Abstract
Although ring finger protein 2 (RNF2) serves an important role in the occurrence, development and regulation of various types of cancer, RNF2 expression in skin squamous cell carcinoma (SCC) remains unknown. The aim of the present study was to investigate the role of RNF2 expression in SCC and adjacent tissues from patients. The protein and gene expression levels of RNF2 in SCC and adjacent tissues were detected by immunohistochemistry (IHC), western blot analysis and semi-quantitative reverse transcription (RT) PCR. Single factor analysis was used to study the association between RNF2 expression level and the clinicopathological characteristics of patients with SCC. Multifactor Cox survival analysis was used to examine the association between RNF2 expression and the overall survival rate of postoperative patients with SCC. The results from IHC staining demonstrated that the positive expression rate of RNF2 was 84.68% (210/248) and 56.05% (139/248) in SCC and in adjacent tissues, respectively. Furthermore, results from western blot analysis demonstrated that RNF2 protein expression in SCC tissues was significantly higher compared with that in the adjacent tissues (P<0.05). The positive rate of RNF2 mRNA in SCC was 81.05% (201/248), which was significantly higher compared with that in the adjacent tissues 54.44% (135/248; P<0.05). Furthermore, RNF2 protein and gene expression levels were associated with tumor diameter, tumor stage, tumor metastasis and the degree of tumor differentiation in patients with SCC. Patients exhibiting higher RNF2 protein expression in SCC tissues had a significantly shorter disease-specific survival rate compared with patients with low RNF2 expression. In addition, RNF2 protein expression, tumor diameter, tumors site and tumor stage were independent factors affecting the overall survival rate of postoperative patients. High protein and gene expression levels of RNF2 in SCC tissues may be associated with the occurrence and development of SCC and prognosis of patients. The results form this study may serve the development of novel therapeutic options and diagnostic strategies for patients with SCC.
Keywords: ring finger protein 2, skin squamous cell carcinoma, protein-coding genes, prognosis
Introduction
Skin squamous cell carcinoma (SCC) is one of the commonest non-melanoma skin cancers worldwide (1,2). SCC is a type of malignant tumor originating from epidermis or adnexal keratinocytes, which often manifests in light exposed areas, such as the scalp, the face and the dorsum of the hand. In Europe and the United States, the incidence rate of SCC ranks second and increases yearly, preceding basal cell carcinoma (3). In China, the incidence rate of SCC is the highest among non-melanoma skin tumors (29.4%) and is slightly higher than that of basal cell carcinoma (28%) (4). At present, the incidence rate of SCC increases annually by 2.6% with the increased population age in China, and 10–15% patients develop metastasis (5,6). Once metastasis occurs or when the disease develops rapidly, the follow-up treatment becomes less efficient and the prognosis of patients is poor. At present, surgical resection remains the best clinical treatment for SCC, followed by radiotherapy and chemotherapy (7). Skin is constantly exposed to chemical stress and ultraviolet radiation, which lead to the overproduction of reactive oxygen species (ROS) (8). Increased oxidative stress can deplete and destroy the skin's non-enzymatic and enzymatic antioxidant defense systems, leading to increased oxidative stress and photosensitivity (9). Increased oxidative stress induces protein oxidation and lipid peroxidation, leading to numerous physiological dysfunctions, including tumor formation and aging (10).
Ring finger protein 2 (RNF2), a member of the polycomb genes family, is an ubiquitinated ligase with finger ring structure (11,12). As one of the core members of the polycomb family, RNF2 gene is highly expressed in a variety of human tumors (13–15), and its expression is associated with the degree of malignancy and the prognosis of patients. In addition, RNF2 expression was reported to promote tumor growth and metastasis (16). Bosch et al (13) demonstrated that RNF2 expression in breast cancer tissues is significantly increased compared with normal breast tissues and promotes breast cancer cell invasive ability. Chen et al (14) reported that RNF2 knockdown inhibits the proliferation and invasive ability of pancreatic cancer cells. Li et al (15) demonstrated that RNF10 may be the central target for the regulation of diabetic vascular remodeling due to its anti-hyperproliferative, pro-apoptotic anti-inflammatory activities. However, RNF2 expression in SCC remains unknown.
Investigating the association between RNF2 expression and the occurrence and development of SCC may therefore provide novel insights for diagnosis, treatment option and prevention of SCC. The present study aimed to evaluate the role of RNF2 expression in SCC by using immunohistochemistry (IHC), western blot analysis and semi-quantitative reverse transcription (RT) PCR, in order to determine a potential prognostic and diagnostic biomarker for patients with SCC.
