Abstract
A number of studies have demonstrated the crucial functions of GINS2 within the GINS complex in various types of cancer. However, the molecular mechanisms and prognostic value of GINS2 in breast cancer remain unknown. The present study used; BC-GenExMiner, COSMIC, UCSC Xena, The Human Protein Atlas, GEPIA, cBioPortal, GeneMANIA, TIMER and Oncomine, in order to investigate gene expression, co-expression, clinical parameters and mutations in GINS2 in patients with breast cancer. Furthermore, the present study assessed the prognostic value of GINS2 in patients with breast cancer via the Kaplan-Meier plotter database. The results of the present study demonstrated that the mRNA levels of GINS2 were significantly higher in breast cancer tissue compared with normal tissue. In addition, high mRNA expression levels of GINS2 were associated with high Scarff-Bloom-Richardson status grades, a basal-like status and age (≤51 years); however, it was not associated with lymph node metastasis. The survival analysis revealed that increased GINS2 mRNA levels were associated with a worse prognosis for relapse-free survival in all patients with breast cancer, particularly in those with estrogen receptor-positive and progesterone receptor-positive subtypes. In addition, a positive association between the GINS2, CENPM and MCM4 genes was confirmed. The results of the present study suggest that GINS2 could be used as a potential prognostic biomarker for breast cancer. Nevertheless, further studies are necessary to confirm the effects of GINS2 on the pathogenesis and development of patients with breast cancer.
Keywords: breast cancer, prognosis values, GINS2
Introduction
In 2018, there were ~2.1 million newly diagnosed female breast cancer cases (1). Although several advancements in early diagnosis and treatment have been made, the prevalence of breast cancer remains a crucial health issue worldwide (2–4). Accurate molecular biomarkers for early diagnosis may be a useful way to improve the current treatments for breast cancer. Therefore, investigating specific and sensitive molecular biomarkers involved in breast cancer has significant clinical value.
DNA helicases unwind or rearrange duplex DNA during replication, recombination and repair, thus playing an essential role in the preservation of genome stability (5). A number of studies have demonstrated that cancer ensues when the activity of DNA helicases is altered, which indicates that a significant number of helicases are tumor-associated (6–10). In addition to its fundamental role within the GINS complex, GINS2 is also a vital component of the CMG complex (11). The CMG complex is the eukaryotic replicative helicase that can unwind double-stranded DNA, including, minichromosome maintenance proteins 2–7 (MCM2-7), cell division cycle 45 (CDC45) and the GINS complex, at replication forks (11,12). Close attention should be paid to the expression of GINS2 in different human malignancies, due to the inextricable linkage between DNA helicases and human cancer types (13). In addition, the upregulation of GINS2 has been reported in various types of human cancer, including glioma, cervical cancer, rectal cancer and lung adenocarcinoma (14–17). Regarding breast cancer, GINS2 expression is also increased in triple-negative breast cancer cells (18). However, to the best of our knowledge, the prognostic value of GINS2 and its role in other types of breast cancer remains unknown.
The present study sought to conduct a more in-depth analysis of GINS2 in order to investigate whether it could be used as a potential prognostic biomarker for breast cancer. The following databases were used; Oncomine, Tumor Immune Estimation Resource (TIMER) and The Human Protein Atlas (HPA), in order to evaluate the differential expression of GINS2 between breast tumor tissue and normal tissue. The association between GINS2 expression and clinical parameters including expression patterns, gene mutations and distinct prognostic values of GINS2 in breast cancer, was also analyzed using the publicly accessible databases. The present study also used the following databases; Oncomine, GeneMANIA, Gene Expression Profiling Interactive Analysis (GEPIA) and University of California Santa Cruz Xena (UCSC Xena), in order to analyze the co-expression and neighboring genes of GINS2.
Materials and methods
The following datasets and patients' information were acquired from the aforementioned publicly accessible online databases. No human or animal specimens were used in the present study.
Oncomine analysis
The Oncomine database (oncomine.org) is a publicly accessible online database containing 715 datasets and 86,733 samples (19). Oncomine was used to compare the mRNA expression of GINS2 in breast cancer tissues with normal tissues. Paired Student's t-test was used in order to analyze the differences in transcriptional expression between datasets derived from cancer specimens and normal controls. The following values were used as thresholds: Gene rank, 10%; fold change, 2; and P<0.0001.
Breast cancer Gene-expression miner v4.1 (bc-GenExMiner v4.1) analysis
bc-GenExMiner v4.1 (bcgenex.centregauducheau.fr) is a statistical mining tool that contains 36 annotated genomic datasets and the data of 5,861 patients with breast cancer (20,21). The association between GINS2 expression levels and different clinical parameters [age, Tumor-Node-Metastasis stage, Scarff-Bloom-Richardson (SBR) grade, Nottingham Prognostic Index (NPI), estrogen receptor status, progesterone receptor status and human epidermal growth factor receptor 2 status] of patients with breast cancer was analyzed using this publicly accessible database (22–24). Welch's t-test was used in order to compare the difference in expression of GINS2 between the groups of patients according to different clinicopathological parameters and Dunnett-Tukey-Kramer test was used for pair-wise comparisons when a significant difference was observed. P<0.05 was considered to indicate a statistically significant difference.
Kaplan-Meier (KM) plotter analysis
The KM plotter database interprets information regarding gene expression and survival analysis of different types of cancer including, breast, liver, ovarian, lung and gastric cancer (25,26). KM plotter was used to analyze the prognostic values of GINS2 in patients with breast cancer and relapse-free survival (RFS). The log-rank P-value was presented on the website, P<0.05 was considered to indicate a statistically significant difference.
Catalogue of somatic mutations in cancer (COSMIC) analysis
The COSMIC database (cancer.sanger.ac.uk/cosmic) is one of the most abundant resources for searching gene mutations in several types of human cancer (27). The COSMIC database was used in order to examine the different types of GINS2 mutations in breast cancer. The following formula was used in order to calculate the percentage of each type of mutation in different types of breast cancer: [(Genetic alternation samples)/(total samples)] ×100%.
UCSC Xena analysis
The UCSC Xena database (xena.ucsc.edu) is a powerful genomic online tool that provides visualization and integration heat maps for analyzing the publicly accessible datasets (28,29). The UCSC Xena database was used to generate a heat map of GINS2, MCM4 and CENPM expression, according to PAM50 breast cancer subtypes in The Cancer Genome Atlas (TCGA) Breast Invasive Carcinoma database (portal.gdc.cancer.gov/).
HPA analysis
The HPA (proteinatlas.org) database aims to map all the human proteins in cells, tissues and organs (30–32). The HPA online tool has already helped thousands of researchers in the fields of biomedicine and disease. The HPA database was used in order to determine GINS2 protein expression level via immunohistochemistry (IHC) staining and the IHC images were obtained from the HPA database.
