulated by androgens and estrogen in an androgen receptor dependent manner (14). Additionally, AGR3 is highly expressed in intrahepatic cholangiocarcinoma compared with its expression levels in hepatocellular carcinoma (15). In breast cancer, AGR3 is positively associated with low histological grade breast tumors (16). Recent studies have demonstrated that extracellular AGR3 can regulate breast cancer cell migration via Src signaling (17), and that AGR3 can promote the proliferative and invasive abilities of breast cancer cells, as well as chemotherapy response (18).
Although differential expression of AGR3 has been identified among different types of cancer, including ovarian, prostate, liver and breast cancers (13–16), the role of AGR3 in breast cancer oncogenesis and development remains unclear. The present study aimed to investigate the association between AGR3 and ER status, and the function of AGR3 in ER-positive breast cancer. It was hypothesized that AGR3 may promote breast cancer development in an ER-dependent manner, and AGR3 may serve as a potential therapeutic target for patients with ER-positive breast cancer.
Materials and methods
Tissue sample collection
A total of 72 breast tumor and paired adjacent normal tissue samples were collected from 72 patients with breast cancer (age range, 30–74 years; median age, 46 years) at The First Affiliated Hospital of Chongqing Medical University (Chongqing, China) between July 2017 and October 2017. Female patients with primary breast cancer, normal cardiopulmonary function and willingness to undergo breast surgery were included, while patients with secondary breast cancer, intolerable or unwilling to undergo breast surgery, and male patients with breast cancer were excluded from the present study. All patients signed informed consent for the retention and analysis of their tissues for research purposes. Breast tumor and paired normal tissue samples (>3 cm from the primary tumor boundary) were collected after surgery and immediately transferred to liquid nitrogen for fast-frozen storage and long-term storage at −80°C. The collection of tumor samples and patient information was approved by the Ethics Committee of The First Affiliated Hospital of Chongqing Medical University (approval no. 2017-16).
Cell culture and drug treatment
MCF7, T47D, ZR75-1, BT549, MDA-MB-231, MDA-MB-468 and SK-BR-3 cell lines were obtained from the American Type Culture Collection. T47D and ZR75-1 cells were cultured at 37°C with 5% CO2 in RPMI-1640 medium, whereas the other cell lines were grown in DMEM, both RPMI-1640 and DMEM medium were supplemented with 10% FBS (all Gibco; Thermo Fisher Scientific, Inc.). Before adding estrogen (MedChemExpress, Inc.), cells in the logarithmic growth phase were seeded in 6-well plates at a confluency of 60–70% and were serum-starved for 24 h in DMEM. RNA was extracted from estrogen-treated cells at different time points (0, 1, 3, 6, 12 and 24 h). For 4-hydroxytamoxifen (4-OH Tam; MedChemExpress, Inc.) treatment, cells in the logarithmic growth phase were seeded in 6-well plates at a confluency of 60–70% and cultured in DMEM supplemented with 10% FBS. Different concentrations of 4-OH Tam (0, 2, 4, 6, 8 and 10 µmol) were added, and RNA and proteins were extracted after 24 h for RT-qPCR and western blot analysis.
siRNA transfection
The siRNAs sequences used to target AGR3 and ESR1 (Shanghai GenePharma Co., Ltd.) were as follows: AGR3-siRNA1 forward, 5′-CAGAUUGUACACAUAUGAG-3′ and reverse, 5′-CUCAUAUGUGUACAAUCUG-3′; AGR3-siRNA2 forward, 5′-AGUUCAUCAUGCUAAACCU-3′ and reverse, 5′-AGGUUUAGCAUGAUGAACU-3′; ESR1-siRNA1 forward, 5′-GGAGGAUGUUGAAACACAATT-3′ and reverse, 5′-UUGUGUUUCAACAUUCUCCTT-3′; and ESR1-siRNA2 forward, 5′-GGAUUUGACCCUCCAUGAUTT-3′ and reverse, 5′-AUCAUGGAGGGUCAAAUCCTT-3′. T47D cells (at a confluency of 60–70%) were transfected with siRNAs (100 nM) using Lipofectamine® RNAiMAX reagent (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The culture medium was changed after 4–6 h, and knockdown efficiencies were determined by RT-qPCR after 24 h.
RNA extraction and qPCR analysis
Total RNA was extracted from breast cancer tissues and cells using TRIzol reagent (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol, and the RNA concentration was measured using a NanoDrop ND-2000 (Thermo Fisher Scientific, Inc.). cDNA was synthesized from 1 µg total RNA with a PrimeScript II cDNA Synthesis kit (37°C for 15 min, 85°C for 5 sec, 4°C for short preservation (usually 30 min prior to qPCR); Takara Bio, Inc.). Targeted genes were detected with a SYBR Premix Ex Taq II qPCR kit (Takara Bio, Inc.) using a primer mixture, and cDNA was amplified by CFX96 fluorescence qPCR (thermocycling conditions: Initial denaturation at 95°C for 2 min; 40 of cycles of denaturation at 95°C for 30 sec, annealing at 58°C for 30 sec, elongation at 72°C for 20 sec; and a final extension at 65°C for 5 sec; Bio-Rad Laboratories, Inc.). Gene-specific primer sequences were designed as follows: AGR3 forward, 5′-AGAGGCCTCCTCAGACACTC-3′ and reverse, 5′-GGCACATATTGCCCATCAGGT-3′; ESR1 forward, 5′-GGTCAGTGCCTTGTTGGATG-3′ and reverse, 5′-CAGGTTGGTGAGTAAGC-3′; GAPDH forward, 5′-TTCCAGGAGCGAGATCCCT-3′ and reverse, 5′-GGCTGTTGTCATACCTTCTCATGG-3′; Growth regulation by estrogen in breast cancer 1 (GREB1) forward, 5′-AAATCGAGGATGTGGAGTG-3′ and reverse, 5′-TCTCACCAAGCAGGAGGA-3′. mRNA abundance was calculated according to the 2−ΔΔCq method (19), relative to the internal control GAPDH.