Materials and methods
Patients and controls
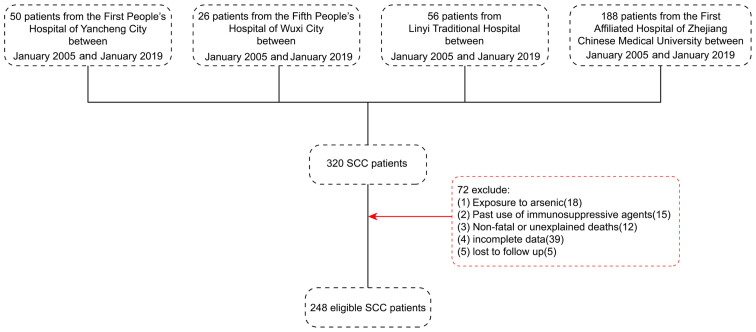
The present study retrospectively analyzed a cross-section of 248 eligible patients with SCC from a total of 320 patients from the First People's Hospital of Yancheng City, The Fifth People's Hospital of Wuxi, Linyi Traditional Hospital and the First Affiliated Hospital of Zhejiang Chinese Medical University between January 2005 and January 2019. The flow chart for patients screening is presented in Fig. 1. All patients underwent surgical resection. The inclusion criteria were as follows: i) Clinicopathological diagnosis of SCC; ii) no radiotherapy or chemotherapy was administered prior to surgery; iii) clinicopathological and follow-up data were complete; and iv) the location was exposed to at least one of the following: Head, face and neck. The exclusion criteria were as follows: i) Exposure to arsenic; ii) prior-use of immunosuppressive agents; iii) non-fatal or unexplained deaths; iv) incomplete clinicopathological and follow-up data; and v) patients lost to follow up.
Figure 1.
Screening flow chart for patients with SCC patients. SCC, skin squamous cell carcinoma.
The 248 patients with SCC included 140 men and 108 women (age range, 25–86 years; mean age, 57 years). The tumor locations were as follows: 119 samples from the head, 77 samples from the face and 52 cases from the neck. Furthermore, the tumor size was <5 cm for 104 cases and >5 cm for 144 cases. A total of 190 cases exhibited poor differentiation and 58 cases showed a high-to-moderate differentiation. According to Broders' pathological grading criteria for SCC (17), 68 cases were grades I and II and 180 cases were grades III and IV. Adjacent tissue specimens were collected by surgical resection from 248 individuals to serve as a control group (age range, 27–81 years; mean age, 55 years).
The follow-up results from the 248 patients enrolled in the present study were obtained via medical records (laboratory data, imaging data and clinical characteristics of the patient during hospitalization) and telephone interviews (follow-up data after discharge: Including survival time, treatment and laboratory data.) Postoperative follow-up was performed every three months during the first year, every four months during the second year, every six months during the third year and every eight months until patient succumbed to the disease. All participants provided oral informed consent. The present study was approved by the Ethics Committee of the First People's Hospital of Yancheng City (approval no. 2017044) and of the Fifth People's Hospital of Wuxi [approval no. HMU (Ethics) 2017-k-133].
IHC
IHC was used to detect RNF2 distribution by using the Envision and DAB chromogenic reagent kit (cat. no. MAX-002 MAX007TM). Sections were fixed with 4% paraformaldehyde for 24 h at room temperature. The dehydration was then carried out under a gradually increasing ethanol gradient at room temperature. Tissues were immersed in 70% ethanol, 80% ethanol, 95% ethanol I, 95% ethanol II, anhydrous ethanol I, anhydrous ethanol II and anhydrous ethanol III for 1 h each. Xylene was subsequently added to make the tissues transparent, then immersed in paraffin and finally embedded into wax blocks. Sections were embedded in paraffin and cut into 5-µm-thick sections. Subsequently, sections were dewaxed with xylene I, xylene II and xylene III for 10 min at room temperature, respectively, and then immersed in anhydrous ethanol I, anhydrous ethanol II respectively for 2 min at room temperature, and rinsed with distilled water three times (30 sec/wash). Antigen repair was performed with 0.01 citrate buffer, pH 6.0 at 90°C for 1 h, and incubated with 0.3% hydrogen peroxide (Sigma-Aldrich; Merck KGaA) at room temperature for 30 min to inhibit peroxidase activity. Sections were incubated with the primary rabbit anti-human RNF2 monoclonal antibody (1:100; cat. no. EPR12245; Upstate, Biotechnology, Inc.) overnight at 4°C, washed 3 times with PBS solution (PH 7.4, 1:100v/v; 3 min/wash) and then incubated with polymer-HRP secondary antibody (1:200; cat. no. KXX0022; Dako; Agilent Technologies, Inc.) for 20 min at room temperature. The sections were washed 3 times with PBS solution (3 min/wash) and subsequently incubated with DAB solution (OriGene Technologies, Inc.) at room temperature for 5 min. Tissues were subsequently stained with Improved Harris Hematoxylin Dye Solution (2 g hematoxylin, 5 g aluminum sulfate, 0.2 g potassium iodate, 250 ml 95% ethanol, 750 ml distilled water, 50.0 ml glycerol and 0.3 g citric acid; BASO Biotechnology Co., Ltd.; http://www.baso.com.cn) for 20 sec at room temperature. The positive staining was observed by using a light microscope (magnification, ×400) and the proportion of positive area was calculated.
