Abstract
N6-methyladenosine (m6A) in messenger RNA (mRNA) is regulated by m6A methyltransferases and demethylases. Modifications of m6A are dynamic and reversible, may regulate gene expression levels and serve vital roles in numerous life processes, such as cell cycle regulation, cell fate decision and cell differentiation. In recent years, m6A modifications have been reported to exhibit functions in human cancers via regulation of RNA stability, microRNA processing, mRNA splicing and mRNA translation, including lung cancer, breast tumor and acute myeloid leukemia. In the present review, the roles of m6A modifications in the onset and progression of cancer were summarized. These modifications display an oncogenic role in certain types of cancer, whereas in other types of cancer they exhibit a tumor suppressor role. Therefore, understanding the biological functions performed by m6A in different types of tumors and identifying pivotal m6A target genes to deduce the potential mechanisms underlying the progression of cancer may assist in the development of novel therapeutics.
Keywords: RNA modification, N6-methyladenosine, cancer, epigenetics, mRNA
1. Introduction
Over 100 different types of post-transcriptional chemical modifications of RNA have been identified in living organisms (1). A total of three types of RNA modifications present in eukaryotic messenger RNA (mRNA) have become prevalent in recent years: N6-methyladenosine (m6A), 5-methylcytosine (m5C) and N1-methyladenosine (m1A) (2). m6A was first identified by Desrosiers et al (3) in 1974 and is the most commonly observed internal modification between long noncoding RNA (lncRNA) and mRNA in eukaryotes. Dominissini et al (4) used high-throughput sequencing to demonstrate that m6A modifications are frequently present at stop codons, 3′-untranslated regions (3′-UTRs) and internal long exons, whereas m6A modifications are typically present at RRm6ACH motifs (5).
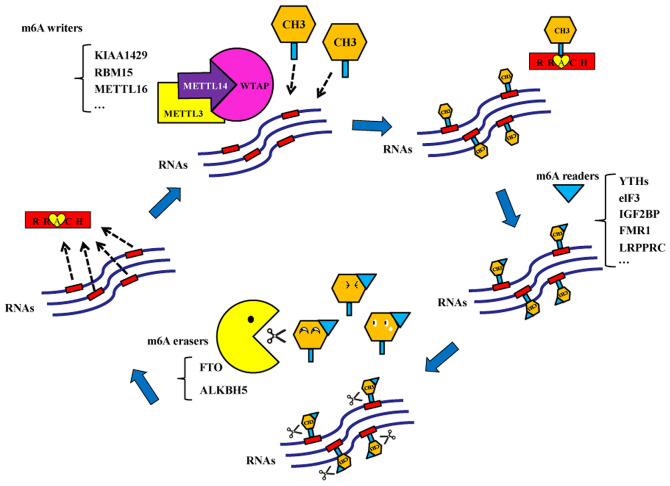
m6A methylation is a methylation modification of adenine (A) at position 6N and is catalyzed by the methyltransferases (6). Bokar et al (7) first discovered that a methyltransferase complex catalyzed the formation of an m6A modification in 1994. m6A methylation modifications are typically present in mRNAs, transport RNAs, ribosomal RNAs and non-coding RNAs (8). It was demonstrated that m6A methylation may be present on microRNAs (miRNAs) (9). There are three types of enzymes that participate in the dynamic and reversible m6A methylation and demethylation modifications. The first type is the methyltransferases, which serve a crucial role in RNA transformation into m6A-modified RNA (7). Their coding genes are called m6A ‘writers’. Methyltransferase-like 3 (METTL3) was the first protein reported to exhibit methyltransferase activity in 1997 (10). Subsequently, additional m6A writers were discovered in mammals, including methyltransferase-like 14 (METTL14), Wilms' tumor 1-associating protein (WTAP), KIAA1429, RNA binding motif protein 15/15B and methyltransferase-like 16 (11–14). The second type of enzymes is the demethylases, which may alter the m6A-modified RNA back to RNA (15). Demethylase coding genes are called m6A ‘erasers’. Fat mass and obesity-associated protein (FTO) and alkB homolog 5 (ALKBH5) may function as m6A erasers (15,16). The last type of enzymes is the m6A ‘readers’, which read the m6A-mediated physiological effects and influence RNA behaviors (17). There are several m6A readers, including YT521-B homology (YTH) domain family-YTHDF (YTHDF1, YTHDF2 and YTHDF3) and YTHDC subtypes (YTHDC1 and YTHDC2) (4), eukaryotic translation initiation factor 3 (eIF3), heterogeneous nuclear ribonucleoprotein A2B1 and C, insulin-like growth factor 2 mRNA-binding proteins (IGF2BP), fragile X mental retardation 1 (FMR1) and leucine rich pentatricopeptide repeat containing (LRPPRC; Fig. 1).
Figure 1.
Mechanism of N6-methyladenine RNA modification. m6A methylation is dynamically and reversibly regulated by methyltransferases (METTL3, METTL14, WTAP, KIAA1429 among others) and demethylases (FTO and ALKBH5). M6A readers (YTHDF1, YTHDF2, YTHDF3, YTHDC1 and YTHDC2) read the m6A-mediated physiological effects and influence RNA behaviors. METTL3, methyltransferase-like 3; METTL14, methyltransferase-like 14; METTL16, methyltransferase-like 16; WTAP, Wilms' tumor 1-associating protein; RBM15, RNA binding motif protein 15; FTO, fat mass and obesity-associated protein; ALKBH5, alkB homolog 5; YTHDF1, YTH domain-containing family protein 1; YTHDF2, YTH domain-containing family protein 2; YTHDF3, YTH domain-containing family protein 3; YTHDC1, YTH domain-containing protein 1; YTHDC2, YTH domain-containing protein 2; eIF3, eukaryotic translation initiation factor 3; IGF2BP, insulin-like growth factor 2 mRNA-binding protein; FMR1, fragile X mental retardation 1; LRPPRC, leucine rich pentatricopeptide repeat containing.
Recently, increasing evidence has demonstrated that aberrantly expressed m6A modifications are associated with human tumors (18–20). In the present review, the roles of m6A regulators in various types of cancer are summarized.
2. m6A methylation and cancer
METTL3
METTL3 was first identified as an m6A writer by Bokar et al (10) in 1997, and was first isolated from HeLa cells as a protein holding an S-adenosylmethionine (SAM)-binding activity. METTL3 catalyzes m6A synthesis and possesses two binding sites for SAM (21). METTL3 is an important subunit of the methyltransferase (10). m6A modifications may occur in the cytoplasm and nucleus, as METTL3 is present in both locations (22).
In 2016, Lin et al (23) first reported that METTL3 may enhance translation independently of methyltransferase activity and methyl reader proteins. METTL3 may directly increase translation of target RNAs via recruiting translation initiation factors. METTL3 increased growth, survival and invasion of lung adenocarcinoma cells via increasing the expression of epidermal growth factor receptor (EGFR) and the activity of the Hippo signaling pathway effector TAZ. Subsequently, numerous studies have focused on METTL3 and its effects on various types of cancer (24–27). Visvanathan et al (24) demonstrated that the expression of METTL3 is increased in glioblastomas (GBM). In addition, METTL3 promoted tumor growth by targeting the 3′-UTR of SOX2 mRNA for m6A methylation. Silencing METTL3 reduced the tumor growth and enhanced the radiosensitivity of tumors, and thus may be used as a potential molecular target for GBM therapy. Barbieri et al (25) discovered that METTL3 may sustain acute myeloid leukemia (AML) via an m6A-dependent translation control. Taketo et al (26) reported that METTL3-depleted cells exhibited higher sensitivity to anticancer agents, which suggested that METTL3 was a potent target for enhancing the efficacy of therapeutics in patients with pancreatic cancer. In 2018, Choe et al (27) demonstrated that METTL3 may enhance translation, oncogenic transformation and the formation of densely packed polyribosomes via interaction with eIF3h in primary lung tumors, and thus may serve as a potential therapeutic target. Li et al (18) demonstrated that METTL3 retained SOX2 expression via an m6A-IGF2BP2-dependent manner in colorectal cancer (CRC) cells. A total of three consecutive studies (19,20,28) indicated that METTL3 may promote progression of gastric cancer (GC) via m6A modifications.