Protein extraction and western blot analysis
Total protein from cell lines was extracted on ice using RIPA lysis buffer (Thermo Fisher Scientific, Inc.). Protein concentration was quantified using a Pierce™ BCA Protein assay kit (Thermo Fisher Scientific, Inc.), mixed with 5X SDS-PAGE loading buffer and double-distilled H2O, boiled at 95°C for 10 min and stored at −20°C. Proteins (30 ug/lane) were separated by 12% SDS-PAGE and transferred onto 0.2 µm immobilon-PSQ polyvinylidene difluoride membranes (EMD Millipore). The membranes were blocked with 5% skimmed powdered milk for 1 h at room temperature and incubated overnight at 4°C with the following primary antibodies: Antibodies against GAPDH (1:1,000; cat. no. 10494-1-AP) and ER (1:1,000; cat. no. 21244-1-AP) were purchased from Wuhan Sanying Biotechnology, and the antibody against AGR3 (1:800; cat. no. AP9424b) was purchased from Abgent Biotech Co., Ltd. After rinsing three times with Tris-buffered saline containing 0.1% Tween-20 (TBST) for 5 min, the membranes were incubated with HRP-conjugated anti-rabbit IgG (cat. no. NEF812001EA) or anti-mouse IgG (cat. no. NEF822001EA) secondary antibodies (both at a dilution ratio of 1:3,000 and from PerkinElmer, Inc.) for 90 min at room temperature, rinsed 3 times with TBST for 5 min and soaked in an enhanced chemiluminescence detection reagent (Bio-Rad Laboratories, Inc.) for 1 min. The membranes were developed using a GeneGnome Chemiluminescence Imaging system (Syngene) to automatically detect protein levels, which were quantified according to the gray value and normalized to that of GAPDH using ImageJ software (version 1.8.0; National Institutes of Health).
Cell Counting Kit-8 (CCK-8) cell viability test
Cells were seeded into 96-well plates at a density of 5×103 cells/well following siRNA transfection, or following the addition of different concentrations of 4-OH Tam (0, 1, 2, 3, 4, 5, 6, 7, 8, 9 and 10 µmol) at 37°C for 24 h. CCK-8 solution (MedChemExpress, Inc.) was used according to the manufacturer's protocol. The cells were incubated for 2 h at 37°C. The mixture of CCK-8 solution and DMEM supplemented with 10% FBS was used as a blank control, and absorbance was measured at 450 nm. The experiment was repeated in triplicate.
Gene Expression Omnibus (GEO) and Gene Set Enrichment Analysis (GSEA)
Datasets from patients with breast cancer were obtained from the GEO data repository (https://www.ncbi.nlm.nih.gov/geo) and The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/). The GEO datasets used in the current study were as follows: Richardson (GSE5460; n=129) (20), Borresen-Dale (GSE19783; n=115) (21) and Wang (GSE19615; n=115) (22). Series matrix files were downloaded from the online GEO datasets and used for correlation analyses. GSEA was used to explore in which pathway AGR3 may be involved. The dataset TCGA-BRCA was downloaded from TCGA using the GDC Data Transfer Tool (23), processed and analyzed using R2 software (http://r2.amc.nl) and normalized with the ‘DESeq2’ package (24), according to the variance stabilizing transformation method (25). A total of 186 Kyoto Encyclopedia of Genes and Genomes (KEGG) gene sets from the Molecular Signatures Database (MSigDB; http://www.broadinstitute.org/gsea/msigdb/) were used in GSEA. GSEA requires ≥2 different phenotypes. TCGA data for 1,096 patients with breast cancer were divided into 2 groups according to AGR3 expression. The top 10% (110) of the data with the highest AGR3 expression were defined as the high expression group, and the bottom 10% (110) of the date with the lowest AGR3 expression were defined as the low expression group. The R script of GSEA from the Broad Institute (https://www.gsea-msigdb.org/gsea) was used for the GSEA analysis of the two groups.
Statistical analysis
All the statistical analyses were performed using GraphPad Prism 7 (GraphPad Software, Inc.) and R 3.4.0 (https://www.r-project.org), for GSEA analysis. The statistical significance of GSEA analysis was determined by unpaired Student's t-test. The statistical significance of paired groups was determined by Wilcoxon matched-pairs signed-rank test. The statistical significance of unpaired two groups was determined by Mann-Whitney test. Pearson's correlation analysis was performed to analyze the correlation of target genes. Multiple groups were statistically compared using one-way ANOVA and Dunnett's multiple comparisons test. The results are presented as the mean ± standard deviation (n≥3). P<0.05 was considered to indicate a statistically significant difference.
Results
AGR3 expression is upregulated in luminal breast cancer tissues and is positively correlated with ESR1 expression
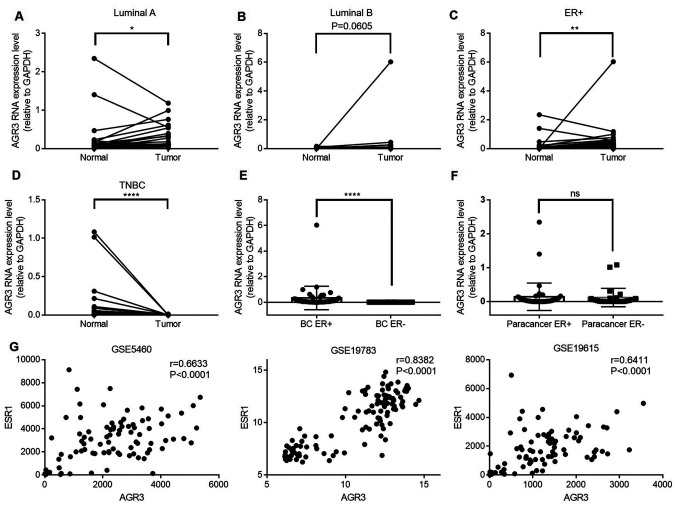
To investigate AGR3 expression in breast cancer, AGR3 expression was analyzed in ER-positive and ER-negative paired cancer tissues and adjacent normal tissues by RT-qPCR. AGR3 expression was significantly upregulated in luminal A breast cancer tissues compared with that in their paired adjacent normal tissues (n=21; Tumor, 0.3335±0.0719 vs. normal, 0.2704±0.1231; P=0.0421; Fig. 1A), and was visibly increased in luminal B breast cancer tissues compared with normal tissues (n=23; Tumor, 0.3344±0.2599 vs. normal, 0.0279±0.0084; P=0.0605; Fig. 1B). Among the 44 pairs of ER-positive breast cancer and adjacent normal tissues, the RNA levels of AGR3 were significantly upregulated in breast cancer tissues compared with that in paired normal tissues (n=44; Tumor, 0.3340±0.1386 vs. normal, 0.1436±0.0610; P=0.0016; Fig. 1C). Additionally, AGR3 RNA expression was detected in patients with TN breast cancer and was significantly downregulated in ER-negative TN breast cancer tissues compared with that in normal adjacent tissues (n=28; Tumor, 0.0017±0.0006 vs. normal, 0.1197±0.0514; P<0.0001; Fig. 1D).