IHC staining scores were determined by two blinded independent pathologists. The positively stained cells in SCC tissues and adjacent tissues were observed in 10 randomly selected high-power fields of view and 100 tumor cells were counted. RNF2 was located in the nucleus under light microscopy. The staining score was determined as follows: Negative (−), no brown-yellow positive staining in tumors or glandular epithelial cells; weakly positive (+), <25% positive cells; positive (++), 25–50% positive cells; strong positive (+++), >50% positive cells. For statistical analysis, negative (−) or weakly positive (+) were defined as low expression, whereas positive (++) or strong positive (+++) were defined as high expression.
Detection of RNF2 expression by western blot analysis
Tissue samples were homogenized, lysed with tissue lysate RIPA lysis buffer (Beijing Solarbio Science & Technology Co., Ltd.), centrifuged at 12,000 × g for 5 min at 4°C, and the supernatan t containing the total proteins was collected. Protein concentration was determined using the BCA kit (Pierce; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. A total of 20 µg protein/lane was separated via SDS-PAGE on a 10% gel and electrotransferred onto polyvinylidene difluoride membranes for 1–2 h. Membranes were blocked with 5% skimmed milk for 1 h at room temperature, and incubated with the following antibodies: Rabbit RNF2 (1:2,000; cat. no. ab101273; Abcam) and rabbit β-actin (1:10,000; cat. no. BS1002; Bioworld Technology, Inc.) overnight at 4°C, followed by incubation with anti-rabbit IgG secondary antibody (1:2,000; cat. no. ab205718; Abcam) at room temperature for 1–2 h. The membranes were analyzed using the Dolphin-Doc Plus gel imaging system (25×25 cm; Wealtec Corp.).
Detection of RNF2 mRNA expression levels by RT-CR
Total RNA was isolated from tissues using TRIzol® and quantified by Nanodrop spectrophotometer. Total RNA (10 µg/sample) was isolated and used to generate complementary DNA. cDNA was amplified by semi-quantitative PCR and normalized to the internal reference gene β-actin. The sequences of primers used were as follows: RNF2, forward 5′-AGCACAATAATCAGCAAGCACTC-3′, reverse 5′-GCTCCACTACCATTTTCAATCTG-3′; and β-actin, forward 5′-TGGCATCCACGAAACTACC3-3′ and reverse 5′-GTGTTGGCGTACAGGTCTT-3′. The thermocycling conditions were as follows: Pre-denaturation at 95°C for 30 sec, 95°C for 5 sec and 60°C for 30 sec, 40 cycles, and final extension at 60°C for 30 sec. Amplification of RNF2 by PCR was examined by 1.0% agarose gel electrophoresis using a Quantity-One electrophoresis apparatus. The absorbance (a) value of the belt and the reference were read, and the results were expressed by the ratio (sample value/reference value). If the ratio of the SCC value was greater than the reference value, it was positively expressed. Otherwise, it was negatively expressed.
Statistical analysis
SPSS 13.0 (SPSS, Inc.) software was used for statistical analysis. The χ2 test was used to compare the association between the expression status of RNF2 mRNA and protein levels and the clinicopathological characteristics of patients in adjacent and cancer tissues. Kaplan-Meier survival analysis was used to determine the disease-specific and disease-free survival rates. The log-rank test was used to analyze the difference in survival curves. Multivariable regression analysis was performed to determine the prognostic factors by using the Cox proportional-hazards model. P<0.05 was considered to indicate a statistically significant difference.
Results
RNF2 protein expression in SCC and adjacent tissues
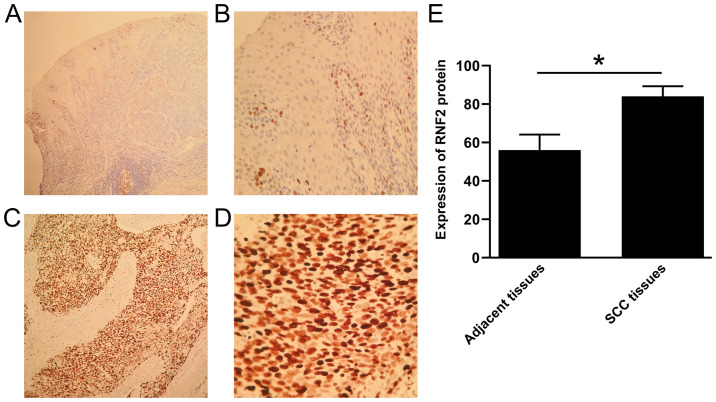
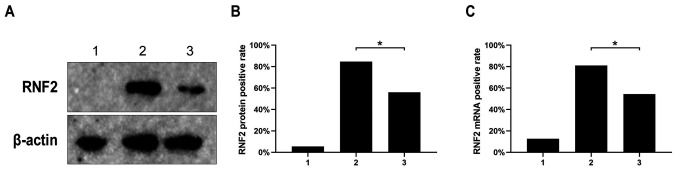
IHC was used to detect the expression of RNF2 in SCC tissues and adjacent tissues. The results demonstrated that RNF2 was mainly expressed in the nucleus of SCC tissue cells. A small amount of RNF2 was found in the cytoplasm (Fig. 2A-D). Furthermore, RNF2 expression in SCC tissues was significantly higher compared with adjacent tissues (P<0.05; Fig. 2). The positive expression rate of RNF2 protein was 84.68% (210/248) and 56.05% (139/248) in SCC tissues and in adjacent tissues, respectively. The results from western blot analysis also revealed that RNF2 expression in SCC tissues was significantly higher compared with that in the adjacent tissues (P<0.05; Fig. 3A-B).
