Abstract
Acute myeloid leukemia (AML) is the most common childhood cancer and is a major cause of morbidity among adults with hematologic malignancies. Several novel genetic alterations, which target critical cellular pathways, including alterations in lymphoid development-regulating genes, tumor suppressors and oncogenes that contribute to leukemogenesis, have been identified. The present study aimed to identify molecular markers associated with the occurrence and poor prognosis of AML. Information on these molecular markers may facilitate prediction of clinical outcomes. Clinical data and RNA expression profiles of AML specimens from The Cancer Genome Atlas database were assessed. Mutation data were analyzed and mapped using the maftools package in R software. Kyoto Encyclopedia of Genes and Genomes, Reactome and Gene Ontology analyses were performed using the clusterProfiler package in R software. Furthermore, Kaplan-Meier survival analysis was performed using the survminer package in R software. The expression data of RNAs were subjected to univariate Cox regression analysis, which demonstrated that the mutation loads varied considerably among patients with AML. Subsequently, the expression data of mRNAs, microRNAs (miRNAs/miR) and long non-coding RNAs (lncRNAs) were subjected to univariate Cox regression analysis to determine the the 100 genes most associated with the survival of patients with AML, which revealed 48 mRNAs and 52 miRNAs. The top 1,900 mRNAs (P<0.05) were selected through enrichment analysis to determine their functional role in AML prognosis. The results demonstrated that these molecules were involved in the transforming growth factor-β, SMAD and fibroblast growth factor receptor-1 fusion mutant signaling pathways. Survival analysis indicated that patients with AML, with high MYH15, TREML2, ATP13A2, MMP7, hsa-let-7a-2-3p, hsa-miR-362-3p, hsa-miR-500a-5p, hsa-miR-500b-5p, hsa-miR-362-5p, LINC00987, LACAT143, THCAT393, THCAT531 and KHCAT230 expression levels had a shorter survival time compared with those without these factors. Conversely, a high KANSL1L expression level in patients was associated with a longer survival time. The present study determined genetic mutations, mRNAs, miRNAs, lncRNAs and signaling pathways involved in AML, in order to elucidate the underlying molecular mechanisms of the development and recurrence of this disease.
Keywords: acute myeloid leukemia, survival analysis, prognosis, bioinformatics analysis
Introduction
Acute myeloid leukemia (AML) is a heterogeneous hematologic malignancy characterized by clonal expansion of myeloblasts in peripheral blood, bone marrow and/or other tissues. AML prognosis is suboptimal and several patients will ultimately experience recurrence (1). The latest National Comprehensive Cancer Network (NCCN) guidelines predicted that 21,450 new cases of AML will be diagnosed in 2019 and 10,920 patients will die from the disease (1). According to the SEER Cancer Statistics Review, the median age at diagnosis of AML is 67 years (2). However, other registries have reported a median age of 71 years (3), with 54% of patients diagnosed at ≥65 years, and ~1/3 diagnosed at ≥75 years (2). Thus, the incidence of AML and myelodysplastic syndromes increases with age.
Currently, the pathogenesis of AML remains unclear. Previous studies have demonstrated that molecular abnormalities, such as chromosomal abnormalities, gene mutations, abnormal myeloid cell proliferation in bone marrow and peripheral blood, and inhibition of normal hematopoietic stem cell (HSC) apoptosis and differentiation, contribute to the progression of AML and clinical outcomes (1,2,4). Recently, considerable progress has been made in risk stratification for AML, based on molecular biology and cytogenetic changes, such as chromosomal abnormalities and somatic mutations (1,4,5). Based on retrospective analyses of data from large cooperative group studies, 40–50% of patients with de novo AML exhibit a normal karyotype, representing the single most important prognostic factor for predicting remission rates, relapse risks, and OS outcomes (6,7). However, clinical outcome is heterogeneous even in patients with normal karyotype AML (1,3,4).
The most frequently used chemotherapy regimen for AML involves combination therapy with continuous infusion of cytarabine for 7 days and intravenous injection of daunorubicin for 3 days (1–4). However, despite the substantial initial sensitivity of AML to chemotherapy in the early stages of treatment, patients eventually develop clinical drug resistance due to relapse (1–3). Thus, identification of effective markers to improve the clinical outcome prediction of AML is crucial.
Gene expression profiling in AML is valuable in the diagnosis of different cytogenetic subtypes, discovery of novel AML subclasses and prediction of prognosis. Furthermore, molecular analysis is a promising source of clinically useful prognostic biomarkers (1–4). The present study aimed to identify prognostic biomarkers for patients with AML, using a gene expression profile dataset from a publicly available database, and set out to construct a gene signature for AML prognostic prediction.
Materials and methods
Data collection and baseline characteristics of patients with AML
Expression of RNA profiles, such as those of mRNAs, microRNAs (miRNAs/miRs) and long non-coding RNAs (lncRNAs), and the corresponding clinical information mutation data of 200 patients with AML in The Cancer Genome Atlas (TCGA) dataset (TCGA, Firehose Legacy Version; http://www.cbioportal.org/study/summary?id=laml_tcga) (8) were downloaded from the cBioPortal online platform (http://www.cbioportal.org) (9). Of the 200 patients assessed in the present study, 109 were male and 91 were female. The median age of the patients was 58 years (P25-P75: 44–67 years), and the median mutation count was nine. The clinical data of patients with AML are listed in Table I.
Table I.
Characteristics of patients with acute myeloid leukemia (n=200).
| Characteristic | Patient, n (%) |
|---|---|
| Median age, years (P25-P75) | 58 (44–67) |
| Age, years | |
| <60 | 108 (54.0) |
| ≥60 | 92 (46.0) |
| Sex | |
| Male | 109 (54.5) |
| Female | 91 (45.5) |
| Race | |
| White | 181 (90.5) |
| Black/African American | 15 (7.5) |
| Asian | 2 (1.0) |
| Missing | 2 (1.0) |
| Median mutation count (P25-P75) | 9 (5–14) |
| Mutation count | |
| <10 | 101 (50.5) |
| ≥10 | 95 (47.5) |
| Missing | 4 (2.0) |
| Ethnicity | |
| Not Hispanic or Latino | 194 (97.0) |
| Hispanic or Latino | 3 (1.5) |
| Missing | 3 (1.5) |
| Median platelet count preresection (P25-P75) | 52 (31–87) |
| Median abnormal lymphocyte (P25-P75) | 0 (0–2) |
P25-P75, 25th percentile to 75th percentile.