Figure 1.
AGR3 expression is upregulated in ER-positive breast cancer and positively correlates with ESR1 expression. (A-C) AGR3 RNA expression was detected in (A) luminal A, (B) luminal B and (C) ER-positive breast cancer tissues and their paired normal tissues, and compared using the Wilcoxon matched-pairs signed-rank test. (D) AGR3 RNA expression was detected in TN breast cancer tissues and their paired normal tissues, and compared using the Wilcoxon matched-pairs signed-rank test. (E) AGR3 RNA expression was compared between ER-positive and ER-negative breast cancer tissues using the Mann-Whitney test. (F) AGR3 RNA expression was compared between ER-positive and ER-negative adjacent normal tissues using the Mann-Whitney test. (G) AGR3 and ESR1 expression values were extracted from GEO dataset series matrix files (GSE5460, GSE19783 and GSE19615), and correlation analyses of AGR3 and ESR1 from each GEO expression dataset were conducted using the Pearson correlation coefficient. *P<0.05, **P<0.01, ****P<0.0001. AGR3, anterior gradient 3; ER, estrogen receptor; ESR1, estrogen receptor 1; TNBC, triple-negative breast cancer; ns, not significant; GEO, Gene Expression Omnibus.
AGR3 expression between ER-positive and ER-negative breast cancer tissues, as well as between their paired ER-positive and ER-negative adjacent normal tissues was analyzed. AGR3 RNA levels were significantly upregulated in ER-positive (n=44) compared with in ER-negative (n=28) breast cancer tissues (BC ER+, 0.3340±0.1386 vs. BC ER-, 0.0017±0.0006; P<0.0001; Fig. 1E), although this difference was not observed between ER-positive (n=44) and ER-negative (n=28) adjacent normal tissues (paracancer ER+, 0.1436±0.0610 vs. paracancer ER-, 0.1197±0.0514; P=0.5994; Fig. 1F), suggesting that AGR3 may be a tumor-specific gene that is upregulated in breast cancer depending on the ER status. To confirm the present findings, the correlation between AGR3 and ESR1 expression was analyzed using the GEO datasets. The correlation coefficients in three GEO datasets were analyzed, and all displayed significant positive correlations between AGR3 and ESR1 expression (GSE5460, r=0.6633; 95% CI: 0.554–0.7502; P<0.0001; GSE19783, r=0.8382; 95% CI: 0.7739–0.8854; P<0.0001; GSE19615, r=0.6411; 95% CI: 0.5189–0.7376; P<0.0001; Fig. 1G). The present results suggested that ESR1 and AGR3 may act in a similar manner and play similar roles in breast cancer.
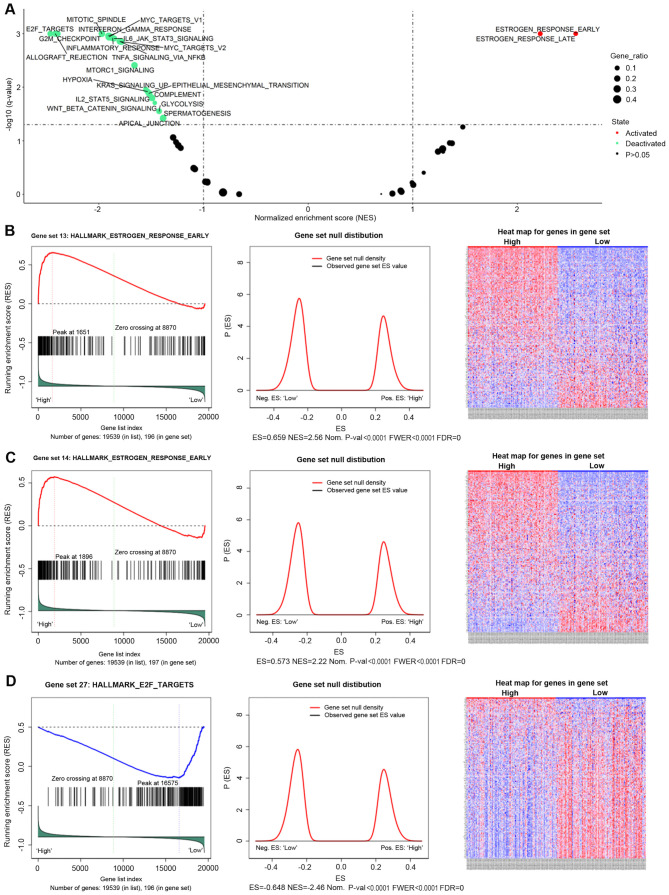
AGR3 is associated with the estrogen response and cancer-associated signaling pathways according to GSEA
To comprehensively understand the role of AGR3 in breast cancer, GSEA was performed to explore the potential pathways associated with AGR3. GSEA is an analytical method that can reveal the biological pathways of candidate genes from gene sets (26). A total of 186 KEGG gene sets from the MSigDB were used for the GSEA, and a number of AGR3-associated gene sets were identified (Fig. 2A and Table I). AGR3 was positively associated with the ‘HALLMARK_ESTROGEN_RESPONSE_EARLY’ [enrichment score (ES)=0.6592; normalized ES (NES)=2.5579; nominal P-value (Nom P<0.0001; false discovery rate q-value (FDR q-val)=0; family-wise error rate P-value (FWER P<0.0001; Fig. 2B] and the ‘HALLMARK_ESTROGEN_RESPONSE_LATE’ (ES=0.57267; NES=2.2178; NOM P<0.0001; FDR q-val=0; FWER P<0.0001; Fig. 2C) gene sets, indicating that AGR3 may function as an estrogen response factor. Additionally, AGR3 was negatively associated with ‘HALLMARK_E2F_TARGETS’ (ES=−0.64839; NES=−2.4647; NOM P<0.0001; FDR q-val=0; FWER P<0.0001; Fig. 2D), allograft rejection, G2/M checkpoint, mitotic spindle, interferon γ response, Myc targets, mTORc1 signaling and other gene sets (Fig. 2A and Table I), indicating that AGR3 may function in different cancer-associated signaling pathways in breast cancer.
