Abstract
With the development of new biochemical and computational methods, circular RNAs (circRNAs) have been identified as microRNA sponges. circRNAs are associated with many diseases, particularly cancer. The present study aimed to investigate the expression profile of circRNAs in irradiated A549 lung cancer cells using high-throughput sequencing. Bioinformatics analyses were used to examine the potential functions of circRNAs. RNA sequencing data demonstrated that 1,875 circRNA targets were differentially expressed in A549 cells in response to irradiation. A total of 30 circRNAs were upregulated and 37 circRNAs were downregulated significantly in irradiation-treated A549 cells (fold change ≥2.0; P<0.05). The top 5 upregulated and downregulated circRNAs were successfully validated by reverse transcription-quantitative PCR. In addition, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis suggested that differentially expressed circRNAs might be pivotal in biological irradiation responses to irradiation. circRNA-microRNA co-expression networks highlighted the biological significance of circRNA_0002174 and circRNA_0036627, which require further study. In conclusion, the present study is, to the best of the authors' knowledge, the first to describe the differentially expressed profile of circRNAs in response to irradiation in A549 cells. These results provide a new perspective to elucidate insight into the molecular mechanisms by which A549 cells respond to radiation, and a basis for a more in-depth analysis of the potential application of circRNAs in the treatment of lung cancer therapy.
Keywords: circular RNA, high-throughput sequencing, irradiation, bioinformatics, lung cancer
Introduction
According to Globocan, a total of 18.1 million new cancer cases and 9.6 million cancer-related deaths occurred in 2018 (1). Both in men and women, lung cancer is the most commonly diagnosed cancer (11.6% of total cases) and the leading cause of cancer-related death (18.4% of all cancer-related deaths) (1). It is estimated that ~80% of lung cancer cases are non-small cell lung cancer (NSCLC), and ~50% of NSCLC are adenocarcinomas (2). In recent years, radiotherapy has been a critical therapeutic approach for lung cancer. However, both recurrence and metastasis after radiotherapy remain life-threatening complications of cancer and are primarily responsible for cancer patient mortality (3). Therefore, elucidating the mechanism of radiation damage is crucial for radiotherapy of lung cancer.
Circular RNA (circRNA) is a broad category of non-coding RNA characterized by a covalently closed ring framework without 5′-3′ polarity (4–7). Unlike other forms of RNA, circRNA not only exists in organisms stably but also has resistance to RNase R treatment (8–10). Although Sanger et al (11) first identified circRNA in viruses in 1976, its functions in various diseases have only been recently reported. With the development of new biochemical and computational methods, circRNA has attracted increasing research attention (5,12). Previous studies demonstrated that circRNAs regulate mRNA transcription via two main mechanisms: Competition with mRNAs leading to inhibition of host gene expression (13), and elimination of the inhibitory effects of microRNA (miRNA/miR) on target genes by acting as miRNA sponges (5,14).
Several previous studies suggested that circRNAs were involved in various human diseases, such as lupus nephritis (15), cardiovascular diseases (16) and systemic neural diseases (17). The role of circRNAs in human cancer was also previously examined. For instance, hsa_circ_002059 could be a potential novel and stable biomarker for the diagnosis of gastric carcinoma (18). circRNA-000911 might regulate the carcinogenesis of breast cancer (19). Furthermore, circRNAs could promote cell proliferation, migration, invasion and metastasis in NSCLC (20–22). Moreover, previous studies have suggested that circRNA may play an importantly regulatory role in radiation-induced esophageal injury (23). circRNA was significantly differentially expressed in radioresistant esophageal cancer cells (24) and radioresistant HeLa cells (25). circRNA_014511 could affect the radiosensitivity of bone marrow mesenchymal stem cells by binding to miR-29b-2-5p (26). However, the circRNA expression profiles of NSCLC cells in response to radiation treatment remain unclear.
The present study, investigated for the first time to the best of the authors' knowledge, the regulation of circRNAs in irradiated and non-irradiated A549 cells using high-throughput RNA sequencing (RNAseq). The expression levels of dysregulated circRNAs were also verified by reverse transcription-quantitative PCR (RT-qPCR), and the application and function of dysregulated circRNAs in disease diagnosis were assessed. Overall, the present study suggested that circRNAs could play an important role in lung cancer radiotherapy.
Materials and methods
Cell culture
The human NSCLC cell lines A549 and NCI-H1299 were purchased from The American Type Culture Collection and were authenticated by STR DNA profiling analysis. NSCLC cells were cultured in RPMI-1640 medium (GE Healthcare) containing 10% FBS (Thermo Fisher Scientific, Inc.) and 5 mg/ml penicillin/streptomycin (Thermo Fisher Scientific, Inc.) at 37°C with 5% CO2. NSCLC cells were digested and passaged every 3 days by using EDTA-containing trypsin (Thermo Fisher Scientific, Inc.).
Irradiation
A549 cells were plated at 1×104 cells/well in 96-well plates. After incubation for 24 h, A549 cells were exposed to various doses (0, 2, 4 and 8 Gy) of irradiation using the X-RAD 160–225 instrument (42 cm; 225 kV/s; 12.4 mA; 2.0 Gy/min; filter, 2 mm aluminum; Precision X-Ray, Inc.) (27,28). Radiosensitivity was assessed after treatment with the irradiation for 24 h or 48 h.
Cell Counting Kit-8 (CCK-8) assay
Following irradiation, cell proliferation was measured by CCK-8 according to the manufacturer's instructions. The CCK-8 kit was purchased from Beyotime Biotechnology Co., Ltd. The 450 nm absorbance (630 nm reference) was determined for each well, using a Multiskan™ FC Microplate Photometer (Thermo Fisher Scientific, Inc.). Each experiment was repeated three times.