Recently, METTL3-mediated m6A modifications were discovered to promote tumor progression via interacting with various miRNAs in numerous types of cancer (29–35). Du et al (29) demonstrated that miR-33a suppressed the proliferation of non-small-cell lung cancer (NSCLC) cells via targeting METTL3 mRNA. Jin et al (30) revealed that METTL3 may initiate m6A modification and promote the translation of the yes-associated protein (YAP) mRNA to increase the resistance of NSCLC to anticancer therapeutics and metastasis via a metastasis associated lung adenocarcinoma transcript 1-miR-1914-3p-YAP axis. Cai et al (31) revealed that Hepatitis B virus X-interacting protein (HBXIP) modulated METTL3 by reducing the expression of miRNA let-7 g, thus promoting breast cancer progression. Wei et al (32) demonstrated that miR-600 inhibited the migration and proliferation of lung cancer cells via downregulation of METTL3 expression. Han et al (33) revealed that METTL3 interacted with DiGeorge Syndrome Critical Region 8 (DGCR8) and upregulated pre-miR-221/222, which restricted PTEN expression, thereby resulting in the proliferation of bladder cell carcinoma. Cheng et al (34) demonstrated that METTL3 enhanced the progression of bladder cell carcinoma via AF4/FMR2 family member 4/NF-κB/MYC signaling. Peng et al (35) revealed that the upregulation of METTL3 promoted CRC metastasis via miR-1246/sprouty-related, EVH1 domain-containing protein 2/mitogen-activated protein kinase signaling.
METTL3 has not only been revealed to promote the proliferation and metastasis of tumor cells, but also to inhibit the progression of tumors. Li et al (36) demonstrated that reduced expression of METTL3 was associated with higher histological grades and larger tumor sizes in nude mice. Furthermore, patients with renal cell carcinoma (RCC) and upregulated expression of METTL3 exhibited longer survival times.
Collectively, the results from these previous studies suggested that METTL3-mediated m6A modifications may promote tumor progression in different types of cancer; however the underlying mechanisms, such as the specific signaling pathways involved, may differ in different types of tumors. METTL3 may also affect tumors independently of its catalytic activity, for example via enhancing the translation of oncogenes. Currently, RCC is the sole tumor type where METTL3 is demonstrated to inhibit the progression of cancer. Additional studies are required to unravel the specific mechanisms by which METTL3-mediated inhibition affects RCC progression.
METTL14
METTL14, which is a homolog of METTL3, is another methyltransferase that catalyzes m6A modifications on RNA (11,12,37). METTL14 interacts with METTL3 and modifies the m6A content, and together, they form the core m6A methylation complex (38). METTL14 has been demonstrated to occupy a degraded, non-functional SAM-binding domain, based on the crystal structure analysis of the METTL3-METTL14 complex (39). On the other hand, METTL14 is important for the positioning of the methyl group and the binding of the RNA substrate to promote the catalytic activity of METTL3 (40,41). Thus, METTL14 is an indispensable factor for METTL3 activity.
METTL14 may inhibit the initiation and progression of tumors. A previous study revealed that the mRNA levels of METTL14 in patients with metastatic HCC were significantly lower compared with that in patients with non-metastatic HCC (42). Further experiments demonstrated that METTL14 interacted with DGCR8 and upregulated the expression of miRNA-126. Overexpression of METTL14 inhibited the metastasis of HCC in an established liver metastasis mouse model. Based on these results, it was hypothesized that METTL14 regulated the expression of miRNA-126 via m6A modifications, thereby regulating its downstream targets to inhibit the metastasis of HCC. Therefore, METTL14 expression may be used as a prognostic factor for HCC (42). Previous studies revealed that low methylation levels of m6A, caused by mutations in the METTL14 gene, were observed in endometrial cancer cells (43,44). Using the CRISPR technology, METTL14 was removed from hec-1-a endometrial cancer cells and a lack of METTL14 was demonstrated to increase the tumor cell proliferation, colony formation and metastasis. It was observed that the levels of m6A methylation in endometrial cancer cells were reduced compared with adjacent normal tissues. Thus, it was deduced that the mutations of METTL14 decreased m6A methylation levels, which served a key role in the development of endometrial cancer. Subsequently, it was demonstrated in patient samples and endometrial cancer cell lines that downregulation of m6A methylation altered the activity of the Akt/protein kinase B signaling pathway, thereby promoting the proliferation and metastasis of cancer cells. Liu et al (45) also discovered that the downregulation of METTL14 decreased PH domain and leucine rich repeat protein phosphatase 2 expression, which is a negative regulator of the AKT signaling pathway. Downregulation of METTL14 also increased the expression of mTOR Complex 2, a positive regulator of the AKT signaling pathway, which resulted in the proliferation of endometrial cancer cells. Gong et al (46) revealed that METTL14 downregulated the translation of P2X purinoceptor 6 (P2RX6), thus reducing the RCC cell invasion and metastasis via ATP-P2RX6-Ca2+-p-ERK1/2-matrix metalloproteinase-9-mediated signaling. METTL14-mediated m6A modifications may also suppress tumor progression in CRC and bladder cancer (47,48).
The METTL14-driven m6A modifications consistently result in the inhibition of tumor progression, including HCC, endometrial cancer, RCC, CRC and bladder cancer. Whether METTL14 promotes tumor progression in an m6A modification-dependent manner remains to be elucidated.
WTAP
WTAP was discovered by Little et al (49) in 2000, using the yeast two-hybrid system. After eight years, Zhong et al (50) demonstrated that WTAP interacted with METTL3. WTAP is a methyltransferase that does not possess a catalytic domain, thus WTAP does not exhibit an independent catalytic capacity on RNA. However, it can form a methyltransferase complex with METTL3 and METTL14, thus facilitating the catalysis of the m6A modification on RNA (11).
Downregulation of WTAP may result in the formation of METTL14 and METTL3 degradation complexes and considerably decrease m6A levels (11). In addition, Schwartz et al (12) reported the WTAP-independent and -dependent m6A modification sites upon WTAP depletion. WTAP-independent sites are located at the cap structure of the transcripts, whereas WTAP-dependent sites are present at internal positions (12), highlighting the complexity of the co-transcriptional regulation. Ping et al (51) revealed that WTAP assisted in the binding of METTL3 and METTL14 with the target RNA, whereas the absence of WTAP resulted in the failure of METTL3 and METTL14 binding with the RNA in the nuclear speckle; however the protein levels of METTL3 and METTL14 remained unaltered. Schöller et al (52) discovered that WTAP bound to a region of METTL3 in the first 150 amino acid region. Even if WTAP was truncated from the C-terminal, it would still bind with METTL3, as long as the remaining structure contained the first 150 amino acids from the N-terminal.
Recently, WTAP was demonstrated to regulate tumor progression in an m6A modification-dependent manner. Chen et al (53) revealed that WTAP expression was upregulated in HCC. WTAP-mediated m6A modifications contributed to the progression of HCC via human antigen R/protein C-ets-1/p21/p27 signaling.
