Abstract
Early studies have indicated that insulin-like growth factor II mRNA binding protein 3 (IGF2BP3/IMP3) may affect the progression of hepatocellular carcinoma (HCC); however, the detailed underlying mechanisms, particularly its linkage to tight junction protein-mediated cell invasion, remain unclear. The present study revealed that IGF2BP3 increased HCC cell invasiveness by suppressing zonula occludens-1 (ZO-1) expression, via direct binding to the 3′ untranslated region (3′-UTR). Analysis of the molecular mechanisms demonstrated that IGF2BP3 binds to the overlapping targets of IGF2BP3-RNA cross-linkage and microRNA (miR)191-5p targeting sites, and promotes the formation of an miR191-5p-induced RNA-induced silencing complex. The knockdown of IGF2BP3 or the addition of a miR-191-5p inhibitor decreased cellular invasiveness and increased ZO-1 expression. Analysis of the human HCC database also confirmed the association between IGF2BP3 and HCC progression. Collectively, these preclinical findings suggest that IGF2BP3 increases HCC cell invasiveness by promoting the miR191-5p-induced suppression of ZO-1 signaling. This newly identified signaling effect on small molecule targeting may aid in the development of novel strategies with which to inhibit HCC progression more effectively.
Keywords: hepatocellular carcinoma, insulin-like growth factor II mRNA binding protein 3, RNA binding protein, microRNA-191-5p, cell invasion, tjp1, zonula occludens-1
Introduction
Hepatocellular carcinoma (HCC) is the most common primary liver cancer with a high recurrence rate and a poor prognosis. Annually, among the newly diagnosed HCC cases, those in China account for approximately >50% of all global cases (1). Although great advances have been made in traditional hepatectomy and other comprehensive therapeutic strategies for HCC, the results remain unsatisfactory (2). Early recurrence and metastasis are the leading causes of poor patient outcome; therefore, in order to improve HCC outcome, the investigation of novel molecular mechanisms underlying HCC recurrence and metastasis is urgently required.
Insulin-like growth factor II mRNA-binding protein 3 (IGF2BP3/IMP3), a member of the IGF2BP family, has been shown to exhibit low or undetectable protein levels in normal adult tissues, whereas higher expression levels are observed in malignant tumors (3–5). Moreover, a number of studies have found that the overexpression of IGF2BP3 is associated with poor outcome in various types of cancer, such as breast cancer, cervical cancer, renal cell carcinoma, neuroblastoma and HCC (6–13). Pre-clinical studies have demonstrated that IGF2BP3 can promote cancer cell motility, invasiveness and migration (14–17). Functional studies have also demonstrated that IGF2BP3, an RNA binding protein, plays a critical physiological role in regulating RNA splicing, stability, localization, modification and translation (18,19). Studies of multiple cancer types have demonstrated that a set of cancer-related pathways, particularly those involved in invasion, may be mediated by IGF2BP3 (13,16). The results of a mechanical study also demonstrated that IGF2BP3 facilitated partial invasion-promoted gene expression and decreased the expression of certain tumor-suppressing genes (20,21). For example, IGF2BP3 has been shown to function as a cytoplasmic ‘safehouse’ and to prevent the mRNA-directed decay of oncogene Hmga2 during tumorigenesis (22). However, IGF2BP3 can also influence the mRNA degradation of tumor-suppressor genes by enhancing micro(mi)RNA-mRNA interactions (23). To date, the detailed molecular pathways underlying the effects of IGF2BP3 on HCC cell invasion ability remain unknown.
Zonula occludens-1 (ZO-1) is a member of the zona occludens protein family, which is associated with the function of tight and adherens junctions, and may serve critical roles in HCC progression and metastasis by promoting cellular migration and invasion (24,25). Previous studies have demonstrated that ZO-1 expression is associated with the prognosis of lung cancer and HCC (26,27). In a previous in vitro study, increasing ZO-1 expression significantly decreased cellular invasiveness; by contrast, the knockdown of ZO-1 markedly increased the invasive capacity of HCC cell lines. The result of IGF2BP3 RIP (RNP immunoprecipitation) sequencing data from pancreatic ductal adenocarcinoma cells also demonstrated that IGF2BP3 could bind ZO-1 mRNA (23). However, whether IGF2BP3 can bind and mediate ZO-1 expression in HCC invasion remains to be elucidated.
In the present study, the potential association between IGF2BP3 expression and poor prognosis in patients with HCC was determined by analyzing data from the Gene Expression Omnibus (GEO), the European Genome-phenome Archive (EGA) and The Cancer Genome Atlas (TCGA). In in vitro experiments, the results demonstrate that the knockdown of IGF2BP3 deceases cell invasiveness by increasing ZO-1 expression; conversely, the overexpression of IGF2BP3 increased invasion capacity by decreasing ZO-1 expression in HCC cell lines. Mechanistic analyses revealed that IGF2BP3 suppressed ZO-1 expression by enhancing miR191-5p-induced ZO-1 mRNA silencing. Taken together, the findings of the present study suggest that directly targeting IGF2BP3 or miR191-5p may improve ZO-1 expression and suppress cell invasiveness.
Materials and methods
Bioinformatics data
Liver cancer RNA-seq data (http://kmplot.com/analysis/index.php?p=background) were extracted from the GEO, the EGA and TCGA databases. IGF2BP3 expression data associated with overall survival (OS) and relapse-free survival (RFS) were analyzed using the Kaplan-Meier plotter online tool (http://kmplot.com).
Cell culture
The Huh-7 and HA22T liver cancer cell lines (both adult hepatocellular carcinoma) were purchased from the American Type Culture Collection (28–30). Cells were maintained in Dulbecco's modified Eagle's medium (Invitrogen; Thermo Fisher Scientific Inc.) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific, Inc.), 1% glutamine and 1% penicillin/streptomycin (Invitrogen; Thermo Fisher Scientific, Inc.) (31,32), and cultured at 37°C in a humidified incubator [5% (v/v) CO2]. PCR detection for mycoplasma contamination yielded negative results.