Total RNA extraction
Total RNA was extracted using the Total RNA extraction kit (Beijing Solarbio Science & Technology Co., Ltd.), according to the manufacturer's instructions. RNA concentration and purity were determined using a NanoDrop™ 2000 spectrophotometer (Thermo Fisher Scientific, Inc.). RNA integrity was assessed by 1% formaldehyde denaturing gel electrophoresis. The total RNA of the control group cells (0 Gy/48 h) and the irradiation group cells (2 Gy/48 h) were extracted separately for RNAseq, three samples per group.
circRNA sequencing analysis
RNA libraries were constructed by CloudSeq Pte Ltd. circRNA sequencing analysis was also performed by CloudSeq Pte Ltd according to the manufacturer's instructions (14). Cluster and TreeView were used to conduct hierarchical clustering analysis using R (v3.6.3) (29) in order to assess differential expression of circRNAs in irradiated and non-irradiated A549 cells. These circRNAs that can be found in the circbase (http://www.circbase.org/) are known circRNAs, and those that cannot be found are novel circRNAs. DCC (detect circRNAs from chimeric reads, v0.4.4) software was used to detect circRNAs from chimeric reads. The functional roles of target genes in biological process (BP), cellular component (CC) and molecular function (MF) were examined through Gene Ontology (GO) (current release 2020-03-24, http://geneontology.org) analysis. The involvement of genes encoding circRNAs in biological pathways was assessed by the Kyoto Encyclopedia of Genes and Genomes (KEGG) using the Database for Annotation, Visualization and Integrated Discovery v6.8 (https://david.ncifcrf.gov/).
RT-qPCR
circRNA expression profiles obtained from high-throughput sequencing were verified by RT-qPCR. Total RNA was extracted from cells using the Total RNA extraction kit (Beijing Solarbio Science & Technology Co., Ltd.). cDNA was synthesized using the iScript cDNA synthesis kit (Bio-Rad Laboratories, Inc.) by RT of 2 µl total RNA. The CFX96 Real-time System (Bio-Rad Laboratories, Inc.) was used to perform RT-qPCR with SYBR Green Supermix (Bio-Rad Laboratories, Inc.). The extraction of total RNA, the synthesis of cDNA, and the steps of RT-qPCR were all performed according to the manufacturers' instructions (27). The thermocycling conditions were: Denaturation at 95°C for 30 sec, annealing at 60°C for 30 sec, extension at 72°C for 60 sec, repeated for 30 cycles. Finally, the product was kept intact at 72°C for 5 min to complete the extension and stored at 4°C. The primer sequences used in the present study are presented in Table SI. The control gene was β-actin and the 2−ΔΔCq method was used to calculate the mRNA expression relative to the control group (30).
miRNA prediction and circRNA-miRNA interaction network analysis
TargetScan (v7.0) and miRanda (v3.3a) were used to identify potential miRNA targets of circRNAs (14). The Cloudseq software was used to analyze interactions between circRNA and miRNA; each circRNA has multiple miRNA binding sites. The construction of circRNA-miRNA networks was performed using Cytoscape (v3.1.0) software.
Statistical analysis
Data analysis was performed using GraphPad Prism 7.0 (GraphPad Software, Inc.). Results are presented as the mean ± standard deviation from at least three independent experiments. A two-tailed Student's t-test was used to analyze the differences between the two groups. P<0.05 was considered to indicate a statistically significant difference.
Results
Overview of circRNA expression profiles
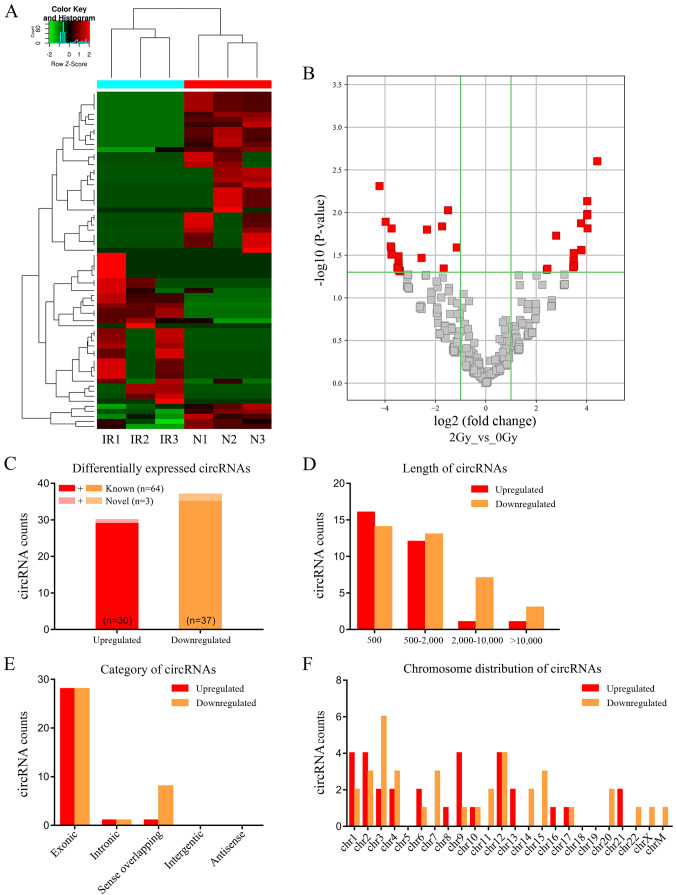
Based on the results of the CCK-8 assay (Fig. S1), after a 2 Gy irradiation treatment for 48 hours, the cell viability of A549 was significantly reduced (P<0.05). The total RNA of the control group A549 cells (0 Gy/48 h) and the irradiation group A549 cells (2 Gy/48 h) were extracted separately for RNAseq, three samples per group. The expression profiles of circRNA were first analyzed in the irradiated and non-irradiated A549 cells. A total of 1,875 circRNA targets was detected by RNAseq. A hierarchical clustering heatmap (Fig. 1A) and volcano plots (Fig. 1B) suggested that the expression levels of circRNA were clustered and distinguishable. A total of 67 significantly differentially expressed circRNAs in irradiated and non-irradiated A549 cells (fold change ≥2.0; P<0.05) were identified. Of these, 30 circRNAs were significantly upregulated (Table I) and 37 were downregulated (Table II) >2-fold in irradiated A549 cells, compared with non-irradiated A549 cells.
Figure 1.