In comparison with METTL3 and METTL14, there is a decreased number of publications reporting the relationship between WTAP and cancer. The primary reason for this may be that WTAP does not exhibit an independent catalytic activity, while it mainly forms methyltransferase complexes with METTL3 and METTL14 to catalyze m6A modifications on RNA.
KIAA1429. KIAA1429 was first identified in 2003 on account of the discovery of its homologs, which exhibited an interaction with WTAP in a sex-specific manner (54). Subsequent studies demonstrated that METTL14, METTL3 and KIAA1429 are required for methylation in mammals (12). Recently, two studies revealed that METTL14, METTL3, WTAP and KIAA1429 form a methyltransferase complex (55,56). The downregulation of KIAA1429 resulted in a higher decrease of the m6A index in the mRNA compared with the downregulation of either METTL14 or METTL3, suggesting that KIAA1429 is required for the complete catalytic activity of the methyltransferase complex.
Cheng et al (57) investigated the role of KIAA1429 in HCC and discovered that the mRNA levels of KIAA1429 were higher in HCC compared with the adjacent normal liver tissues, based on the analysis of the cancer genome atlas (TCGA) database. KIAA1429 was demonstrated to regulate the invasion and migration of HCC cells via upregulation of the m6A modifications on ID2 mRNA, thereby inhibiting ID2. Qian et al (58) revealed that KIAA1429 promoted the progression of breast cancer via modulating CDK1 activity in an m6A-independent manne.
As in the case of WTAP, KIAA1429 also requires the formation of a methyltransferase complex with other m6A writers to perform its function. KIAA1429 may affect tumorigenesis in both an m6A-dependent and -independent manner.
3. m6A demethylation and cancer
FTO
In 2007, Frayling et al (59) discovered a cluster of single-nucleotide polymorphisms in the first intron of the FTO gene, which were associated with obesity. Subsequently, the gene was officially named FTO. FTO belongs to the Fe (II)- and α-ketoglutarate-dependent dioxygenase AlkB protein family and demethylates the 3-methylthymine in single-stranded DNA and the 3-methyluracil in single-stranded RNA (60). Jia et al (15) revealed that FTO exhibited a powerful oxidative demethylation activity, targeting the m6A residues in RNA in vitro. It was demonstrated that m6A in nuclear RNA was a physiological substrate of FTO, via the partial colocalization of FTO with nuclear speckles. In 2017, Mauer et al (61) presented a novel perspective on the actual substrate of FTO. It was hypothesized that the demethylation affinity of FTO was higher for m6Am, which was the first nucleotide following the 7-methylguanosine cap of certain mRNAs (62), compared with m6A, increasing mRNA stability. The demethylation activity of FTO on m6Am was ~100× higher compared with that on m6A. After the depletion of FTO, the levels of m6A were not notably altered, whereas the levels of m6Am increased considerably. These results suggested that the substrate of FTO may be m6Am, a finding which was different from previous studies. Subsequently, other researchers demonstrated that the distribution of FTO differed in the nucleus and cytoplasm of different cell lines. FTO had a higher affinity for m6A in the nucleus and m6Am in the cytoplasm (63,64).
In recent years, it has been demonstrated that FTO is closely associated with the onset and progression of various malignant tumors, such as breast cancer (65), lung cancer (66,67), gastric cancer (68,69) and AML (70–72).
Niu et al (65) discovered that FTO was highly expressed in breast cancer tissues and that FTO expression was associated with a worse prognosis. FTO promoted the proliferation and metastasis of breast cancer cells via inhibiting Bcl-2 interacting protein 3. Li et al (66) discovered that FTO promoted the proliferation of NSCLC via increasing the expression of ubiquitin carboxyl-terminal hydrolase 7. Liu et al (67) revealed that overexpression of FTO may increase the stability of myeloid zinc finger 1 (MZF1) mRNA and promote the expression of MZF1 via downregulating the m6A levels in the MZF1 mRNA transcripts, leading to the proliferation and invasion of lung squamous cell carcinoma cells. Xu et al (68) reported that the expression levels of FTO were closely associated with reduced tumor differentiation, lymph node metastasis and advanced tumor-node-metastasis stage in gastric cancer. Via the analysis of the Kaplan-Meier plotter database and TCGA database, Li et al (69) discovered that high expression of FTO and ALKBH1 mRNAs was associated with a poor prognosis in gastric cancer. A previous study also revealed that FTO serves a crucial role in the progression of AML (70). FTO enhanced leukemia oncogene-mediated cell transformation and the occurrence of leukemia, inhibited the all-transretinoic acid-mediated differentiation of AML cells and regulated the synthesis of target genes, such as ankyrin repeat and SOCS box containing 2 and retinoic acid receptor alpha, via downregulating the levels of m6A. Su et al (71) reported that R-2HG inhibited the activity of FTO, increased the m6A modifications on RNA, reduced the stability of the MYC/CCAAT/enhancer-binding protein alpha transcripts and inhibited the proliferation of leukemia cells, thus resulting in cell cycle arrest. Huang et al (72) revealed that FTO promoted leukemogenesis. An FTO inhibitor, termed FB23-2, was developed, which may directly bind to and selectively inhibit the m6A demethylase activity of FTO. It was demonstrated that FB23-2 may suppress the proliferation and promote the differentiation/apoptosis of AML cells, and thus may be used as a potential druggable target for treating AML. Zhou et al (73) discovered that FTO enhanced the resistance to chemo-radiotherapy in cervical squamous cell carcinoma via reducing the m6A demethylation in β-catenin mRNA, thereby regulating its expression. Li et al (74) revealed that FTO promoted HCC tumorigenesis by mediating pyruvate kinase M2 demethylation. Yang et al (75) found that FTO increased m6A demethylation and enhanced melanoma tumorigenesis and anti-programmed cell death protein 1 resistance.
FTO is an important m6A demethylation enzyme. At present, it has been demonstrated that FTO promotes tumorigenesis. However, whether FTO inhibits tumor progression requires further investigation.
ALKBH5
ALKBH5 is an additional m6A demethylase. ALKBH5 is a 2-oxoglutarate- and ferrous iron-dependent nucleic acid oxygenase that catalyzes the demethylation of m6A on RNA (76). ALKHB5 downregulation is required to increase m6A levels, whereas its overexpression decreases the m6A modification on mRNA in human cell lines. ALKBH5 is upregulated under hypoxic conditions and serves a role in spermatogenesis (16,77).
ALKBH5 has been associated with the progression of human tumors. Nettersheim et al (78) discovered that ALKBH5 may serve a role in the development of testicular germ cell tumors, via using immunohistochemistry, mass spectrometry and ELISA. Zhang et al (77) revealed that ALKBH5 mediated m6A demethylation of NANOG mRNA (an embryonic stem cell transcription factor), which increased the expression of NANOG in breast cancer. The deletion of ALKBH5 increased the m6A methylation of NANOG mRNA, thus reducing the expression levels of NANOG, and significantly inhibited the development of lung metastasis of breast cancer. Subsequently, it was demonstrated that hypoxia-inducible factors inhibited the methylation and the ALKBH5-induced demethylation by modulating zinc finger protein 217 to promote the invasion and metastasis of breast cancer cells (79). ALKBH5 was also highly expressed in GBMs, while silencing ALKBH5 inhibited the proliferation of GBMs. ALKBH5 induced the demethylation of forkhead box protein M1 (FOXM1), which is a transcription factor, and stimulated cell proliferation, leading to an increase in FOXM1 expression (80).