Reagents and materials
Mouse anti-ZO-1 (1:1,000; sc-33725), mouse anti-GAPDH (1:1,000; sc-47724) and mouse anti-IGF2BP3 (1:1,000; sc-365641) antibodies were purchased from Santa Cruz Biotechnology, Inc. Rabbit polyclonal argonaute-2 antibody (1:100; ab32381) was purchased from Abcam. Anti-rabbit/mouse secondary antibodies (1:5,000; A10547 and A10668, respectively) were purchased from Invitrogen; Thermo Fisher Scientific, Inc. Rabbit IgG (1:100; sc-69786) was also obtained from Santa Cruz Biotechnology, Inc. The miRNA-191-5p inhibitor was purchased from Shanghai GenePharma Co., Ltd.
Lentiviral expression plasmids and virus infection
The lentivirus system and standard calcium chloride transfection method were applied to generate the virus. The pWPI–IGF2BP3, pLKO.1 pLKO.1-shIGF2BP3#1 or pLKO.1-shIGF2BP3#2, pWPI-ZO-1 and pLKO.1-oemiR191-5p/pLKO.1/pLKO.1-oemiR429, or the corresponding empty control plasmids (EMD Millipore) were co-transfected into 293 cells with the pMD2G envelope plasmid and psPAX2 packaging plasmid (12259 and 12260, respectively; both Addgene, Inc.), using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.). After 8 h, the medium was replaced with fresh warm medium. The cells were then cultured in a virus room incubator for virus generation as previously decreased (33). After 48 h, the virus-containing supernatants were harvested, used immediately or stored at −80°C for later use. Huh-7 or HA22T cells were seeded into a 6-well plate (~1×106/well) and infected with virus (MOI=2). Green fluorescence protein was used to monitor gene overexpression, and 1 µg/ml puromycin was to select the cells with gene knockdown or miR-191-5p overexpression. In order to downregulate miR-191-5p expression, ~1×106 cells/well were seeded into a 6-well plate, and transfected with 50 pmol miR-191-5p inhibitor or NC inhibitor using Lipofectamine® 3000, and incubated for 24 h. The expression level of miR-191-5p was monitored by reverse transcription-quantitative (RT-q) PCR to determine whether upregulation or downregulation was successful. The shRNA sequences were as follows: shIGF2BP3#1 targeting sequence, 5′-GCAGGAATTGACGCTGTATAA-3′; shIGF2BP3#2 targeting sequence, 5′-TCTGCGGCTTGTAAGTCTAT-3′; miR-191-5p inhibitor, 5′-CAGCUGCUUUUGGGAUUCCGUUG-3′; and NC inhibitor, 5′-CAGUACUUUUGUGUAGUACAA-3′.
RNA extraction and RT-qPCR analysis
Total RNA was extracted from Huh-7 and HA22T cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). Total RNA (1 µg) was reverse transcribed into cDNA using Superscript III transcriptase (Invitrogen; Thermo Fisher Scientific, Inc.) at 42°C for 60 min and 95°C for 5 min. qPCR was conducted using a Bio-Rad CFX96 system (Applied Biosystems) with SYBR-Green to detect the mRNA expression level of a gene of interest using the following thermocycling conditions: 95°C for 3 min, followed by 35 cycles at 95°C for 15 sec, 60°C for 30 sec and 68°C for 1 min. Expression levels were normalized to the GAPDH levels using the 2−ΔΔCq method (34). The primer sequences were as follows: miR-191-5p forward, 5′-CAACGGAATCCCAAAAGCAGCTG-3′ and reverse, 5′-TGTCGTGGAGTCGGC-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′; IGF2BP3 forward, 5′-ACTGCACGGGAAACCCATAG-3′ and reverse, 5′-ACTATCCAGCACCTCCCACT-3′; ZO-1 forward, 5′-GTGGGTAACGCCATCCTCTG-3′ and reverse, 5′-TCCGGGATTTCACCAGTGTG-3′; and GAPDH forward, 5′-TGCACCACCAACTGCTTAGC-3′ and reverse, 5′-GGCATGGACTGTGGTCATGAG-3′. All primers were purchased from Integrated DNA Technologies, Inc.
Invasion assay
As previously described, a Transwell assay was used to determine the invasive capacity of HCC cells. Following transfection, 5×104 cells/well were seeded with serum-free medium into the upper chambers of the Transwell inserts, which had been pre-coated with diluted Matrigel (Corning Inc.) at 37°C for 4 h. Subsequently, 750 µl media supplemented with 10% FBS was pipetted into the lower chambers, and the cells were incubated at 37°C [5% (v/v) CO2] for 18 h. After being fixed with methanol at room temperature for 20 min, the invasive cells were stained with 0.1% (w/v) crystal violet at room temperature for 15 min. The average number of cells from three randomly selected microscopic areas using an inverted light microscope (magnification, ×100; Olympus Corporation) were counted and for quantification, and the number of invasive cells was calculated using ImageJ software 1.8 (National Institutes of Health).
Western blot analysis
Cells were lysed to extract total proteins using RIPA buffer (Wuhan Boster Biological Technology, Ltd.,). The protein concentration was measured using a BCA Protein assay kit (ab102536; Abcam) and ultraviolet spectrophotometry. Proteins (30 µg/well) were separated using 6–10% SDS/PAGE gels and then transferred onto PVDF membranes (EMD Millipore). After blocking with 5% milk at room temperature for 1.5 h, the membranes were incubated with primary antibodies at 4°C overnight, and subsequently with HRP-conjugated secondary antibodies (detail in the Reagents and materials part) at room temperature for 2 h. An enhanced chemiluminescent substrate (35055; Thermo Fisher Scientific, Inc.) was then used to detect and visualize the signals. The relative expression levels were quantified using the Image Lab soft 4.1 (Bio-Rad Laboratories, Inc.) with GAPDH as the internal reference.
RNA immunoprecipitation (RIP)
Following transfection, cells were lysed in ice-cold lysis buffer supplemented with RNase inhibitor. Following centrifugation, 500 µl of the supernatant was clarified using protein A/G beads for 1 h and incubated with an argonaute-2 antibody at 4°C overnight. RNA was extracted using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol and subjected to RT-qPCR analysis.