GEPIA analysis
The GEPIA database (gepia.cancer-pku.cn) is an online website that can be used to analyze the RNA expression data, based on TCGA and the Genotype-Tissue Expression (GTEx) Projects (33,34). Pearson's correlation test was used in order to assess the association between GINS2, MCM4 and CENPM in breast cancer via the GEPIA database.
cBioPortal for cancer genomics analysis
The cBioPortal for Cancer Genomics database (http://www.cbioportal.org) is an online website that allows for the visualization, analysis and download of large-scale cancer genomic datasets (35–37). The cBioPortal for Cancer Genomics database was used to analyze the expression of and mutations in GINS2. The breast cancer dataset (TCGA Cell 2015) was used for further analysis within the database (38). Kaplan-Meier analysis was used in order to assess the overall survival (OS) and disease-free survival (DFS) rates of GINS2, using the database.
GeneMANIA analysis
GeneMANIA (genemania.org) is an online database that identifies other associated with a set of input genes. The GeneMANIA database was used to identify genes that are associated with GINS2 at the genetic level. Furthermore, the associations between pathways, shared protein domains, and the co-localization and co-expression of GINS2 were determined using GeneMANIA.
TIMER analysis
TIMER (cistrome.shinyapps.io/timer) is an online database used for the systematic analysis of immune infiltrates and gene expression across different types of cancer (39). Wilcoxon signed-rank test was used to analyze GINS2 expression levels between breast cancer tissue and normal breast tissue via the TIMER database.
Results
Expression of GINS2 in human breast cancer
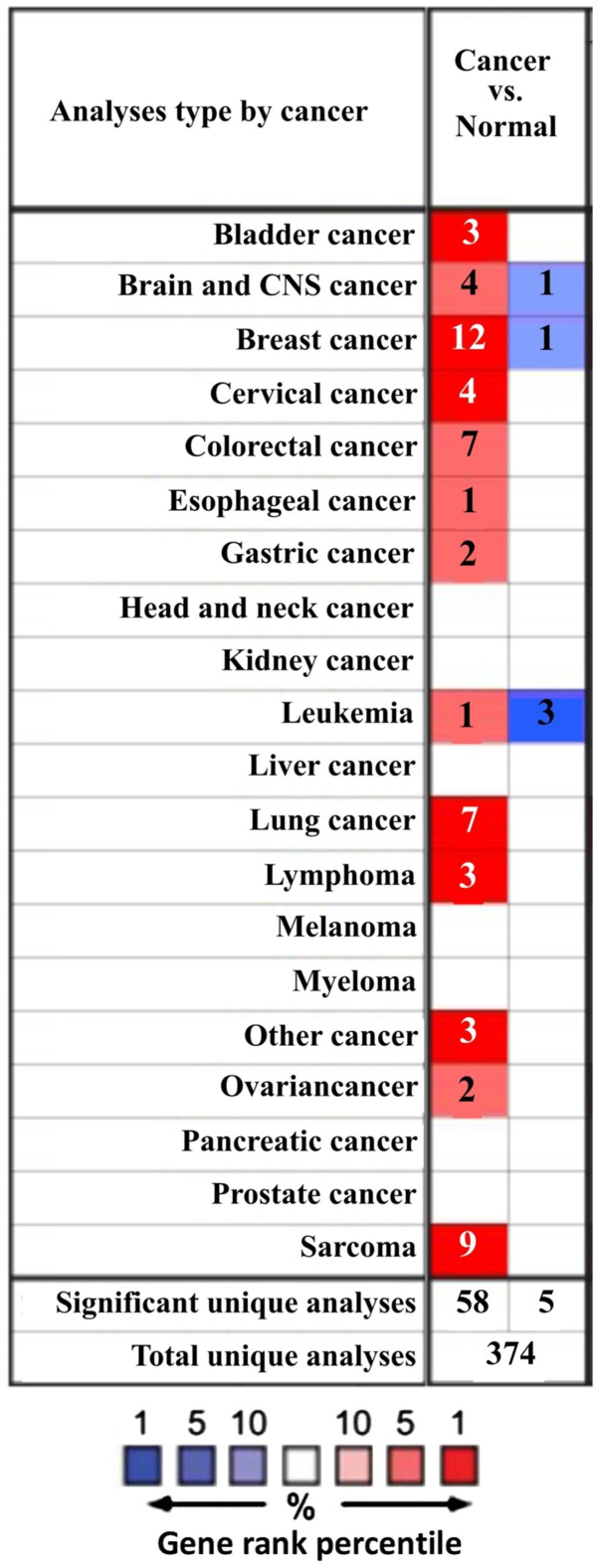
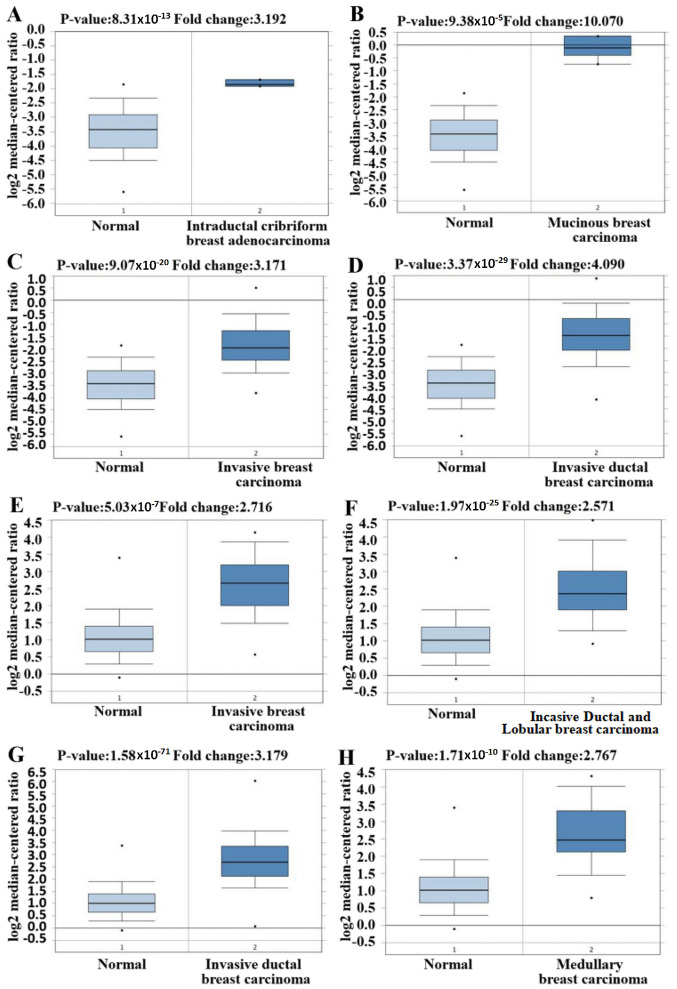
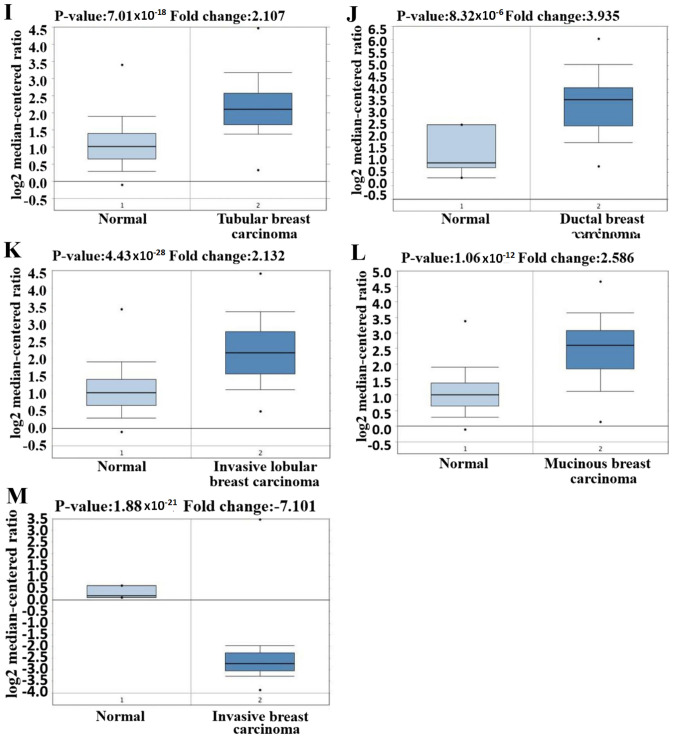
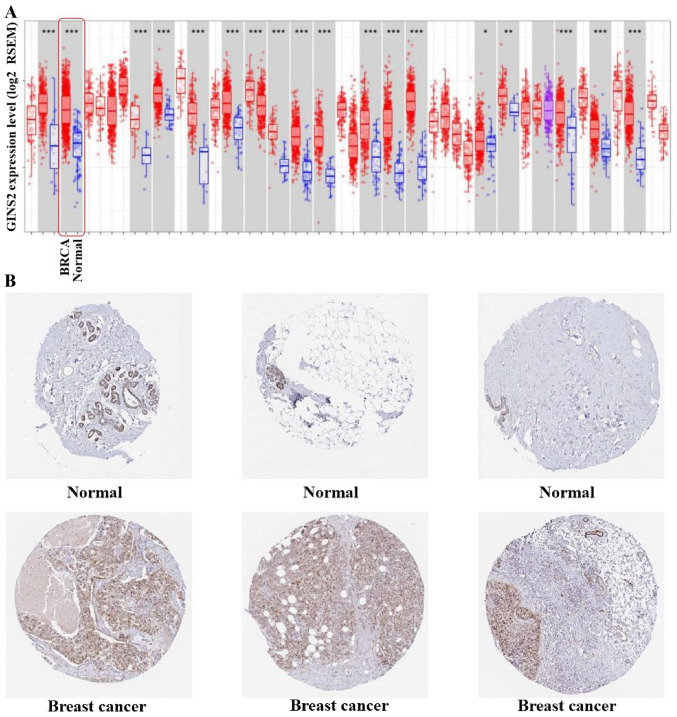
The difference in mRNA expression of GINS2 between tumor and normal tissues, in multiple types of cancer, was analyzed in the present study using the online Oncomine database. The Oncomine database contained a total of 374 uniquely analyzed GINS2. In the breast cancer analysis, the mRNA expression of GINS2 was significantly upregulated and downregulated in 12 and one studies, respectively (Figs. 1 and 2). In these 13 studies, Finak et al (40) is the only study to have demonstrated the downregulation of GINS2 in invasive breast carcinoma, (P=1.88×10−21; fold change, −7.101; Fig. 2M). In contrast, 12 different datasets revealed that GINS2 had higher mRNA expression in breast cancer compared with normal breast tissues (41,42). Different types of breast cancer, including: Invasive breast carcinoma (P=5.03×10−7; fold change, 2.716; Fig. 2E); Invasive Ductal and Invasive Lobular Breast Carcinoma (P=1.97×10−25; fold change, 2.571; Fig. 2F); invasive ductal breast carcinoma (P=1.58×10−71; fold change, 3.179; Fig. 2G); medullary breast carcinoma (P=1.71×10−10; fold change, 2.767; Fig. 2H); invasive lobular breast carcinoma (P=4.43×10−28; fold change, 2.132; Fig. 