Figure 2.
Staining results of Envision immunohistochemistry for RNF2 in SCC and adjacent tissues. (A) Weak positive staining of RNF2 in adjacent tissues (magnification, ×100). (B) Weak positive staining of RNF2 in adjacent tissues (magnification, ×400). (C) Strong positive staining of RNF2 in SCC tissues (magnification, ×100). (D) Strongly positive staining of RNF2 in SCC tissues (magnification, ×400). (E) Expression of RNF2 protein in SCC tissues significantly higher compared with adjacent tissues. *P<0.05. RNF2, ring finger protein 2; SCC, skin squamous cell carcinoma.
Figure 3.
Expression of RNF2 protein and mRNA in SCC and adjacent tissues. (A and B) The positive expression rate of RNF2 protein in SCC and adjacent tissues following western blot analysis. (C) The positive expression rate of mRNA in SCC and adjacent tissues. 1, negative control group; 2, SCC tissues; 3, adjacent tissues. *P<0.05. RNF2, ring finger protein 2; SCC, skin squamous cell carcinoma.
RNF2 mRNA expression in SCC and adjacent tissues
RNF2 expression level in SCC and adjacent tissues was evaluated by semi-quantitative RT-PCR. The results demonstrated that the positive rate of RNF2 mRNA in SCC tissues was 81.05% (201/248), which was significantly higher compared with adjacent tissues 54.44% (135/248; P<0.05; Fig. 3C).
Association between RNF2 mRNA and protein expression and the clinicopathological characteristics in patients with SCC
The expression of RNF2 mRNA and protein were both higher in SCC tissues compared with adjacent tissues. The results from single factor analysis demonstrated that RNF2 mRNA and protein expression was not associated with sex, age or tumor site (P>0.05). However, they were significantly associated with tumor diameter, degree of tumor differentiation, tumor stage and tumor metastasis (P<0.05; Table I).
Table I.
Association between RNF2 protein and mRNA expression and the clinicopathological characteristics of patients with SCC.
| Characteristic | n | RNF2 protein positive rate | χ2 | P-value | RNF2 mRNA positive rate | χ2 | P-value |
|---|---|---|---|---|---|---|---|
| Sex | |||||||
| Male | 140 | 119 (85.00) | 0.155 | 0.586 | 115 (82.14) | 0.022 | 0.878 |
| Female | 108 | 91 (84.26) | 86 (79.63) | ||||
| Age (years) | |||||||
| <40 | 49 | 41 (83.67) | 0.192 | 0.675 | 39 (79.59) | 0.039 | 0.943 |
| ≥40 | 199 | 169 (84.92) | 162 (81.41) | ||||
| Tumor diameter (cm) | |||||||
| <5 | 104 | 80 (76.92) | 0.145 | 0.010 | 75 (72.12) | 0.113 | 0.021 |
| ≥5 | 144 | 130 (90.28) | 126 (87.50) | ||||
| Degree of tumor differentiation | |||||||
| High and moderate differentiation | 58 | 28 (48.28) | 0.499 | 0.002 | 27 (46.55) | 0.441 | 0.003 |
| Poor differentiation | 190 | 182 (95.79) | 174 (91.58) | ||||
| Tumor stage | |||||||
| I–II | 68 | 41 (60.29) | 7.131 | 0.004 | 37 (54.41) | 6.928 | 0.005 |
| III–IV | 180 | 169 (93.89) | 164 (91.11) | ||||
| Lymph node metastasis | |||||||
| Yes | 86 | 81 (94.19) | 0.219 | 0.032 | 79 (91.86) | 0.201 | 0.031 |
| No | 162 | 129 (79.63) | 122 (75.31) | ||||
| Tumor site | |||||||
| Head | 119 | 103 (86.55) | 0.209 | 0.912 | 99 (83.19) | 0.129 | 0.372 |
| Face | 77 | 64 (83.12) | 61 (79.22) | ||||
| Neck | 52 | 43 (82.69) | 41 (78.85) |
RNF2, ring finger protein 2; SCC, skin squamous cell carcinoma.