Characteristics of differentially expressed circRNAs were detected by high-throughput sequencing of IR and N A549 cells. (A) Expression heatmap of distinguishable and clustered circRNAs in IR and N cells. (B) Volcano plots of differentially expressed circRNAs in IR and N cells. Vertical line indicates a 2.0-fold (log2 scale) change; the horizontal line represents the P-value of 0.05 (−log10 scale). Red dots indicate the differentially expressed circRNAs with statistical significance. (C) Counts of upregulated and downregulated circRNAs. In total, 67 circRNAs were significantly differentially expressed between IR and N A549 cells (fold change ≥2.0; P<0.05); of these, 30 circRNAs were significantly upregulated (red), 37 circRNAs were downregulated (yellow), and three circRNAs were novel (light red or light yellow). (D) Counts of differentially expressed circRNAs based on nucleotide length. (E) Counts of differentially expressed circRNAs based on five categories of circRNAs. (F) Counts of differentially expressed circRNAs based on chromosomal location. IR, irradiated; N, non-irradiated; circRNAs, circular RNAs; chr, chromosome.
Table I.
Upregulated circRNAs in irradiated A549 cells, compared with non-irradiated A549 cells.
| circRNA | Log FC | P-value | Chromosomal location | Gene name |
|---|---|---|---|---|
| hsa_circ_0008278 | 4.423 | 0.003 | Chr2: 120885264−120932580+ | EPB41L5 |
| hsa_circ_0003187 | 4.035 | 0.015 | Chr4: 146767108−146770713− | ZNF827 |
| hsa_circ_0002174 | 4.029 | 0.01 | Chr3: 195686054−195686957− | GSE61474_XLOC_045249 |
| hsa_circ_0070039 | 4.023 | 0.007 | Chr4: 77055328−77065626− | NUP54 |
| hsa_circ_0004270 | 4.020 | 0.011 | Chr1: 108690901−108703915− | SLC25A24 |
| hsa_circ_0005591 | 3.794 | 0.028 | Chr12: 110566755−110570441+ | IFT81 |
| hsa_circ_0029605 | 3.789 | 0.028 | Chr13: 20235838−20244503+ | MPHOSPH8 |
| hsa_circ_0001865 | 3.787 | 0.028 | Chr9: 86292642−86293514− | UBQLN1 |
| hsa_circ_0002874 | 3.780 | 0.013 | Chr9: 4286038−4286523− | GLIS3 |
| hsa_circ_0011218 | 3.506 | 0.042 | Chr1: 31413998−31414970− | PUM1 |
| hsa_circ_0030632 | 3.499 | 0.042 | Chr13: 96636058−96651561− | UGGT2 |
| hsa_circ_0005579 | 3.499 | 0.042 | Chr2: 36623757−36691798+ | CRIM1 |
| hsa_circ_0088239 | 3.497 | 0.043 | Chr9: 119093523−119097353+ | PAPPA |
| hsa_circ_0105573 | 3.497 | 0.043 | Chr16: 53472928−53504834+ | RBL2 |
| hsa_circ_0099569 | 3.497 | 0.043 | Chr12:95663815-95681633+ | VEZT |
| Novel | 3.497 | 0.043 | Chr6:154749262-154763424− | CNKSR3 |
| hsa_circ_0044158 | 3.490 | 0.036 | Chr17:43197692-43198739− | PLCD3 |
| hsa_circ_0005238 | 3.490 | 0.043 | Chr21:17205667-17214859+ | USP25 |
| hsa_circ_0136108 | 3.490 | 0.043 | Chr8:19442677-19459376− | CSGALNACT1 |
| hsa_circ_0003677 | 3.488 | 0.03 | Chr12:117365828-117402659+ | FBXW8 |
| hsa_circ_0066776 | 3.485 | 0.043 | Chr3:110830877-110845182+ | PVRL3 |
| hsa_circ_0009061 | 3.485 | 0.043 | Chr1:23356962-23377013+ | KDM1A |
| hsa_circ_0001603 | 3.482 | 0.043 | Chr6:42559889-42562042+ | UBR2 |
| hsa_circ_0000199 | 3.479 | 0.037 | Chr1:243708812-243736350− | AKT3 |
| hsa_circ_0004820 | 3.475 | 0.044 | Chr2:183993015-183995273+ | NUP35 |
| hsa_circ_0002224 | 3.475 | 0.044 | Chr10:51374370-51387763+ | TIMM23B |
| hsa_circ_0053317 | 3.468 | 0.044 | Chr2:28460069-28464260+ | BRE |
| hsa_circ_0000378 | 2.792 | 0.019 | Chr12:12397196-12397589− | LRP6 |
| hsa_circ_0001190 | 2.446 | 0.046 | Chr21:38792601-38794168+ | DYRK1A |
| hsa_circ_0005456 | 2.422 | 0.047 | Chr9:118949433-118950495+ | PAPPA |
circRNA, circular RNA; FC, fold change; Chr, chromosome.
Table II.
Downregulated circRNAs in irradiated A549 cells, compared with non-irradiated A549 cells.