ALKBH5 also regulates lncRNA demethylation to affect tumor progression. He et al (81) revealed that ALKBH5 expression was downregulated in pancreatic cancer cells and inhibited the cell motility via demethylating the lncRNA KCNK15-AS1, which may be used as a potential therapeutic target for pancreatic cancer. Zhang et al (82) demonstrated that ALKBH5 promoted GC invasion and metastasis via demethylating the lncRNA nuclear enriched abundant transcript 1.
Although ALKBH5 and FTO are both demethylases, they display opposing effects on tumor progression. ALKBH5 inhibits tumor progression, whereas FTO promotes tumorigenesis. The elucidation of the mechanisms underlying the differences in their effects requires further investigation.
4. m6A-binding proteins and cancer
YTH domain-containing proteins
The YTH domain- containing proteins include five members: YTHDF1, YTHDF2, YTHDF3, YTHDC1 and YTHDC2. Although YTHDF proteins share high similarity, their functions may differ. In the cytoplasm, YTHDF1 assists in the translation of m6A-modified mRNAs, whereas YTHDF2 expedites the recycling of m6A-modified transcripts (83). YTHDF3 facilitates the protein synthesis in synergy with YTHDF1 and promotes the recycling of methylated mRNA, mediated by YTHDF2 (84). All three YTHDF proteins function in a coordinated manner to influence biological processes, associated with m6A RNA methylation. YTHDC1 regulates mRNA splicing (85), whereas YTHDC2 binds to certain noncoding RNAs to perform its function (86). The binding sites of each protein are known; all the YTH domain-containing proteins, except YTHDC2, bind to m6A, which was identified using the individual-nucleotide-resolution UV crosslinking and immunoprecipitation method (87).
YTHDF1
Using bioinformatics analysis, Zhao et al (88) discovered that YTHDF1 expression was significantly upregulated in HCC and was positively correlated with the pathological stage. Lower YTHDF1 expression levels were associated with improved survival in patients with HCC.
Han et al (89) described a novel role of YTHDF1 in regulating the cytotoxic T lymphocyte (CTL) response to tumor antigens. It was discovered that YTHDF1-deficient mice exhibited an elevated antigen-specific CD8+ T cell anticancer response. Loss of YTHDF1 in classical dendritic cells (DCs) enhanced the cross-presentation of tumor antigens and the cross-priming of CD8+ T cells, which indicated YTHDF1 as a potential therapeutic target in anticancer immunotherapy. Kim et al (90) revealed that YTHDF1 regulated the cross-presentation of extracellular antigens to DCs. Via promoting the translation of the m6A-modified cathepsin mRNA, YTHDF1 enhanced the antigen degradation in the phagosome and limited the cross-presentation of neoantigens to DCs. Therefore, it may be possible to combine an anti-YTHDF1 therapy with the immune checkpoint blockade to increase the number of CTLs in the tumor.
A recent study revealed that YTHDF1 promoted the proliferation of CRC cells and protected the tumor cells from antitumor drugs (91). In addition, c-Myc, which is an oncogenic transcription factor, is an upstream gene of YTHDF1 and is associated with its expression. Knocking out YTHDF1 inhibited the proliferation of tumor cells, whereas knocking out c-Myc has been indicated to reduce the expression of YTHDF1, thus restricting the proliferation of CRC cells. Bai et al (92) obtained similar results when YTHDF1 was overexpressed in CRC. Silencing YTHDF1 significantly reduced Wnt/β-catenin signaling in CRC cells, thus highlighting a YTHDF1 as a potential therapeutic target for the treatment of CRC.
YTHDF2
YTHDF2 has been known to be involved in the development of AML for a considerable amount of time (93). In 2014, Wang et al (94) first reported that m6A was selectively recognized by the YTHDF2 protein to regulate mRNA degradation. It was demonstrated that reversible m6A deposition dynamically tuned the stability and localization of the target RNAs via YTHDF2. YTHDF2 may specifically interact with m6A-modified mRNAs via its C-terminal YTH domain and recruit the mRNAs to cytoplasmic foci to control the mRNA degradation via its N-terminal region.
Numerous studies verified that YTHDF2 may promote tumor progression. Chen et al (95) revealed that METTL3 promoted the progression of HCC cells via a YTHDF2-dependent posttranscriptional silencing of suppressor of cytokine signaling 2 (SOCS2). The downregulation of YTHDF2 reduced the proliferation and invasion of prostate cancer cells (96). Additionally, a negative correlation was indicated between YTHDF2 and miR-493-3p. Knocking out YTHDF2 increased the levels of m6A and inhibited the proliferation and invasion of DU-145 and PC3 cells. A dual-luciferase reporter assay confirmed that YTHDF2 was a direct target of miR-493-3p. The upregulation of miR-493-3p suppressed the expression of YTHDF2, thereby inhibiting the proliferation and invasion of prostate cancer cells. Paris et al (97) discovered that YTHDF2 was overexpressed in AML and was required for the disease initiation and propagation. It was revealed that targeting YTHDF2 extended the half-life of the m6A-modified transcripts to selectively compromise the initiation and propagation of AML without dysregulating physiological hematopoiesis.
YTHDF2 also acts as a tumor suppressor protein in addition to acting as an oncogene. Hypoxia resulted in a decrease of YTHDF2 expression in HCC (98). It was demonstrated that the overexpression of YTHDF2 inhibited the proliferation of HCC cells and activated the mitogen-activated protein kinase kinase- and ERK-mediated signaling. YTHDF2 directly targeted the m6A modification site in the 3′-UTR of the EGFR mRNA, leading to the degradation of the EGFR mRNA in HCC cells. In addition, hypoxia-induced phosphorylation of ERK was also inhibited by YTHDF2. Therefore, YTHDF2 may inhibit the ERK/MAPK signaling via reducing the stability of the EGFR mRNA in HCC cells to reduce the proliferation of HCC (98).
eIF3
Dysregulation of translation initiation results in abnormal gene expression, leading to altered cell growth and potentially cancer (99). Translation initiation is regulated by many eIFs and one of these, eIF3, is a critical factor controlling translation, with a molecular weight of 550–700 kDa in mammalian cells. eIF3 consists of ~13 subunits ranging in mass from 35–170 kDa, which associate with the 40S ribosome in an early stage of translation initiation and facilitate the interaction of mRNA with methionyl-tRNAi (99). Several subunits, known as eIF3a-eIF3m, have previously been revealed to serve vital roles in modulating the translation of specific mRNAs encoding proteins important for cell growth and oncogenesis (100). The abnormal expression of eIF3 may result in alterations in the translation efficiency of certain mRNAs, including those involved in angiogenesis, cellular growth and malignancy (101,102).
Meyer et al (103) discovered that mRNAs containing m6A in the 5′-UTR were translated in a cap-independent manner. eIF3 directly bound to m6A in the 5′-UTR and initiated an eIF4E-independent translation. Wang et al (83) revealed that eIF3 was recruited to the YTHDF1 5′-UTR and bound to m6A near a stop codon.
Previous studies demonstrated that eIF3 expression was upregulated in various types of cancer. Bachmann et al (104) discovered that the expression of eIF3 was increased in breast cancer compared with adjacent normal tissues. Subsequently, Dellas et al (105) revealed that eIF3 expression was upregulated in cervical cancer and it was higher during the earlier stages of cancer.
5. Conclusion
m6A modifications have been investigated for >40 years. However, this dynamic and reversible modification is still garnering increasing attention, particularly in relation to cancer research. m6A writers perform the modification, whereas m6A erasers reverse it. m6A readers are responsible for reading the modification to regulate the mRNA fate. Thus, m6A modifications serve as another level of regulation of RNAs in addition to histones and DNA modifications (106).
