Abstract
Colorectal cancer (CRC) is one of the most common malignancies, and the fourth most common cause of cancer-associated mortality globally. The epithelial to mesenchymal transition (EMT) serves an important function in metastatic dissemination and determines the aggressiveness of CRC. However, the regulatory mechanism of EMT in CRC has not yet been elucidated. γ-glutamylcyclotransferase (GGCT) is an important enzyme in glutathione metabolism and highly expressed in numerous forms of cancer, making it a promising therapeutic target. In the present study, GGCT was demonstrated to be highly expressed in CRC tissues, and patients with CRC with a higher expression of GGCT exhibited a worse prognosis compared with patients exhibiting a lower expression of GGCT. This result suggests that GGCT may serve as a novel prognostic marker for CRC. Furthermore, GGCT was indicated to promote CRC cell migration and invasion through regulating EMT-associated genes, including N-cadherin, Vimentin, snail family transcriptional repressor 2 and snail family transcriptional repressor 1. In conclusion, the present study provides novel insights into the mechanism governing CRC migration and invasion, and identified GGCT as a promising therapeutic target that may be used in the treatment of CRC.
Keywords: colorectal cancer, γ-glutamylcyclotransferase, migration and invasion, epithelial-mesenchymal transition
Introduction
Colorectal cancer (CRC) is one of the most common malignancies and the second most common cause of cancer-associated mortality globally (1). The incidence and mortality of CRC has increased in previous years, particularly in developing Asian countries, including China (2). Despite advances in the prevention, diagnosis and treatment of CRC, to the best of our knowledge, no effective treatment strategy for this disease has yet been developed. Metastasis and recurrence are the main reasons why CRC treatment may fail (3,4). Therefore, it is vital to elucidate the molecular mechanisms governing CRC progression and migration and invasion.
γ-glutamylcyclotransferase (GGCT) is an important enzyme in glutathione metabolism, which catalyzes the reaction of the γ-glutamyl peptide to produce 5-oxyproline and free amino acids (5). The GGCT gene is highly preserved in a number of species, including in bacteria, plants and nematodes, and a number of other higher organisms (6). GGCT has been reported to accumulate in a variety of cancer types, including breast, ovarian, cervical, lung, bladder, prostate, colon, osteosarcoma and glioma, indicating that it may serve as an oncogene in these tumor types (7–12). The depletion of GGCT has been indicated to inhibit the proliferation of bladder cancer cells and induce the cytotoxicity of McF-7/ADR cells in vitro and in vivo (13). In previous years, one study has identified the suppression of cancer cell proliferation by disrupting GGCT, highlighting the potential for treatment of malignant tumor types through inhibiting GGCT (14). However, the underlying mechanism as to how GGCT regulates CRC progression remains yet to be determined. Therefore, investigation into the precise function of GGCT in human CRC is urgently required.
The present study demonstrated the association between GGCT expression level and the prognosis of patients with CRC. Further results indicated that GGCT promoted CRC cells migration and invasion via the epithelial to mesenchymal transition (EMT). In summary, these data support that GGCT may be a novel therapeutic target for use in the treatment of CRC.
Materials and methods
Microarray and database analysis
A microarray of mRNA from 286 colorectal tumor samples was downloaded from The Cancer Genome Atlas (TCGA) Colorectal Cancer database (https://www.cancer.gov/tcga). These samples were separated into a low and high group based on the cut-off value of GGCT median expression (cut-off value=39.1), which was obtained from the online TCGA Colorectal Cancer database. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of this microarray was performed using KEGG PathwayFinder with a gene correlation that was based on GGCT expression using the R2 web application (15–17). The absolute r cutoff was set at r=0.5, P<0.05. Gene Set Enrichment Analysis (GSEA; http://www.broadinstitute.org/gsea/index.jsp) was conducted using the KEGG_CELL_ADHENSION_MOLECULES_CAMS, Gene Ontology (GO)_EXTRACELLULAR_MATRIX, KEGG_FOCAL_ADHENSION and HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION gene sets (18,19). A group of 594 patient specimens from the TCGA Colorectal Cancer database (http://www.cbioportal.org/datasets) was used to evaluate the expression and prognostic value of GGCT in patients with CRC.
Cell culture and patient specimens
CRC cell lines HCT-116 (ATCC® CCL-247) and SW620 (ATCC® CCL-227) were purchased from American Type Culture Collection. These cells were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS), digested using trypsin every 2 days and cultured at 37°C in a humidified incubator with 5% CO2. A total of 6 paired CRC surgical specimens and corresponding adjacent normal colon specimens were obtained from the Department of General Surgery, Renhe Hospital (Shanghai, China) once written informed consent was obtained from patients or their guardians. The present study was ethically approved by the Ethic Committee of Renhe Hospital.