| circRNA | Log FC | P-value | Chromosomal location | Gene name |
|---|---|---|---|---|
| hsa_circ_0001009 | −4.213 | 0.005 | Chr2: 58449077−58459247− | FANCL |
| hsa_circ_0004815 | −3.972 | 0.013 | Chr12: 42745687−42792796+ | PPHLN1 |
| hsa_circ_0096360 | −3.769 | 0.025 | Chr11: 70505933−70507783− | SHANK2 |
| hsa_circ_0036627 | −3.767 | 0.025 | Chr15: 85656608−85669605+ | PDE8A |
| hsa_circ_0004524 | −3.750 | 0.026 | Chr3: 138289160−138291774− | CEP70 |
| hsa_circ_0006834 | −3.740 | 0.031 | Chr2: 120885264−120932576+ | EPB41L5 |
| hsa_circ_0006284 | −3.740 | 0.031 | Chr11: 12883797−12886447+ | TEAD1 |
| hsa_circ_0001345 | −3.736 | 0.015 | Chr3: 142455221−142467302+ | TRPC1 |
| hsa_circ_0127359 | −3.736 | 0.031 | Chr4: 95796982−95820201+ | BMPR1B |
| hsa_circ_0098537 | −3.736 | 0.031 | Chr12: 41408030−41423021+ | CNTN1 |
| hsa_circ_0005925 | −3.503 | 0.044 | Chr7: 99952766−99953427+ | PILRB |
| hsa_circ_0002658 | −3.479 | 0.045 | Chr3: 15726733−15731727− | ANKRD28 |
| hsa_circ_0116480 | −3.479 | 0.045 | Chr22: 32664275−32669494+ | G053490 |
| hsa_circ_0124903 | −3.479 | 0.045 | Chr4: 103644028−103651893− | MANBA |
| hsa_circ_0000606 | −3.479 | 0.045 | Chr15: 60734615−60737990− | ICE2 |
| hsa_circ_0001977 | −3.466 | 0.046 | Chr20: 17928130−17937681− | SNX5 |
| Novel | −3.464 | 0.040 | ChrM: 13298−13449+ | MTND5 |
| Novel | −3.464 | 0.046 | Chr20: 45801355−45809584+ | EYA2 |
| hsa_circ_0004844 | −3.460 | 0.033 | ChrX: 2871184−2876476− | ARSE |
| hsa_circ_0005062 | −3.455 | 0.047 | Chr9: 710804−713464+ | KANK1 |
| hsa_circ_0002714 | −3.453 | 0.047 | Chr7: 80418622−80435074− | SEMA3C |
| hsa_circ_0058495 | −3.453 | 0.046 | Chr2: 227729320−227779067+ | RHBDD1 |
| hsa_circ_0003692 | −3.453 | 0.046 | Chr3: 171969050−172028671+ | FNDC3B |
| hsa_circ_0078299 | −3.453 | 0.041 | Chr6: 151669846−151674887+ | AKAP12 |
| hsa_circ_0101802 | −3.442 | 0.047 | Chr14: 39648295−39648666+ | PNN |
| hsa_circ_0000532 | −3.440 | 0.047 | Chr14: 45564424−45566208+ | PRPF39 |
| hsa_circ_0006501 | −3.431 | 0.048 | Chr7: 101870647−101870949+ | CUX1 |
| hsa_circ_0006021 | −3.418 | 0.048 | Chr1: 235582788−235597595+ | TBCE |
| hsa_circ_0123217 | −3.418 | 0.048 | Chr3: 196214270−196215554− | RNF168 |
| hsa_circ_0004532 | −3.418 | 0.048 | Chr17: 12011107−12016677+ | MAP2K4 |
| hsa_circ_0006668 | −3.407 | 0.049 | Chr12: 51442817−51451911+ | LETMD1 |
| hsa_circ_0008153 | −2.549 | 0.034 | Chr15: 63845914−63855207+ | USP3 |
| hsa_circ_0093688 | −2.333 | 0.016 | Chr10: 4872867−4950612+ | AKR1E2 |
| hsa_circ_0000443 | −1.733 | 0.015 | Chr12: 116668338−116675510− | MED13L |
| hsa_circ_0001460 | −1.669 | 0.045 | Chr4:178274462-178281831+ | NEIL3 |
| hsa_circ_0112879 | −1.497 | 0.009 | Chr1:247319708-247323115− | ZNF124 |
| hsa_circ_0005332 | −1.152 | 0.026 | Chr3:114069121-114070725− | ZBTB20 |
circRNA, circular RNA; FC, fold change; Chr, chromosome.
Of the 67 differentially expressed circRNAs, 64 were known circRNAs listed in circbase, and 3 were novel (Fig. 1C). Most of the 67 identified circRNAs were <2,000 nucleotides in length (Fig. 1D). Based on structure, five categories of circRNAs were identified: i) Exonic circRNAs; ii) intronic circRNAs; iii) sense overlapping circRNAs; iv) intergenic circRNAs; and v) antisense circRNAs. Exonic circRNAs represented 83.6% (56/67) of all identified circRNAs, sense overlapping circRNAs were at 13.4% (9/67), and other circRNAs at 3% (2/67; Fig. 1E). Sixty-six circRNAs were located on chromosomes, while one downregulated circRNA was located in the mitochondrial genome (Fig. 1F).
Predicted functions and pathways of differentially expressed circRNAs
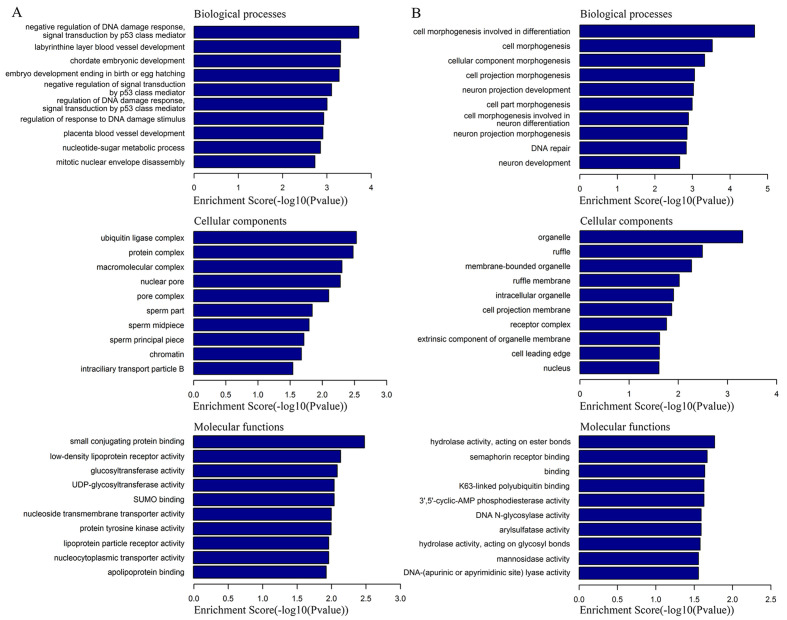
The special RNA analysis software (DCC) was used to obtain the genomic location of circRNA target genes. In combination with the gene annotation in the circbase, the target genes of circRNAs were obtained. The functions and pathways of differentially expressed circRNAs were predicted by GO and KEGG analysis based on their corresponding target genes. GO enrichment analysis predicted the top 10 functional roles of target host genes according to BP, CC and MF (Fig. 2). GO analysis suggested the target genes participated in multiple BPs related to human tumorigenesis, including ‘cell morphogenesis involved in differentiation’, ‘neuron development’ and ‘chordate embryonic development’.
Figure 2.