Survival analysis
Samples with incomplete lncRNA, mRNA or miRNA data were excluded, while patients with AML, with complete lncRNA-sequence (seq, n=20), mRNA-seq (n=150) and miRNA-seq data (n=164), and survival information were included in the survival analysis. Univariate Cox regression analysis was performed to determine the associations between survival and lncRNAs, miRNAs and mRNAs. Kaplan-Meier plots were generated for the top five lncRNAs, miRNAs and mRNAs, using the survminer package [version 0.4.6; (10)] within R software version 3.3.3 (11).
Bioinformatics analysis
The statistically significant mRNAs (P<0.05) identified during univariate Cox regression analysis were included in the Kyoto Encyclopedia of Genes and Genomes (KEGG), Reactome and Gene Ontology (GO) analyses; Gene set enrichment analysis (GSEA) was performed using the clusterProfiler package [version 3.14.3; (12)] within R software, in order to identify significantly enriched signaling pathways. The transcripts were ranked by the log2 (fold change) between high and low expression groups, according to the median expression, and GSEA was performed. A protein-protein interaction (PPI) network was constructed and modular analyses were performed using Metascape (http://metascape.org/gp/index.html) (13). The ggplot2 R package was used to visualize the correlation heatmap (14). The ggplot2 R package within R software was also used to visualize the results of KEGG and GO analyses, and P<0.05 was set as the cut-off value. Spearmen's correlation analysis was performed to assess the association between expression levels of the five mRNAs (MYH15, TREML2, ATP13A2, MMP7 and KANSL1L).
Statistical analysis
Statistical analysis was performed using SPSS software (version 24.0; IBM Corp). R software was used to plot figures. Kaplan-Meier analysis was used to assess survival outcomes and a log-rank test was used to compare differences between survival curves. The ComplexHeatmap package [version 2.2.0; (15)] within R software was used to analyze mutation data and plot gene mutation landscapes. P<0.05 (two-tailed) was considered to indicate a statistically significant difference.
Results
Mutation profiles of patients with AML
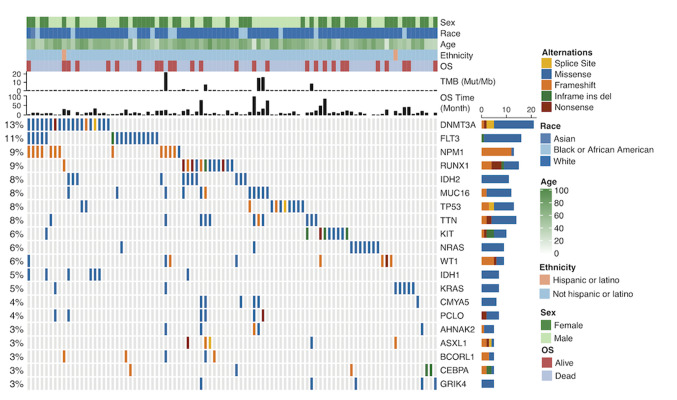
Analysis of somatic mutation data using the ComplexHeatmap package within R software demonstrated that among the mutation landscapes of patients with AML (Fig. 1), the genes with the highest mutation frequencies were: DNMT3A (13%), FLT3 (11%), NPM1 (9%), RUNX1 (9%), IDH2 (8%), MUC16 (8%) and TP53 (8%) (Fig. 1). The mutation rates varied among patients. In TCGA cohort, the median age of patients with AML at diagnosis was 58 years (Table I).
Figure 1.
Mutation landscape and clinical information of patients with acute myeloid leukemia. Alternation type, OS status, age, race, ethnicity, OS time and TMB are annotated. OS, overall survival; TMB, tumor mutation burden; Mut, mutation; Mb, megabase; ins, insertion; del, deletion. The genes with the highest mutation frequencies were: DNMT3A, FLT3, NPM1, RUNX1, IDH2, MUC16 and TP53. Most of the gene mutations were missense mutations. The mutation rates varied among patients.
Survival analysis of the prognosis of patients with AML
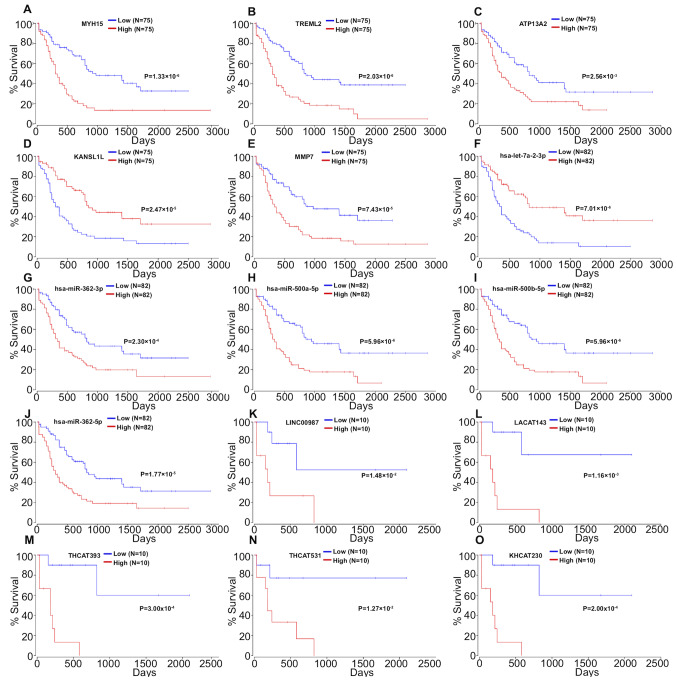
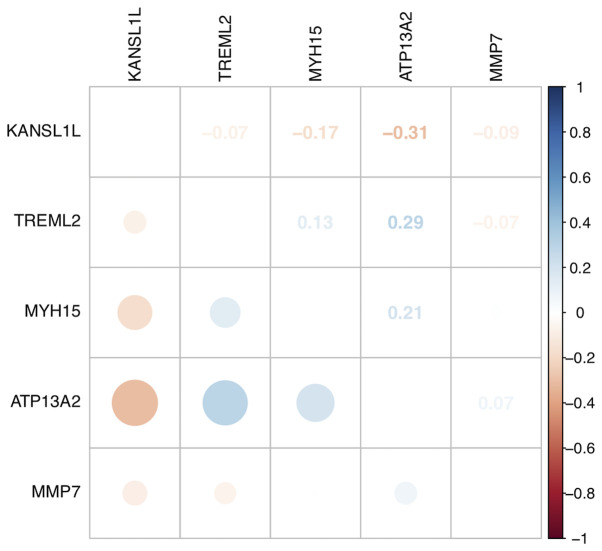
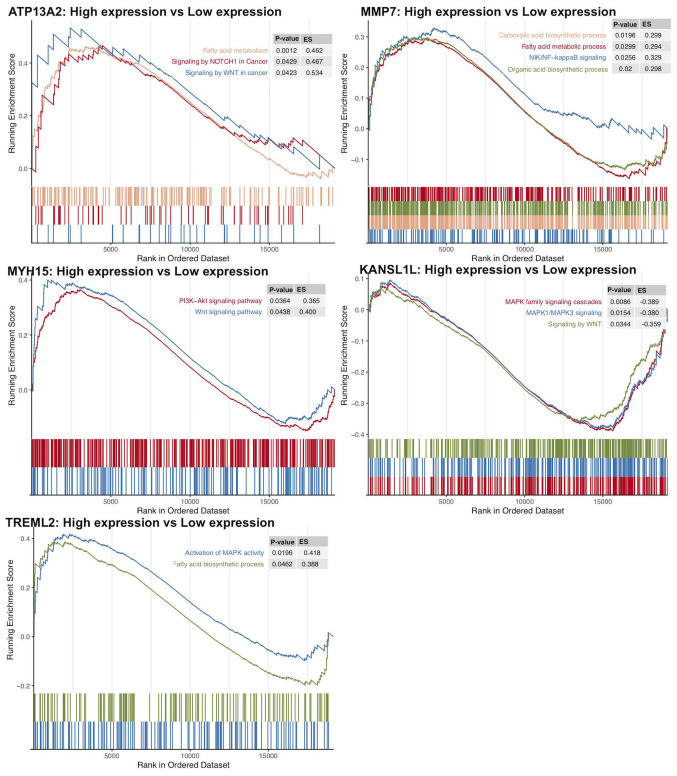