Luciferase reporter assay
A 500 bp fragment of the ZO-1 3′ untranslated region (UTR), with wild-type or mutant IGF2BP3 or miRNA-responsive elements, was cloned into the psiCHECK-2 vector (Promega Corporation) downstream of the Renilla luciferase open reading frame. HA22T and Huh-7 cells were seeded into 24-well plates, and the cDNA was transfected with Lipofectamine® 3000 transfection reagent according to the manufacturer's instructions. PRL-TK was used as an internal control that served as the baseline control response. Luciferase activity was measured at 48 h post-transfection using a dual-luciferase reporter assay (Promega Corporation) according to the manufacturer's protocol. The relative luciferase activity was normalized to Renilla luciferase activity.
Statistical analysis
Statistical analysis was performed using SPSS software, version 23.0 (IBM Inc.). The Mann-Whitney U test was conducted to compare two groups of continuous data. The Kruskal-Wallis test with the Dunn-Bonferroni post hoc test was applied to multiple groups of continuous data. All cellular experiments were conducted in three technical replicates. Quantitation Results are presented as the mean ± SD. Statistical significance for cell experiments was determined using the independent-sample t-test. P<0.05 was considered to indicate a statistically significant difference.
Results
Higher IGF2BP3 expression may indicate a lower survival rate in patients with HCC
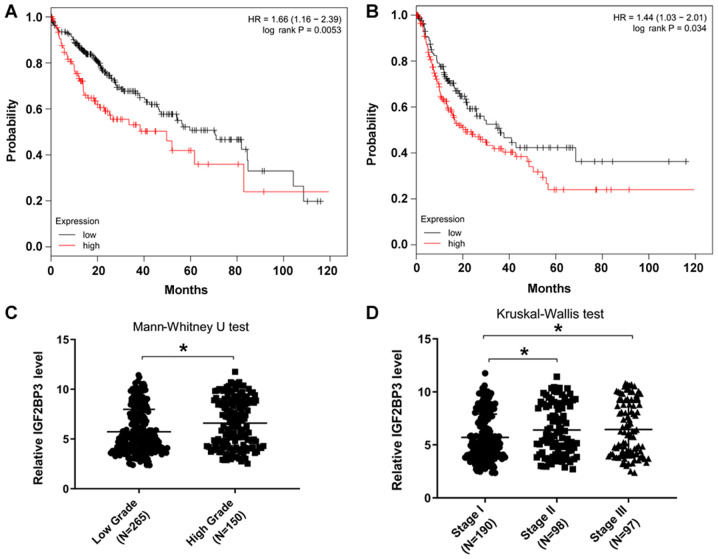
The datasets, which included gene expression associated with OS and RFS, were retrieved from the GEO, EGA and TCGA, and were analyzed online (http://kmplot.com). The results revealed that patients with a high IGF2BP3 expression level (50% of the top IGF2BP3 expression samples) exhibited lower OS and RFS rates than those with low IGF2BP3 expression (50% of the lower IGF2BP3 expression samples) (Fig. 1A and B), suggesting that IGF2BP3 may be involved in HCC progression. Furthermore, IGF2BP3 expression was determined in patients at different grades or stages of HCC using data from TCGA database. The results demonstrated that patients with higher grades or stages of the disease possessed higher IGF2BP3 expression levels (Fig. 1C and D). Taken together, these data are consistent with those previously published, which suggest that IGF2BP3 promotes HCC progression (13).
Figure 1.
Patients with HCC with higher IGF2BP3 expression levels have a lower survival rate. (A) OS and (B) RFS results from online databases (kmplot, http://kmplot.com), including IGF2BP3 expression data from 364 primary HCC samples. (C) IGF2BP3 expression at different pathological grades from 415 primary HCC samples from TCGA database (P=0.015). (D) IGF2BP3 expression at different stages from 385 primary HCC samples from the TCGA database. IGF2BP3 expression was significantly higher in the stage II (P=0.024) group or stage III (P=0.027) group than that in the stage I group. *P<0.05. HCC, hepatocellular carcinoma; IGF2BP3, insulin-like growth factor II mRNA-binding protein 3; OS, overall survival; RFS, relapse-free survival; TCGA, The Cancer Genome Atlas; HR, hazard ratio.
IGF2BP3 may promote HCC cell invasiveness by suppressing ZO-1 expression
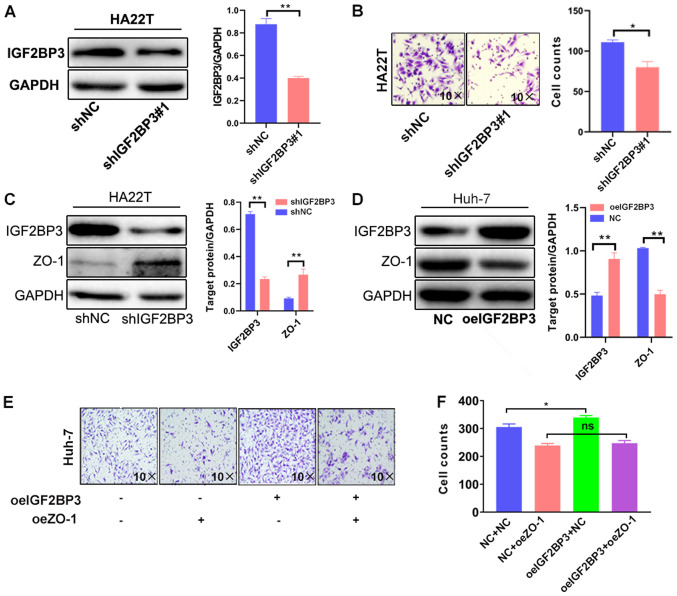
To further demonstrate the function of IGF2BP3 in HCC progression, its effects on cell invasiveness were assessed by IGF2BP3 shRNA knockdown or overexpression with IGF2BP3-cDNA in HA22T and Huh-7 cells, respectively. As shown in Fig. 2A and B, IGF2BP3 knockdown with IGF2BP3-shRNA#1 markedly decreased the invasive ability of HA22T cells. To avoid the off-target effect of IGF2BP3-shRNA, a second shIGF2BP3#2 plasmid was constructed to repeat the experiment, and the results were consistent with those obtained with IGF2BP3-shRNA#1 (Fig. S1A and B). By contrast, the overexpression of IGF2BP3 markedly increased the invasive capacity of Huh-7 cells (Fig. S1C and D).
Figure 2.