2K); mucinous breast carcinoma (P=1.06×10−12; fold change, 2.586; Fig. 2L) and tubular breast carcinoma (P=7.01×10−18; fold change, 2.107; Fig. 2I) demonstrated upregulated mRNA expression of GINS2 in the Curtis Breast dataset (41). Furthermore, increased mRNA expression of GINS2 in: Intraductal cribriform breast adenocarcinoma (P=8.31×10−13; fold change, 3.192; Fig. 2A); mucinous breast carcinoma (P=9.38×10−5; fold change, 10.070; Fig. 2B); invasive breast carcinoma (P=9.07×10−20; fold change, 3.171; Fig. 2C) and invasive ductal breast carcinoma (P=3.37×10−29; fold change, 4.090; Fig. 2D) was observed in TCGA datasets (43) Increased mRNA expression of GINS2 in ductal breast carcinoma (P=8.32×10−6; fold change, 3.935; Fig. 2J) were demonstrated in the Richardson Breast2 dataset (42). These data all indicate that GINS2 expression is markedly higher in breast cancer samples compared with in normal breast tissues. In order to further evaluate the upregulation of GINS2 in breast cancer, the present study examined GINS2 expression using the TIMER database. mRNA expression levels of GINS2 between breast cancer and normal breast tissues were compared. The results of the present study indicate that the expression level of GINS2 is higher in BRCA compared with in normal breast tissue (Fig. 3A).
Figure 1.
mRNA expression levels of GINS2 in different types of cancer based on data obtained from the Oncomine database. Red or blue represents the numbers of datasets which statistically significant upregulation or downregulation of GINS2 expression levels, respectively.
Figure 2.
GINS2 expression in different studies and different types of breast cancer using the Oncomine database. (A-M) Box plots of GINS2 expression comparing different subtypes of breast cancer and normal tissues using the Oncomine database. Upregulated mRNA expression of GINS2 in TCGA datasets of (A) intraductal cribriform breast adenocarcinoma (P=8.31×10−13); (B) mucinous breast carcinoma (P=9.38×10−5); (C) invasive breast carcinoma (P=9.07×10−20); (D) invasive ductal breast carcinoma (P=3.37×10−29). Upregulated mRNA expression of GINS2 in the Curtis breast datasets of (E) invasive breast carcinoma (P=5.03×10−7); (F) invasive ductal and invasive lobular breast carcinoma (P=1.97E-25); (G) invasive ductal breast carcinoma (P=1.58×10−71); (H) medullary breast carcinoma (P=1.71×10−10). GINS2 expression in different studies and different types of breast cancer using the Oncomine database. (A-M) Box plots of GINS2 expression comparing different subtypes of breast cancer and normal tissues using the Oncomine database. Upregulated mRNA expression of GINS2 in the Curtis Breast dataset of (I) tubular breast carcinoma (P=7.01×10−18); (J) upregulated mRNA expression of GINS2 in the Richardson Breast2 dataset of ductal breast carcinoma (P=8.32×10−6); (K) invasive lobular breast carcinoma (P=4.43×10−28); (L) mucinous breast carcinoma (P=1.06×10−12); (M) downregulation mRNA expression of GINS2 in the Finak dataset of invasive breast carcinoma, (P=1.88×10−21).