Figure 2.
GSEA results of hallmark gene set analysis of AGR3 in breast cancer. (A) Volcano plot of the AGR3 GSEA results; enriched hallmark gene sets with an FDR q-value <0.01 were considered statistically significant. GSEA enrichment scores (left), gene set null distribution (middle) for gene sets and heat maps (right) for genes in the hallmark gene sets: (B) ‘HALLMARK_ESTROGEN_RESPONSE_EARLY’, (C) ‘HALLMARK_ESTROGEN_RESPONSE_LATE’ and (D) ‘HALLMARK_E2F_TARGETS’. The gene set null distribution was drawn using 1,000 permutations, and the gene labels were rearranged each time. ES, enrichment score; NES, normalized ES; NOM p-val, nominal P-value; FDR, false discovery rate; FWER p-val: Familywise error rate P-value; GSEA, Gene Set Enrichment Analysis; AGR3, anterior gradient 3.
Table I.
Gene Set Enrichment Analysis results of hallmark gene sets of anterior gradient 3 in breast cancer.
| Gene set | Size | ES | NES | NOM P-value | FDR q-value | FWER P-value | Gene ratio |
|---|---|---|---|---|---|---|---|
| HALLMARK_ESTROGEN_RESPONSE_EARLY | 196 | 0.65919 | 2.5579 | <0.0001 | 0.00000000 | <0.0001 | 0.0845 |
| HALLMARK_ESTROGEN_RESPONSE_LATE | 197 | 0.57267 | 2.2178 | <0.0001 | 0.00000000 | <0.0001 | 0.0970 |
| HALLMARK_PEROXISOME | 103 | 0.41390 | 1.4776 | 0.0113100 | 0.05422300 | 0.1878 | 0.1120 |
| HALLMARK_E2F_TARGETS | 196 | −0.64839 | −2.4647 | <0.0001 | 0.00000000 | <0.0001 | 0.1520 |
| HALLMARK_ALLOGRAFT_REJECTION | 199 | −0.63245 | −2.4169 | <0.0001 | 0.00000000 | <0.0001 | 0.2010 |
| HALLMARK_G2M_CHECKPOINT | 194 | −0.63038 | −2.3917 | <0.0001 | 0.00000000 | <0.0001 | 0.1380 |
| HALLMARK_MITOTIC_SPINDLE | 198 | −0.51817 | −1.9722 | <0.0001 | 0.00000000 | <0.0001 | 0.2300 |
| HALLMARK_INTERFERON_GAMMA_RESPONSE | 198 | −0.50047 | −1.9088 | <0.0001 | 0.00009480 | 0.0004 | 0.2280 |
| HALLMARK_MYC_TARGETS_V1 | 197 | −0.49912 | −1.9013 | <0.0001 | 0.00013949 | 0.0007 | 0.3410 |
| HALLMARK_INFLAMMATORY_RESPONSE | 196 | −0.49297 | −1.8694 | <0.0001 | 0.00020188 | 0.0012 | 0.2130 |
| HALLMARK_IL6_JAK_STAT3_SIGNALING | 87 | −0.54467 | −1.8545 | <0.0001 | 0.00023175 | 0.0016 | 0.1880 |
| HALLMARK_TNFA_SIGNALING_VIA_NFKB | 198 | −0.47236 | −1.7999 | <0.0001 | 0.00039442 | 0.0031 | 0.2300 |
| HALLMARK_MYC_TARGETS_V2 | 58 | −0.56107 | −1.7859 | 0.0003642 | 0.00040505 | 0.0035 | 0.2310 |
| HALLMARK_MTORC1_SIGNALING | 197 | −0.43572 | −1.6602 | <0.0001 | 0.00288310 | 0.0266 | 0.2200 |
| HALLMARK_KRAS_SIGNALING_UP | 195 | −0.40799 | −1.5502 | 0.0003493 | 0.01011000 | 0.0989 | 0.1380 |
ES, enrichment score; NES, normalized ES; NOM, nominal; FDR, false discovery rate; FWER, family-wise error rate.
AGR3 is regulated by estrogen in T47D cells
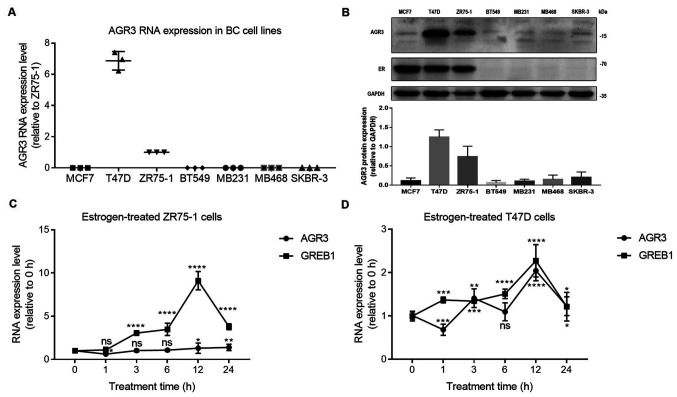
To explore the possible association between AGR3 and ESR1, the RNA expression levels of AGR3 in different breast cancer cell lines, including the ER-positive breast cancer MCF7, ZR75-1 and T47D cell lines and the ER-negative breast cancer MB231, MB468, BT549 and SKBR-3 cell lines, were detected. AGR3 RNA expression was markedly upregulated in T47D cells and was relatively abundant in the ZR75-1 cell line, whereas very low levels were observed in the other ER-negative breast cancer cell lines (MCF7, 0.0009±0.0001; T47D, 6.8700±0.3437; ZR75-1, 1.0000±0.0000 (for the convenience of comparing with other cell lines, ZR75-1 was normalized to 1); BT549, NA (not available value that was too low to detect); MB231, NA; MB468, 0.0013±0.0001; and SKBR-3, 5.443×10−5±5.862×10−6; Fig. 3A). To confirm AGR3 protein expression in breast cancer cell lines, AGR3 protein levels were detected using an AGR3 antibody. AGR3 protein was highly expressed in T47D and ZR75-1 cells compared with the other cell lines (MCF7, BT549, MB231, MB468 and SKBR-3). However, the AGR3 protein was lowly expressed in ER-negative breast cancer cell lines, including BT549, MB231, MB468 and SKBR-3 cells (Fig. 3B), consistent with the findings at the RNA level. ER protein was also expressed in all the ER-positive breast cancer cell lines; however, no ER protein was detected in ER-negative BT549, MB231, MB468 and SKBR-3 breast cancer cell lines (Fig. 3B).