IGF2BP3 promotes HCC cell invasiveness by suppressing ZO-1 expression. (A) Western blot analysis of IGF2BP3 knockdown in HA22T cells. (B) Transwell invasion assays were performed using HA22T cells transfected with pLKO and IGF2BP3-shRNA. The invasive cells were counted and averaged from 3 randomly selected microscopic fields (magnification, ×10). Each sample was run in triplicate. Western blot analysis was performed on (C) HA22T cells transfected with pLKO and IGF2BP3-shRNA and (D) Huh-7 cells transfected with pWPI and IGF2BP3-cDNA. (E) Transwell invasion assays were performed using Huh-7 cells transfected with pWPI+pWPI, pWPI+oeZO-1, oeIGF2BP3+pWPI or oeIGF2BP3+oeZO-1. (F) All quantifications are shown, and are presented as the mean ± SD. *P≤0.05 and **P≤0.01. ns, not significant; IGF2BP3, insulin-like growth factor II mRNA-binding protein 3; HCC, hepatocellular carcinoma; ZO-1, zonula occludens-1; sh, short hairpin (RNA); oe, overexpression; NC, negative control.
To elucidate the mechanisms responsible for the effects of IGF2BP3 on HCC cell invasiveness, the expression of ZO-1 was examined, a protein which is associated with cellular invasiveness in a variety of cancers, such as breast, pancreatic and lung cancer (27,35,36). IGF2BP3 knockdown in HA22T cells resulted in increased expression of ZO-1 (Fig. 2C). Conversely, the overexpression of IGF2BP3 in Huh-7 cells decreased the expression of ZO-1 (Fig. 2D). To verify whether increased levels of ZO-1 could reduce cellular invasion capacity, a lentivirus system was used to overexpress ZO-1 and western blot analysis was used confirm expression in Huh-7 cells (Fig. S1E). The results of Transwell assay revealed that ZO-1 overexpression markedly decreased Huh-7 cell invasiveness (Fig. 2E). These results suggest that IGF2BP3 affects HCC cell invasiveness by altering ZO-1 expression.
A reverse assay was then employed to examine the roles of ZO-1 in the IGF2BP3-induced increase in HCC cell invasiveness. The results revealed that the ZO-1 overexpression partially blocked the IGF2BP3-induced increase in HCC cell invasiveness (Fig. 2E-F). Collectively, the results shown in Fig. 2A-F demonstrate that IGF2BP3 may increase HCC cell invasiveness by suppressing ZO-1 expression.
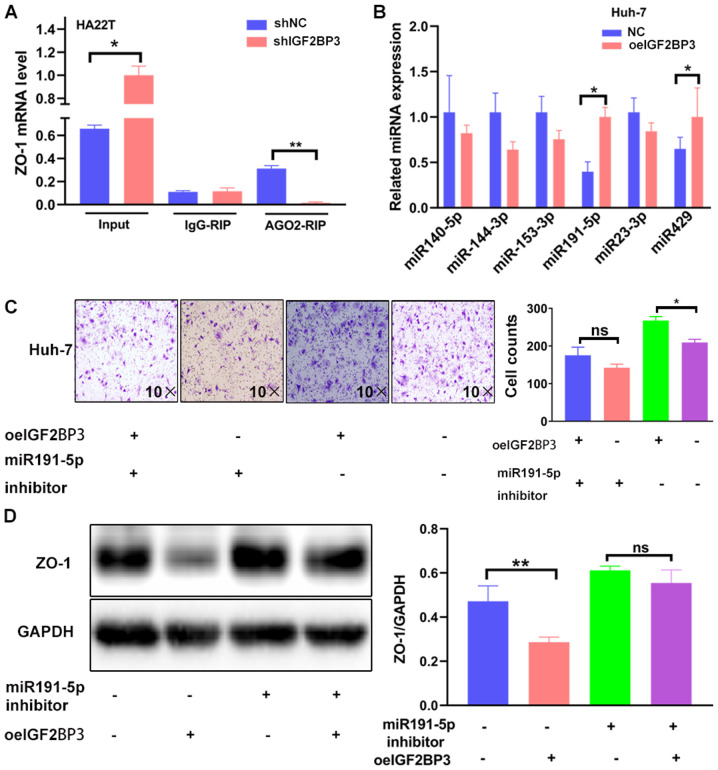
Mechanistic analysis of the mechanisms through which IGF2BP3 suppresses ZO-1 expression: By synergizing with miR191-5p through the RNA-induced silencing complex (RISC)
The present study then aimed to reveal the mechanisms responsible for the suppressive effects of IGF2BP3 on ZO-1 expression. It has been previously reported that IGF2BP3 acts as an RNA-binding protein by recognizing specific motifs to determine targeted RNA fate (37), and that IGF2BP3 can modulate invasion-associated gene transcripts by promoting Ago2-mRNA interactions (23). Therefore, the potential regulation of ZO-1 by Ago2-mRNA interactions was investigated by detecting ZO-1 mRNA in the Ago2 complex using an RIP assay. ZO-1 mRNA co-precipitation with Ago2 was decreased in the IGF2BP3-depleted cells (Fig. 3A). Subsequently, to determine which miRNAs mediated ZO-1 expression through the IGF2BP3-Ago2 complex, online miRNA software (http://www.targetscan.org) was used to screen six potential candidates and detect miRNA expression from the Ago2-antibody pull-down complex between the negative control (NC) cells and IGF2BP3-cDNA cells. The results revealed that the miR-191-5p and miR429 levels were significantly higher in the IGF2BP3-overexpressing cells than in the NC cells in the Ago2 complex (Fig. 3B). The roles of the two miRNA candidates in ZO-1 reduction were then investigated; an increase in miR191-5p alone decreased ZO-1 expression (Fig. S2A and B). However, overexpressing miR429 in HA22T cells (Fig. S2C) had no impact on ZO-1 protein expression (Fig. S2D). It was thus hypothesized that the miR191-5p regulatory effects were associated with the 3′UTR overlap of the IGF2BP3 binding sites, and that IGF2BP3 synergized miR191-5p via the RISC. To further investigate this hypothesis, a rescue experiment was performed to determine the role of miR191-5p in the IGF2BP3-induced increase in HCC cell invasiveness. The findings revealed that miR191-5p knockdown with an miR191-5p inhibitor (Fig. S2E) partially reversed the IGF2BP3-induced increase in HCC cell invasion and decreased ZO-1 expression (Fig. 3C-D). Taken together, the data shown in Fig. 3A-D suggest that IGF2BP3 decreases ZO-1 expression by cooperating with the miR191-5p-Ago2 complex via the RISC.