GO enrichment analysis. (A) Top 10 GO terms of upregulated circRNAs. (B) Top 10 GO terms of downregulated circRNAs. GO, Gene Ontology; circRNAs, circular RNAs.
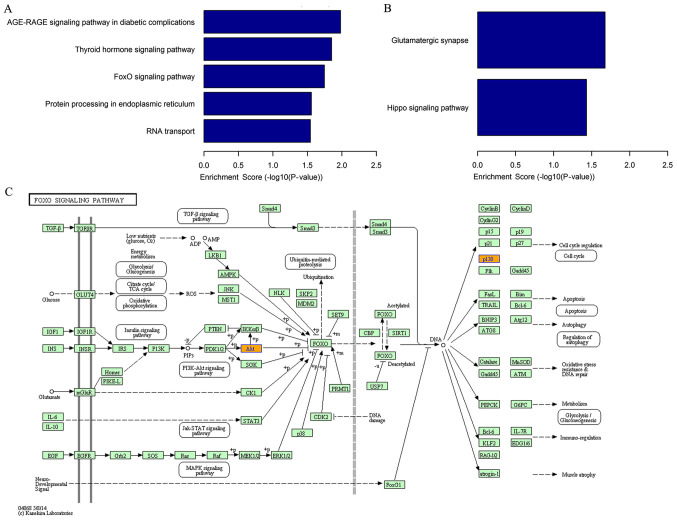
The KEGG enrichment analysis identified five pathways related to the upregulated circRNAs, the ‘AGE-RAGE signaling pathway in diabetic complications’, ‘thyroid hormone signaling pathway’, ‘FoxO signaling pathway’, ‘protein processing in the endoplasmic reticulum’ and ‘RNA transport’ (Fig. 3A). The pathway analysis also indicated two pathways associated with downregulated circRNAs, namely the ‘glutamatergic synapse’ and the ‘Hippo signaling pathway’ (Fig. 3B). The FOXO signaling pathway is presented as an example (Fig. 3C). The FOXO signaling pathway was associated with several BPs, including ‘cell cycle’, ‘apoptosis’ and ‘regulation of autophagy’.
Figure 3.
Kyoto Encyclopedia of Genes and Genomes enrichment analysis. (A) Pathways associated with upregulated circRNAs. (B) Pathways associated with downregulated circRNAs. (C) Representation of the FOXO signaling pathway. Orange nodes represent upregulated target genes. circRNAs, circular RNAs.
Validation of the accuracy of circRNA sequencing data by RT-qPCR
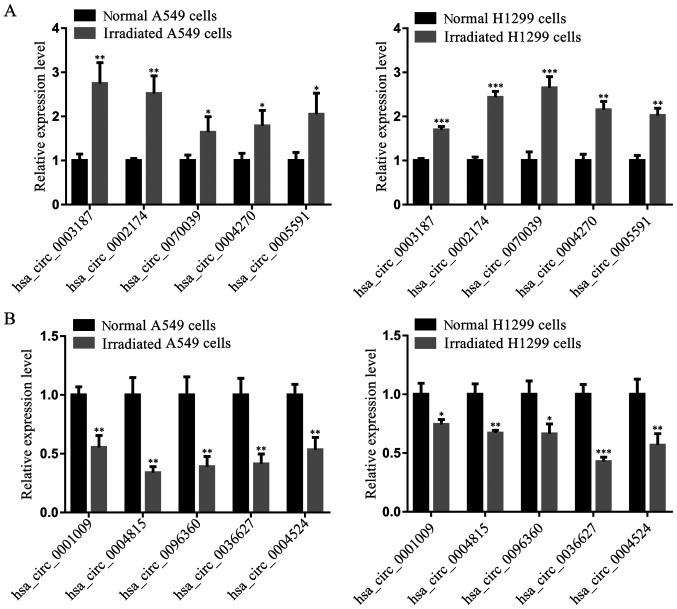
To validate the RNAseq data, the top 5 upregulated (excluding hsa_circ_0008278 due to its sequence length >40,000 nucleotides) and downregulated circRNAs were selected based on P-value and fold change, and subjected to RT-qPCR. RT-qPCR demonstrated that hsa_circ_0003187, hsa_circ_0002174, hsa_circ_0070039, hsa_circ_0004270 and hsa_circ_0005591 expressions were significantly upregulated in irradiated cells compared with corresponding non-irradiated cells (Fig. 4A). Moreover, the expression of hsa_circ_0001009, hsa_circ_0004815, hsa_circ_0096360, hsa_circ_0036627 and hsa_circ_0004524 were significantly downregulated in irradiated cells (Fig. 4B). These 10 circRNA expression levels detected by RT-qPCR were consistent with RNAseq data, demonstrating the accuracy of high-throughput RNAseq results.
Figure 4.
Differentially expressed circRNAs verified by reverse transcription-quantitative PCR. (A) Expression levels of the top 5 upregulated circRNAs in two NSCLC cell lines. (B) Expression levels of the top 5 downregulated circRNAs in two NSCLC cell lines. Each assay was performed at least in triplicate. Differences between groups were analyzed using two-tailed Student's t-test. *P<0.05, **P<0.01 and ***P<0.001 vs. respective normal cells. NSCLC, non-small cell lung cancer; circRNAs, circular RNAs.