Figure 3.
AGR3 is upregulated in estrogen receptor-positive breast cancer cell lines and regulated by estrogen in T47D breast cancer cells. (A) Reverse transcription-quantitative PCR analysis of AGR3 RNA expression in breast cancer MCF7, T47D, ZR75-1 (normalized to 1 for the convenience of comparing with other cell lines), BT549, MB231, MB468 and SKBR-3 cell lines. (B) Western blot analysis of AGR3 protein levels in breast cancer MCF7, T47D, ZR75-1, BT549, MB231, MB468 and SKBR-3 cell lines (top) and the bar graph of AGR3 protein levels (bottom). AGR3 and GREB1 RNA expression was analyzed in (C) ZR75-1 cells and (D) T47D cells after treatment with 10 nmol estrogen for 0, 1, 3, 6, 12 and 24 h. The RNA expression level of 0 h was normalized to 1 in (C) and (D), and GREB1 was used as an estrogen-regulated positive control. Each experiment was repeated ≥3 times. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001 vs. 0 h. AGR3, anterior gradient 3; BC, breast cancer; GREB1, Growth regulating estrogen receptor binding 1; ns, not significant.
Estrogen is an important factor in breast cancer, and it can directly activate the ER (27). To examine whether estrogen regulates AGR3 expression, estrogen was added to T47D and ZR75-1 cells at a final concentration of 10 nmol, and the RNA level of AGR3 at different time intervals after treatment was detected. Although the RNA levels of GREB1, an estrogen response molecule (28), were gradually increased and reached the highest level after 12 h of estrogen treatment in ZR75-1 cells (12 h, 9.1120±0.2447 vs. 0 h, 1.0000±0.0000 (0 h was normalized to 1); P<0.0001; one-way ANOVA, F=512.4 and P<0.0001), AGR3 did not appear to be significantly increased in ZR75-1 cells treated with estrogen for 12 h (12 h, 1.3008±0.1390 vs. 0 h, 1.0000±0.0000 (0 h was normalized to 1); P=0.0368) or 24 h (24 h, 1.3900±0.08388 vs. 0 h, 1.0000±0.0000; P=0.0085; one-way ANOVA, F=14.8 and P<0.0001; Fig. 3C). However, the RNA expression levels of AGR3 and GREB1 were increased in a similar manner in T47D cells, and both reached their highest levels after estrogen treatment for 12 h (AGR3: 12 h, 2.043±0.05334 vs. 0 h, 1.0000±0.0000 (0 h was normalized to 1); P<0.0001; one-way ANOVA, F=110.6 and P<0.0001; GREB1: 12 h, 2.274±0.08661 vs. 0 h, 1.0000±0.0000 (0 h was normalized to 1); P<0.0001; one-way ANOVA, F=77.2 and P<0.0001; Fig. 3D), suggesting that AGR3 may be regulated by estrogen in ER-positive breast cancer cells.
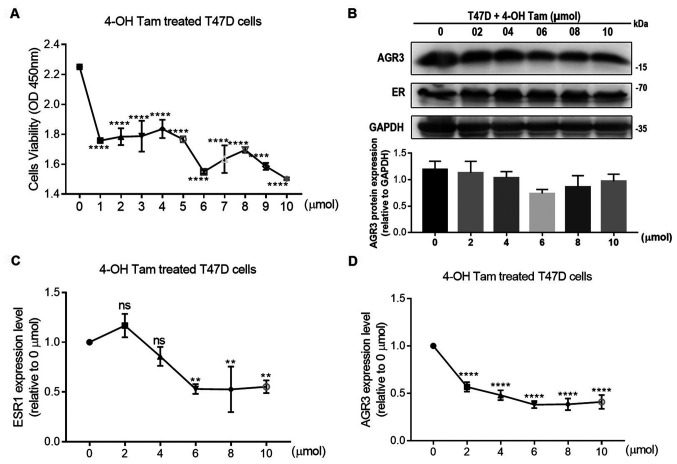
To further investigate the association between estrogen and AGR3, the present study assessed whether 4-OH Tam may influence the viability of T47D cells and the expression levels of AGR3. When different concentrations of 4-OH Tam were added to T47D cells, cell viability was significantly decreased with increasing 4-OH Tam concentrations, and was the lowest after the addition of 10 µmol 4-OH Tam (10 µmol, 1.5030±0.0052 vs. 0 µmol, 2.2510±0.0092; P<0.0001; one-way ANOVA, F=48.2 and P<0.0001; Fig. 4A). Additionally, the protein levels of AGR3 and ER were analyzed; these appeared to decrease following 4-OH Tam treatment, with protein levels of AGR3 in T47D cells reaching their lowest level using 6 µmol 4-OH Tam, and ER protein levels also had a visible decline following addition of 6 µmol 4-OH Tam (Fig. 4B). Additionally, AGR3 and ESR1 RNA expression was significantly decreased in T47D cells following treatment with different concentrations of 4-OH Tam. ESR1 and AGR3 RNA expression reached their lowest levels in T47D cells using 6 µmol 4-OH Tam (ESR1: 6 µmol, 0.5301±0.0282 vs. 0 µmol, 1.0000±0.0000 (0 µmol was normalized to 1); P=0.0015; one-way ANOVA, F=16.9 and P<0.0001; AGR3: 6 µmol, 0.3810±0.0212 vs. 0 µmol, 1.0000±0.0000 (0 µmol was normalized to 1); P<0.0001; one-way ANOVA, F=65.8 and P<0.0001; Fig. 4C and D). Therefore, both ESR1 and AGR3 expression may be regulated by 4-OH Tam, which further confirmed the effect of estrogen on AGR3 RNA and protein expression.