Figure 3.
GINS2 expression at the mRNA and protein levels. (A) GINS2 expression levels between breast tumor tissue and normal tissue in different types of tumors from The Cancer Genome Atlas database were determined using the Tumor Immune Estimation Resource database. *P<0.05; **P<0.01; ***P<0.001 vs. respective normal tissue. (B) GINS2 protein expression was determined using the Human Protein Atlas database. mRNA, messenger RNA.
However, IHC analysis obtained from the HPA database contradicts this observation. GINS2 protein was identified across both instances following staining with the HPA057285 antibody in the glandular and myoepithelial cells in normal breast tissue and the breast cancer tissues (all samples; Fig. 3B).
GINS2 mutation in breast cancer
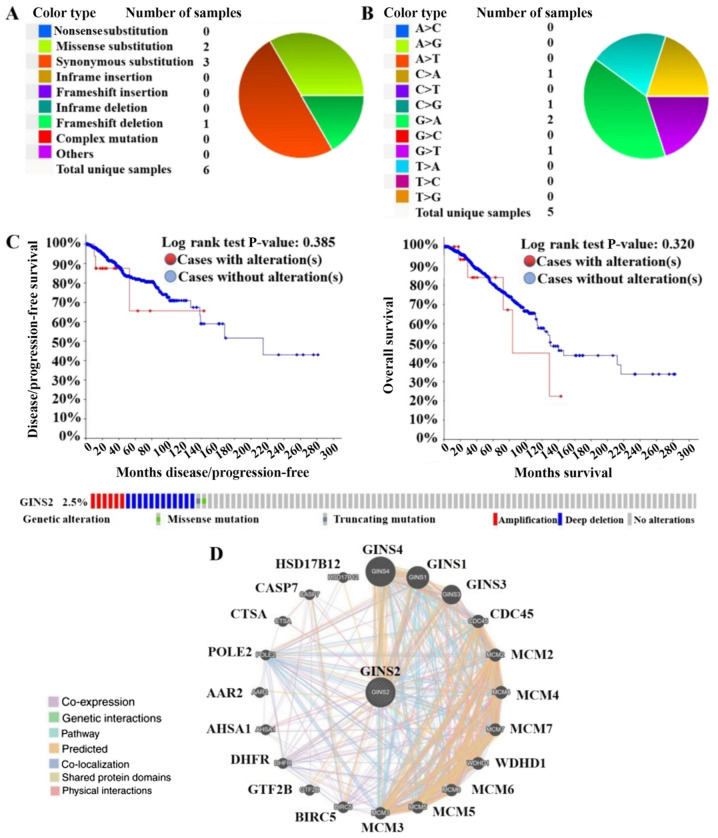
The genetic alterations affecting GINS2 in breast cancer samples were analyzed using the COSMIC online database. The GINS2 mutations were tested in 2,522 samples from patients with breast cancer. Of the five point mutations in the GINS2 gene, three were synonymous and two were a missense mutation (Table I). A total of nine copy number variations (CNV) were identified in breast cancer including, two CNV gains and seven CNV losses (Table I). A summary of the types of mutations in GINS2 are presented on the pie chart in Fig. 4A. The occurrence of a particular type of mutation is presented in Table I, with the number next to it indicating the percentage of mutations in all samples. The most common type of mutation was synonymous substitution, which accounts for 50% of all mutations. The mutations of the coding strand in GINS2 are presented on the pie chart in Fig. 4B and there was 40% G>A mutations in the GINS2 coding strand.
Table I.
Genetic alterations affecting GINS2 in 2,522 breast cancer samples Catalogue of Somatic Mutations in Cancer database.
| Genetic alteration | No. | Percentage, % |
|---|---|---|
| Substitution missense | 2 | 0.08 |
| Substitution synonymous | 3 | 0.12 |
| Substitution nonsense | 0 | 0.00 |
| Copy number gain | 2 | 0.08 |
| Copy number loss | 7 | 0.28 |
| Insertion | 0 | 0.00 |
| Deletion | 1 | 0.04 |
Figure 4.
Network of alterations and neighboring genes of GINS2 using the COSMIC, cBioPortal and GeneMANIA databases. (A) Summary of the types of GINS2 mutations in breast cancer according to the COSMIC database. (B) Summary of the mutations of the coding strand in GINS2 according to the COSMIC database. (C) Kaplan-Meier analysis was used in order to assess the disease-free survival and overall survival rates, with or without GINS2 alterations in breast cancer. (D) Gene-gene interaction network of GINS2. COSMIC, Catalogue of Somatic Mutations in Cancer.
In order to investigate the association between survival time and gene alterations of GINS2 in breast cancer, genetic alterations affecting GINS2 in breast cancer samples were analyzed in the present study using the cBioPortal for Cancer Genomics database. Mutations of GINS2 were tested in 816 samples from patients with breast cancer in TCGA (38). Gene alterations of GINS2 were demonstrated to have occurred in 20 of the 816 (2.5%) queried samples. However, the results of the present study demonstrated that there were no statistically significant difference between OS/DFS and patients with breast cancer, with or without GINS2 alterations (P=0.320 and P=0.385, respectively; Fig. 4C).