Aberrant m6A modifications are involved in the onset and progression of various types of cancer, including HCC, breast cancer, RCC, endometrial cancer, AML and lung cancer (45,46,65–72,74). Numerous genes are regulated by m6A modifications and participate in tumorigenesis, such as SOX2, HBXIP, MZF1, NANOG and SOCS2 (24,31,67,77,95). m6A modifications also participate in the modulation of lncRNAs at the post-transcriptional level (81,82). Furthermore, m6A may alter the local structure of mRNAs and lncRNAs to regulate the gene expression and RNA maturation (107,108). These findings may facilitate the discovery of novel therapeutic strategies.
Although considerable progress has been made in understanding the relevance of m6A modifications on RNA, several domains remain still unknown. Firstly, certain m6A enzymes have been investigated extensively in relation to tumorigenesis, whereas studies on the involvement of other m6A enzymes is limited, such as KIAA1429, RBM15, FMR1 and LRPPRC. Additional studies are required to elucidate the complex pathological processes of tumors. Second, whether m6A modifications interact with other RNA modifications, such as m5C and m1A, and whether these interactions affect tumorigenesis remains to be explored. Finally, additional clinical trials are required to determine the potential diagnostic and therapeutic functions of m6A modifications in human tumors.
Acknowledgements
Not applicable.
Funding
This study was funded by National Natural Science Foundation of China (grant. no. 81502208), Department of Education of Liaoning Province (grant. no. L2014310), Natural Science Foundation of Liaoning Province (grant. no. 2019-MS-360), Shenyang Science and Technology Bureau Plan Projects (grant. no. F16-206-9-09) and 345 Talent Project of Shengjing Hospital of China Medical University, to K Wang.
Availability of data and materials
Not applicable.
Authors' contributions
KW, JY and JC conceived of the review. KW, XF and XW wrote the manuscript. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, WireckI TK, de Crécy-Lagard V, Ross R, Limbach PA, Kotter A, et al. MODOMICS: A database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018;46(D1):D303–D307. doi: 10.1093/nar/gkx1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu ZX, Li LM, Sun HL, Liu SM. Link between m6A modification and cancers. Front Bioeng Biotechnol. 2018;6:89. doi: 10.3389/fbioe.2018.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci USA. 1974;71:3971–3975. doi: 10.1073/pnas.71.10.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon- Divon M, Ungar L, Osenberg L, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 5.Kane SE, Beemon K. Precise localization of m6A in Rous sarcoma virus RNA reveals clustering of methylation sites: Implications for RNA processing. Mol Cell Biol. 1985;5:2298–2306. doi: 10.1128/MCB.5.9.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li YL, Yu J, Song SH. Recent progresses in RNA N6-methyladenosine research. Yi Chuan. 2013;35:1340–1351. doi: 10.3724/SP.J.1005.2013.01340. (In Chinese) [DOI] [PubMed] [Google Scholar]
- 7.Bokar JA, Rath-Shambaugh ME, Ludwiczak R, Narayan P, Rottman F. Caracterization and partial purification of mRNA N6-adenosine methyltransferase from Hela cell nuclei. Internal mRNA methylation requires a multisubunit complex. J Biol Chem. 1994;269:17697–17704. [PubMed] [Google Scholar]
- 8.Wu X, Sang L, Gong Y. N6-methyladenine RNA modification and cancers. Am J Cancer Res. 2018;8:1957–1966. [PMC free article] [PubMed] [Google Scholar]
- 9.Alarcón CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015;519:482–485. doi: 10.1038/nature14281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bokar JA, Shambaugh ME, Polayes D, Matera AG, Rottman FM. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA. 1997;3:1233–1247. [PMC free article] [PubMed] [Google Scholar]
- 11.Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10:93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwartz S, Mumbach MR, Jovanovic M, Wang T, Maciag K, Bushkin GG, Mertins P, Ter-Ovanesyan D, Habib N, Cacchiarelli D, et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep. 2014;8:284–296. doi: 10.1016/j.celrep.2014.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Visvanathan A, Somasundaram K. mRNA traffic control reviewed: N6-methyladenosine (M6A) takes the driver's seat. Bioessays. 2018;40 doi: 10.1002/bies.201700093. [DOI] [PubMed] [Google Scholar]
- 14.Pendleton KE, Chen B, Liu K, Hunter OV, Xie Y, Tu BP, Conrad NK. The U6 snRNA m6A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell. 2017;169:824–835.e14. doi: 10.1016/j.cell.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, He C. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vagbo CB, Shi Y, Wang WL, Song SH, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li A, Chen YS, Ping XL, Yang X, Xiao W, Yang Y, Sun HY, Zhu Q, Baidya P, Wang X, et al. Cytoplasmic m6A reader YTHDF3 promotes mRNA translation. Cell Res. 2017;27:444–447. doi: 10.1038/cr.2017.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li T, Hu PS, Zuo Z, Lin JF, Li X, Wu QN, Chen ZH, Zeng ZL, Wang F, Zheng J, et al. METTL3 facilitates tumor progression via an m6A-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol Cancer. 2019;18:112. doi: 10.1186/s12943-019-1038-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Q, Chen C, Ding QQ, Zhao Y, Wang ZD, Chen JJ, Jiang ZR, Zhang Y, Xu GF, Zhang JJ, et al. METTL3-mediated m6A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut. 2019 Oct 3; doi: 10.1136/gutjnl-2019-319639. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 20.Yue B, Song C, Yang L, Cui R, Cheng X, Zhang Z, Zhao G. METTL3-mediated N6-methyladenosine modification is critical for epithelial-mesenchymal transition and metastasis of gastric cancer. Mol Cancer. 2019;18:142. doi: 10.1186/s12943-019-1065-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leach RA, Tuck MT. Expression of the mRNA (N6-adenosine)-methyltransferase S-adenosyl-L-methionine binding subunit mRNA in cultured cells. Int J Biochem Cell Biol. 2001;33:984–999. doi: 10.1016/S1357-2725(01)00071-1. [DOI] [PubMed] [Google Scholar]
- 22.Chen T, Hao YJ, Zhang Y, Li MM, Wang M, Han W, Wu Y, Lv Y, Hao J, Wang L, et al. m(6)A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem Cell. 2015;16:289–301. doi: 10.1016/j.stem.2015.01.016. [DOI] [PubMed] [Google Scholar]
- 23.Lin S, Choe J, Du P, Triboulet R, Gregory RI. The m(6)A Methyltransferase METTL3 promotes translation in human cancer cells. Mol Cell. 2016;62:335–345. doi: 10.1016/j.molcel.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Visvanathan A, Patil V, Arora A, Hegde AS, Arivazhagan A, Santosh V, Somasundaram K. Essential role of METTL3- mediated m6A modification in glioma stem-like cells maintenance and radioresistance. Oncogene. 2018;37:522–533. doi: 10.1038/onc.2017.351. [DOI] [PubMed] [Google Scholar]