Figure 4.
AGR3 and ER are negatively regulated by 4-OH Tam in T47D cells. (A) The viability of T47D cells treated with 0–10 µmol 4-OH Tam was detected using the Cell Counting Kit-8 assay. (B) The protein levels of AGR3 and ER in T47D cells treated with 0, 2, 4, 6, 8 and 10 µmol 4-OH Tam for 24 h were detected by western blotting. (C and D) RNA expression levels of (C) ESR1 and (D) AGR3 were detected in T47D cells after the addition of different concentrations of 4-OH Tam for 24 h. The RNA expression level of 0 µmol was normalized to 1 in (C) and (D). Each experiment was repeated ≥3 times. **P<0.01 and ****P<0.0001 vs. 0 µmol. 4-OH Tam, 4-hydroxytamoxifen; AGR3, anterior gradient 3; ER, estrogen receptor; ESR1, estrogen receptor 1; ns, not significant.
AGR3 promotes T47D cell proliferation in an estrogen- dependent manner
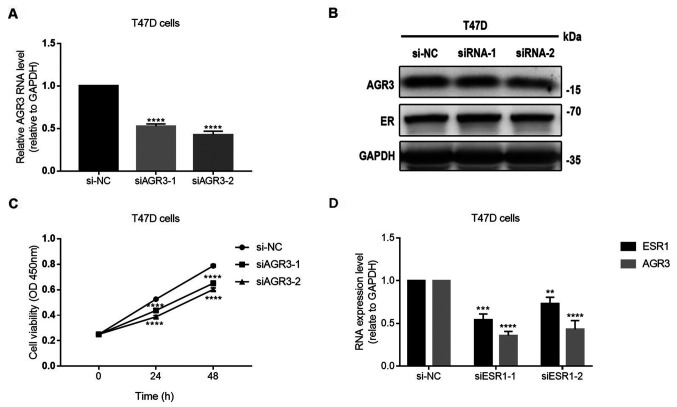
To further explore the role of AGR3 in ER-positive breast cancer, the present study analyzed whether AGR3 expression may influence the ER status by knocking down AGR3 expression in T47D cells using two siRNAs. Both siRNAs significantly decreased AGR3 RNA levels (siAGR3-1, 0.5265±0.0150 vs. si-NC, 1.0000±0.0000; P<0.0001; siAGR3-2, 0.4257±0.0243 vs. si-NC, 1.0000±0.0000; P<0.0001; one-way ANOVA, F=345.2 and P<0.0001; Fig. 5A). The protein levels of AGR3 decreased after transfecting T47D cells with the AGR3 siRNAs, but there was no notable change in ER protein expression (Fig. 5B). After knocking down AGR3 using siRNAs, the viability of T47D cells was significantly decreased compared with that of the negative control cells 24 h post-transfection (siAGR3-1, 0.4383±0.007 vs. si-NC 0.5259±0.002963; P<0.0001; siAGR3-2, 0.3883±0.006245 vs. si-NC, 0.5259±0.002963; P<0.0001; one-way ANOVA, F=150.5 and P<0.0001) and 48 h post-transfection (siAGR3-1, 0.6512±0.0103 vs. si-NC, 0.7879±0.0062; P<0.0001; siAGR3-2, 0.6032±0.0067 vs. si-NC, 0.7879±0.0062; P<0.0001; one-way ANOVA, F=145.5 and P<0.0001) (Fig. 5C). Additionally, siRNAs against ESR1 significantly decreased the expression levels of ESR1 (siESR1-1, 0.7283±0.0456 vs. si-NC, 1.0000±0.0000; P=0.0026; siESR1-2, 0.5435±0.0385 vs. si-NC, 1.0000±0.0000; P=0.0002; one-way ANOVA, F=44.4 and P=0.0003) and AGR3 (siESR1-1, 0.4302±0.0604 vs. si-NC, 1.0000±0.0000; P<0.0001; siESR1-2, 0.3576±0.0285 vs. si-NC, 1.0000±0.0000; P<0.0001; one-way ANOVA, F=83.3 and P<0.0001) in T47D cells (Fig. 5D). In conclusion, the present results suggest that AGR3, which is regulated by ESR1, may promote the proliferation of ER-positive breast cancer cells in an estrogen-dependent manner.
Figure 5.
AGR3 promotes T47D cell proliferation in an estrogen-dependent manner. (A) RNA levels of AGR3 were detected by RT-qPCR after knocking down AGR3 in T47D cells. (B) Protein levels of AGR3 and ER were detected by western blotting after knocking down AGR3 in T47D cells. (C) Cell viability was detected using the Cell Counting Kit-8 assay after knocking down AGR3 in T47D cells for 2 consecutive days. (D) RNA levels of AGR3 and ESR1 were detected by RT-qPCR after knocking down ESR1 in T47D cells. Each experiment was repeated ≥3 times. **P<0.01; ***P<0.001; ****P<0.0001 vs. si-NC. RT-qPCR, reverse transcription-quantitative PCR; siRNA, small interfering RNA; si-NC, small interfering negative control; AGR3, anterior gradient 3; ER, estrogen receptor; ESR1, estrogen receptor 1.
Discussion
AGR2 and AGR3 both belong to the PDI family, and they are similarly expressed in breast cancer due to the 71% similarity in their protein sequences and adjacent positions at 7p21 (29). AGR2 is a widely accepted oncogene in breast cancer and is regulated by estrogen in normal breast tissue and ER-positive breast cancer cells (30,31). Although AGR2 and AGR3 share sequence homology, they are not identical in function, and AGR3 may serve a different role in breast cancer (12). Recent studies have revealed the oncogenic functions of AGR3 in breast cancer. For example, extracellular AGR3 was demonstrated to regulate breast cancer cell migration via Src signaling, which provided an exogenous mode of expression (17), whereas the present study focused on endogenous AGR3 and its function in ER-positive breast cancer cells. In addition, AGR3 promotes the proliferative and invasive abilities of breast cancer cells and chemotherapy response (18), which further confirmed the oncogenic role of AGR3 in breast cancer. In order to fully understand the function of AGR3 in breast cancer, the present study aimed to explore the function of AGR3 in ER-positive breast cancer, and focused on the direct association between AGR3 and ER.