Western blot analysis
Western blot analysis was performed as previously described (20). A total of 5×105 CRC cells were washed three times in cold phosphate buffered saline (PBS), and total protein extracts were obtained using RIPA buffer [50 mM Tris (pH 8.0), 150 mM NaCl, 0.5% sodium deoxycholate, 0.1% SDS, 1% NP40, 1 mM EDTA and a mix of protease inhibitors]. Total proteins were quantified using a bicinchoninic acid assay (Pierce; Thermo Fisher Scientific, Inc.). Lysates were subsequently collected using centrifugation at 12,000 × g for 10 min at room temperature, and 20 µg proteins were separated by 10% SDS-PAGE and transferred to polyvinylidene difluoride membranes. The membranes were blocked with 5% non-fat dried milk in TBS and Tween-20 for 1 h at room temperature. The membranes were then incubated overnight at 4°C with the following specific primary antibodies: Anti-GGCT (1:1,000; cat. no. PA5-54263; Invitrogen; Thermo Fisher Scientific), anti-N-Cadherin (1:1,000; cat. no. 13116; Cell Signaling Technology, Inc.), anti-vimentin (1:1,000; cat. no. 5741; Cell Signaling Technology, Inc.), anti-snail family transcriptional repressor 2 (Slug; 1:1,000; cat. no. 9585; Cell Signaling Technology, Inc.), anti-snail family transcriptional repressor 1 (Snail; 1:1,000; cat. no. 3879; Cell Signaling Technology, Inc.) and anti-β-Actin (1:1,000; cat. no. 3700; Cell Signaling Technology, Inc.). Following the primary incubation, membranes were incubated for 2 h at room temperature with either of the following horseradish peroxidase-conjugated secondary antibodies: Goat anti-mouse (1:2,000; cat. no. sc-2005) and goat anti-rabbit IgG (1-,000; cat. no. sc-2004) (all from Santa Cruz Biotechnlogy).
Lentiviral vectors construction
GGCT-short-hairpin (sh)RNA lentiviral vectors and a non-targeting control shRNA (shNT; SHC002) vector were purchased from Hanbio Biotechnology Co., Ltd. The shRNAs used were as follows; shCtrl forward, 5′-GCTCAGCGGGAGGAGGCTATATATTGCAAGAGAACGTTT-3′ and reverse, 5′-ACTTGAACAAAGCACGCGGAGGCACAGTGTCCTCT-3′; shGGCT#1 forward, 5′-GACACGACGTCCCGTCTAGGCTGTCAGTCAAGCGGCCTCGGCAT-3′ and reverse, 5′-ACGCGCACCCGTGCGACTTGTGCATACACAC-3′; shGGCT#2 forward, 5′-GCTCTGTATCAGATGTGTCTCGTATACATGAGCTT-3′; and reverse, 5′-ACTTGGATAAGTTACGACCTATCCGTCATCTCTTGAAT-3′. A total of 8×104 CRC cells were plated overnight in 500 µl growth medium, in a single well of a 24-well plate. Cells were transiently transfected with 15 pmol shRNAs against GGCT or a non-targeting control using 1.5 µl Lipofectamine 3000 reagent for 48 h at 37°C in a humidified incubator with 5% CO2 according to the manufacturer's protocol (cat. no. L3000015; Thermo Fisher Scientific, Inc.).
RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR) assay
Total RNA of GGCT-knockdown and control HCT-116 and SW620 cells were extracted using a total RNA extracting kit purchased from Fastagen Biotechnology according to the manufacturer's protocol. For RT-qPCR, mRNAs mixture was incubated at 42°C for 60 min and 70°C for 15 min to be reverse transcribed to cDNA using the Reverse Transcription kit (Takara Biotechnology Co., Ltd.). RT-qPCR was performed using SYBR Premix Ex Taq (Takara Biotechnology Co., Ltd.). The thermocycling conditions were as follows: Pre-denaturation at 94°C for 5 min; 33 cycles of 94°C for 30 sec, 64°C for 30 sec and 72°C for 45 sec; and 72°C for 10 min. Results were normalized to the expression of β-actin using the 2−ΔΔCq method (21). The sequence of the primers used were as follows: β-actin forward, 5′-CATGTACGTTGCTATCCAGGC-3′ and reverse, 5′-CTCCTTAATGTCACGCACGAT-3′; GGCT forward, 5′-TGGCAATTCCCAAGGCAAAAC-3′ and reverse, 5′-CCCCTTCTTGCTCATCCAGAG-3′; N-cadherin forward, 5′-TCAGGCGTCTGTAGAGGCTT-3′ and reverse, 5′-ATGCACATCCTTCGATAAGACTG-3′; Vimentin forward 5′-GACGCCATCAACACCGAGTT-3′ and reverse, 5′-CTTTGTCGTTGGTTAGCTGGT-3′; Slug forward, 5′-CGAACTGGACACACATACAGTG-3′ and reverse, 5′-CTGAGGATCTCTGGTTGTGGT-3′; Snail forward, 5′-TCGGAAGCCTAACTACAGCGA-3′ and reverse, 5′-AGATGAGCATTGGCAGCGAG-3′.