The expression data of mRNAs, miRNAs and lncRNAs were subjected to univariate Cox regression analysis to determine the top 100 genes associated with the survival of patients with AML. Age and sex were used as covariates. The top 100 genes (P<0.05) for each gene type are listed in Table II. A total of 48 mRNAs, 52 miRNAs, and 73 lncRNAs were identified as risk factors for poor prognosis, whereas 52 mRNAs, 48 miRNAs and 27 lncRNAs were identified as protective factors for prognosis. Subsequently, the top five miRNAs, mRNAs and lncRNAs in the cox regression model were analyzed via Kaplan-Meier analysis. The 5 mRNAs (MYH15, TREML2, ATP13A2, KANSL1L and MMP7; Fig. 2A-E), 5 miRNAs (hsa-let-7a-2-3p, hsa-miR-362-3p, hsa-miR-500a-5p, hsa-miR-500b-5p and hsa-miR-362-5p; Fig. 2F-J) and 5 lncRNAs (LINC00987, LACAT143, THCAT393, THCAT531 and KHCAT230; Fig. 2K-O) were significantly associated with overall survival time (all P<0.05). KANSL1L expression was negatively correlated with the other four mRNAs, while ATP13A2 expression was positively correlated with MYH15 and TREML2 expression levels (Fig. 3).
Table II.
Univariate Cox regression analysis of top 100 miRNA, mRNA and lncRNA in patients with acute myeloid leukemia.
| Gene type | Risk factor | Protective factor |
|---|---|---|
| mRNAa | MYH15, TREML2, ATP13A2, MMP7, SPINK2, FAM207A, IL2RA, CLNK, FAM83G, F12, MAP4K1, FBXW4, RPS6KA1, NUP210, PRR7, RHOF, STK10, CBR1, TGFB1, LSP1, CKLF, SH3BP1, PNPLA6, PRICKLE4, IL12RB1, SPAG1, ETFB, UNC45A, BCKDK, DAXX, KBTBD8, ZNF296, SELPLG, LSM4, RAC2, TOMM40L, ANKEF1, C10orf128, GRK6, FERMT3, TAGLN2, PARVB, PPCDC, UPF1, TBCC, PTP4A3, PDE7B, FIBP | KANSL1L, HSDL1, ZFYVE16, MYB, NMT2, GALNT1, MAP3K1, MBNL1-AS1, C14orf37, DCBLD2, GFPT1, POU5F2, SUZ12P1, TM6SF1, PWWP2A, ANKRD50, DCP2, MIB1, PDE3B, ADAMTS7, ADSS, TVP23A, ZNF333, CCSAP, MMP14, DHRSX, ZNF124, SERINC5, SNRK, CANX, CYP4F2, CCNJL, PLXNB1, SMA4, PRRC1, TAS2R43, ARL15, LOC100130264, CENPC, SLC35F5, LRRC37A6P, EPHX1, ZSCAN23, LCORL, LOXL4, TGIF1, DDX59, SRPK2, CLINT1, LRRTM2, GABRE, H2AFV |
| miRNAa | hsa-miR-362-3p, hsa-miR-500a-5p, hsa-miR-500b-5p, hsa-miR-362-5p, hsa-miR-532-5p, hsa-miR-502-3p, hsa-miR-660-5p, hsa-miR-155-3p, hsa-miR-20b-5p, hsa-let-7b-5p, hsa-miR-10a-5p, hsa-miR-501-3p, hsa-miR-500a-3p, hsa-miR-141-5p, hsa-miR-20b-3p, hsa-miR-501-5p, hsa-miR-532-3p, hsa-miR-20a-5p, hsa-miR-363-3p, hsa-miR-188-5p, hsa-miR-339-5p, hsa-miR-196b-5p, hsa-miR-155-5p, hsa-miR-107, hsa-miR-196a-5p, hsa-let-7a-3p, hsa-let-7b-3p, hsa-miR-339-3p, hsa-miR-29c-5p, hsa-miR-18a-5p, hsa-miR-9-5p, hsa-miR-769-5p, hsa-miR-1284, hsa-miR-185-3p, hsa-miR-548d-3p, hsa-miR-194-5p, hsa-miR-15b-3p, hsa-miR-29b-2-5p, hsa-miR-10a-3p, hsa-miR-17-5p, hsa-miR-15a-3p, hsa-miR-451a, hsa-miR-106a-5p, hsa-miR-200c-3p, hsa-miR-629-3p, hsa-miR-301a-3p, hsa-miR-33b-5p, hsa-miR-151a-5p, hsa-miR-15a-5p, hsa-miR-338-3p, hsa-miR-1-3p, hsa-miR-1976 | hsa-let-7a-2-3p, hsa-miR-100-5p, hsa-miR-3913-5p, hsa-miR-181b-3p, hsa-miR-181c-5p, hsa-miR-452-5p, hsa-miR-181a-5p, hsa-miR-181a-3p, hsa-miR-224-5p, hsa-miR-181b-5p, hsa-miR-181a-2-3p, hsa-miR-195-5p, hsa-miR-203a-3p, hsa-miR-199a-3p, hsa-miR-199b-3p, hsa-miR-3607-3p, hsa-miR-193a-5p, hsa-miR-125b-5p, hsa-miR-574-3p, hsa-miR-3605-3p, hsa-miR-1287-3p, hsa-miR-3653-3p, hsa-miR-193b-3p, hsa-miR-25-3p, hsa-miR-10b-5p, hsa-miR-142-5p, hsa-miR-365a-3p, hsa-miR-424-3p, hsa-miR-181c-3p, hsa-miR-32-5p, hsa-miR-335-5p, hsa-miR-148a-5p, hsa-miR-98-3p, hsa-miR-127-3p, hsa-miR-450b-5p, hsa-miR-143-3p, hsa-miR-146a-5p, hsa-miR-340-3p, hsa-miR-148a-3p, hsa-miR-181b-2-3p, hsa-miR-335-3p, hsa-miR-30a-3p, hsa-miR-106b-3p, hsa-miR-1468-5p, hsa-miR-219a-1-3p, hsa-miR-30e-3p, hsa-miR-125a-5p, hsa-miR-424-5p |