circRNA-miRNA co-expression network
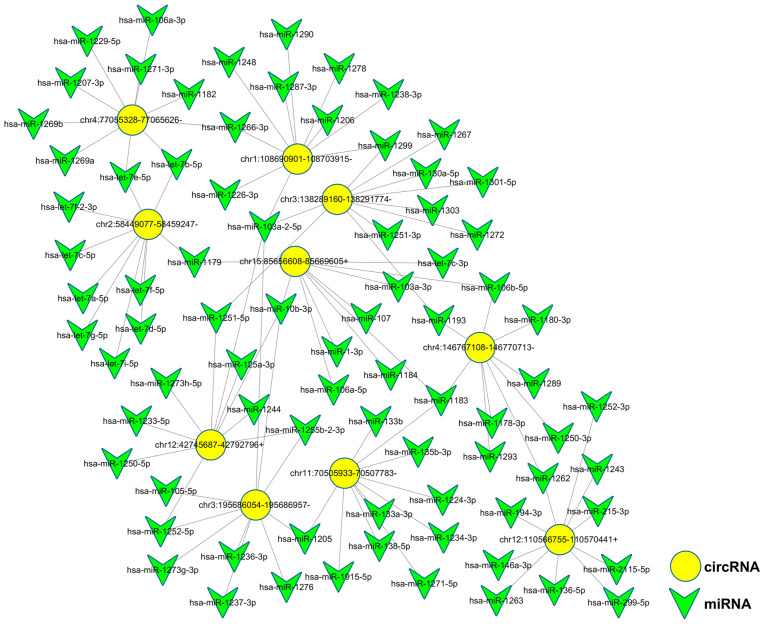
Potential interactions between the 10 validated circRNAs and microRNAs were theoretically predicted through conserved binding sites using miRanda and TargetScan analyses. All 10 circRNAs had complementary miRNA response elements. circRNA_0002174 and circRNA_0036627 were the two circRNAs with the largest number of predicted miRNA-binding sites among upregulated circRNAs and downregulated circRNAs, respectively (Fig. 5; Table III). These two circRNAs could be investigated to help resolve the molecular mechanisms of radiotherapy.
Figure 5.
circRNA-miRNA interaction network. Network was constructed using the top 5 upregulated and downregulated circRNAs (yellow nodes) and their first 10 complementary binding miRNAs (green nodes) using Cytoscape software. circRNAs, circular RNAs; miRNA, microRNA.
Table III.
A total of 10 validated circRNA and their miRNA targets.
| circRNA | Chromosomal location | No. of miRNA targets |
|---|---|---|
| hsa_circ_0036627 | Chr15: 85656608−85669605+ | 378 |
| hsa_circ_0002174 | Chr3: 195686054−195686957− | 276 |
| hsa_circ_0004815 | Chr12: 42745687−42792796+ | 274 |
| hsa_circ_0070039 | Chr4: 77055328−77065626− | 229 |
| hsa_circ_0004270 | Chr1: 108690901−108703915− | 164 |
| hsa_circ_0004524 | Chr3: 138289160−138291774− | 121 |
| hsa_circ_0003187 | Chr4: 146767108−146770713− | 114 |
| hsa_circ_0001009 | Chr2: 58449077−58459247− | 103 |
| hsa_circ_0005591 | Chr12: 110566755−110570441+ | 66 |
| hsa_circ_0096360 | Chr11: 70505933−70507783− | 60 |
circRNA, circular RNA; Chr, chromosome, miRNA, microRNA.
Discussion
With the development of new computational methods and high-throughput sequencing, circRNA has been recognized as a stable and abundant form of RNA, and is presently regarded as an essential part of the non-coding RNA family (31). Previously, several studies demonstrated that circRNAs played a vital role in the pathophysiology of various diseases, with particular relevance in different types of cancer (18,32,33). Furthermore, circRNAs could promote cell proliferation, migration, invasion and metastasis in NSCLC (20–22). Moreover, previous studies have found that circRNA may play a major role in response to radiation (23). circRNA was significantly differently expressed in radioresistant esophageal cancer cells (24) and radioresistant HeLa cells (25). circRNA_014511 could affect the radiosensitivity of bone marrow mesenchymal stem cells by binding to miR-29b-2-5p (26). However, the functions and biological features of circRNAs in lung cancer radiotherapy remain poorly understood. In the present study, the expression levels of circRNAs in irradiated and non-irradiated A549 cells were determined by circRNA sequencing, to examine the possible underlying involvement of differentially expressed circRNAs in the mechanisms of lung cancer radiotherapy.
To the best of the authors' knowledge, the present study was the first to identify differentially expressed circRNAs in irradiated A549 cells by examining junction reads. A549 cells received a 2 Gy irradiation dose and total RNA was collected after 48 h. This time interval was chosen because most of the radiation-related genes with high expression were strongly inhibited after 48 h, based on previous studies (25,34). Among the total 1,875 identified circRNAs, 30 significantly upregulated circRNAs and 37 significantly downregulated circRNAs were detected in irradiated A549 cells. The expression levels of the top 5 upregulated and downregulated circRNAs were further validated by RT-qPCR, which were consistent with high-throughput sequencing data.
Differentially expressed circRNAs may regulate the transcription of several target genes in irradiated lung cancer cells. In order to further investigate the potential functions of circRNAs in irradiated A549 cells, GO and KEGG analyses were conducted. GO analysis was used to identify the BP, CC and MF of target genes. Previous studies demonstrated that irradiation can cause DNA strand breaks via different mechanisms (35,36). Notably, irradiation induces DNA damage (37). The results of the present study suggested that the target genes associated with dysregulated circRNAs might be related to the regulation of DNA damage response (‘signal transduction by p53 class mediator’). In addition, the present study also indicated that ‘labyrinthine layer blood vessel development’ was a notable BP term in response to irradiation. Similarly, Lee et al (38) had previously identified that vascular endothelial growth factor was highly expressed following irradiation, which might lead to rapid blood vessel development. ‘Small conjugating protein binding’ was also an important GO term. KEGG analysis identified several pathways associated with the upregulated and downregulated circRNAs identified in the present study. Among these pathways, the FOXO signaling pathway was identified. The FOXO signaling pathway was associated with several cellular functions in previous studies, including proliferation, apoptosis, inflammation migration, antitumor activities and metabolism through regulation of numerous transcriptional targets (39,40). A further understanding of the function of these differentially expressed circular RNAs will help to understand the mechanisms associated with radiation damage.
Previous studies demonstrated that circRNAs could negatively regulate the effects of miRNAs, and promote the effects of endogenous mRNAs (15,41). A single circRNA may regulate multiple miRNAs, thereby affecting several pathways. Zhao et al (42) suggested that ciRS-7 had >60 well-known miR-7 binding sites, which is far more than any known linear RNA sponge. In the present study, a circRNA-miRNA interaction network was constructed to examine the biological role of circRNAs following irradiation. The network analysis suggested that two potential circRNAs (circRNA_0002174 and circRNA_0036627) might play an important role in the regulation of target miRNAs. Based on the characteristics of circRNAs, including structural stability, stable expression and tissue specificity, circRNAs could serve as a novel biomarker and therapeutic target for cancer treatment. Chen et al (43) suggested that circRNA epithelial stromal interaction 1 played a vital role as a prognostic marker in patients with triple-negative breast cancer. Nair et al (44) suggested that circRNAs might function as markers of cell proliferation in breast cancer. circPRKCI may serve as a potential therapeutic target for patients with lung adenocarcinoma (45). The present study demonstrated that differential expression of circRNAs in irradiated and non-irradiated A549 cells could be a critical marker of radiation-induced damage.