Co-expression and neighboring genes of GINS2 gene
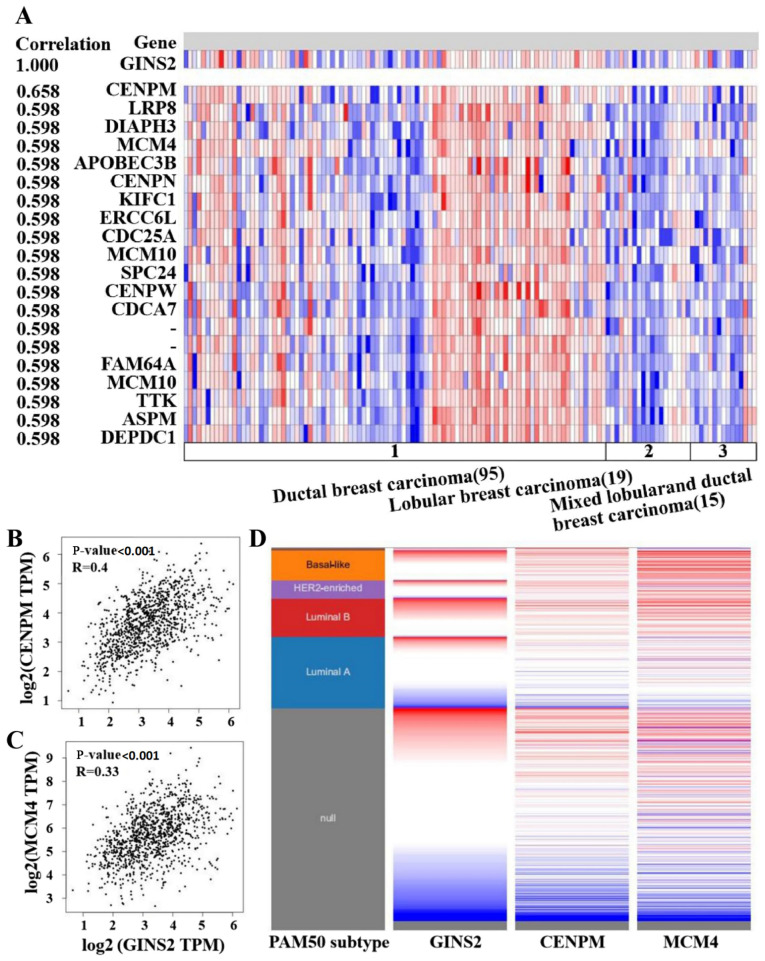
Co-expression of the GINS2 gene in breast cancer was performed using the Oncomine database and genes co-expressed with GINS2 were analyzed by Lu et al (44). The present study demonstrated that GINS2 was associated with CENPM, LRP8, DIAPH3, MCM4, APOBEC3B, CENPN, KIFC1, ERCC6L, CDC25A, MCM10, SPC24, CENPW, CDCA7, FAM64A, MCM10, TTK, ASPM and DEPDC1 (Fig. 5A). Subsequently, the present study constructed a network analysis of the neighboring genes of GINS2 using the GeneMANIA database. The results of the present study demonstrated that the following 20 genes; GINS4, GINS1, GINS3, CDC45, MCM2, MCM4, MCM7, WDHD1, MCM6, MCM5, MCM3, BIRC5, GTF2B, DHFR, AHSA1, AAR2, POLE2, CTSA, CASP7 and HSD17B12 were closely associated with GINS2 (Fig. 4D). In the Oncomine co-expression analysis, CENPM ranked first with a score of 0.658. Furthermore, the MCM4 also ranked with a high score of 0.598 in the Oncomine co-expression analysis. Thus, heat maps derived from the UCSC Xena database were used in order to compare GINS2, CENPM and MCM4 expression. The results of the present study demonstrated that GINS2 was upregulated when the expression level of CENPM and MCM4 were increased, which was determined using TCGA database (Fig. 5D). Furthermore, data mining in GEPIA also revealed a positive association between GINS2, CENPM and MCM4. However, the coefficients were relatively low between GINS2 and CENPM (0.4) or MCM4 (0.33) (Fig. 5B and C). Collectively, the results of the present study suggest that in breast cancer, GINS2 may be associated with the CENPM and MCM4 signaling pathways.
Figure 5.
Co-expression genes of GINS2. (A) Co-expression genes of GINS2 were obtained using the Oncomine database. Red or blue represent the upregulation or the downregulation of GINS2 gene, respectively. (B) The association between GINS2 and CENPM expression in breast cancer was analyzed using the Gene Expression Profiling Interactive Analysis database. P<0.001. (C) Association between GINS2 and MCM4 expression in breast cancer was analyzed using the Gene Expression Profiling Interactive Analysis database. P<0.001. (D) Heat maps of GINS2, CENPM and MCM4 expression according to the PAM50 breast cancer subtypes using the University of California Santa Cruz Xena web-based tool. Red or blue represent the upregulation or the downregulation of GINS2, respectively.
GINS2 expression and clinical parameters of patients with breast cancer
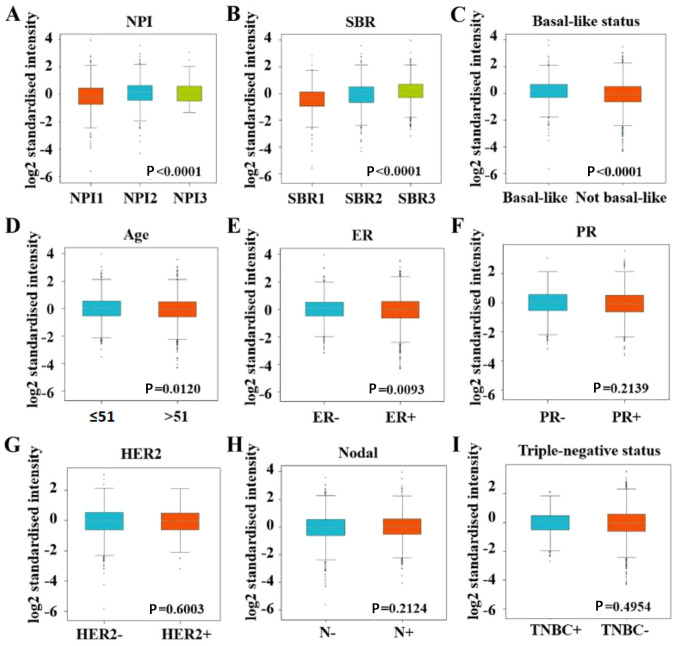
Using the bc-GenExMiner database, the present study also evaluated GINS2 expression in different groups, such as age and ER expression status. Regarding age, GINS2 expression was demonstrated to be significantly elevated in patients ≤51 years (P=0.012; Fig. 6D). Patients with estrogen receptor (ER)-negative breast cancer were demonstrated to have significantly increased GINS2 gene expression compared with ER-positive patients (P=0.0093; Fig. 6E). Regarding progesterone receptor (PR) status, human epidermal growth factor receptor 2 (HER2) and triple-negative status, there were no statistically significant differences in GINS2 expression between the positive and negative groups (Fig. 6F, G and I). Similarly, there was no statistically significant difference in GINS2 expression between patients with positive and negative lymph nodes in breast cancer (Fig. 6H). However, GINS2 expression was significantly elevated in those patients with the basal-like subtype of breast cancer compared with those with the non-basal-like subtype (P<0.0001; Fig. 6C). The SBR grade and the NPI are commonly accepted prognostic factors for histological grade and tumor grade in breast cancer, respectively (22,23). The results from the bc-GenExMiner database in the present study demonstrated that an advanced SBR grade was associated with higher GINS2 expression (Fig. 6B; Table II). As for NPI, GINS2 expression was demonstrated to be higher in NPI2 compared with NPI1 (Fig. 6A; Table II).
Figure 6.