- 25.Barbieri I, Tzelepis K, Pandolfini L, Shi J, Millán-Zambrano G, Robson SC, Aspris D, Migliori V, Bannister AJ, Han N, et al. Promoter-bound METTL3 maintains myeloid leukaemia by m6A-dependent translation control. Nature. 2017;522:126–131. doi: 10.1038/nature24678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taketo K, Konno M, Asai A, Koseki J, Toratani M, Satoh T, Doki Y, Mori M, Ishii H, Ogawa K. The epitranscriptome m6A writer METTL3 promotes chemo- and radioresistance in pancreatic cancer cells. Int J Oncol. 2018;52:621–629. doi: 10.3892/ijo.2017.4219. [DOI] [PubMed] [Google Scholar]
- 27.Choe J, Lin S, Zhang W, Liu Q, Wang L, Ramirez-Moya J, Du P, Kim W, Tang S, Sliz P, et al. mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis. Nature. 2018;561:556–560. doi: 10.1038/s41586-018-0538-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu T, Yang S, Sui J, Xu SY, Cheng YP, Shen B, Zhang Y, Zhang XM, Yin LH, Pu YP, Liang GY. Dysregulated N6-methyladenosine methylation writer METTL3 contributes to the proliferation and migration of gastric cancer. J Cell Physiol. 2020;235:548–562. doi: 10.1002/jcp.28994. [DOI] [PubMed] [Google Scholar]
- 29.Du M, Zhang Y, Mao Y, Mou J, Zhao J, Xue Q, Wang D, Huang J, Gao S, Gao Y. MiR-33a suppresses proliferation of NSCLC cells via targeting METTL3 mRNA. Biochem Biophys Res Commun. 2017;482:582–589. doi: 10.1016/j.bbrc.2016.11.077. [DOI] [PubMed] [Google Scholar]
- 30.Jin D, Guo J, Wu Y, Du J, Yang L, Wang X, Di W, Hu B, An J, Kong L, et al. m6A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1-miR-1914-3p-YAP axis to induce NSCLC drug resistance and metastasis. J Hematol Oncol. 2019;12:135. doi: 10.1186/s13045-019-0830-6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Cai X, Wang X, Cao C, Gao Y, Zhang S, Yang Z, Liu Y, Zhang X, Zhang W, Ye L. HBXIP-elevated methyltransferase METTL3 promotes the progression of breast cancer via inhibiting tumor suppressor let-7 g. Cancer Lett. 2018;415:11–19. doi: 10.1016/j.canlet.2017.11.018. [DOI] [PubMed] [Google Scholar]
- 32.Wei W, Huo B, Shi X. miR-600 inhibits lung cancer via downregulating the expression of METTL3. Cancer Manag Res. 2019;11:1177–1187. doi: 10.2147/CMAR.S181058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han J, Wang JZ, Yang X, Yu H, Zhou R, Lu HC, Yuan WB, Lu JC, Zhou ZJ, Lu Q, et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol Cancer. 2019;18:110. doi: 10.1186/s12943-019-1036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng M, Sheng L, Gao Q, Xiong Q, Zhang H, Wu M, Liang Y, Zhu F, Zhang Y, Zhang X, et al. The m6A methyltransferase METTL3 promotes bladder cancer progression via AFF4/NF-κB/MYC signaling network. Oncogene. 2019;38:3667–3680. doi: 10.1038/s41388-019-0683-z. [DOI] [PubMed] [Google Scholar]
- 35.Peng W, Li J, Chen R, Gu Q, Yang P, Qian W, Ji D, Wang Q, Zhang Z, Tang J, Sun Y. Upregulated METTL3 promotes metastasis of colorectal cancer via miR-1246/SPRED2/MAPK signaling pathway. J Exp Clin Cancer Res. 2019;38:393. doi: 10.1186/s13046-019-1408-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li X, Tang J, Huang W, Wang F, Li P, Qin C, Qin Z, Zou Q, WeI J, Hua L, et al. The M6A methyltransferase METTL3: Acting as a tumor suppressor in renal cell carcinoma. Oncotarget. 2017;8:96103–96116. doi: 10.18632/oncotarget.21726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol. 2014;16:191–198. doi: 10.1038/ncb2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tong J, Flavell RA, Li HB. RNA m6A modification and its function in diseases. Front Med. 2018;12:481–489. doi: 10.1007/s11684-018-0654-8. [DOI] [PubMed] [Google Scholar]
- 39.Śledź P, Jinek M. Structural insights into the molecular mechanism of the m(6)A writer complex. Elife. 2016;5:e18434. doi: 10.7554/eLife.18434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang P, Doxtader KA, Nam Y. Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol Cell. 2016;63:306–317. doi: 10.1016/j.molcel.2016.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang X, Feng J, Xue Y, Guan Z, Zhang D, Liu Z, Gong Z, Wang Q, Huang J, Tang C, et al. Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature. 2016;534:575–578. doi: 10.1038/nature18298. [DOI] [PubMed] [Google Scholar]
- 42.Ma JZ, Yang F, Zhou CC, Liu F, Yuan JH, Wang F, Wang TT, Xu QG, Zhou WP, Sun SH. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N6 methyladenosine-dependent primary MicroRNA processing. Hepatology. 2017;65:529–543. doi: 10.1002/hep.28885. [DOI] [PubMed] [Google Scholar]
- 43.Manning BD, Toker A. AKT/PKB signaling: Navigating the network. Cell. 2017;169:381–405. doi: 10.1016/j.cell.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 45.Liu J, Eckert MA, Harada BT, Liu SM, Lu Z, Yu K, Tienda SM, Chryplewicz A, Zhu AC, Yang Y, et al. m6A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat Cell Biol. 2018;20:1074–1083. doi: 10.1038/s41556-018-0174-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gong D, Zhang J, Chen Y, Xu Y, Ma J, Hu G, Huang Y, Zheng J, ZhaI W, Xue W. The m6A-suppressed P2RX6 activation promotes renal cancer cells migration and invasion through ATP-induced Ca2+ Influx Modulating ERK1/2 phosphorylation and MMP9 signaling pathway. J Exp Clin Cancer Res. 2019;38:233. doi: 10.1186/s13046-019-1223-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen X, Xu M, Xu X, Zeng K, Liu X, Sun L, Pan B, He B, Pan Y, Sun H, et al. METTL14 suppresses crc progression via regulating N6-methyladenosine-dependent primary miR-375 processing. Mol Ther. 2020;28:599–612. doi: 10.1016/j.ymthe.2019.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 48.Gu C, Wang Z, Zhou N, Li G, Kou Y, Luo Y, Wang Y, Yang J, Tian F. Mettl14 inhibits bladder TIC self-renewal and bladder tumorigenesis through N6-methyladenosine of Notch1. Mol Cancer. 2019;18:168. doi: 10.1186/s12943-019-1084-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Little NA, Hastie ND, Davies RC. Identification of WTAP, a novel Wilms' tumour 1-associating protein. Hum Mol Genet. 2000;9:2231–2239. doi: 10.1093/oxfordjournals.hmg.a018914. [DOI] [PubMed] [Google Scholar]
- 50.Zhong S, Li H, Bodi Z, Button J, Vespa L, Herzog M, Fray RG. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell. 2008;20:1278–1288. doi: 10.1105/tpc.108.058883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177–189. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schöller E, Weichmann F, Treiber T, Ringle S, Treiber N, Flatley A, Feederle R, Bruckmann A, Meister G. Interactions, localization, and phosphorylation of the m6A generating METTL3-METTL14-WTAP complex. RNA. 2018;24:499–512. doi: 10.1261/rna.064063.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen Y, Peng C, Chen J, Chen D, Yang B, He B, Hu W, Zhang Y, Liu H, Dai L, et al. WTAP facilitates progression of hepatocellular carcinoma via m6A-HuR-dependent epigenetic silencing of ETS1. Mol Cancer. 2019;18:127. doi: 10.1186/s12943-019-1053-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ortega A, Niksic M, Bachi A, Wilm M, Sánchez L, Hastie N, Valcárcel J. Biochemical function of female-lethal(2)D/Wilms' tumor suppressor-1-associated proteins in alternative pre-mRNA splicing. J Biol Chem. 2003;278:3040–3047. doi: 10.1074/jbc.M210737200. [DOI] [PubMed] [Google Scholar]