Transwell invasion and migration assay
For the transwell invasion assay, GGCT-knockdown and control HCT-116 cells were trypsinized and resuspended with serum-free DMEM. Cells were then seeded into suspension cell culture inserts (8.0 µm; EMD Millipore), which were firstly coated with 1 mg/ml Matrigel (BD Biosciences), at a density of 4×104 cells/well in 200 µl DMEM with 6 replicate wells, and 500 µl DMEM with 10% FBS was added to the lower chambers of the 24-well plate. Subsequent to incubation at 37°C with 5% CO2 for 24 h, inserts were washed with PBS and fixed with 4% paraformaldehyde for 15 min at room temperature. The migration ability of cells was examined by using 8.0 µm cell culture inserts without coating Matrigel. Following 12 h incubation at 37°C, the upper inserts were removed and then washed and fixed as described above. All inserts were subsequently wiped and cells were stained using a crystal violet solution (Beyotime Institute of Biotechnology) for 15 min at room temperature. Stained cells were counted and images were acquired using a microscope (Olympus Corporation).
Wound-healing assay
GGCT-knockdown and control HCT-116 cells were seeded at a density of 2×103 cells/well in 24-well plates with serum-free DMEM, and a wound was created using a 10 µl plastic pipette tip in three replicate wells. The migration distance was observed and measured after 24 h incubation at 37°C. The images were acquired using a light microscope (Olympus) at a magnification of ×100. A total of 9 areas were selected randomly in each well at ×100 magnification and the migration distance was measured using the software program HMIAS-2000 version 2.0 (Wuhan Qianping Imaging Technology Co., Ltd.) as previously described (22).
Statistical analysis
All experiments were performed at least three times with triplicate samples. The survival curves were created using Kaplan-Meier analysis, and P values were calculated using a Log-rank test. Statistical analysis was performed using SPSS statistical software (SPSS16.0; SPSS, Inc.) and GraphPad Prism 6 software (GraphPad Software, Inc.) The unpaired two-group comparison and multiple comparisons were made using the unpaired Student's t-test or one-way analysis of variance with Student-Keuls-Neuman post hoc test, respectively. Multiple comparison between the groups was performed using method. Data were presented as the mean ± standard deviation. P<0.05 was considered to indicate a statistically significant difference.
Results
GGCT is upregulated in human CRC tissues and is associated with a poor prognosis
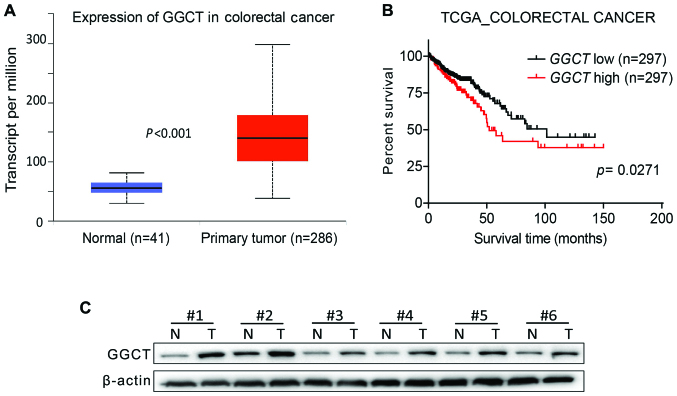
As GGCT has been previously demonstrated to be overexpressed in a variety of cancer types (5), the effect of GGCT expression on mRNA levels in human CRC was assessed by analyzing TCGA Colorectal Cancer database, and the results indicated that GGCT expression was increased significantly in CRC tissues compared with normal colon tissues (P<0.001; Fig. 1A). Furthermore, the prognostic value of GGCT was evaluated, and the Kaplan-Meier survival curve revealed that the overall survival time for patients with CRC with a higher GGCT expression was significantly decreased compared with patients exhibiting a lower GGCT expression (P=0.0271; Fig. 1B). To evaluate the expression of GGCT on the protein expression in human CRC, six pairs of tissue samples were examined using western blot analysis. The pathological features of the CRC specimens were summarized in Table I. The results demonstrated that GGCT expression was increased in the tissues of patients with CRC compared with corresponding normal colon tissues (Fig. 1C). In conclusion, the results revealed that GGCT was upregulated in human CRC tissues and was associated with a poor patient prognosis.
Figure 1.