| lncRNAa | LINC00987, HICLINC36.3, LACAT143, THCAT393, THCAT531, KHCAT230, LINC00511.12, HICLINC365.1, CAT74, AMAT158, LSCAT43, CAT58.2, PRCAT231, KCCAT685, BRCAT407, BRCAT320, KCCAT44, DDX11-AS1.3, SNHG12.4, LACAT91, OVAT85, THCAT614, CAT1620, KCCAT539, THCAT283.1, SNHG9, CAT1764.1, OVAT60, ESAT58, KCCAT680.2, BRCAT104, KHCAT392, LINC00152.2, CAT990, CAT2214, KPCAT17, CAT18.1, HICLINC36.2, CAT862.2, THCAT591, CAT461, CAT1560.2, OVAT76, LSCAT160, STCAT14, GBAT5, THCAT220, HICLINC36.1, HICLINC285, CAT1010, THCAT422.1, CAT43, CAT122, BRCAT392, CAT2313, CAT277.2, THCAT546, CAT993, CAT1358.2, CAT1424.3, KCCAT499, PRKCQ-AS1.3, CAT295.1, LSCAT277, TMCC1-AS1.1, KCCAT52, LINC00621.3, SMAT11, LSCAT218, PRKCQ-AS1.2, CAT292, CAT1405, CAT318 | CAT121.2, CAT313.1, FOXD2-AS1.1, CAT1824, CAT1631, CAT1569.1, HNCAT240, CAT2189, PRCAT188.3, CAT1950.1, CAT2155, KCCAT349, CAT1966.1, CAT541, WAC-AS1, KCCAT448, MPAT11, CAT2031, CAT353.3, CAT1966.2, HICLINC283.5, CAT1849, CAT439, CAT1319.2, CAT1140, CAT2062, CAT1569.2 |
miRNA, microRNA; lncRNA, long non-coding RNA.
Figure 2.
Kaplan-Meier survival curves for (A) MYH15, (B) TREML2, (C) ATP13A2, (D) KANSL1L and (E) MMP7, (F) hsa-let-7a-2-3p, (G) hsa-miR-362-3p, (H) has-miR-500a-5p, (I) hsa-miR-500b-5p, (J) hsa-miR-362-5p, (K) LINC00987, (L) LACAT143, (M) THCAT393, (N) THCAT531 and (O) KHCAT230 associated with overall survival time of patients with acute myeloid leukemia.
Figure 3.
Correlation between different the gene expression levels of ATP13A2, MMP7, TREML2, MYH15 and KANSL1L. Dot size and color represent the associations between the five gene expression levels of patients with acute myeloid leukemia in The Cancer Genome Atlas cohort. Blue dots indicate a positive correlation, while red dots indicate a negative correlation. Both the values, and dot sizes correspond to Pearson's correlation coefficients.
Function analysis
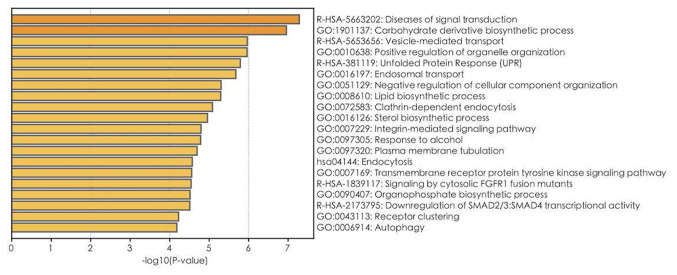
GO, Reactome and KEGG pathway enrichment analyses on 1,900 mRNAs with P<0.05 in the univariate Cox regression model were performed (Table III). The top 20 results of GO, Reactome and KEGG analyses are presented in Fig. 4. A total of 112 GO terms were enriched in biological mechanisms, including ‘SMAD protein complex assembly’, ‘transmembrane receptor tyrosine kinase signaling pathway’, ‘cell proliferation’, ‘apoptosis’, ‘autophagy’ and ‘protein processing’ (Fig. 4; Table III). Pathway enrichment analysis demonstrated that different genes are involved in multiple signaling pathways, such as signaling by TGF-beta family members, interleukin-2 family signaling, loss of function of SMAD4 in cancer and signaling by cytosolic FGFR1 fusion mutants (Table III). Furthermore, using GSEA, gene sets, such as signaling by Wnt in cancer, signaling by NOTCH1 in cancer, and fatty acid metabolism were demonstrated to be enriched in the ATP13A2 high expression group (Fig. 5). Oncogenic signaling pathways and metabolism pathways were differentially enriched in MMP7 (NF-κB-inducing kinase/NF-κB and the fatty acid metabolic process), TREML2 (the fatty acid metabolic process and MAPK signaling) and MYH15 (phosphoinositide-3-kinase-protein kinase B/Akt, WNT signaling) high expression phenotypes. Conversely, Wnt and MAPK1/MAPK3 signaling pathways were differentially enriched in the KANSL1L low expression phenotype.