Association between mRNA expression of GINS2 and different clinical parameters using the bc-GenExMiner database. Analysis is demonstrated for: (A) NPI (P<0.001); (B) SBR (P<0.001); (C) basal-like status (P<0.001); (D) age (P=0.012); (E) ER (P=0.0093); (F) PR (P=0.2139); (G) HER2 (P=0.6003); (H) Nodal (P=0.2124); and (I) triple-negative status (P=0.4954). NPI, Nottingham prognostic index; SBR, Scarff-Bloom-Richardson; ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2.
Table II.
GINS2 expression levels according to the SBR grade status and NPI values.
| Group comparison (GINS2) | P-value | Group comparison (GINS2) | P-value |
|---|---|---|---|
| SBR2>SBR1 | P<0.0001 | NPI2>NPI1 | P<0.0001 |
| SBR3>SBR1 | P<0.0001 | NPI3=NPI2 | P>0.10 |
| SBR3>SBR2 | P<0.0001 | NPI3=NPI1 | P>0.10 |
SBR, Scarff-Bloom-Richardson; NPI, Nottingham Prognostic Index.
Prognostic value of GINS2 in patients with breast cancer
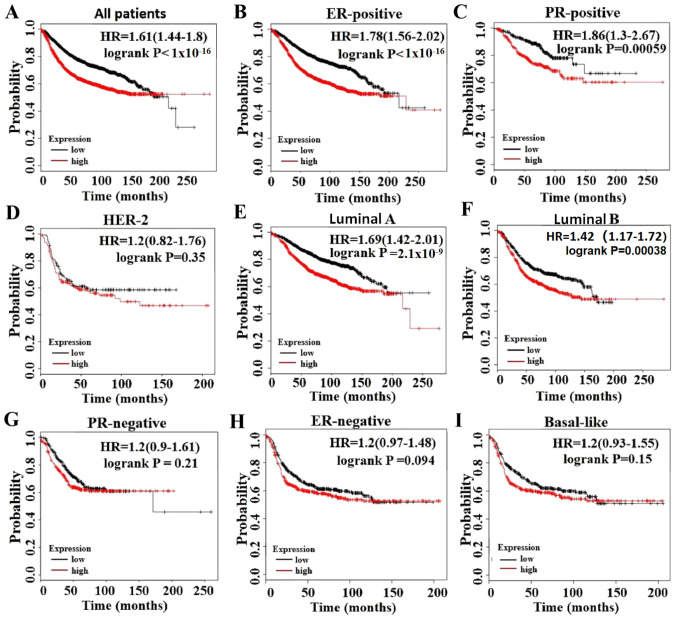
The RFS rate was analyzed for GINS2 using the KM plotter database. The present study demonstrated that elevated GINS2 expression was associated with a poor prognosis of RFS in patients with breast cancer [hazard ratio (HR), 1.61; P<1×10−16; Fig. 7A]. In particular, the sub-analysis revealed that elevated GINS2 mRNA expression was significantly associated with a poor prognosis of RFS in the ER-positive (HR, 1.78; P<1×10−16; Fig. 7B) and PR-positive (HR, 1.86; P=0.00059; Fig. 7C) subgroups of patients with breast cancer, but not in the ER-negative (HR, 1.2; P=0.094; Fig. 7H) and PR-negative (HR, 1.2; P=0.21; Fig. 7G) subgroups of patients. Furthermore, increased GINS2 mRNA expression was significantly associated with a poor prognosis of RFS in patients with unique molecular subtypes in Luminal-A (HR, 1.69; P=2.1×10−9; Fig. 7E) and Luminal-B (HR, 1.42; P=0.00038; Fig. 7F), but not in other molecular subtypes, including basal-like type (HR, 1.2; P=0.15; Fig. 7I) and HER2 type (HR, 1.2; P=0.35; Fig. 7D).
Figure 7.
Prognostic values of GINS2 in breast cancer. Analysis is demonstrated for (A) All patients (P<1×10−16); (B) ER-positive (P<1×10−16); (C) PR-positive (P=0.00059); (D) HER-2 (P=0.35); (E) Luminal A (P=2.1×10−9); (F) Luminal B (P=0.00038); (G) PR-negative (P=0.21); (H) ER-negative (P=0.094); and (I) Basal-like (P=0.15). HR, hazard ratio; ER, estrogen receptor; PR, progesterone receptor; HER-2, human epidermal growth factor receptor 2.
Discussion
Breast cancer comprises different subtypes that are associated with unique biological and clinical features (45,46). Previously, critical molecular biomarkers, such as ER, PR and HER-2, have been identified and well characterized in breast cancer (47,48). Therefore, understanding the role of biomarkers in determining the prognosis and identifying appropriate therapies in patients with breast cancer is critical.
The GINS complex is a protein complex that is composed of SLD5, PSF1, PSF2 and PSF3 subunits, which correspond to GINS4, GINS1, GINS2 and GINS3 in the human genome, respectively (5). These four subunits are structurally similar and are likely to have derived from a single protein through gene duplication and a subsequent domain swap (49). Previous studies have demonstrated that GINS1 expression is elevated in bladder cancer and GINS4 is upregulated in lung adenocarcinoma (50,51). Furthermore, upregulation of GINS2 has also been reported in different types of human cancer, including glioma, cervical cancer, rectal cancer and lung adenocarcinoma (14–17). Unlike GINS1 and GINS4, GINS2 is the only GINS subunit that has previously been reported to be upregulated in patients with breast cancer (18). Therefore, the role of GINS2 in breast cancer requires further analyses.
GINS2 is a vital component of the CMG complex (11). The CMG complex is the eukaryotic replicative helicase and when the activity of DNA helicases is altered diseases, such as cancer may occur, suggesting that the CMG complex is tumor-associated (6–10). Therefore, as a critical component of the CMG complex, the upregulation of GINS2 may contribute to breast cancer. Furthermore, the GINS complex plays an essential role in multiple biological processes, such as cell cycle regulation, cell proliferation and apoptosis; thus, as a significant part of the GINS complex, GINS2 may be closely associated with tumorigenesis (52,53). However, to the best of our knowledge, the function of GINS2 as a valid biomarker for poor prognosis in breast cancer remains unknown.
In the present study, analyses of the TIMER, bc-GenExMiner and Oncomine databases were performed in order to determine the expression of GINS2 in breast cancer. In the Oncomine and TIMER analyses, the present study demonstrated that upregulated GINS2 expression was observed in different types of cancer. Regarding breast cancer, the results of the present study indicated that GINS2 expression was markedly higher in breast cancer tissues compared with normal breast tissues. Furthermore, the results of the bc-GenExMiner database in the present study demonstrated that increased GINS2 expression was also associated with a basal-like status, age ≤51 years and high SBR grade status in patients.