- 55.Guo J, Tang HW, Li J, Perrimon N, Yan D. Xio is a component of the Drosophila sex determination pathway and RNA N6-methyladenosine methyltransferase complex. Proc Natl Acad Sci USA. 2018;115:3674–3679. doi: 10.1073/pnas.1720945115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Robinson M, Shah P, Cui YH, He YY. The role of dynamic m6A RNA methylation in photobiology. Photochem Photobiol. 2019;95:95–104. doi: 10.1111/php.12930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cheng X, Li M, Rao X, Zhang W, Li XP, Wang L, Huang G. KIAA1429 regulates the migration and invasion of hepatocellular carcinoma by altering m6A modification of ID2 mRNA. Onco Targets Ther. 2019;12:3421–3428. doi: 10.2147/OTT.S180954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qian JY, Gao J, Sun X, Cao MD, Shi L, Xia TS, Zhou WB, Wang S, Ding Q, Wei JF. KIAA1429 acts as an oncogenic factor in breast cancer by regulating CDK1 in an N6-methyladenosine-independent Manner. Oncogene. 2019;38:6123–6141. doi: 10.1038/s41388-019-0861-z. [DOI] [PubMed] [Google Scholar]
- 59.Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, Perry JRB, Elliott KS, Lango H, Rayner NW, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jia G, Yang CG, Yang S, Jian X, Yi C, Zhou Z, He C. Oxidative demethylation of 3-methylthymine and 3-methyluracil in single-stranded DNA and RNA by mouse and human FTO. FEBS Lett. 2008;582:3313–3319. doi: 10.1016/j.febslet.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mauer J, Luo X, Blanjoie A, Jiao X, Grozhik AV, Patil DP, Linder B, Pickering BF, Vasseur JJ, Chen Q, et al. Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature. 2017;541:371–375. doi: 10.1038/nature21022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, Jaffrey SR. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods. 2015;12:767–772. doi: 10.1038/nmeth.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wei J, Liu F, Lu Z, Fei Q, Ai Y, He PC, Shi H, Cui X, Su R, Klungland A, et al. Differential m6A, m6Am, and m1A demethylation mediated by FTO in the cell nucleus and cytoplasm. Mol Cell. 2018;71:973–985 e5. doi: 10.1016/j.molcel.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu WC, Feng J, Jiang D, Zhou X, Jiang Q, Cai M, Wang X, Shan T, Wang Y. AMPK regulates lipid accumulation in skeletal muscle cells through FTO-dependent demethylation of N6-methyladenosine. Sci Rep. 2017;7:41606. doi: 10.1038/srep41606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Niu Y, Lin Z, Wan A, Chen H, Liang H, Sun L, Wang Y, Li X, Xiong XF, Wei B, et al. RNA N6-methyladenosine demethylase FTO promotes breast tumor progression through inhibiting BNIP3. Mol Cancer. 2019;18:46. doi: 10.1186/s12943-019-1004-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li J, Han Y, Zhang H, Qian Z, Jia W, Gao Y, Zheng H, Li B. The m6A demethylase FTO promotes the growth of lung cancer cells by regulating the m6A level of USP7 mRNA. Biochem Biophys Res Commun. 2019;512:479–485. doi: 10.1016/j.bbrc.2019.03.093. [DOI] [PubMed] [Google Scholar]
- 67.Liu J, Ren D, Du Z, Wang H, Zhang H, Jin Y. m6A demethylase FTO facilitates tumor progression in lung squamous cell carcinoma by regulating MZF1 expression. Biochem Biophys Res Commun. 2018;502:456–464. doi: 10.1016/j.bbrc.2018.05.175. [DOI] [PubMed] [Google Scholar]
- 68.Xu D, Shao W, Jiang Y, Wang X, Liu Y, Liu X. FTO expression is associated with the occurrence of gastric cancer and prognosis. Oncol Rep. 2017;38:2285–2292. doi: 10.3892/or.2017.5904. [DOI] [PubMed] [Google Scholar]
- 69.Li Y, Zheng D, Wang F, Xu Y, Yu H, Zhang H. Expression of demethylase gene, FTO and ALKBH1, is associated with prognosis of gastric cancer. Dig Dis Sci. 2019;64:1503–1513. doi: 10.1007/s10620-018-5452-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li Z, Weng H, Su R, Weng X, Zuo Z, Li C, Huang H, Nachtergaele S, Dong L, Hu C, et al. FTO plays an oncogenic role in acute myeloid leukemia as a N6-methyladenosine RNA demethylase. Cancer Cell. 2017;31:127–141. doi: 10.1016/j.ccell.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Su R, Dong L, Li C, Nachtergaele S, Wunderlich M, Qing Y, Deng X, Wang Y, Weng X, Hu C, et al. R-2HG exhibits anti-tumor activity by targeting FTO/m6A/MYC/CEBPA signaling. Cell. 2018;172:90–105.e23. doi: 10.1016/j.cell.2017.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang Y, Su R, Sheng Y, Dong L, Dong Z, Xu H, Ni T, Zhang ZS, Zhang T, Li C, et al. Small-molecule targeting of oncogenic Fto demethylase in acute myeloid leukemia. Cancer Cell. 2019;35:677–691.e10. doi: 10.1016/j.ccell.2019.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhou S, Bai ZL, Xia D, Zhao ZJ, Zhao R, Wang YY, Zhe H. FTO regulates the chemo-radiotherapy resistance of cervical squamous cell carcinoma (CSCC) by targeting β-catenin through mRNA demethylation. Mol Carcinog. 2018;57:590–597. doi: 10.1002/mc.22782. [DOI] [PubMed] [Google Scholar]
- 74.Li J, Zhu L, Shi Y, Liu J, Lin L, Chen X. m6A demethylase FTO promotes hepatocellular carcinoma tumorigenesis via mediating PKM2 demethylation. Am J Transl Res. 2019;11:6084–6092. [PMC free article] [PubMed] [Google Scholar]
- 75.Yang S, Wei J, Cui YH, Park G, Shah P, Deng Y, Aplin AE, Lu Z, Hwang S, He C, He YY. m6A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat Commun. 2019;10:2782. doi: 10.1038/s41467-019-10669-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Aik W, Scotti JS, Choi H, Gong L, Demetriades M, Schofield CJ, McDonough MA. Structure of human RNA N6-methyladenine demethylase ALKBH5 provides insights into its mechanisms of nucleic acid recognition and demethylation. Nucleic Acids Res. 2014;42:4741–4754. doi: 10.1093/nar/gku085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang C, Samanta D, Lu H, Bullen JW, Zhang H, Chen I, He X, Semenza GL. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m6A-demethylation of NANOG mRNA. Proc Natl Acad Sci USA. 2016;113:E2047–E2056. doi: 10.1073/pnas.1602883113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nettersheim D, Berger D, Jostes S, Kristiansen G, Lochnit G, Schorle H. N6-Methyladenosine detected in RNA of testicular germ cell tumors is controlled by METTL3, ALKBH5, YTHDC1/F1/F2, and HNRNPC as writers, erasers, and readers. Andrology. 2019;7:498–506. doi: 10.1111/andr.12612. [DOI] [PubMed] [Google Scholar]