Table III.
Significantly enriched pathways (GO, KEGG and Reactome) of 1,900 mRNAs in univariate Cox regression model.
| Term | Pathway | P-value | Count | Gene symbol |
|---|---|---|---|---|
| R-HSA-5663202 | Diseases of signal transduction | <0.001 | 62 | AREG, CREB1, CSK, CTBP1, CUX1, GSK3A, HGF, LRP6, SMAD2, SMAD3, SMAD4, MAP3K11, PHB, PIK3CA, PIK3R1, POLR2E, POLR2I, PPP2R5B, PSMA7, PSMB10, PSMC4, PSMD2, PSMD3, PSMD8, RAC2, SEL1L, STAT5B, TGFA, TGFB1, TLN1, VAV1, VCL, VCP, ZMYM2, FXR1, CUL1, IRS2, TRIM24, NEURL1, ZFYVE9, QKI, PSMF1, NCOR2, GAB2, BCL2L11, AKAP9, CPSF6, CDC37, RASA3, NCBP2, ERLEC1, TRAT1, HDAC7, MIB1, KIAA1549, MAPKAP1, HDAC11, KDM7A, APH1B, NRG4, SPRED2, RICTOR |
| R-HSA-1839117 | Signaling by cytosolic FGFR1 fusion mutants | <0.001 | 8 | CUX1, PIK3CA, PIK3R1, STAT5B, ZMYM2, TRIM24, GAB2, CPSF6 |
| R-HSA-2173795 | Downregulation of SMAD2/3: SMAD4 transcriptional activity | <0.001 | 9 | PARP1, SMAD2, SMAD3, SMAD4, PPM1A, SKIL, TGIF1, NCOR2, SMURF2 |
| R-HSA-3304349 | Loss of function of SMAD2/3 in cancer | <0.001 | 5 | SMAD2, SMAD3, SMAD4, TGFB1, ZFYVE9 |
| R-HSA-9006934 | Signaling by receptor tyrosine kinases | <0.001 | 59 | AREG, AP2M1, COL2A1, COL3A1, COL4A2, COL6A3, CREB1, CSK, CTNNA1, DOCK1, DUSP7, ELK1, PTK2B, FLT4, HGF, INSR, ITGB1, ITPR2, LAMA3, LAMB1, NCF4, PDE3B, PIK3CA, PIK3R1, PLAT, POLR2E, POLR2I, PRKCD, PTPN1, PTPN6, PTPRS, PXN, RALGDS, RPS6KA1, STAT5B, TGFA, THBS2, VAV1, SOCS1, IRS2, NRP2, SOCS6, GAB2, SPINT2, CDC37, NCBP2, TAB2, NELFB, VRK3, ATP6V1H, TLR9, ERBIN, MAPKAP1, APH1B, NRG4, ATP6V0E2, SPRED2, ATP6V1C2, RICTOR |
| R-HSA-9006936 | Signaling by TGF-beta family members | <0.001 | 20 | ACVR1B, PARP1, SMAD2, SMAD3, SMAD4, SMAD5, PPM1A, PPP1CA, SKIL, TGFB1, TGIF1, FOXH1, MTMR4, ZFYVE9, NCOR2, ZFYVE16, FSTL1, BAMBI, PARD3, SMURF2 |
| R-HSA-3304351 | Signaling by TGF-beta Receptor Complex in Cancer | <0.001 | 5 | SMAD2, SMAD3, SMAD4, TGFB1, ZFYVE9 |
| R-HSA-170834 | Signaling by TGF-beta Receptor Complex | <0.001 | 15 | PARP1, SMAD2, SMAD3, SMAD4, PPM1A, PPP1CA, SKIL, TGFB1, TGIF1, MTMR4, ZFYVE9, NCOR2, BAMBI, PARD3, SMURF2 |
| R-HSA-3304347 | Loss of function of SMAD4 in cancer | <0.001 | 3 | SMAD2, SMAD3, SMAD4 |
| R-HSA-3311021 | SMAD4 MH2 domain mutants in cancer | <0.001 | 3 | SMAD2, SMAD3, SMAD4 |
| R-HSA-3315487 | SMAD2/3 MH2 domain mutants in cancer | <0.001 | 3 | SMAD2, SMAD3, SMAD4 |
| R-HSA-3304356 | SMAD2/3 phosphorylation motif mutants in cancer | <0.001 | 4 | SMAD2, SMAD3, TGFB1, ZFYVE9 |
| R-HSA-3656532 | TGFBR1 KD mutants in cancer | <0.001 | 4 | SMAD2, SMAD3, TGFB1, ZFYVE9 |
| R-HSA-2173788 | Downregulation of TGF-beta receptor signaling | 0.001 | 8 | SMAD2, SMAD3, PPP1CA, TGFB1, MTMR4, ZFYVE9, BAMBI, SMURF2 |
| R-HSA-3656534 | Loss of function of TGFBR1 in cancer | 0.001 | 4 | SMAD2, SMAD3, TGFB1, ZFYVE9 |
| R-HSA-451927 | Interleukin-2 family signaling | 0.002 | 10 | PTK2B, IL2RA, IL3RA, IL15RA, PIK3CA, PIK3R1, PTPN6, SOS2, STAT5B, GAB2 |
| R-HSA-1839124 | FGFR1 mutant receptor activation | 0.002 | 8 | CUX1, PIK3CA, PIK3R1, STAT5B, ZMYM2, TRIM24, GAB2, CPSF6 |
| R-HSA-1655829 | Regulation of cholesterol biosynthesis by SREBP (SREBF) | 0.003 | 11 | FDPS, LSS, MVD, MVK, NFYA, NFYB, SREBF2, NCOA1, MBTPS1, SEC24A, MBTPS2 |
| R-HSA-8952158 | RUNX3 regulates BCL2L11 (BIM) transcription | 0.004 | 3 | SMAD3, SMAD4, BCL2L11 |
| R-HSA-381038 | XBP1(S) activates chaperone genes | 0.005 | 10 | DDX11, GFPT1, GSK3A, DNAJB9, PPP2R5B, DNAJC3, SSR1, TLN1, PDIA6, SEC31A |
| R-HSA-2173793 | Transcriptional activity of SMAD2/SMAD3:SMAD4 heterotrimer | 0.006 | 9 | PARP1, SMAD2, SMAD3, SMAD4, PPM1A, SKIL, TGIF1, NCOR2, SMURF2 |
| R-HSA-5655302 | Signaling by FGFR1 in disease | 0.008 | 8 | CUX1, PIK3CA, PIK3R1, STAT5B, ZMYM2, TRIM24, GAB2, CPSF6 |
| R-HSA-1226099 | Signaling by FGFR in disease | 0.009 | 11 | CUX1, PIK3CA, PIK3R1, POLR2E, POLR2I, STAT5B, ZMYM2, TRIM24, GAB2, CPSF6, NCBP2 |
Figure 4.