GGCT is upregulated in human CRC tissues and associated with a poor prognosis. (A) Expression of GGCT in CRC tissues compared with corresponding normal colon tissues that were obtained from the TCGA Colorectal Cancer database. (B) Kaplan-Meier analyses for the association between GGCT and the prognosis of patients with CRC derived from TCGA Colorectal Cancer database. These samples were separated into a low and high group based on the median expression of GGCT. (C) Western blot assay assessed the expression of GGCT in six paired CRC tissues and adjacent normal colon tissues. T, tumor; N, normal tissue; GGCT, γ-glutamylcyclotransferase; CRC, colorectal cancer; TCGA, The Cancer Genome Atlas.
Table I.
Basic clinicopathological characteristics of the colorectal cancer specimens used in the present study.
| Characteristic | Number (%) |
|---|---|
| Sex | |
| Male | 4 (66.7) |
| Female | 2 (33.3) |
| Tumor size, cm | |
| <5 | 3 (50.0) |
| ≥5 | 3 (50.0) |
| Age, years | |
| <65 | 4 (66.7) |
| ≥65 | 2 (33.3) |
| Lymphatic metastasis | |
| No | 4 (66.7) |
| Yes | 2 (33.3) |
GGCT is associated with migration and invasion according to GSEA enrichment analysis
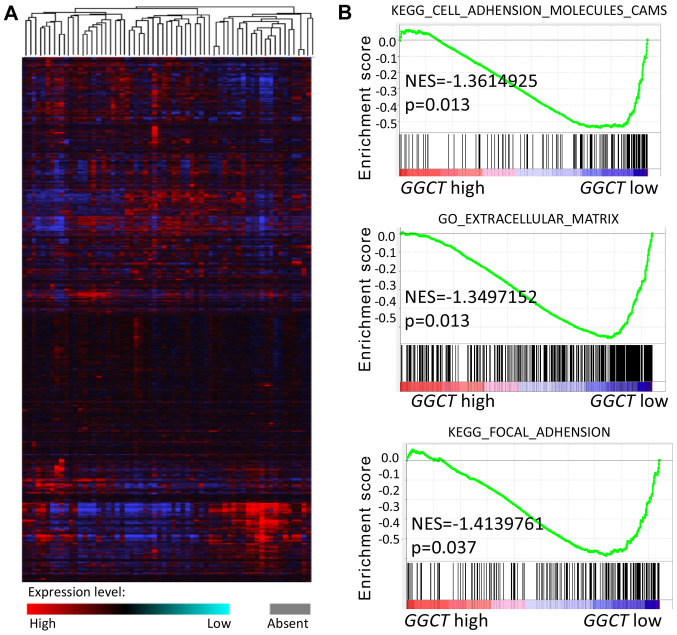
As aforementioned, GGCT was demonstrated to be upregulated in CRC tissues and was associated with a poor prognosis. Therefore, the function of GGCT in CRC was further examined. A microarray of mRNA from 286 colorectal tumor samples was downloaded from the TCGA Colorectal Cancer database (Fig. 2A). GSEA enrichment analysis was subsequently performed and the results indicated that GGCT was significantly associated with CRC migration and invasion in the context of the KEGG_CELL_ADHENSION_MOLECULES_CAMS, GO_EXTRACELLULAR_MATRIX and KEGG_FOCAL_ADHENSION gene sets (P<0.05). Patients with CRC, which exhibited a high GGCT expression, were indicated to exhibit an increased number of EMT-associated genes in the context of the HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION gene set (Fig. 2B). These results revealed that GGCT may regulate the migration and invasion of CRC cells via EMT.
Figure 2.
GSEA of gene expression profiles in patients with colorectal cancer for the expression of GGCT. (A) Heat map of expression values for differentially expressed genes from 286 colorectal tumor samples downloaded from TCGA Colorectal_Cancer Database. (B) RNA-seq data from TCGA was analyzed with GSEA enrichment plots for KEGG_CELL_ADHENSION_MOLECULES_CAMS, GO_EXTRACELLULAR_MATRIX, KEGG_FOCAL_ADHENSION and HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION gene sets. NES, normalized enrichment score; KEGG, Kyoto Encyclopedia of Genes and Genomes; GGCT, γ-glutamylcyclotransferase; GO, Gene Ontology; TCGA, The Cancer Genome Atlas; GSEA, Gene Set Enrichment Analysis.