In the present study, AGR3 expression was significantly upregulated in ER-positive compared with ER-negative breast cancer tissues. Considering the close association between AGR3 and ER status, and that ER is a key molecule in distinguishing between low- and high-grade breast cancer, AGR3 RNA expression was detected in ER-positive and ER-negative breast cancer tissues and their paired adjacent normal breast tissues. AGR3 expression was significantly upregulated in luminal breast cancer tissues compared with that in paired adjacent normal tissues, and was significantly downregulated in TN breast cancer tissues compared with that in paired adjacent normal tissues. The results of the present study suggest that there may be different modes of AGR3 expression between ER-positive and ER-negative breast cancer. However, further studies are required to determine whether differential expression tendencies of AGR3 have the same or different function in ER-positive and ER-negative breast cancers. Of note, no differences were observed in AGR3 expression between ER-positive and ER-negative adjacent normal breast tissues, indicating that the differential expression tendency only occurred in breast cancer samples. Therefore, AGR3 may be a tumor-specific gene, and its expression may depend on the ER status of the breast.
To fully understand the role of AGR3 in breast cancer, GSEA of AGR3 was conducted using a breast cancer TCGA dataset; the results revealed that AGR3 was positively associated with estrogen response gene sets, including early and late steps. Additionally, when estrogen was added to T47D cells, AGR3 RNA expression was increased by indicating the peak time, and AGR3 was confirmed to be an estrogen response molecule in ER-positive breast cancer. Furthermore, AGR3 was also negatively associated with several cancer-associated gene sets, such as ‘E2F_TARGETS’, ‘G2M_CHECKPOINT’ and ‘MITOTIC_SPINDLE’, which are associated with the cell cycle regulation; as well as ‘ALLOGRAFT_REJECTION’, ‘IL6_JAK_STAT3_SIGNALING’, ‘TNFA_SIGNALING_VIA_NFKB’, which are associated with immunity and inflammation; ‘EPITHELIAL_MESENCHYMAL_TRANSITION’ and ‘WNT_BETA_CATENIN_SIGNALING’, which are associated with epithelial-mesenchymal transition. Therefore, the function of AGR3 in breast cancer requires further investigation.
In conclusion, the results of the present study demonstrated that AGR3 was an estrogen response gene that was closely associated with ER status in breast cancer. In ER-positive breast cancer, AGR3 expression was positively correlated with ESR1 expression levels, and in ER-negative breast cancer, AGR3 expression was significantly downregulated compared with that in paired adjacent normal tissues. The differential expression modes of AGR3 according to ER status may lead to different functions in breast cancer. In ER-positive breast cancer cells, AGR3 was responsive to estrogen, and ESR1-regulated AGR3 promoted the proliferation of ER-positive T47D breast cancer cells. Thus, AGR3 may serve an important function in estrogen-mediated cell proliferation in breast cancer, whereas the role of AGR3 in ER-negative breast cancer requires further investigation. Knockdown of AGR3 may be a potential therapeutic strategy for ER-positive breast cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science Foundation of China (grant no. 81772979).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
LJ, JX, HY, KT, CY and KL performed the experiments and analyzed the data. SL provided the reagents, designed the experiments and critically revised the manuscript. LJ, SG and RC designed the study and drafted the initial manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The collection of tumor samples and patient information was approved by the Ethics Committee of The First Affiliated Hospital of Chongqing Medical University (approval no. 2017-16; Chongqing, China). All patients signed informed consent for the retention and analysis of their tissues for research purposes.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Coates AS, Winer EP, Goldhirsch A, Gelber RD, Gnant M, Piccart-Gebhart M, Thürlimann B, Senn HJ, Panel M. Tailoring therapies-improving the management of early breast cancer: St gallen international expert consensus on the primary therapy of early breast cancer 2015. Ann Oncol. 2015;26:1533–1546. doi: 10.1093/annonc/mdv221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allred DC. Issues and updates: Evaluating estrogen receptor- alpha, progesterone receptor, and HER2 in breast cancer. Mod Pathol. 2010;23(Suppl 2):S52–S59. doi: 10.1038/modpathol.2010.55. [DOI] [PubMed] [Google Scholar]
- 4.Bai Z, Gust R. Breast cancer, estrogen receptor and ligands. Arch Pharm (Weinheim) 2009;342:133–149. doi: 10.1002/ardp.200800174. [DOI] [PubMed] [Google Scholar]
- 5.Yue W, Yager JD, Wang JP, Jupe ER, Santen RJ. Estrogen receptor-dependent and independent mechanisms of breast cancer carcinogenesis. Steroids. 2013;78:161–170. doi: 10.1016/j.steroids.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 6.Jia M, Dahlman-Wright K, Gustafsson JA. Estrogen receptor alpha and beta in health and disease. Best Pract Res Clin Endocrinol Metab. 2015;29:557–568. doi: 10.1016/j.beem.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 7.Jia M, Andreassen T, Jensen L, Bathen TF, Sinha I, Gao H, Zhao C, Haldosen LA, Cao Y, Girnita L, et al. Estrogen receptor α promotes breast cancer by reprogramming choline metabolism. Cancer Res. 2016;76:5634–5646. doi: 10.1158/0008-5472.CAN-15-2910. [DOI] [PubMed] [Google Scholar]