Figure 3.
IGF2BP3 decreases ZO-1 expression by synergizing with miR191-5p via the RISC. (A) ZO-1 mRNA expression was detected in the argonaute 2 complex using an RIP assay in HA22T cells transfected with pLKO and shIGF2BP3. (B) Reverse transcription-quantitative PCR was used to screen 6 potential miRNAs which may be able to regulate ZO-1 from the argonaute 2 complex in Huh-7 cells transfected with pWPI and IGF2BP3-cDNA. (C) Transwell invasion assays were performed using Huh-7 cells transfected with pWPI+pWPI, pWPI+oeIGF2BP3, oemiR-191-5p+pWPI or oemiR-191-5p+oeIGF2BP3. (D) Western blotting was performed to detect ZO-1 expression in Huh-7 cells transfected with pWPI+pWPI, pWPI+oeIGF2BP3, oemiR-191-5p+ pWPI or oemiR-191-5p+oeIGF2BP3. For C and D, the quantifications are shown on the right, and are presented as the mean ± SD. *P≤0.05 and **P≤0.01. ns, not significant; IGF2BP3, insulin-like growth factor II mRNA-binding protein 3; ZO-1, zonula occludens-1; miRNA/miR, microRNA; RISC, RNA-induced silencing complex; RIP, RNA interaction-precipitation; sh, short hairpin (RNA); oe, overexpression; NC, negative control.
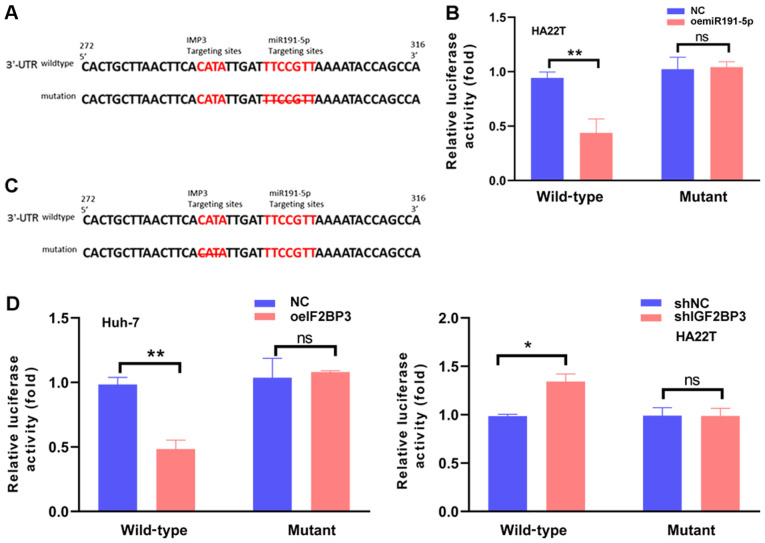
Molecular mechanistic analysis of the mechanisms by which IGF2BP3 enhances RISC function: By promoting Ago2-mRNA interactions
To further determine the mechanisms of the regulatory effects of the miR191-5p-IGF2BP3-Ago2 complex on ZO-1 mRNA degradation. The 3′UTRs of ZO-1 mRNA were first analyzed with a consensus sequence of ‘AAUGU’ and IGF2BP3-RNA was found to crosslink the overlapping miR191-5p target sites in the of ZO-1 mRNA 3′UTRs (23). A luciferase assay was then applied to determine whether IGF2BP3-miR191-5p directly targeted the overlap of ZO-1 mRNA 3′UTR to downregulate its expression. The wild-type and mutant miRNA-target sites were first cloned into the psicheck2 vector to construct the reporter plasmids (Fig. 4A). As expected, increasing miR191-5p resulted in decreased luciferase activity in HA22T cells transfected with the wild-type, but not the mutant ZO-1 3′UTR (Fig. 4B). Subsequently, other reporter plasmids were constructed with psicheck2 vectors carrying the wild-type and mutant IGF2BP3-target sites (Fig. 4C). The luciferase assay revealed that increasing IGF2BP3 expression resulted in decreased luciferase activity in Huh-7 cells transfected with the wild-type ZO-1 3′UTR, but not in the mutant 3′UTR. In addition, attenuating IGF2BP3 significantly increased the luciferase activity in HA22T cells transfected with the wild-type, but not in the mutant ZO-1 3′UTR (Fig. 4D). Taken together, the results presented in Fig. 4A-D suggest that IGF2BP3 can promote the function of miR191-5p by binding the overlap target sites and enhancing Ago2-mRNA interactions.
Figure 4.
IGF2BP3 improves miR191-5p-induced silencing complex function by binding the 3′-UTR of ZO-1 mRNA and promoting Ago2-mRNA interactions. (A) Sequence alignment of the ZO-1 3′UTR with wild-type vs. mutant potential miR191-5p targeting sites. The left red sequence represents the potential IGF2BP3 binding site of the ZO-1 mRNA 3′UTR. The right red sequence represents the potential miR191-5p binding site of the ZO-1 mRNA 3′UTR, and the crossed sequence represents deletion in the mutant ZO-1 3′UTR. (B) Luciferase reporter activity after transfection of wild-type and mutant ZO-1 3′UTR reporter constructs in HA22T cells with/without miR191-5p. (C) Sequence alignment of the ZO-1 3′UTR with wild-type vs. mutant potential IGF2BP3 targeting sites. The left red sequence represents the potential IGF2BP3 binding site of ZO-1 mRNA 3′UTR, and the crossed sequence represents deletion in the mutant ZO-1 3′UTR. The right red sequence represents the potential miR191-5p binding site of ZO-1 mRNA 3′UTR. (D) Luciferase reporter activity following transfection with wild-type or mutant ZO-1 3′UTR reporter constructs in Huh-7 cells with/without IGF2BP3-cDNA, and HA22T cells treated with/without IGF2BP3-shRNA, compared with the control cells. All quantifications are presented as the mean ± SD. *P≤0.05 and **P≤0.01. ns, not significant; IGF2BP3/IMP3, insulin-like growth factor II mRNA-binding protein 3; ZO-1, zonula occludens-1; UTR, untranslated region; miR, microRNA; sh, short hairpin (RNA); oe, overexpression; NC, negative control.