GGCT promotes the migration and invasion of CRC cells in vitro
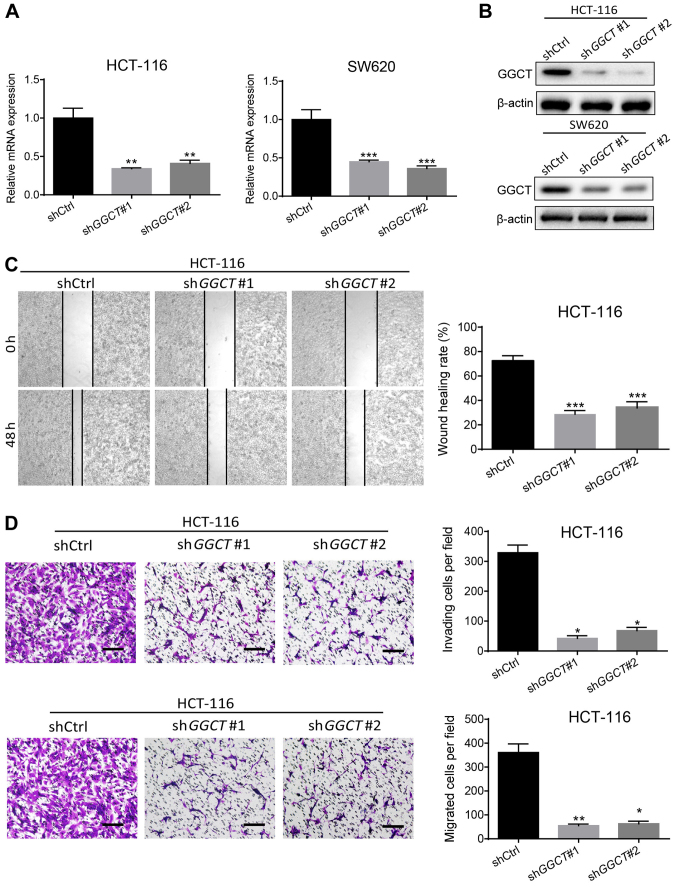
GSEA enrichment analysis revealed that GGCT may regulate the migration and invasion of CRC; therefore, to assess the function of GGCT in CRC malignancy, GGCT-knockdown colon cancer cell lines HCT-116 and SW620 were established using a lentiviral system. RT-qPCR and western blot analysis demonstrated that the expression of GGCT significantly decreased in infected HCT-116 and SW620 cells compared with corresponding control cells (P<0.01), indicating that the interference effects of knocking down GGCT were consistent (Fig. 3A and B). The migratory ability of GGCT-knockdown cells was detected and analyzed using a wound healing assay. In the GGCT-knockdown groups, a significantly lower wound healing rate was observed compared with the control cells (P<0.001; Fig. 3C). Furthermore, a transwell invasion and migration assay was performed, which indicated that the disruption of GGCT significantly inhibited the invasion and migration capability of GGCT-knockdown HCT-116 cells compared with the control cells (P<0.05; Fig. 3D). In conclusion, the results demonstrated that GGCT promoted CRC migration and invasion in vitro.
Figure 3.
Disruption of GGCT inhibited the migration and invasion of HCT-116 cells. (A) Reverse transcription-quantitative polymerase chain reaction assay assessed the expression of GGCT in GGCT-knockdown HCT-116 and SW620 cells compared with corresponding control cells. (B) Western blot assay assessed the expression of GGCT in GGCT-knockdown HCT-116 and SW620 cells compared with corresponding control cells. (C) Representative images of wound healing assay with GGCT-knockdown HCT-116 cells and corresponding control cells. Right panel shows the quantified results. (D) Representative images of transwell invasion and migration assays with GGCT-knockdown HCT-116 cells and corresponding control cells. Right panel shows the quantified results. Scale bar, 100 µm. Data are presented as the means ± standard deviation. *P<0.05, **P<0.01 and ***P<0.001 vs. shCtrl. sh-, short hairpin RNA; Ctrl, control; GGCT, γ-glutamylcyclotransferase.
GGCT induces EMT in the colon cancer cell line HCT-116
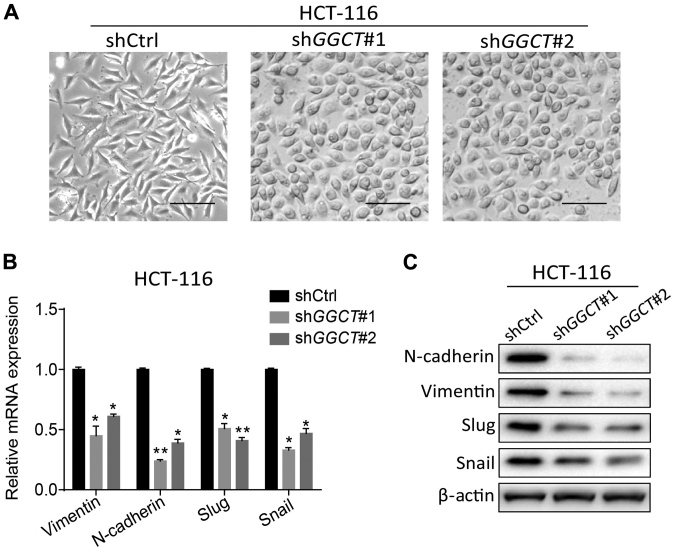
To determine the mechanism in which GGCT promoted the migration and invasion of CRC cells, the present study assessed whether the depletion of GGCT inhibited EMT in HCT-116 cells. The cellular morphology was observed using a light microscope, and the cell morphology of GGCT-knockdown HCT-116 cells was indicated to have notably changed into the epithelial cell type from the original HCT-116 mesenchymal-like cell type (Fig. 4A). EMT-associated biomarkers were subsequently assessed in GGCT-knockdown HCT-116 cells, and it was revealed that the depletion of GGCT significantly decreased the expression of N-cadherin, Vimentin, Slug and Snail compared with control cells (P<0.05; Fig. 4B and C). Overall, these data demonstrated that GGCT induced EMT in the colon cancer cell line HCT-116.