Top 20 GO, Reactome and KEGG results for the 1,900 mRNAs with P<0.05 in the univariate Cox regression model. The bar plot indicates the enrichment scores of significant GO, Reactome and KEGG terms. P<0.05; gene count ≥3. GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Figure 5.
Transcriptome traits of biological function in Gene Set Enrichment Analysis. Each run was performed with 1,000 permutations. ES, enrichment score.
Protein interaction network visualization and analysis
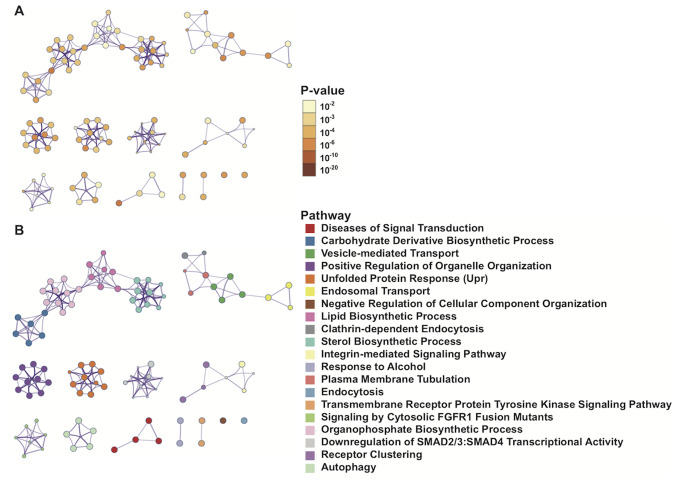
Metascape was used to visualize the PPIs of 1,900 mRNAs with significant differences in the univariate Cox regression model. The interactions among enrichment pathways are presented in Fig. 6. The metabolic pathways, including ‘carbohydrate derivative biosynthetic processing’, ‘vesicle-mediated transport’ and ‘lipid biosynthetic processing’ were significantly associated and highly interactive.
Figure 6.
Network of enriched Gene Ontology, Reactome and Kyoto Encyclopedia of Gene and Genomes pathways. (A) Colored based on significance, where terms containing more genes tend to have a more significant P-value. (B) Colored based on pathways, where nodes that share the same pathways are typically closer together.
Discussion
AML is a heterogeneous clonal disorder characterized by immature myeloid cell proliferation and bone marrow failure. Currently, 7 + 3 induction therapy is the standard treatment for young adults and fit elderly patients (<60 years). Under the 7 + 3 regimen, 65–73% of patients with AML achieve complete remission (CR). However, only 38–62% of patients with AML, >60 years achieve CR (1–4). Elderly patients are unable to tolerate the toxicity of high-dose chemotherapy drugs (1–4). The 5-year survival rate of patients with AML is ~30% (5,16). Advancements in genetics and molecular research, as well as improved understanding of AML biology, certain cytogenetic abnormalities, and specific gene mutations and/or changes, have been achieved (17). Furthermore, novel targets that can considerably improve therapeutic efficacy in some patients with specific molecular genetics have been developed (1,2,4). The presence or absence of specific gene mutations and/or changes in gene expression, such as P53 or FLT3 gene mutations, can be used to further classify AML cases and affect the prognosis of patients with somatic mutations (18–22). Thus, the NCCN has added NPM1, FLT3, CEBPA, IDH1/2, DNMT3A, KIT, TP53, RUNX1 and ASXL1 gene mutations to the AML prognostic stratification system. However, ~50% of all patients with AML exhibit normal karyotypes and lack specificity (1,3,4). Novel molecular biomarkers based on somatic mutations can improve the risk stratification of patients with normal karyotypes (1). Research on novel biomarkers and treatment methods, as well as improvement of current chemotherapy methods, represent breakthroughs through which the quality of life of patients with AML can be improved. The present study analyzed the AML dataset from TCGA database through bioinformatics analysis. Univariate Cox's regression analysis demonstrated that mRNAs, miRNAs and lncRNAs gene mutations are significantly associated with survival prognosis. Enrichment analysis of 1,900 mRNAs with P<0.05 in the univariate Cox regression model was also performed.
The TGF-β and SMAD signaling pathways are enriched, and FGFR1 fusion gene mutations are significantly associated with poor survival in patients with AML (23,24). Previous studies have demonstrated that the TGF-β and SMAD signaling pathways are dysregulated in leukemia (23,24). De Visser and Kast (25) reported that loss of TGF-β function is involved in the occurrence, progression and metastasis of AML. Furthermore, Lin et al (26) demonstrated that missense and frameshift mutations of SMAD4 disrupt gene function in the TGF-β signaling pathway, and ultimately block the TGF-β signaling pathway in AML. These studies suggest that the TGF-β and SMAD signaling pathways may be closely associated with the occurrence, development and poor prognosis of patients with AML (23–26).