Discussion
IGF2BP3 is an oncofetal protein that is detected in a number of malignant tumors (5). Clinical research has indicated that IGF2BP3 can be used as a biomarker to distinguish tumors from normal tissue. In addition, according to previous studies, IGF2BP3 can also be used as an independent prognostic indicator (11,38,39). Notably, using data from online databases, the findings of the present study indicate that higher IGF2BP3 expression levels may be associated with the poor survival rates of patients with HCC, which is in agreement with the findings of a previous study demonstrating that IGF2BP3 may be involved in tumorigenesis, and that its expression was significantly upregulated in HCC tissues (13). Thus, increased IGF2BP3 expression may contribute to HCC progression. By contrast, decreasing the expression of IGF2BP3 may result in decreased HCC cell invasiveness.
miRNAs maturing from hairpin miRNA precursors act as critical post-transcriptional regulators involved in tumorigenesis, tumor progression and metastasis (40–44). The RISC is the primary component of post-transcriptional regulation. miRNAs recruit Ago2, a member of the argonaute protein family, to guide the targeting of mRNA cleavage or translation inhibition through the RISC (45). A number of proteins have also been reported to enhance Ago2 activation to improve RISC formation (46). One of these proteins is IGF2BP3, which can influence the expression of invasion- and migration-associated genes by strengthening the function of Ago2 (23).
The present study demonstrated that IGF2BP3 promotes cell invasiveness by downregulating ZO-1 expression, which seems to oppose the suggestion that IGF2BP3 facilitates mRNA translation by binding to and maintaining mRNA stability (37,47). Thus, from the investigation of the mechanisms responsible for the regulatory effects of IGF2BP3 on the invasion of HCC, and analysis of related gene expression, the previous study identified that IGF2BP3 promoted RISC formation (23). In the present study, the results of Ago2 antibody pulldown assays confirm that IGF2BP3 enhances the function of Ago2 (Fig. 3A), but also suggest that miRNAs were targeting ZO-1 mRNA.
The present study demonstrated that miR191-5p increases cell invasiveness. However, decreasing IGF2BP3 expression partly reversed this miR191-5p-induced invasiveness. The combination of other published and those of the present study suggests that targeting IGF2BP3 blocks cell invasiveness via the miR191-5p-Ago2 complex.
ZO-1 is a member of the membrane-associated guanylate kinase homolog family. It interacts with transmembrane proteins and links tight junction components to the cortical actin cytoskeleton to maintain epithelial tight junction integrity (48,49). Increasing evidence indicates that the downregulation of ZO-1 expression is associated with the enhancement of cancer dissemination and metastasis (27,50). The present study revealed that miR191-5p and IGF2BP3 increases cellular invasiveness by binding to the overlapping targeting sites of ZO-1 mRNA. The overexpression of miR-191-5p or IGF2BP3 decreased the ZO-1 protein expression level and promoted HCC cell invasiveness. Conversely, the inhibition of miR191-5p or the knockdown of IGF2BP3 restored ZO-1 expression and decreased invasiveness.
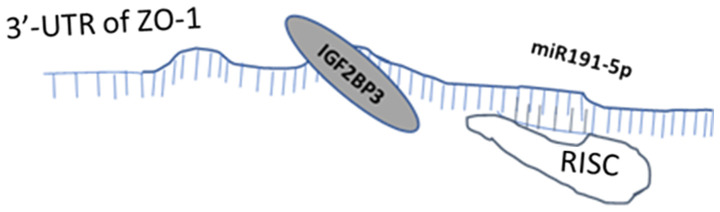
Collectively, the findings of the present study demonstrate that suppressing IGF2BP3 expression may inhibit HCC cell invasiveness by altering IGF2BP3/miR191-5P/ZO-1 signaling (Fig. 5), and that targeting this signaling molecule with small molecules may aid in the development of novel therapeutic strategies with which to better retard HCC progression.
Figure 5.
Schematic diagram illustrating IGF2BP3 function. IGF2BP3 can decrease ZO-1 mRNA stability by enhancing miR191-5p expression and inducing the RISC-mRNA interaction. ZO-1, zonula occludens-1; IGF2BP3, insulin-like growth factor II mRNA-binding protein 3; RISC, RNA-induced silencing complex; miR, microRNA; RISC, RNA-induced silencing complex; UTR, untranslated region.