- 8.Persson S, Rosenquist M, Knoblach B, Khosravi-Far R, Sommarin M, Michalak M. Diversity of the protein disulfide isomerase family: Identification of breast tumor induced Hag2 and Hag3 as novel members of the protein family. Mol Phylogenet Evol. 2005;36:734–740. doi: 10.1016/j.ympev.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 9.Ivanova AS, Tereshina MB, Ermakova GV, Belousov VV, Zaraisky AG. Agr genes, missing in amniotes, are involved in the body appendages regeneration in frog tadpoles. Sci Rep. 2013;3:1279. doi: 10.1038/srep01279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Z, Wu Z, Chen H, Zhu Q, Gao G, Hu L, Negi H, Kamle S, Li D. Induction of anterior gradient 2 (AGR2) plays a key role in insulin-like growth factor-1 (IGF-1)-induced breast cancer cell proliferation and migration. Med Oncol. 2015;32:577. doi: 10.1007/s12032-015-0577-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verma S, Salmans ML, Geyfman M, Wang H, Yu Z, Lu Z, Zhao F, Lipkin SM, Andersen B. The estrogen-responsive Agr2 gene regulates mammary epithelial proliferation and facilitates lobuloalveolar development. Dev Biol. 2012;369:249–260. doi: 10.1016/j.ydbio.2012.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Obacz J, Takacova M, Brychtova V, Dobes P, Pastorekova S, Vojtesek B, Hrstka R. The role of AGR2 and AGR3 in cancer: Similar but not identical. Eur J Cell Biol. 2015;94:139–147. doi: 10.1016/j.ejcb.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 13.King ER, Tung CS, Tsang YT, Zu Z, Lok GT, Deavers MT, Malpica A, Wolf JK, Lu KH, Birrer MJ, et al. The anterior gradient homolog 3 (AGR3) gene is associated with differentiation and survival in ovarian cancer. Am J Surg Pathol. 2011;35:904–912. doi: 10.1097/PAS.0b013e318212ae22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bu H, Schweiger MR, Manke T, Wunderlich A, Timmermann B, Kerick M, Pasqualini L, Shehu E, Fuchsberger C, Cato AC, Klocker H. Anterior gradient 2 and 3-two prototype androgen-responsive genes transcriptionally upregulated by androgens and by oestrogens in prostate cancer cells. FEBS J. 2013;280:1249–1266. doi: 10.1111/febs.12118. [DOI] [PubMed] [Google Scholar]
- 15.Brychtova V, Zampachova V, Hrstka R, Fabian P, Novak J, Hermanova M, Vojtesek B. Differential expression of anterior gradient protein 3 in intrahepatic cholangiocarcinoma and hepatocellular carcinoma. Exp Mol Pathol. 2014;96:375–381. doi: 10.1016/j.yexmp.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 16.Obacz J, Brychtova V, Podhorec J, Fabian P, Dobes P, Vojtesek B, Hrstka R. Anterior gradient protein 3 is associated with less aggressive tumors and better outcome of breast cancer patients. Onco Targets Ther. 2015;8:1523–1532. doi: 10.2147/OTT.S82235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Obacz J, Sommerova L, Sicari D, Durech M, Avril T, Iuliano F, Pastorekova S, Hrstka R, Chevet E, Delom F, Fessart D. Extracellular AGR3 regulates breast cancer cells migration via Src signaling. Oncol Lett. 2019;18:4449–4456. doi: 10.3892/ol.2019.10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu Q, Shao Y, Zhang J, Zhang H, Zhao Y, Liu X, Guo Z, Chong W, Gu F, Ma Y. Anterior gradient 3 promotes breast cancer development and chemotherapy response. Cancer Res Treat. 2020;52:218–245. doi: 10.4143/crt.2019.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Lu X, Lu X, Wang ZC, Iglehart JD, Zhang X, Richardson AL. Predicting features of breast cancer with gene expression patterns. Breast Cancer Res Treat. 2008;108:191–201. doi: 10.1007/s10549-007-9596-6. [DOI] [PubMed] [Google Scholar]
- 21.Enerly E, Steinfeld I, Kleivi K, Leivonen SK, Aure MR, Russnes HG, Rønneberg JA, Johnsen H, Navon R, Rødland E, et al. miRNA-mRNA integrated analysis reveals roles for miRNAs in primary breast tumors. PLoS One. 2011;6:e16915. doi: 10.1371/journal.pone.0016915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y, Zou L, Li Q, Haibe-Kains B, Tian R, Li Y, Desmedt C, Sotiriou C, Szallasi Z, Iglehart JD, et al. Amplification of LAPTM4B and YWHAZ contributes to chemotherapy resistance and recurrence of breast cancer. Nat Med. 2010;16:214–218. doi: 10.1038/nm.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grossman RL, Heath AP, Ferretti V, Varmus HE, Lowy DR, Kibbe WA, Staudt LM. Toward a shared vision for cancer genomic data. N Engl J Med. 2016;375:1109–1112. doi: 10.1056/NEJMp1607591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Durbin BP, Hardin JS, Hawkins DM, Rocke DM. A variance-stabilizing transformation for gene-expression microarray data. Bioinformatics. 2002;18(Suppl 1):S105–S110. doi: 10.1093/bioinformatics/18.suppl_1.S105. [DOI] [PubMed] [Google Scholar]
- 26.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yager JD, Davidson NE. Estrogen carcinogenesis in breast cancer. N Engl J Med. 2006;354:270–282. doi: 10.1056/NEJMra050776. [DOI] [PubMed] [Google Scholar]
- 28.Mohammed H, D'Santos C, Serandour AA, Ali HR, Brown GD, Atkins A, Rueda OM, Holmes KA, Theodorou V, Robinson JL, et al. Endogenous purification reveals GREB1 as a key estrogen receptor regulatory factor. Cell Rep. 2013;3:342–349. doi: 10.1016/j.celrep.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fletcher GC, Patel S, Tyson K, Adam PJ, Schenker M, Loader JA, Daviet L, Legrain P, Parekh R, Harris AL, Terrett JA. hAG-2 and hAG-3, human homologues of genes involved in differentiation, are associated with oestrogen receptor-positive breast tumours and interact with metastasis gene C4.4a and dystroglycan. Br J Cancer. 2003;88:579–585. doi: 10.1038/sj.bjc.6600740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hrstka R, Nenutil R, Fourtouna A, Maslon MM, Naughton C, Langdon S, Murray E, Larionov A, Petrakova K, Muller P, et al. The pro-metastatic protein anterior gradient-2 predicts poor prognosis in tamoxifen-treated breast cancers. Oncogene. 2010;29:4838–4847. doi: 10.1038/onc.2010.228. [DOI] [PubMed] [Google Scholar]
- 31.Wilson CL, Sims AH, Howell A, Miller CJ, Clarke RB. Effects of oestrogen on gene expression in epithelium and stroma of normal human breast tissue. Endocr Relat Cancer. 2006;13:617–628. doi: 10.1677/erc.1.01165. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.