TGF-β signaling plays an important role in the extracellular microenvironment and several cellular processes, including cell proliferation, differentiation, apoptosis and migration (27). TGF-β is a potent inhibitor of hematopoietic stem cell proliferation, which plays an important role in the hematopoietic stem and progenitor cell maintenance at rest, hyperproliferation inhibition, differentiation induction and apoptosis promotion. TGF-β, TGF-β receptor (TGF-βR) and SMAD proteins, and their target genes constitute a tumor-suppressor pathway (27,28). Thus, any component defect can lead to loss of the growth inhibition function of TGF-β, thereby encouraging cell malignant proliferation in several types of solid tumors (28). TGF-β/SMADs act as tumor suppressor signals, whereby mutational inactivation or downregulation inhibits TGF-β signaling in solid tumors, including colon, lung, breast, pancreatic and gastric cancer (28). TGF-β/SMAD signaling is a negative regulatory signal for HSC proliferation in the blood system. In vitro experiments have demonstrated that exogenous TGF-β can decrease the number of HSCs that enter the cell cycle (27–29). Furthermore, antibodies against TGF-β1 may allow HSCs to stay in the S phase for a long period of time (29). Patients with AML lose their TGF-βR function, which is considered to induce insensitivity to TGF-β signaling and TGF-β-mediated growth inhibition (22). The combination treatment with TGF-β and activated vitamin D3 may enhance therapeutic effects on patients through the reduction of SMAD2/3 phosphorylation levels and nuclear translocation (30).
The extracellular receptor binding domain of FGFR1 binds to its corresponding ligand and activates downstream signals, such as PLC-γ, STAT5, STAT1, PI3K and RAS/MAP kinase (31,32). Although FGFR1 plays an important role in controlling cell proliferation, differentiation, migration and cell phenotype transformation, to the best of our knowledge, the function of this gene in HSCs is not yet fully understood (31). A novel protein composed of FGFR1 rearrangements activates the receptor tyrosine kinase (RTK) activity of FGFR1 in a non-ligand-dependent manner, continues to activate downstream signaling pathways and inhibits apoptosis (32). Jackson et al (33) reported that the ZNF198-FGFR1 fusion gene mutation retains the RTK domain of FGFR1 and causes the proline-rich domain of ZNF198 to mediate the ligand-free intracellular partial dimerization of FGFR1. Furthermore, ZNF198-FGFR1 fusion gene mutation further promotes autophosphorylation of tyrosine residues, thereby activating multiple downstream signaling pathways, inhibiting apoptosis and promoting proliferation. These effects result in tumorigenic transformation. Thus, it is speculated that FGFR1 fusion gene mutation may play a role in AML occurrence and development, and in the poor prognosis of patients with this disease.
In the present study, GSEA demonstrated that high expression levels of ATP13A2, MMP7, TREML2 and MYH15 were associated with oncogenic signaling pathways and metabolism pathways, including the NIK/NF-κB, PI3K-Akt, Wnt and MAPK signaling pathways, and the fatty acid metabolic process. Yu et al (34) reported that the Wnt signaling pathway promotes tumor cell proliferation and metastasis in chronic lymphocytic leukemia. Furthermore, Agarwal et al (35) demonstrated that the PI3K/AKT/IkB kinase signaling pathway is abnormally activated in colorectal cancer, eventually leading to tumor growth.
High expression levels of the following markers: MYH15, TREML2, ATP13A2, MMP7, hsa-let-7a-2-3p, hsa-miR-362-3p, hsa-miR-500a-5p, hsa-miR-500b-5p, hsa-miR-362-5p, LINC00987, LACAT143, THCAT393, THCAT531 and KHCAT230 are associated with poor prognosis of patients with AML. Furthermore, the TGF-β and SMAD signaling pathways, and FGFR1 fusion gene mutations may be closely associated with AML occurrence, development and poor prognosis. However, further investigation on the functions of these genes is required to determine their molecular mechanism in the progression of AML.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the cBioPortal database (https://www.cbioportal.org/study/summary?id=laml_tcga).
Authors' contributions
AL and ZJ conceived and designed the present study. GL, YG, KL, AL and ZJ acquired, analyzed and interpreted the data. KL performed the statistical analysis. GL, YG and KL drafted the initial manuscript, in consultation with AL and ZJ. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Tallman MS, Wang ES, Altman JK, Appelbaum FR, Bhatt VR, Bixby D, Coutre SE, De Lima M, Fathi AT, Fiorella M, et al. Acute myeloid leukemia, version 3.2019, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2019;17:721–749. doi: 10.6004/jnccn.2019.0028. [DOI] [PubMed] [Google Scholar]
- 2.National Cancer Institute, corp-author. SEER cancer statistics review, 1975–2015: Overview, median age at diagnosis. https://seer.cancer.gov/csr/1975_2015/ [Sep 10;2018 ]; [Google Scholar]
- 3.Juliusson G. Older patients with acute myeloid leukemia benefit from intensive chemotherapy: An update from the Swedish acute leukemia registry. Clin Lymphoma Myeloma Leuk. 2011;11(Suppl 1):S54–S59. doi: 10.1016/j.clml.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 4.DiNardo C, Lachowiez C. Acute myeloid leukemia: From mutation profiling to treatment decisions. Curr Hematol Malig Rep. 2019;14:386–394. doi: 10.1007/s11899-019-00535-7. [DOI] [PubMed] [Google Scholar]
- 5.Stuani L, Sabatier M, Sarry JE. Exploiting metabolic vulnerabilities for personalized therapy in acute myeloid leukemia. BMC Biol. 2019;17:57. doi: 10.1186/s12915-019-0670-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pagana L, Pulsoni A, Tosti ME, Avvisati G, Mele L, Mele M, Martino B, Visani G, Cerri R, Di Bona E, et al. Clinical and biological features of acute myeloid leukaemia occurring as second malignancy: GIMEMA archive of adult acute leukaemia. Br J Haematol. 2001;112:109–117. doi: 10.1046/j.1365-2141.2001.02527.x. [DOI] [PubMed] [Google Scholar]