Supplementary Material
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science Foundation of China (grant no. 81672469).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YG and XQ conceived and designed the study. TL, XO, CZ and JZ made substantial contributions to the design of the study, acquisition of data, interpretation of data and revising the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Miller KD, Siegel RL, Lin CC, Mariotto AB, Kramer JL, Rowland JH, Stein KD, Alteri R, Jemal A. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66:271–289. doi: 10.3322/caac.21349. [DOI] [PubMed] [Google Scholar]
- 3.Nielsen J, Christiansen J, Lykke-Andersen J, Johnsen AH, Wewer UM, Nielsen FC. A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Mol Cell Biol. 1999;19:1262–1270. doi: 10.1128/MCB.19.2.1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yaniv K, Yisraeli JK. The involvement of a conserved family of RNA binding proteins in embryonic development and carcinogenesis. Gene. 2002;287:49–54. doi: 10.1016/S0378-1119(01)00866-6. [DOI] [PubMed] [Google Scholar]
- 5.Mueller F, Bommer M, Lacher U, Ruhland C, Stagge V, Adler G, Gress TM, Seufferlein T. KOC is a novel molecular indicator of malignancy. Br J Cancer. 2003;88:699–701. doi: 10.1038/sj.bjc.6600790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohashi R, Sangen M, Namimatsu S, Takei H, Naito Z. IMP3 contributes to poor prognosis of patients with metaplastic breast carcinoma: A clinicopathological study. Ann Diagn Pathol. 2017;31:30–35. doi: 10.1016/j.anndiagpath.2017.05.015. [DOI] [PubMed] [Google Scholar]
- 7.Samanta S, Sun H, Goel HL, Pursell B, Chang C, Khan A, Greiner DL, Cao S, Lim E, Shultz LD, Mercurio AM. IMP3 promotes stem-like properties in triple-negative breast cancer by regulating SLUG. Oncogene. 2016;35:1111–1121. doi: 10.1038/onc.2015.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu D, Yang X, Jiang NY, Woda BA, Liu Q, Dresser K, Mercurio AM, Rock KL, Jiang Z. IMP3, a new biomarker to predict progression of cervical intraepithelial neoplasia into invasive cancer. Am J Surg Pathol. 2011;35:1638–1645. doi: 10.1097/PAS.0b013e31823272d4. [DOI] [PubMed] [Google Scholar]
- 9.Li C, Rock KL, Woda BA, Jiang Z, Fraire AE, Dresser K. IMP3 is a novel biomarker for adenocarcinoma in situ of the uterine cervix: An immunohistochemical study in comparison with p16(INK4a) expression. Mod Pathol. 2007;20:242–247. doi: 10.1038/modpathol.3800735. [DOI] [PubMed] [Google Scholar]
- 10.Tschirdewahn S, Panic A, Püllen L, Harke NN, Hadaschik B, Riesz P, Horváth A, Szalontai J, Nyirády P, Baba HA, et al. Circulating and tissue IMP3 levels are correlated with poor survival in renal cell carcinoma. Int J Cancer. 2019;145:531–539. doi: 10.1002/ijc.32124. [DOI] [PubMed] [Google Scholar]
- 11.Jiang Z, Chu PG, Woda BA, Rock KL, Liu Q, Hsieh CC, Li C, Chen W, Duan HO, McDougal S, Wu CL. Analysis of RNA-binding protein IMP3 to predict metastasis and prognosis of renal-cell carcinoma: A retrospective study. Lancet Oncol. 2006;7:556–564. doi: 10.1016/S1470-2045(06)70732-X. [DOI] [PubMed] [Google Scholar]
- 12.Chen ST, Jeng YM, Chang CC, Chang HH, Huang MC, Juan HF, Hsu CH, Lee H, Liao YF, Lee YL, et al. Insulin-like growth factor II mRNA-binding protein 3 expression predicts unfavorable prognosis in patients with neuroblastoma. Cancer Sci. 2011;102:2191–2198. doi: 10.1111/j.1349-7006.2011.02100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeng YM, Chang CC, Hu FC, Chou HY, Kao HL, Wang TH, Hsu HC. RNA-binding protein insulin-like growth factor II mRNA-binding protein 3 expression promotes tumor invasion and predicts early recurrence and poor prognosis in hepatocellular carcinoma. Hepatology. 2008;48:1118–1127. doi: 10.1002/hep.22459. [DOI] [PubMed] [Google Scholar]
- 14.Yan J, Wei Q, Jian W, Qiu B, Wen J, Liu J, Fu B, Zhou X, Zhao T. IMP3 predicts invasion and prognosis in human lung adenocarcinoma. Lung. 2016;194:137–146. doi: 10.1007/s00408-015-9829-0. [DOI] [PubMed] [Google Scholar]
- 15.Li W, Liu D, Chang W, Lu X, Wang YL, Wang H, Zhu C, Lin HY, Zhang Y, Zhou J, Wang H. Role of IGF2BP3 in trophoblast cell invasion and migration. Cell Death Dis. 2014;5:e1025. doi: 10.1038/cddis.2013.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taniuchi K, Furihata M, Hanazaki K, Saito M, Saibara T. IGF2BP3-mediated translation in cell protrusions promotes cell invasiveness and metastasis of pancreatic cancer. Oncotarget. 2014;5:6832–6845. doi: 10.18632/oncotarget.2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pasiliao CC, Chang CW, Sutherland BW, Valdez SM, Schaeffer D, Yapp DT, Ng SS. The involvement of insulin-like growth factor 2 binding protein 3 (IMP3) in pancreatic cancer cell migration, invasion, and adhesion. BMC Cancer. 2015;15:266. doi: 10.1186/s12885-015-1251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jens M, Rajewsky N. Competition between target sites of regulators shapes post-transcriptional gene regulation. Nat Rev Genet. 2015;16:113–126. doi: 10.1038/nrg3853. [DOI] [PubMed] [Google Scholar]
- 19.Bell JL, Wächter K, Mühleck B, Pazaitis N, Köhn M, Lederer M, Hüttelmaier S. Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs): Post-transcriptional drivers of cancer progression? Cell Mol Life Sci. 2013;70:2657–2675. doi: 10.1007/s00018-012-1186-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang W, Cheng X, Wang T, Song X, Zheng Y, Wang L. LINC00467 promotes cell proliferation and metastasis by binding with IGF2BP3 to enhance the mRNA stability of TRAF5 in hepatocellular carcinoma. J Gene Med. 2020;22:e3134. doi: 10.1002/jgm.3134. [DOI] [PubMed] [Google Scholar]
- 21.Mizutani R, Imamachi N, Suzuki Y, Yoshida H, Tochigi N, Oonishi T, Suzuki Y, Akimitsu N. Oncofetal protein IGF2BP3 facilitates the activity of proto-oncogene protein eIF4E through the destabilization of EIF4E-BP2 mRNA. Oncogene. 2016;35:3495–3502. doi: 10.1038/onc.2015.410. [DOI] [PubMed] [Google Scholar]
- 22.Jønson L, Christiansen J, Hansen TVO, Vikeså J, Yamamoto Y, Nielsen FC. IMP3 RNP safe houses prevent miRNA-directed HMGA2 mRNA decay in cancer and development. Cell Rep. 2014;7:539–551. doi: 10.1016/j.celrep.2014.03.015. [DOI] [PubMed] [Google Scholar]