In conclusion, the preset study identified differentially expressed circRNAs and their target genes in irradiated A549 cells. The results of the present study suggested that circRNAs might play an important role in response to irradiation. The top 5 upregulated and downregulated circRNAs were validated; however, further investigation is required in order to examine the molecular functions of circRNAs in radiotherapy. Several potential circRNA-miRNA networks were predicted through bioinformatics analysis. These results warrant further investigation of the potential biological functions of circRNAs in lung cancer radiotherapy.
Supplementary Material
Acknowledgements
The authors would like to thank Cloud-Seq Biotech Ltd. Co. (Shanghai, China) for the circRNA sequencing service and the subsequent bioinformatics analysis.
Funding
The present study was funded by The National Natural Science Foundation of China (grant nos. 81802955 and 81972977), The Foundation of Sichuan Science and Technology Agency (grant nos. 2018JY0648 and 2019YJ0589), The Foundation of The First Affiliated Hospital of Chengdu Medical College (grant nos. CYFY2017ZD03 and CYFY2018ZD02) and The Foundation of Chengdu Medical College (grant no. CYCG16-04).
Availability of data and materials
The datasets supporting the conclusions of this article are available in the Gene Expression Omnibus repository (GSE124396; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE1243 96).
Authors' contributions
TZ, DW and YX conceived and designed the experiments. TZ and DW performed the experiments and wrote drafts of the paper; TZ, SD and RH analyzed the data; DW, TL and JL acquired the data; YX reviewed the drafts of the paper. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Zhu W, Li Z, Xiong L, Yu X, Chen X, Lin Q. FKBP3 Promotes proliferation of non-small cell lung cancer cells through regulating Sp1/HDAC2/p27. Theranostics. 2017;7:3078–3089. doi: 10.7150/thno.18067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiu LY, Hsin IL, Yang TY, Sung WW, Chi JY, Chang JT, Ko JL, Sheu GT. The ERK-ZEB1 pathway mediates epithelial-mesenchymal transition in pemetrexed resistant lung cancer cells with suppression by vinca alkaloids. Oncogene. 2017;36:242–253. doi: 10.1038/onc.2016.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haimovich G, Medina DA, Causse SZ, Garber M, Millán-Zambrano G, Barkai O, Chávez S, Pérez-Ortín JE, Darzacq X, Choder M. Gene expression is circular: Factors for mRNA degradation also foster mRNA synthesis. Cell. 2013;153:1000–10011. doi: 10.1016/j.cell.2013.05.012. [DOI] [PubMed] [Google Scholar]
- 5.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 6.Wilusz JE, Sharp PA. Molecular biology. A circuitous route to noncoding RNA. Science. 2013;340:440–441. doi: 10.1126/science.1238522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, Sun W, Dou K, Li H. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015;365:141–148. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 8.Rybak-Wolf A, Stottmeister C, Glažar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58:870–885. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 9.Szabo L, Morey R, Palpant NJ, Wang PL, Afari N, Jiang C, Parast MM, Murry CE, Laurent LC, Salzman J. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol. 2015;16:126. doi: 10.1186/s13059-015-0690-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qiu X, Ke X, Ma H, Han L, Chen Q, Zhang S, Da P, Wu H. Profiling and bioinformatics analyses reveal differential expression of circular RNA in tongue cancer revealed by high-throughput sequencing. J Cell Biochem. 2018;120:4102–4112. doi: 10.1002/jcb.27695. [DOI] [PubMed] [Google Scholar]
- 11.Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci USA. 1976;73:3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381–388. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salzman J. Circular RNA expression: Its potential regulation and function. Trends Genet. 2016;32:309–316. doi: 10.1016/j.tig.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luan J, Jiao C, Kong W, Fu J, Qu W, Chen Y, Zhu X, Zeng Y, Guo G, Qi H, et al. circHLA-C plays an important role in lupus nephritis by sponging miR-150. Mol Ther Nucl Acids. 2018;10:245–253. doi: 10.1016/j.omtn.2017.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tan WL, Lim BT, Anene-Nzelu CG, Ackers-Johnson M, Dashi A, See K, Tiang Z, Lee DP, Chua WW, Luu TD. A landscape of circular RNA expression in the human heart. Cardiovasc Res. 2017;113:298–309. doi: 10.1093/cvr/cvw250. [DOI] [PubMed] [Google Scholar]
- 17.Shao Y, Chen Y. Roles of circular RNAs in neurologic disease. Front Mol Neurosci. 2016;9:25. doi: 10.3389/fnmol.2016.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li P, Chen S, Chen H, Mo X, Li T, Shao Y, Xiao B, Guo J. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta. 2015;444:132–136. doi: 10.1016/j.cca.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 19.Wang H, Xiao Y, Wu L, Ma D. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-000911/miR-449a pathway in breast carcinogenesis. Int J Oncol. 2018;52:743–754. doi: 10.3892/ijo.2018.4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu K, Liao X, Gong Y, He J, Zhou JK, Tan S, Pu W, Huang C, Wei YQ, Peng Y. Circular RNA F-circSR derived from SLC34A2-ROS1 fusion gene promotes cell migration in non-small cell lung cancer. Mol Cancer. 2019;18:98. doi: 10.1186/s12943-019-1028-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li XY, Liu YR, Zhou JH, Li W, Guo HH, Ma HP. Enhanced expression of circular RNA hsa_circ_000984 promotes cells proliferation and metastasis in non-small cell lung cancer by modulating Wnt/β-catenin pathway. Eur Rev Med Pharmacol Sci. 2019;23:3366–3374. doi: 10.26355/eurrev_201904_17700. [DOI] [PubMed] [Google Scholar]