Prognostic value of RNF2 expression status for the overall survival of postoperative patients with SCC
The follow-up examination of patients with SCC was performed up until 80 months. Patients with high RNF2 expression in SCC tissues had a significantly shorter disease-specific survival rate compared with patients with low RNF2 expression. Kaplan-Meier survival analysis demonstrated that there was a significant difference between RNF2 high and RNF2 low patients (P<0.0001; Fig. 4). High RNF2 expression in SCC tissues was associated with a significantly shorter disease-specific survival rate than those with low RNF2 expression. Among the 210 patients exhibiting high RNF2 protein expression in SCC tissues, 195 patients succumbed to the disease and 15 patients survived. Among the 38 patients with low RNF2 protein expression, 16 patients succumbed and 22 patients survived. The median survival time of patients with higher and lower expression of RNF2 was 14.5±1.3 and 25.5±7.5 months, respectively. These findings indicated that high RNF2 expression may affect the prognosis of patients with SCC.
Figure 4.
RNF2 expression and survival analysis of patients with SCC. RNF2, ring finger protein 2; SCC, skin squamous cell carcinoma.
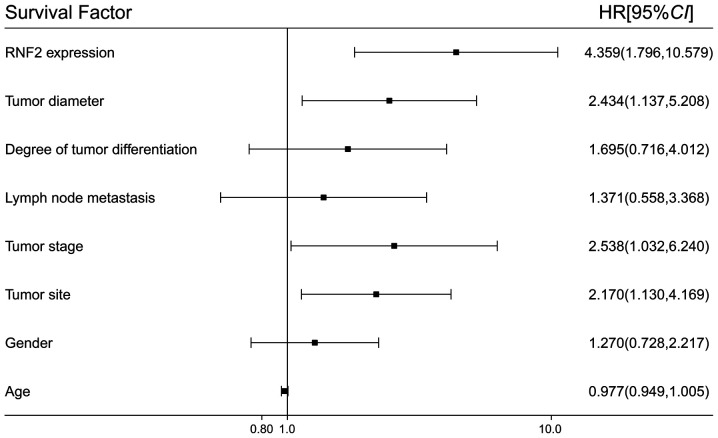
The results from multifactor Cox survival analysis demonstrated that RNF2 protein expression, tumor diameter, tumor site and tumor stage were independent prognostic factors affecting the overall survival of patients with SCC following surgery. However, sex, age, tumor differentiation degree and tumor metastasis were not independent prognostic factors affecting the overall survival of patients with SCC (Fig. 5).
Figure 5.
Association between RNF2 expression and the prognosis of patients with SCC by Multifactor Cox survival analysis. RNF2, ring finger protein 2; SCC, skin squamous cell carcinoma.
Discussion
RNF2 is a member of the protein-coding genes (PCGs) also known as RING1B (18). PCGs were first discovered in Drosophila melanogaster and are reported to maintain the expression level of growth regulators such as Hox, promote histone deacetylation and block ATP-dependent chromosome remodeling, inhibiting therefore gene transcription (19,20). Furthermore, PCGs are involved in numerous biological processes, including cell cycle regulation, cancer progression, aging, X chromosome inactivation, cell fate and stem cell differentiation (21). In mammals, PCGs consist of two core components: Polycomb inhibitory complexes (PRC) 1 and PRC2. PCGs mainly inhibit the activity of target genes through epigenetic events, thus participating in the occurrence, development and regulation of cancer via DNA methylation, histone demethylation, macrohistone H2A linkage and histone deacetylation (22). An important member of PRC1, RNF2, has a ring finger domain (23). This ring finger has E3 ligase, which helps RNF2 catalyze the universalization of histone H2A 119 lysine, then silence genes by compressing chromatin and inhibiting the extension of transcription (24). Due to its E3 ligase, RNF2 catalyzes the universalization of histone H2A 119 lysine, which can silence genes by compressing chromatin and therefore inhibit the extension of transcription (25). RNF2 serves a role in the proliferation and differentiation of stem cells, maintaining their ability of self-renewal and the occurrence and development of tumors (26). Curatti et al (27) demonstrated that RNF1 serves a crucial role in the existence of a redox regulatory mechanism in A. vinelandii, and controls the rate of expression and maturation of nitrogenase by the activity of the Rnf protein complexes.