- 23.Ennajdaoui H, Howard JM, Sterne-Weiler T, Jahanbani F, Coyne DJ, Uren PJ, Dargyte M, Katzman S, Draper JM, Wallace A, et al. IGF2BP3 modulates the interaction of invasion-associated transcripts with RISC. Cell Rep. 2016;15:1876–1883. doi: 10.1016/j.celrep.2016.04.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.González-Mariscal L, Betanzos A, Avila-Flores A. MAGUK proteins: Structure and role in the tight junction. Semin Cell Dev Biol. 2000;11:315–324. doi: 10.1006/scdb.2000.0178. [DOI] [PubMed] [Google Scholar]
- 25.Nagai T, Arao T, Nishio K, Matsumoto K, Hagiwara S, Sakurai T, Minami Y, Ida H, Ueshima K, Nishida N, et al. Impact of tight junction protein ZO-1 and twist expression on postoperative survival of patients with hepatocellular carcinoma. Dig Dis. 2016;34:702–707. doi: 10.1159/000448860. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X, Wang L, Zhang H, Tu F, Qiang Y, Nie C. Decreased expression of ZO-1 is associated with tumor metastases in liver cancer. Oncol Lett. 2019;17:1859–1864. doi: 10.3892/ol.2018.9765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsu YL, Hung JY, Chang WA, Lin YS, Pan YC, Tsai PH, Wu CY, Kuo PL. Hypoxic lung cancer-secreted exosomal miR-23a increased angiogenesis and vascular permeability by targeting prolyl hydroxylase and tight junction protein ZO-1. Oncogene. 2017;36:4929–4942. doi: 10.1038/onc.2017.105. [DOI] [PubMed] [Google Scholar]
- 28.Aden DP, Fogel A, Plotkin S, Damjanov I, Knowles BB. Controlled synthesis of HBsAg in a differentiated human liver carcinoma-derived cell line. Nature. 1979;282:615–616. doi: 10.1038/282615a0. [DOI] [PubMed] [Google Scholar]
- 29.Knowles BB, Howe CC, Aden DP. Human hepatocellular carcinoma cell lines secrete the major plasma proteins and hepatitis B surface antigen. Science. 1980;209:497–499. doi: 10.1126/science.6248960. [DOI] [PubMed] [Google Scholar]
- 30.López-Terrada D, Cheung SW, Finegold MJ, Knowles BB. Hep G2 is a hepatoblastoma-derived cell line. Hum Pathol. 2009;40:1512–1515. doi: 10.1016/j.humpath.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 31.Chang C, Lin Y, O-Lee TW, Chou CK, Lee TS, Liu TJ, P'eng FK, Chen TY, Hu CP. Induction of plasma protein secretion in a newly established human hepatoma cell line. Mol Cell Biol. 1983;3:1133–1137. doi: 10.1128/MCB.3.6.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakabayashi H, Taketa K, Yamane T, Miyazaki M, Miyano K, Sato J. Phenotypical stability of a human hepatoma cell line, HuH-7, in long-term culture with chemically defined medium. Gan. 1984;75:151–158. [PubMed] [Google Scholar]
- 33.Stepanenko AA, Dmitrenko VV. HEK293 in cell biology and cancer research: Phenotype, karyotype, tumorigenicity, and stress-induced genome-phenotype evolution. Gene. 2015;569:182–190. doi: 10.1016/j.gene.2015.05.065. [DOI] [PubMed] [Google Scholar]
- 34.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 35.Chen X, Zhao M, Huang J, Li Y, Wang S, Harrington CA, Qian DZ, Sun XX, Dai MS. MicroRNA-130a suppresses breast cancer cell migration and invasion by targeting FOSL1 and upregulating ZO-1. J Cell Biochem. 2018;119:4945–4956. doi: 10.1002/jcb.26739. [DOI] [PubMed] [Google Scholar]
- 36.Liu M, Yang J, Zhang Y, Zhou Z, Cui X, Zhang L, Fung KM, Zheng W, Allard FD, Yee EU, et al. ZIP4 promotes pancreatic cancer progression by repressing ZO-1 and claudin-1 through a ZEB1-dependent transcriptional mechanism. Clin Cancer Res. 2018;24:3186–3196. doi: 10.1158/1078-0432.CCR-18-0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lederer M, Bley N, Schleifer C, Hüttelmaier S. The role of the oncofetal IGF2 mRNA-binding protein 3 (IGF2BP3) in cancer. Semin Cancer Biol. 2014;29:3–12. doi: 10.1016/j.semcancer.2014.07.006. [DOI] [PubMed] [Google Scholar]
- 38.Li D, Yan D, Tang H, Zhou C, Fan J, Li S, Wang X, Xia J, Huang F, Qiu G, Peng Z. IMP3 is a novel prognostic marker that correlates with colon cancer progression and pathogenesis. Ann Surg Oncol. 2009;16:3499–3506. doi: 10.1245/s10434-009-0648-5. [DOI] [PubMed] [Google Scholar]
- 39.Li S, Cha J, Kim J, Kim KY, Kim HJ, Nam W, Cha IH. Insulin-like growth factor II mRNA-binding protein 3: A novel prognostic biomarker for oral squamous cell carcinoma. Head Neck. 2011;33:368–374. doi: 10.1002/hed.21457. [DOI] [PubMed] [Google Scholar]
- 40.Farazi TA, Spitzer JI, Morozov P, Tuschl T. miRNAs in human cancer. J Pathol. 2011;223:102–115. doi: 10.1002/path.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 42.Farazi TA, Hoell JI, Morozov P, Tuschl T. MicroRNAs in human cancer. In: Schmitz U, Wolkenhauer O, Vera J, editors. MicroRNA cancer regulation. Dordrecht Springer Netherlands; 2013. pp. 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Di Leva G, Croce CM. Roles of small RNAs in tumor formation. Trends Mol Med. 2010;16:257–267. doi: 10.1016/j.molmed.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 45.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 46.Jing Q, Huang S, Guth S, Zarubin T, Motoyama A, Chen J, Di Padova F, Lin SC, Gram H, Han J. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120:623–634. doi: 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
- 47.Glisovic T, Bachorik JL, Yong J, Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008;582:1977–1986. doi: 10.1016/j.febslet.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anderson JM. Cell signalling: MAGUK magic. Curr Biol. 1996;6:382–384. doi: 10.1016/S0960-9822(02)00501-8. [DOI] [PubMed] [Google Scholar]
- 49.Gottardi CJ, Arpin M, Fanning AS, Louvard D. The junction-associated protein, zonula occludens-1, localizes to the nucleus before the maturation and during the remodeling of cell-cell contacts. Proc Natl Acad Sci USA. 1996;93:10779–10784. doi: 10.1073/pnas.93.20.10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hoover KB, Liao SY, Bryant PJ. Loss of the tight junction MAGUK ZO-1 in breast cancer: Relationship to glandular differentiation and loss of heterozygosity. Am J Pathol. 1998;153:1767–1773. doi: 10.1016/S0002-9440(10)65691-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.