The positive association between increased GINS2 expression and the poor outcome of patients with breast cancer was confirmed using the KM plotter database. In particular, the sub-analysis revealed that elevated GINS2 mRNA expression was associated with a poor prognosis of RFS in the patients with ER-positive and PR-positive subgroups of breast cancer. In summary, high GINS2 expression may serve as a useful prognostic biomarker in patients with breast cancer. The present study also analyzed the co-expression and neighboring genes of GINS2 using the Oncomine, GeneMANIA, GEPIA and UCSC Xena databases, and confirmed that the CENPM and MCM4 genes were positively associated with GINS2 expression. The results of the present study indicate that GINS2 could be associated with the CENPM and MCM4 signaling pathways in breast cancer.
Overall, the present study demonstrated that increased GINS2 expression may be a useful and predictive biomarker for poor prognosis in patients with breast cancer. In addition, GINS2 may be an effective predictive biomarker for prognosis with co-expressed CENPM and MCM4 genes in breast cancer. The results of the present study provide insight into the current understanding of GINS2 in breast cancer. Nevertheless, further experiments and clinical trials are required in order to clarify the involvement of GINS2 in breast cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
SY, SJ and LZ designed the present study. SY, KW and PX wrote the initial manuscript and performed data analysis. YR, JH and YL interpreted data, reviewed and edited the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Denkert C, Loibl S, Noske A, Roller M, Müller BM, Komor M, Budczies J, Darb-Esfahani S, Kronenwett R, Hanusch C, et al. Tumor-associated lymphocytes as an independent predictor of response to neoadjuvant chemotherapy in breast cancer. J Clin Oncol. 2010;28:105–113. doi: 10.1200/JCO.2009.23.7370. [DOI] [PubMed] [Google Scholar]
- 3.Filipits M, Rudas M, Jakesz R, Dubsky P, Fitzal F, Singer CF, Dietze O, Greil R, Jelen A, Sevelda P, et al. A new molecular predictor of distant recurrence in ER-positive, HER2-negative breast cancer adds independent information to conventional clinical risk factors. Clin Cancer Res. 2011;17:6012–6020. doi: 10.1158/1078-0432.CCR-11-0926. [DOI] [PubMed] [Google Scholar]
- 4.Park BW, Oh JW, Kim JH, Park SH, Kim KS, Kim JH, Lee KS. Preoperative CA 15-3 and CEA serum levels as predictor for breast cancer outcomes. Ann Oncol. 2008;19:675–681. doi: 10.1093/annonc/mdm538. [DOI] [PubMed] [Google Scholar]
- 5.Lian YF, Li SS, Huang YL, Wei H, Chen DM, Wang JL, Huang YH. Up-regulated and interrelated expressions of GINS subunits predict poor prognosis in hepatocellular carcinoma. Biosci Rep. 2018;38(pii):BSR20181178. doi: 10.1042/BSR20181178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernstein KA, Gangloff S, Rothstein R. The RecQ DNA helicases in DNA repair. Annu Rev Genet. 2010;44:393–417. doi: 10.1146/annurev-genet-102209-163602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Brabant AJ, Stan R, Ellis NA. DNA helicases, genomic instability and human genetic disease. Annu Rev Genomics Hum Genet. 2000;1:409–459. doi: 10.1146/annurev.genom.1.1.409. [DOI] [PubMed] [Google Scholar]
- 8.Kim H, D'Andrea AD. Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway. Genes Dev. 2012;26:1393–1408. doi: 10.1101/gad.195248.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu Y, Shin-Ya K, Brosh RM., Jr FANCJ helicase defective in Fanconia anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol Cell Biol. 2008;28:4116–4128. doi: 10.1128/MCB.02210-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walne AJ, Vulliamy T, Kirwan M, Plagnol V, Dokal I. Constitutional mutations in RTEL1 cause severe dyskeratosis congenita. Am J Hum Genet. 2013;92:448–453. doi: 10.1016/j.ajhg.2013.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Onesti S, MacNeill SA. Structure and evolutionary origins of the CMG complex. Chromosoma. 2013;122:47–53. doi: 10.1007/s00412-013-0397-x. [DOI] [PubMed] [Google Scholar]
- 12.Kamada K. The GINS complex: Structure and function. Subcell Biochem. 2012;62:135–156. doi: 10.1007/978-94-007-4572-8_8. [DOI] [PubMed] [Google Scholar]
- 13.Brosh RM., Jr DNA helicases involved in DNA repair and their roles in cancer. Nat Rev Cancer. 2013;13:542–558. doi: 10.1038/nrc3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng M, Zhou Y, Yang X, Tang J, Wei D, Zhang Y, Jiang JL, Chen ZN, Zhu P. High GINS2 transcript level predicts poor prognosis and correlates with high histological grade and endocrine therapy resistance through mammary cancer stem cells in breast cancer patients. Breast Cancer Res Treat. 2014;148:423–436. doi: 10.1007/s10549-014-3172-7. [DOI] [PubMed] [Google Scholar]
- 15.Ouyang F, Liu J, Xia M, Lin C, Wu X, Ye L, Song L, Li J, Wang J, Guo P, He M. GINS2 is a novel prognostic biomarker and promotes tumor progression in early-stage cervical cancer. Oncol Rep. 2017;37:2652–2662. doi: 10.3892/or.2017.5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fischer U, Kim E, Keller A, Meese E. Specific amplifications and copy number decreases during human neural stem cells differentiation towards astrocytes, neurons and oligodendrocytes. Oncotarget. 2017;8:25872–25884. doi: 10.18632/oncotarget.15980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu M, Pan H, Zhang F, Zhang Y, Zhang Y, Xia H, Zhu J, Fu W, Zhang X. Identification of TNM stage-specific genes in lung adenocarcinoma by genome-wide expression profiling. Oncol Lett. 2013;6:763–768. doi: 10.3892/ol.2013.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peng L, Song Z, Chen D, Linghu R, Wang Y, Zhang X, Kou X, Yang J, Jiao S. GINS2 regulates matrix metallopeptidase 9 expression and cancer stem cell property in human triple negative Breast cancer. Biomed Pharmacother. 2016;84:1568–1574. doi: 10.1016/j.biopha.2016.10.032. [DOI] [PubMed] [Google Scholar]
- 19.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/S1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jézéquel P, Campone M, Gouraud W, Guérin-Charbonnel C, Leux C, Ricolleau G, Campion L. bc-GenExMiner: An easy-to-use online platform for gene prognostic analyses in breast cancer. Breast Cancer Res Treat. 2012;131:765–775. doi: 10.1007/s10549-011-1457-7. [DOI] [PubMed] [Google Scholar]