- 79.Zhang C, Zhi WI, Lu H, Samanta D, Chen I, Gabrielson E, Semenza GL. Hypoxia-inducible factors regulate pluripotency factor expression by ZNF217- and ALKBH5-mediated modulation of RNA methylation in breast cancer cells. Oncotarget. 2016;7:64527–64542. doi: 10.18632/oncotarget.11743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z, Chen Y, Sulman EP, Xie K, Bögler O, et al. m6A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017;31:591–606 e6. doi: 10.1016/j.ccell.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.He Y, Hu H, Wang Y, Yuan H, Lu Z, Wu P, Liu D, Tian L, Yin J, Jiang K, Miao Y. ALKBH5 inhibits pancreatic cancer motility by decreasing long non-coding RNA KCNK15-AS1 methylation. Cell Physiol Biochem. 2018;48:838–846. doi: 10.1159/000491915. [DOI] [PubMed] [Google Scholar]
- 82.Zhang J, Guo S, Piao HY, Wang Y, Wu Y, Meng XY, Yang D, Zheng ZC, Zhao Y. ALKBH5 promotes invasion and metastasis of gastric cancer by decreasing methylation of the lncRNA NEAT1. J Physiol Biochem. 2019;75:379–389. doi: 10.1007/s13105-019-00690-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161:1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, Liu C, He C. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 2017;27:315–328. doi: 10.1038/cr.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xu C, Wang X, Liu K, Roundtree IA, Tempel W, Li Y, Lu Z, He C, Min J. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat Chem Biol. 2014;10:927–929. doi: 10.1038/nchembio.1654. [DOI] [PubMed] [Google Scholar]
- 86.Tanabe A, Tanikawa K, Tsunetomi M, Takai K, Ikeda H, Konno J, Torigoe T, Maeda H, Kutomi G, Okita K, et al. RNA helicase YTHDC2 promotes cancer metastasis via the enhancement of the efficiency by which HIF-1α mRNA is translated. Cancer Lett. 2016;376:34–42. doi: 10.1016/j.canlet.2016.02.022. [DOI] [PubMed] [Google Scholar]
- 87.Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M, Jaffrey SR. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537:369–373. doi: 10.1038/nature19342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhao X, Chen Y, Mao Q, Jiang X, Jiang W, Chen J, Xu W, Zhong L, Sun X. Overexpression of YTHDF1 is associated with poor prognosis in patients with hepatocellular carcinoma. Cancer Biomark. 2018;21:859–868. doi: 10.3233/CBM-170791. [DOI] [PubMed] [Google Scholar]
- 89.Han D, Liu J, Chen C, Dong L, Liu Y, Chang R, Huang X, Liu Y, Wang J, Dougherty U, et al. Anti-tumour immunity controlled through mRNA m6A methylation and YTHDF1 in dendritic cells. Nature. 2019;566:270–274. doi: 10.1038/s41586-019-0916-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kim DJ, Iwasaki A. YTHDF1 control of dendritic cell cross-priming as a possible target of cancer immunotherapy. Biochemistry. 2019;58:1945–1946. doi: 10.1021/acs.biochem.9b00200. [DOI] [PubMed] [Google Scholar]
- 91.Nishizawa Y, Konno M, Asai A, Koseki J, Kawamoto K, Miyoshi N, Takahashi H, Nishida N, Haraguchi N, Sakai D, et al. Oncogene c-Myc promotes epitranscriptome m6A reader YTHDF1 expression in colorectal cancer. Oncotarget. 2017;9:7476–7486. doi: 10.18632/oncotarget.23554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bai Y, Yang C, Wu R, Huang L, Song S, Li W, Yan P, Lin C, Li D, Zhang Y. YTHDF1 regulates tumorigenicity and cancer stem cell-like activity in human colorectal carcinoma. Front Oncol. 2019;9:332. doi: 10.3389/fonc.2019.00332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nguyen TT, Ma LN, Slovak ML, Bangs CD, Cherry AM, Arber DA. Identification of novel Runx1 (AML1) translocation partner genes SH3D19, YTHDf2, and ZNF687 in acute myeloid leukemia. Genes Chromosomes Cancer. 2006;45:918–932. doi: 10.1002/gcc.20355. [DOI] [PubMed] [Google Scholar]
- 94.Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chen M, Wei L, Law CT, Tsang FH, Shen J, Cheng CL, Tsang LH, Ho DW, Chiu DK, Lee JM, et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67:2254–2270. doi: 10.1002/hep.29683. [DOI] [PubMed] [Google Scholar]
- 96.Li J, Meng S, Xu M, Wang S, He L, Xu X, Wang X, Xie L. Downregulation of N6-methyladenosine binding YTHDF2 protein mediated by miR-493-3p suppresses prostate cancer by elevating N6-methyladenosine levels. Oncotarget. 2017;9:3752–3764. doi: 10.18632/oncotarget.23365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Paris J, Morgan M, Campos J, Spencer GJ, Shmakova A, Ivanova I, Mapperley C, Lawson H, Wotherspoon DA, Sepulveda C, et al. Targeting the RNA m6A reader YTHDF2 selectively compromises cancer stem cells in acute myeloid leukemia. Cell Stem Cell. 2019;25:137–148.e6. doi: 10.1016/j.stem.2019.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhong L, Liao D, Zhang M, Zeng C, Li X, Zhang R, Ma H, Kang T. YTHDF2 suppresses cell proliferation and growth via destabilizing the EGFR mRNA in hepatocellular carcinoma. Cancer Lett. 2019;442:252–261. doi: 10.1016/j.canlet.2018.11.006. [DOI] [PubMed] [Google Scholar]
- 99.Asano K, Kinzy TG, Merrick WC, Hershey JW. Conservation and diversity of eukaryotic translation initiation factor eIF3. J Biol Chem. 1997;272:1101–1109. doi: 10.1074/jbc.272.2.1101. [DOI] [PubMed] [Google Scholar]
- 100.Dong Z, Zhang JT. Initiation factor eIF3 and regulation of mRNA translation, cell growth, and cancer. Crit Rev Oncol Hematol. 2006;59:169–180. doi: 10.1016/j.critrevonc.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 101.De Benedetti A, Harris AL. eIF4E expression in tumors: Its possible role in progression of malignancies. Int J Biochem Cell Biol. 1999;31:59–72. doi: 10.1016/S1357-2725(98)00132-0. [DOI] [PubMed] [Google Scholar]
- 102.De Benedetti A, Graff JR. eIF-4E expression and its role in malignancies and metastases. Oncogene. 2004;23:3189–3199. doi: 10.1038/sj.onc.1207545. [DOI] [PubMed] [Google Scholar]
- 103.Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, Pestova TV, Qian SB, Jaffrey SR. 5′UTR m(6)A promotes cap-independent translation. Cell. 2015;163:999–1010. doi: 10.1016/j.cell.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bachmann F, Banziger R, Burger MM. Cloning of a novel protein overexpressed in human mammary carcinoma. Cancer Res. 1997;57:988–994. [PubMed] [Google Scholar]
- 105.Dellas A, Torhorst J, Bachmann F, Bänziger R, Schultheiss E, Burger MM. Expression of p150 in cervical neoplasia and its potential value in predicting survival. Cancer. 1998;83:1376–1383. doi: 10.1002/(SICI)1097-0142(19981001)83:7<1376::AID-CNCR15>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 106.Wang S, Sun C, Li J, Zhang E, Ma Z, Xu W, Li H, Qiu M, Xu Y, Xia W, et al. Roles of RNA methylation by means of N6-methyladenosine (m6A) in human cancers. Cancer Lett. 2017;408:112–120. doi: 10.1016/j.canlet.2017.08.030. [DOI] [PubMed] [Google Scholar]
- 107.Yang D, Qiao J, Wang G, Lan Y, Li G, Guo X, Xi J, Ye D, Zhu S, Chen W, et al. N6-methyladenosine modification of lincRNA 1281 is critically required for mESC differentiation potential. Nucleic Acids Res. 2018;46:3906–3920. doi: 10.1093/nar/gky130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560–564. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.