Recent studies reported that RNF2 is highly expressed in malignant tumors and serves a role in promoting cancer (28,29). For example, previous studies demonstrated that RNF2 protein is highly expressed in esophageal, pancreatic, bladder, breast and ovarian cancers, and that RNF2 is manly located in tumor cell nucleus (11,30,31). Yang et al (16) reported a significantly higher RNF2 protein expression in esophageal cancer compared with adjacent tissue, which was associated with tumor size, tumor-node-metastasis (TNM) stage and lymph node metastasis; however, RNF2 expression is not associated with the prognosis of patients. Chen et al (30) demonstrated that the high RNF2 protein and mRNA expression in invasive ovarian cancer tissues following IHC and fluorescence in situ hybridization is associated with TNM stage, histological grade, FIGO stage and Ki-67 proliferation index. Li et al (31) also reported that RNF2 protein and gene expression in bladder urothelial carcinoma is significantly increased compared with that in bladder mucosa, and high RNF2 expression is associated with tumor TNM stage and lymph node metastasis. However, no association was indicated with poor prognosis of patients with bladder mucosa (30). Chen et al (15) reported that RNF2 protein is localized in the nucleus of pancreatic cancer tissues where it is highly expressed. However, low RNF2 expression in normal pancreatic tissues adjacent to cancer, and high RNF2 expression in cancer tissues are negatively correlated with the 5-year survival rate of patients.
The present study confirmed that RNF2 protein was localized in the nucleus of SCC tissues cells. The results from the present study demonstrated that RNF2 expression in SCC was significantly higher compared with adjacent tissue, suggesting an association between the occurrence and development of tumor and a higher expression of RNF2. In the present study, the RNF2 mRNA expression in SCC tissues was significantly higher compared with that in adjacent tissues. The association between RNF2 expression and the clinicopathological characteristics of patients with SCC was consistent, whereby the protein level was consistent with the gene level. The present study demonstrated that RNF2 protein and gene expression in SCC tissues were not associated with sex, age or tumor site. However, RNF2 protein and gene expression were associated with tumor diameter, tumor stage, tumor metastasis and the degree of tumor differentiation in SCC tissues. In addition, the protein and gene expression of RNF2 in the poor differentiation group and the lymph node metastasis group was significantly higher compared with the high and moderate differentiation group and the non-metastasis lymph node group. These findings suggested that higher expression of RNF2 may serve a crucial role in the development, invasion and metastasis of SCC.
The present study also examined the association between the expression of RNF2 and overall survival rate of postoperative patients with SCC. The median survival time for patients with higher RNF2 expression was significantly shorter compared with patients with low RNF2 expression in SCC. Multifactor Cox survival stage demonstrated that RNF2 protein expression, tumor diameter, tumors site and tumor stage were independent prognostic factors affecting the overall survival of postoperative patients with SCC. All these results revealed that high expression of RNF2 was associated with the prognosis of patients with SCC, which were also approved the results of studies by Choi et al (32) and Li et al (31).
The present study had some limitations. Firstly, although this study was, to the best of our knowledge, one of the largest-scale studies on RNF2 protein expression in SCC, further validation is required through recruitment of a larger sample size. Secondly, the underlying mechanism of RNF2 as a predictor for the prognosis of patients with SCC requires further investigation through large samples and multi centers.
In summary, the present study demonstrated that the protein and gene expression of RNF2 in SCC tissues may be associated with the occurrence, development and prognosis of SCC, which may further assist in the development of novel therapeutic and diagnostic strategies for patients with SCC.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- IHC
immunohistochemistry
- RT-PCR
reverse transcription polymerase chain reaction
- SCC
skin squamous cell carcinoma
- RNF2
ring finger protein 2
- PCGs
protein-coding genes
Funding
The present study was supported by the Youth Medical Talent of Jiangsu Province (grant no. QNRC2016163) granted to JL.
Availability of data and materials
All data generated or analyzed during this study are included in the published article.
Authors' contributions