- 7.Pulsoni A, Pagano L, Lo Coco F, Avvisati G, Mele L, Di Bona E, Invernizzi R, Leoni F, Marmont F, Mele A, et al. Clinicobiological features and outcome of acute promyelocytic leukemia occurring as a second tumor: The GIMEMA experience. Blood. 2002;100:1972–1976. doi: 10.1182/blood-2001-12-0312. [DOI] [PubMed] [Google Scholar]
- 8.Tomczak K, Czerwińska P, Wiznerowicz M. The cancer genome atlas (TCGA): An immeasurable source of knowledge. Contemp Oncol (Pozn) 2015;19:A68–A77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alboukadel K, Marcin K, Przemyslaw B. Drawing Survival Curves using ‘ggplot2’. R package version 0.4.6. https://cran.r-project.org/web/packages/survminer/survminer.pdf. [Sep 9;2019 ]; [Google Scholar]
- 11.Chambers J. Software for data analysis. Springer-Verlag; New York, NY: 2008. Programming with R. [DOI] [Google Scholar]
- 12.Yu G, Wang LG, Han Y, He QY. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, Chanda SK. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wickham H. ggplot2. Wiley Interdisciplinary Reviews: Computational Statistics. 2011;3:180–185. doi: 10.1002/wics.147. [DOI] [Google Scholar]
- 15.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 16.Ilyas AM, Ahmad S, Faheem M, Naseer MI, Kumosani TA, Al-Qahtani MH, Gari M, Ahmed F. Next generation sequencing of acute myeloid leukemia: Influencing prognosis. BMC Genomics. 2015;16(Suppl 1):S5. doi: 10.1186/1471-2164-16-S1-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, Ritchey JK, Young MA, Lamprecht T, McLellan MD, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481:506–510. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martelli MP, Sportoletti P, Tiacci E, Martelli MF, Falini B. Mutational landscape of AML with normal cytogenetics: Biological and clinical implications. Blood Rev. 2013;27:13–22. doi: 10.1016/j.blre.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 19.Tomasson MH, Xiang Z, Walgren R, Zhao Y, Kasai Y, Miner T, Ries RE, Lubman O, Fremont DH, McLellan MD, et al. Somatic mutations and germline sequence variants in the expressed tyrosine kinase genes of patients with de novo acute myeloid leukemia. Blood. 2008;111:4797–4808. doi: 10.1182/blood-2007-09-113027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindsley RC, Mar BG, Mazzola E, Grauman PV, Shareef S, Allen SL, Pigneux A, Wetzler M, Stuart RK, Erba HP, et al. Acute myeloid leukemia ontogeny is defined by distinct somatic mutations. Blood. 2015;125:1367–1376. doi: 10.1182/blood-2014-11-610543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cancer Genome Atlas Research Network, corp-author. Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, Robertson A, Hoadley K, Triche TJ, Jr, Laird PW, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marcucci G, Haferlach T, Döhner H. Molecular genetics of adult acute myeloid leukemia: Prognostic and therapeutic implications. J Clin Oncol. 2011;29:475–486. doi: 10.1200/JCO.2010.30.2554. [DOI] [PubMed] [Google Scholar]
- 23.Wu H, Li P, Shao N, Ma J, Ji M, Sun X, Ma D, Ji C. Aberrant expression of Treg-associated cytokine IL-35 along with IL-10 and TGF-β in acute myeloid leukemia. Oncol Lett. 2012;3:1119–1123. doi: 10.3892/ol.2012.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen J, Mu Q, Li X, Yin X, Yu M, Jin J, Li C, Zhou Y, Zhou J, Suo S, et al. Homoharringtonine targets Smad3 and TGF-β pathway to inhibit the proliferation of acute myeloid leukemia cells. Oncotarget. 2017;8:40318–40326. doi: 10.18632/oncotarget.16956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Visser KE, Kast WM. Effects of TGF-beta on the immune system: Implications for cancer immunotherapy. Leukemia. 1999;13:1188–1199. doi: 10.1038/sj.leu.2401477. [DOI] [PubMed] [Google Scholar]
- 26.Lin HK, Bergmann S, Pandolfi PP. Deregulated TGF-beta signaling in leukemogenesis. Oncogene. 2005;24:5693–5700. doi: 10.1038/sj.onc.1208923. [DOI] [PubMed] [Google Scholar]
- 27.Massagué J. TGFβ signalling in context. Nat Rev Mol Cell Biol. 2012;13:616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Massagué J. TGFbeta in cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruscetti FW, Akel S, Bartelmez SH. Autocrine transforming growth factor-beta regulation of hematopoiesis: Many outcomes that depend on the context. Oncogene. 2005;24:5751–5763. doi: 10.1038/sj.onc.1208921. [DOI] [PubMed] [Google Scholar]
- 30.Matsunawa M, Ishii Y, Kasukabe T, Tomoyasu S, Ota H, Honma Y. Cotylenin A-induced differentiation is independent of the transforming growth factor-beta signaling system in human myeloid leukemia HL-60 cells. Leuk Lymphoma. 2006;47:733–740. doi: 10.1080/10428190500375839. [DOI] [PubMed] [Google Scholar]
- 31.Macdonald D, Reiter A, Cross NC. The 8p11 myeloproliferative syndrome: A distinct clinical entity caused by constitutive activation of FGFR1. Acta Haematol. 2002;107:101–107. doi: 10.1159/000046639. [DOI] [PubMed] [Google Scholar]
- 32.Ren M, Li X, Cowell JK. Genetic fingerprinting of the development and progression of T-cell lymphoma in a murine model of atypical myeloproliferative disorder initiated by the ZNF198-fibroblast growth factor receptor-1 chimeric tyrosine kinase. Blood. 2009;114:1576–8154. doi: 10.1182/blood-2009-03-212704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jackson CC, Medeiros LJ, Miranda RN. 8p11 myeloproliferative syndrome: A review. Hum Pathol. 2010;41:461–476. doi: 10.1016/j.humpath.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 34.Yu J, Chen L, Cui B, Widhopf GF, II, Shen Z, Wu R, Zhang L, Zhang S, Briggs SP, Kipps TJ. Wnt5a induces ROR1/ROR2 heterooligomerization to enhance leukemia chemotaxis and proliferation. J Clin Invest. 2016;126:585–598. doi: 10.1172/JCI83535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Agarwal A, Das K, Lerner N, Sathe S, Cicek M, Casey G, Sizemore N. The AKT/IκB kinase pathway promotes angiogenic/metastatic gene expression in colorectal cancer by activating nuclear factor-κB and β-catenin. Oncogene. 2005;24:1021–1031. doi: 10.1038/sj.onc.1208296. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the cBioPortal database (https://www.cbioportal.org/study/summary?id=laml_tcga).