Figure 4.
Disruption of GGCT inhibited the EMT of HCT-116 cells. (A) Representative images of cellular morphology of GGCT-knockdown HCT-116 cells and corresponding control cells obtained using a light microscope. Scale bar, 100 µM. (B) Reverse transcription-quantitative PCR assay assessed the expression of EMT-associated genes in GGCT-knockdown HCT-116 cells and corresponding control cells. (C) Western blot assay assessed the expression of EMT-associated genes in GGCT-knockdown HCT-116 cells and corresponding control cells. Data are presented as the mean ± standard deviation. *P<0.05 and **P<0.01 vs. shCtrl. sh-, short hairpin RNA; Ctrl, control; GGCT, γ-glutamylcyclotransferase; EMT, epithelial-to-mesenchymal transition; Slug, snail family transcriptional repressor 2; Snail, snail family transcriptional repressor 1.
Discussion
CRC is the third most common malignancy, and the fourth most common cause of cancer-associated mortality globally (2). Despite substantial advances in modern medicine, including the development of novel treatment methods, the mortality of patients with CRC remains high, which may be due to the lack of specific biomarkers and effective treatments for the disease (23,24). Therefore the identification of novel prognostic markers and therapeutic targets for use in CRC treatment is urgently required.
EMT is essential for metastatic dissemination (25). During EMT, tumor cells with epithelial characteristics are transformed into tumor cells with mesenchymal characteristics, which contributes to cancer progression (26). It has recently been accepted that EMT is an important and complex phenomenon that determines the aggressiveness of colon cancer (27). Zhang et al (20) demonstrated that Prostate Transmembrane Protein, Androgen Induced 1 induced EMT via a non-canonical transforming growth factor-β signaling pathway in CRC, and Wei et al (28) indicated that FAT Atypical Cadherin 4 modulated CRC tumorigenesis by regulating the status of EMT and autophagy (28). Sun et al (29) demonstrated that Tripartite Motif Containing 29 facilitated the EMT of CRC via the activation of the Wnt/β-catenin signaling pathway. The aforementioned results indicated that elucidating the molecular mechanisms of EMT may aid in the progression of CRC research and the identification of novel therapeutic targets.
In the present study, GGCT was revealed to be increased in CRC tissues compared with normal colon tissues, and patients with CRC that had higher GGCT expression exhibited a worse prognosis compared with patients with a lower expression of GGCT, indicating the prognostic value of GGCT. The dysregulation of GGCT has been reported in certain cancer types, for example Li et al (13) demonstrated that GGCT suppressed the progression of prostate cancer, indicating that GGCT may serve as a tumor suppressor gene. However, Zhang et al (30) indicated that the disruption of GGCT inhibited proliferation and induced late apoptosis in human gastric cancer, demonstrating that GGCT may serve as an oncogene. The present study demonstrated that GGCT was highly expressed in CRC and may function as a novel prognostic marker for the disease. Furthermore, GGCT was indicated to promote CRC cell migration and invasion through regulating EMT. The disruption of GGCT in HCT-116 cells markedly decreased the EMT associated markers and changed the morphology of GGCT-knockdown HCT-116 cells from mesenchymal-like cell type to the epithelial cell type, compared with the control cells. Disrupting GGCT was also observed to notably decrease the cell migration and invasion of HCT-116 cells, which was consistent with previous reports that indicated that GGCT influenced the migration and invasion of cancer cells (5,31).
GGCT has previously been described to serve an important function in the proliferation of gastric, breast and lung cancer cells (12,30,32). In the present study, the proliferation of GGCT-knockdown CRC cells was not assessed, nevertheless this part is worthy of further work.
As GGCT has been reported to regulate the penultimate step in glutathione catabolism and serve a critical function in glutathione homeostasis, biochemical factors were not assessed following the disruption of GGCT in CRC cells. However, assessing how the depletion of GGCT influences EMT in CRC cells requires investigation in future studies.
In the present study, GGCT was demonstrated to serve a novel function as an oncogene in CRC, was demonstrated to promote CRC migration and invasion via EMT and was identified as a novel prognostic marker for this disease. In conclusion, the present study provides novel insight into the mechanisms responsible for CRC migration and invasion and identifies GGCT as a promising therapeutic target for use in the treatment of CRC.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Science and Technology Committee of Baoshan District, Shanghai City Special Funds for Science, Technology and Innovation (grant no. 16-E-21).