BaL, MY and BiL contributed to the conception and design of the study, acquisition of data, analysis and interpretation of data, and drafting of the manuscript. JL, BJ and WW contributed to the statistical analysis. GL contributed to the conception and supervision of the study, and the critical revision of the manuscript. BaL and BJ contributed to the study conception and design, study supervision and critical revision of the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of the First People's Hospital of Yancheng City (approval no. 2017044) and of the Fifth People's Hospital of Wuxi [approval no. HMU (Ethics) 2017-k-133], and was performed in accordance with the Declaration of Helsinki.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Drucker AM, Adam GP, Rofeberg V, Gazula A, Smith B, Moustafa F, Weinstock MA, Trikalinos TA. Treatments for primary squamous cell carcinoma and squamous cell carcinoma in situ of the skin: A systematic review and network meta-analysis: Summary of an Agency for Healthcare Research and Quality Comparative Effectiveness Review. J Am Acad Dermatol. 2020;82:479–482. doi: 10.1016/j.jaad.2019.06.030. [DOI] [PubMed] [Google Scholar]
- 2.Wu J, Lu WY, Cui LL. Clinical significance of STAT3 and MAPK phosphorylation, and the protein expression of cyclin D1 in skin squamous cell carcinoma tissues. Mol Med Rep. 2015;12:8129–8134. doi: 10.3892/mmr.2015.4460. [DOI] [PubMed] [Google Scholar]
- 3.Bashline B. Skin cancer: Squamous and basal cell carcinomas. FP Essent. 2019;481:17–22. [PubMed] [Google Scholar]
- 4.Wu J, Guo NZ, Cui LL, Wang W, Xiong CQ, Zhang XY. Correlation between tripartite motif-containing protein 44 protein expression and the prognosis of postoperative patients exhibiting skin squamous cell carcinoma. Medicine (Baltimore) 2018;97:e13021. doi: 10.1097/MD.0000000000013021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang L, Shan X, Chen Q, Xu D, Fan X, Yu M, Yan Q, Liu J. Downregulation of HDAC3 by ginsenoside Rg3 inhibits epithelial-mesenchymal transition of cutaneous squamous cell carcinoma through c-Jun acetylation. J Cell Physiol. 2019;234:22207–22219. doi: 10.1002/jcp.28788. [DOI] [PubMed] [Google Scholar]
- 6.Lansbury L, Bath-Hextall F, Perkins W, Stanton W, Leonardi-Bee J. Interventions for non-metastatic squamous cell carcinoma of the skin: Systematic review and pooled analysis of observational studies. BMJ. 2013;347:f6153. doi: 10.1136/bmj.f6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lailheugue A, Gibier JB, Lassailly G, Truant S, Pruvot FR, El Amrani M. Primary squamous cell carcinoma of the peristomal skin of gastrostomy in a transplant patient: A first case report. J Gastrointest Oncol. 2019;10:573–576. doi: 10.21037/jgo.2019.01.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Emmert H, Patel H, Brunton VG. Kindlin-1 protects cells from oxidative damage through activation of ERK signalling. Free Radic. Biol Med. 2017;108:896–903. doi: 10.1016/j.freeradbiomed.2017.05.013. [DOI] [PubMed] [Google Scholar]
- 9.Zhang QL, Li XM, Lian DD, Zhu MJ, Yim SH, Lee JH, Jiang RH, Kim CD. Tumor Suppressive function of NQO1 in cutaneous squamous cell carcinoma (SCC) cells. Biomed Res Int. 2019;2019:2076579. doi: 10.1155/2019/2076579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ciani F, Tafuri S, Troiano A, Cimmino A, Fioretto BS, Guarino AM, Pollice A, Vivo M, Evidente A, Carotenuto D, Calabrò V. Anti-proliferative and pro-apoptotic effects of Uncaria tomentosa aqueous extract in squamous carcinoma cells. J Ethnopharmacol. 2018;211:285–294. doi: 10.1016/j.jep.2017.09.031. [DOI] [PubMed] [Google Scholar]
- 11.Yang J, Yu F, Guan J, Wang T, Liu C, Wang Y, Liu G, Zhu S. Knockdown of RNF2 enhances the radiosensitivity of squamous cell carcinoma in lung. Biochem Cell Biol. 2019;97:589–599. doi: 10.1139/bcb-2018-0252. [DOI] [PubMed] [Google Scholar]
- 12.An R, Cheng L, Chen L, Du J. Plk1 interacts with RNF2 and promotes its ubiquitin-dependent degradation. Oncol Rep. 2018;39:2358–2364. doi: 10.3892/or.2018.6326. [DOI] [PubMed] [Google Scholar]
- 13.Bosch A, Panoutsopoulou K, Corominas JM, Gimeno R, Moreno-Bueno G, Martín-Caballero J, Morales S, Lobato T, Martínez-Romero C, Farias EF, et al. The Polycomb group protein RING1B is overexpressed in ductal breast carcinoma and is required to sustain FAK steady state levels in breast cancer epithelial cells. Oncotarget. 2014;5:2065–2076. doi: 10.18632/oncotarget.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen S, Chen JZ, Zhang JQ, Chen HX, Yan ML, Huang L, Tian YF, Chen YL, Wang YD. Hypoxia induces TWIST-activated epithelial-mesenchymal transition and proliferation of pancreatic cancer cells in vitro and in nude mice. Cancer Lett. 2016;383:73–84. doi: 10.1016/j.canlet.2016.02.043. [DOI] [PubMed] [Google Scholar]
- 15.Li S, Yu G, Huang W, Wang R, Pu P, Chen M. RING finger protein 10 is a potential drug target for diabetic vascular complications. Mol Med Rep. 2019;20:931–938. doi: 10.3892/mmr.2019.10358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang XX, Ma M, Sang MX, Wang XX, Song H, Liu ZK, Zhu SC. Radiosensitization of esophageal carcinoma cells by knockdown of RNF2 expression. Int J Oncol. 2016;48:1985–1996. doi: 10.3892/ijo.2016.3404. [DOI] [PubMed] [Google Scholar]