- 21.Jézéquel P, Frénel JS, Campion L, Guérin-Charbonnel C, Gouraud W, Ricolleau G, Campone M. bc-GenExMiner 3.0: New mining module computes breast cancer gene expression correlation analyses. Database (Oxford) 2013;2013:bas060. doi: 10.1093/database/bas060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bansal C, Singh US, Misra S, Sharma KL, Tiwari V, Srivastava AN. Comparative evaluation of the modified Scarff-Bloom-Richardson grading system on breast carcinoma aspirates and histopathology. Cytojournal. 2012;9:4. doi: 10.4103/1742-6413.92550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee AH, Ellis IO. The Nottingham prognostic index for invasive carcinoma of the breast. Pathol Oncol Res. 2008;14:113–115. doi: 10.1007/s12253-008-9067-3. [DOI] [PubMed] [Google Scholar]
- 24.Cserni G, Chmielik E, Cserni B, Tot T. The new TNM-based staging of breast cancer. Virchows Arch. 2018;472:697–703. doi: 10.1007/s00428-018-2301-9. [DOI] [PubMed] [Google Scholar]
- 25.Lánczky A, Nagy Á, Bottai G, Munkácsy G, Szabó A, Santarpia L, Győrffy B. miRpower: A web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res Treat. 2016;160:439–446. doi: 10.1007/s10549-016-4013-7. [DOI] [PubMed] [Google Scholar]
- 26.Nagy A, Lánczky A, Menyhárt O, Győrffy B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep. 2018;8:9227. doi: 10.1038/s41598-018-29514-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Forbes SA, Beare D, Boutselakis H, Bamford S, Bindal N, Tate J, Cole CG, Ward S, Dawson E, Ponting L, et al. COSMIC: Somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017;45:D777–D783. doi: 10.1093/nar/gkw1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hinrichs AS, Raney BJ, Speir ML, Rhead B, Casper J, Karolchik D, Kuhn RM, Rosenbloom KR, Zweig AS, Haussler D, Kent WJ. UCSC data integrator and variant annotation integrator. Bioinformatics. 2016;32:1430–1432. doi: 10.1093/bioinformatics/btv766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hung JH, Weng Z. Visualizing genomic annotations with the UCSC genome browser. Cold Spring Harb Protoc. 2016 Nov 1; doi: 10.1101/pdb.prot093062. (Epub ahead of print). doi: 10.1101/pdb.prot093062. [DOI] [PubMed] [Google Scholar]
- 30.Uhlen M, Zhang C, Lee S, Sjöstedt E, Fagerberg L, Bidkhori G, Benfeitas R, Arif M, Liu Z, Edfors F, et al. A pathology atlas of the human cancer transcriptome. Science. 2017;357:eaan2507. doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- 31.Thul PJ, Åkesson L, Wiking M, Mahdessian D, Geladaki A, Ait Blal H, Alm T, Asplund A, Björk L, Breckels LM, et al. A subcellular map of the human proteome. Science. 2017;356:eaal3321. doi: 10.1126/science.aal3321. [DOI] [PubMed] [Google Scholar]
- 32.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 33.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.GTEx Consortium. The genotype-tissue expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu P, Heins ZJ, Muller JT, Katsnelson L, de Bruijn I, Abeshouse AA, Schultz N, Fenyö D, Gao J. Integration and analysis of CPTAC proteomics data in the context of cancer genomics in the cBioPortal. Mol Cell Proteomics. 2019;18:1893–1898. doi: 10.1074/mcp.TIR119.001673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ciriello G, Gatza ML, Beck AH, Wilkerson MD, Rhie SK, Pastore A, Zhang H, McLellan M, Yau C, Kandoth C, et al. Comprehensive molecular portraits of invasive lobular breast cancer. Cell. 2015;163:506–519. doi: 10.1016/j.cell.2015.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, Li B, Liu XS. TIMER: A web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77:e108–e110. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H, Chen H, Omeroglu G, Meterissian S, Omeroglu A, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14:518–527. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- 41.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Richardson AL, Wang ZC, De Nicolo A, Lu X, Brown M, Miron A, Liao X, Iglehart JD, Livingston DM, Ganesan S. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell. 2006;9:121–132. doi: 10.1016/j.ccr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 43.Cancer Genome Atlas Network: Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu X, Lu X, Wang ZC, Iglehart JD, Zhang X, Richardson AL. Predicting features of breast cancer with gene expression patterns. Breast Cancer Res Treat. 2008;108:191–201. doi: 10.1007/s10549-007-9596-6. [DOI] [PubMed] [Google Scholar]
- 45.Prat A, Pineda E, Adamo B, Galván P, Fernández A, Gaba L, Díez M, Viladot M, Arance A, Muñoz M. Clinical implications of the intrinsic molecular subtypes of breast cancer. Breast. 2015;24(Suppl 2):S26–S35. doi: 10.1016/S0960-9776(15)70012-3. [DOI] [PubMed] [Google Scholar]
- 46.Taherian-Fard A, Srihari S, Ragan MA. Breast cancer classification: Linking molecular mechanisms to disease prognosis. Brief Bioinform. 2015;16:461–474. doi: 10.1093/bib/bbu020. [DOI] [PubMed] [Google Scholar]
- 47.Mar-Aguilar F, Mendoza-Ramírez JA, Malagón-Santiago I, Espino-Silva PK, Santuario-Facio SK, Ruiz-Flores P, Rodríguez-Padilla C, Reséndez-Pérez D. Serum circulating microRNA profiling for identification of potential breast cancer biomarkers. Dis Markers. 2013;34:163–169. doi: 10.1155/2013/259454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nicolini A, Ferrari P, Duffy MJ. Prognostic and predictive biomarkers in breast cancer: Past, present and future. Semin Cancer Biol. 2018;52:56–73. doi: 10.1016/j.semcancer.2017.08.010. [DOI] [PubMed] [Google Scholar]
- 49.Carroni M, De March M, Medagli B, Krastanova I, Taylor IA, Amenitsch H, Araki H, Pisani FM, Patwardhan A, Onesti S. New insights into the GINS complex explain the controversy between existing structural models. Sci Rep. 2017;7:40188. doi: 10.1038/srep40188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamane K, Naito H, Wakabayashi T, Yoshida H, Muramatsu F, Iba T, Kidoya H, Takakura N. Regulation of SLD5 gene expression by miR-370 during acute growth of cancer cells. Sci Rep. 2016;6:1–12. doi: 10.1038/srep30941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tane S, Sakai Y, Hokka D, Okuma H, Ogawa H, Tanaka Y, Uchino K, Nishio W, Yoshimura M, Maniwa Y. Significant role of Psf3 expression in non-small-cell lung cancer. Cancer Sci. 2015;106:1625–1634. doi: 10.1111/cas.12770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mohri T, Ueno M, Nagahama Y, Gong ZY, Asano M, Oshima H, Oshima M, Fujio Y, Takakura N. Requirement of SLD5 for early embryogenesis. PLoS One. 2013;8:e78961. doi: 10.1371/journal.pone.0078961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang J, Wu Q, Wang Z, Zhang Y, Zhang G, Fu J, Liu C. Knockdown of PSF1 expression inhibits cell proliferation in lung cancer cells in vitro. Tumour Biol. 2015;36:2163–2168. doi: 10.1007/s13277-014-2826-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author upon reasonable request.