Availability of data and materials
All data generated and/or analyzed during this study are included in this published article.
Authors' contributions
QH and YZ completed all trial procedures and data analysis in writing the manuscript. QH, YZ and YL performed the experiments. ZL designed the overall idea of the experiment and provided theoretical guidance throughout the process. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was ethically approved by the Ethic Committee of Renhe Hospital (Shanghai, China). All subjects provided written informed consent to participate in the present study.
Patient consent for publication
Not applicable.
Competing interests
The authors have declared that they have no competing interests.
References
- 1.Braillon A. Screening for colorectal cancer at earlier ages: Putting the cart before the horse. Gastroenterology. 2019;156:1532. doi: 10.1053/j.gastro.2018.12.034. [DOI] [PubMed] [Google Scholar]
- 2.Young GP, Rabeneck L, Winawer SJ. The global paradigm shift in screening for colorectal cancer. Gastroenterology. 2019;156:843–851. doi: 10.1053/j.gastro.2019.02.006. [DOI] [PubMed] [Google Scholar]
- 3.de Andrea CE, Schalper KA, Sanmamed MF, Melero I. Immunodivergence in metastatic colorectal cancer. Cancer Cell. 2018;34:876–878. doi: 10.1016/j.ccell.2018.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huyghe JR, Bien SA, Harrison TA, Kang HM, Chen S, Schmit SL, Conti DV, Qu C, Jeon J, Edlund CK, et al. Discovery of common and rare genetic risk variants for colorectal cancer. Nat Genet. 2019;51:76–87. doi: 10.1038/s41588-018-0286-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kageyama S, Hanada E, Ii H, Tomita K, Yoshiki T, Kawauchi A. Gamma-glutamylcyclotransferase: A novel target molecule for cancer diagnosis and treatment. Biomed Res Int. 2015;2015:345219. doi: 10.1155/2015/345219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Y, Hyde AS, Simpson MA, Barycki JJ. Emerging regulatory paradigms in glutathione metabolism. Adv Cancer Res. 2014;122:69–101. doi: 10.1016/B978-0-12-420117-0.00002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amano T, Eishi Y, Yamada T, Uchida K, Minegishi K, Tamura T, Kobayashi D, Hiroshi K, Suzuki T, Board PG. Widespread expression of gamma-glutamyl cyclotransferase suggests it is not a general tumor marker. J Histochem Cytochem. 2012;60:76–86. doi: 10.1369/0022155411428468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kageyama S, Isono T, Iwaki H, Wakabayashi Y, Okada Y, Kontani K, Yoshimura K, Terai A, Arai Y, Yoshiki T. Identification by proteomic analysis of calreticulin as a marker for bladder cancer and evaluation of the diagnostic accuracy of its detection in urine. Clin Chem. 2004;50:857–866. doi: 10.1373/clinchem.2003.027425. [DOI] [PubMed] [Google Scholar]
- 9.Uejima D, Nishijo K, Kajita Y, Ishibe T, Aoyama T, Kageyama S, Iwaki H, Nakamura T, Iida H, Yoshiki T, Toguchida J. Involvement of cancer biomarker C7orf24 in the growth of human osteosarcoma. Anticancer Res. 2011;31:1297–1305. [PubMed] [Google Scholar]
- 10.Takemura K, Kawachi H, Eishi Y, Kitagaki K, Negi M, Kobayashi M, Uchida K, Inoue J, Inazawa J, Kawano T, Board P. γ-Glutamylcyclotransferase as a novel immunohistochemical biomarker for the malignancy of esophageal squamous tumors. Hum Pathol. 2014;45:331–341. doi: 10.1016/j.humpath.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 11.Shen SH, Yu N, Liu XY, Tan GW, Wang ZX. Gamma-glutamylcyclotransferase promotes the growth of human glioma cells by activating Notch-Akt signaling. Biochem Biophys Res Commun. 2016;471:616–620. doi: 10.1016/j.bbrc.2016.01.165. [DOI] [PubMed] [Google Scholar]
- 12.Matsumura K, Nakata S, Taniguchi K, Ii H, Ashihara E, Kageyama S, Kawauchi A, Yoshiki T. Depletion of gamma-glutamylcyclotransferase inhibits breast cancer cell growth via cellular senescence induction mediated by CDK inhibitor upregulation. BMC Cancer. 2016;16:748. doi: 10.1186/s12885-016-2779-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H, Yoshiya T, Nakata S, Taniguchi K, Hidaka K, Tsuda S, Mochizuki M, Nishiuchi Y, Tsuda Y, Ito K, et al. A Novel Prodrug of a γ-Glutamylcyclotransferase Inhibitor Suppresses Cancer Cell Proliferation in vitro and Inhibits Tumor Growth in a Xenograft Mouse Model of Prostate Cancer. ChemMedChem. 2018;13:155–163. doi: 10.1002/cmdc.201700660. [DOI] [PubMed] [Google Scholar]