- 17.Vasconcelos L, Melo JC, Miot HA, Marques ME, Abbade LP. Invasive head and neck cutaneous squamous cell carcinoma: Clinical and histopathological characteristics, frequency of local recurrence and metastasis. An Bras Dermatol. 2014;89:562–568. doi: 10.1590/abd1806-4841.20142810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chrispijn ND, Elurbe DM, Mickoleit M, Aben M, de Bakker DEM, Andralojc KM, Huisken J, Bakkers J, Kamminga LM. Loss of the Polycomb group protein Rnf2 results in derepression of tbx-transcription factors and defects in embryonic and cardiac development. Sci Rep. 2019;9:4327. doi: 10.1038/s41598-019-40867-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sui Y, Ju C, Shao B. A lymph node metastasis-related protein-coding genes combining with long noncoding RNA signature for breast cancer survival prediction. J Cell Physiol. 2019;234:20036–20045. doi: 10.1002/jcp.28600. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, Tao Y, Ji H, Li W, Guo X, Ng DM, Haleem M, Xi Y, Dong C, Zhao J, et al. Genome-wide identification of the essential protein-coding genes and long noncoding RNAs for human pan-cancer. Bioinformatics. 2019;35:4344–4349. doi: 10.1093/bioinformatics/btz230. [DOI] [PubMed] [Google Scholar]
- 21.Guo JC, Li CQ, Wang QY, Zhao JM, Ding JY, Li EM, Xu LY. Protein-coding genes combined with long non-coding RNAs predict prognosis in esophageal squamous cell carcinoma patients as a novel clinical multi-dimensional signature. Mol Biosyst. 2016;12:3467–3477. doi: 10.1039/C6MB00585C. [DOI] [PubMed] [Google Scholar]
- 22.Dempsey J, Zhang A, Cui JY. Coordinate regulation of long non-coding RNAs and protein-coding genes in germ-free mice. BMC Genomics. 2018;19:834. doi: 10.1186/s12864-018-5235-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu S, Zhao D, Yan L, Jiang W, Kim JS, Gu B, Liu Q, Wang R, Xia B, Zhao JC, et al. BMI1 regulates androgen receptor in prostate cancer independently of the polycomb repressive complex 1. Nat Commun. 2018;9:500. doi: 10.1038/s41467-018-02863-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang J, Sun Z, Han Y, Yao R, Yue L, Xu Y, Zhang J. Rnf2 knockdown reduces cell viability and promotes cell cycle arrest in gastric cancer cells. Oncol Lett. 2017;13:3817–3822. doi: 10.3892/ol.2017.5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koike H, Ueno Y, Naito T, Shiina T, Nakata S, Ouchi R, Obana Y, Sekine K, Zheng YW, Takebe T, et al. Ring1B promotes hepatic stem/progenitor cell expansion through simultaneous suppression of Cdkn1a and Cdkn2a in mice. Hepatology. 2014;60:323–333. doi: 10.1002/hep.27046. [DOI] [PubMed] [Google Scholar]
- 26.Román-Trufero M, Méndez-Gómez HR, Pérez C, Hijikata A, Fujimura Y, Endo T, Koseki H, Vicario-Abejón C, Vidal M. Maintenance of undifferentiated state and self-renewal of embryonic neural stem cells by Polycomb protein Ring1B. Stem Cells. 2009;27:1559–1570. doi: 10.1002/stem.82. [DOI] [PubMed] [Google Scholar]
- 27.Curatti L, Brown CS, Ludden PW, Rubio LM. Genes required for rapid expression of nitrogenase activity in Azotobacter vinelandii. Proc Natl Acad Sci USA. 2005;102:6291–6296. doi: 10.1073/pnas.0501216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Q, Li S, Yang X, Zhang X, Song C, Zhu S. Association between RNF2+P-AKT expression in pretreatment biopsy specimens, and poor survival following radiotherapy in patients with esophageal squamous cell carcinoma. Oncol Lett. 2019;18:3734–3742. doi: 10.3892/ol.2019.10727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu J, Wang H, Li Q, Guo QY, Tao SQ, Shen YX, Wu ZS. The oncogenic impact of RNF2 on cell proliferation, invasion and migration through EMT on mammary carcinoma. Pathol Res Pract. 2019;215:152523. doi: 10.1016/j.prp.2019.152523. [DOI] [PubMed] [Google Scholar]
- 30.Chen Y, Cao XY, Li YN, Qiu YY, Li YN, Li W, Wang H. Reversal of cisplatin resistance by microRNA-139-5p-independent RNF2 downregulation and MAPK inhibition in ovarian cancer. Am J Physiol Cell Physiol. 2018;315:C225–C235. doi: 10.1152/ajpcell.00283.2017. [DOI] [PubMed] [Google Scholar]
- 31.Li XD, Chen SL, Dong P, Chen JW, Wang FW, Guo SJ, Jiang LJ, Zhou FJ, Xie D, Liu ZW, et al. Overexpression of RNF2 is an independent predictor of outcome in patients with urothelial carcinoma of the bladder undergoing radical cystectomy. Sci Rep. 2016;6:20894. doi: 10.1038/srep20894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi D, Lee SJ, Hong S, Kim IH, Kang S. Prohibitin interacts with RNF2 and regulates E2F1 function via dual pathways. Oncogene. 2008;27:1716–1725. doi: 10.1038/sj.onc.1210806. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in the published article.