- 14.Ii H, Yoshiya T, Nakata S, Taniguchi K, Hidaka K, Tsuda S, Mochizuki M, Nishiuchi Y, Tsuda Y, Ito K, et al. A novel prodrug of a γ-glutamylcyclotransferase inhibitor suppresses cancer cell proliferation in vitro and inhibits tumor growth in a xenograft mouse model of prostate cancer. Chemmedchem. 2018;13:155–163. doi: 10.1002/cmdc.201700660. [DOI] [PubMed] [Google Scholar]
- 15.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanehisa M, Sato Y, Furumichi M, Morishima K, Tanabe M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019;47:D590–D595. doi: 10.1093/nar/gky962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019;28:1947–1951. doi: 10.1002/pro.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.The Gene Ontology Consortium, corp-author. The Gene Ontology resource: 20 years and still GOing strong. Nucleic Acids Res. 2019;47:D330–D338. doi: 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang L, Wang X, Lai C, Zhang H, Lai M. PMEPA1 induces EMT via a non-canonical TGF-β signalling in colorectal cancer. J Cell Mol Med. 2019;23:3603–3615. doi: 10.1111/jcmm.14261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Yang R, Hoang BH, Kubo T, Kawano H, Chou A, Sowers R, Huvos AG, Meyers PA, Healey JH, Gorlick R. Over-expression of parathyroid hormone Type 1 receptor confers an aggressive phenotype in osteosarcoma. Int J Cancer. 2007;121:943–954. doi: 10.1002/ijc.22749. [DOI] [PubMed] [Google Scholar]
- 23.Ganesh K, Stadler ZK, Cercek A, Mendelsohn RB, Shia J, Segal NH, Diaz LJ., Jr Immunotherapy in colorectal cancer: Rationale, challenges and potential. Nat Rev Gastroenterol Hepatol. 2019;16:361–375. doi: 10.1038/s41575-019-0126-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valle L, de Voer RM, Goldberg Y, Sjursen W, Försti A, Ruiz-Ponte C, Caldés T, Garré P, Olsen MF, Nordling M, et al. Update on genetic predisposition to colorectal cancer and polyposis. Mol Aspects Med. 2019;69:10–26. doi: 10.1016/j.mam.2019.03.001. [DOI] [PubMed] [Google Scholar]
- 25.Pastushenko I, Blanpain C. EMT transition states during tumor progression and metastasis. Trends Cell Biol. 2019;29:212–226. doi: 10.1016/j.tcb.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 26.Singh M, Yelle N, Venugopal C, Singh SK. EMT: Mechanisms and therapeutic implications. Pharmacol Ther. 2018;182:80–94. doi: 10.1016/j.pharmthera.2017.08.009. [DOI] [PubMed] [Google Scholar]
- 27.Zhu Y, Wang C, Becker SA, Hurst K, Nogueira LM, Findlay VJ, Camp ER. miR-145 antagonizes SNAI1-mediated stemness and radiation resistance in colorectal cancer. Mol Ther. 2018;26:744–754. doi: 10.1016/j.ymthe.2017.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei R, Xiao Y, Song Y, Yuan H, Luo J, Xu W. FAT4 regulates the EMT and autophagy in colorectal cancer cells in part via the PI3K-AKT signaling axis. J Exp Clin Cancer Res. 2019;38:112. doi: 10.1186/s13046-019-1043-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun J, Zhang T, Cheng M, Hong L, Zhang C, Xie M, Sun P, Fan R, Wang Z, Wang L, Zhong J. TRIM29 facilitates the epithelial-to-mesenchymal transition and the progression of colorectal cancer via the activation of the Wnt/β-catenin signaling pathway. J Exp Clin Cancer Res. 2019;38:104. doi: 10.1186/s13046-019-1098-y. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Zhang W, Chen L, Xiang H, Hu C, Shi W, Dong P, Lv W. Knockdown of GGCT inhibits cell proliferation and induces late apoptosis in human gastric cancer. BMC Biochem. 2016;17:19. doi: 10.1186/s12858-016-0075-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kageyama S, Ii H, Taniguchi K, Kubota S, Yoshida T, Isono T, Chano T, Yoshiya T, Ito K, Yoshiki T, et al. Mechanisms of tumor growth inhibition by depletion of γ-glutamylcyclotransferase (GGCT): A novel molecular target for anticancer therapy. Int J Mol Sci. 2018;19:E2054. doi: 10.3390/ijms19072054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y, Zhao W, Zhao Z, Wu J, Chen L, Ma Y, Li Q, Lu D, Jin L, Wang J. IL1B gene polymorphisms, age and the risk of non-small cell lung cancer in a Chinese population. Lung Cancer. 2015;89:232–237. doi: 10.1016/j.lungcan.2015.06.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated and/or analyzed during this study are included in this published article.