- 22.Chang H, Qu J, Wang J, Liang X, Sun W. Circular RNA circ_0026134 regulates non-small cell lung cancer cell proliferation and invasion via sponging miR-1256 and miR-1287. Biomed Pharmacother. 2019;112:108743. doi: 10.1016/j.biopha.2019.108743. [DOI] [PubMed] [Google Scholar]
- 23.Luo J, Zhang C, Zhan Q, An F, Zhu W, Jiang H, Ma C. Profiling circRNA and miRNA of radiation-induced esophageal injury in a rat model. Sci Rep. 2018;8:14605. doi: 10.1038/s41598-018-33038-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Su H, Lin F, Deng X, Shen L, Fang Y, Fei Z, Zhao L, Zhang X, Pan H, Xie D, et al. Profiling and bioinformatics analyses reveal differential circular RNA expression in radioresistant esophageal cancer cells. J Transl Med. 2016;14:225. doi: 10.1186/s12967-016-0977-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu D, Li Y, Ming Z, Wang H, Dong Z, Qiu L, Wang T. Comprehensive circular RNA expression profile in radiation-treated HeLa cells and analysis of radioresistance-related circRNAs. Peer J. 2018;6:e5011. doi: 10.7717/peerj.5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, Zhang J, Li J, Gui R, Nie X, Huang R. CircRNA_014511 affects the radiosensitivity of bone marrow mesenchymal stem cells by binding to miR-29b-2-5p. Bosn J Basic Med Sci. 2019;19:155–163. doi: 10.17305/bjbms.2019.3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu D, Han R, Deng S, Liu T, Zhang T, Xie H, Xu Y. Protective effects of flagellin A N/C against radiation-induced NLR pyrin domain containing 3 inflammasome-dependent pyroptosis in intestinal cells. Int J Radiat Oncol. 2018;101:107–117. doi: 10.1016/j.ijrobp.2018.01.035. [DOI] [PubMed] [Google Scholar]
- 28.Wang M, Meng B, Liu Y, Yu J, Chen Q, Liu Y. MiR-124 inhibits growth and enhances radiation-induced apoptosis in non-small cell lung cancer by inhibiting STAT3. Cell Physiol Biochem. 2017;44:2017–2028. doi: 10.1159/000485907. [DOI] [PubMed] [Google Scholar]
- 29.R Core Team, corp-author. R Foundation for Statistical Computing; Vienna, Austria: 2012. R: A language and environment for statistical computing. ISBN 3-900051-07-0, URL http://www.R-project.org/ [Google Scholar]
- 30.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 31.Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR, Piechotta M, Levanon EY, Landthaler M, Dieterich C, Rajewsky N. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015;10:170–177. doi: 10.1016/j.celrep.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 32.Qin M, Liu G, Huo X, Tao X, Sun X, Ge Z, Yang J, Fan J, Liu L, Qin W. Hsa_circ_0001649: A circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark. 2016;16:161–169. doi: 10.3233/CBM-150552. [DOI] [PubMed] [Google Scholar]
- 33.Su H, Jin X, Zhang X, Zhao L, Lin B, Li L, Fei Z, Shen L, Fang Y, Pan H, Xie C. FH535 increases the radiosensitivity and reverses epithelial-to-mesenchymal transition of radioresistant esophageal cancer cell line KYSE-150R. J Transl Med. 2015;13:104. doi: 10.1186/s12967-015-0464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang H, Yue J, Jiang Z, Zhou R, Xie R, Xu Y, Wu S. CAF-secreted CXCL1 conferred radioresistance by regulating DNA damage response in a ROS-dependent manner in esophageal squamous cell carcinoma. Cell Death Dis. 2017;8:e2790. doi: 10.1038/cddis.2017.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Samstein RM, Riaz N. The DNA damage response in immunotherapy and radiation. Adv Radiat Oncol. 2018;3:527–533. doi: 10.1016/j.adro.2018.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moreno-Villanueva M, Feiveson AH, Krieger S, Kay Brinda A, von Scheven G, Bürkle A, Crucian B, Wu H. Synergistic effects of weightlessness, isoproterenol, and radiation on DNA damage response and cytokine production in immune cells. Int J Mol Sci. 2018;19:E3689. doi: 10.3390/ijms19113689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goldstein M, Kastan MB. The DNA damage response: Implications for tumor responses to radiation and chemotherapy. Annu Rev Med. 2015;66:129–143. doi: 10.1146/annurev-med-081313-121208. [DOI] [PubMed] [Google Scholar]
- 38.Lee C, Shim S, Jang H, Myung H, Lee J, Bae CH, Myung JK, Kim MJ, Lee SB, Jang WS, et al. Human umbilical cord blood-derived mesenchymal stromal cells and small intestinal submucosa hydrogel composite promotes combined radiation-wound healing of mice. Cytotherapy. 2017;19:1048–1059. doi: 10.1016/j.jcyt.2017.06.007. [DOI] [PubMed] [Google Scholar]
- 39.Alikhani M, Alikhani Z, Graves DT. FOXO1 functions as a master switch that regulates gene expression necessary for tumor necrosis factor-induced fibroblast apoptosis. J Biol Chem. 2005;280:12096–12102. doi: 10.1074/jbc.M412171200. [DOI] [PubMed] [Google Scholar]
- 40.Wang Y, Zhou Y, Graves DT. FOXO Transcription factors: Their clinical significance and regulation. Biomed Res Int. 2014;2014:925350. doi: 10.1155/2014/925350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang K, Long B, Liu F, Wang JX, Liu CY, Zhao B, Zhou LY, Sun T, Wang M, Yu T, et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J. 2016;37:2602–2611. doi: 10.1093/eurheartj/ehv713. [DOI] [PubMed] [Google Scholar]
- 42.Zhao ZJ, Shen J. Circular RNA participates in the carcinogenesis and the malignant behavior of cancer. RNA Biol. 2017;14:514–521. doi: 10.1080/15476286.2015.1122162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen B, Wei W, Huang X, Xie X, Kong Y, Dai D, Yang L, Wang J, Tang H, Xie X. circEPSTI1 as a prognostic marker and mediator of triple-negative breast cancer progression. Theranostics. 2018;8:4003–4015. doi: 10.7150/thno.24106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nair AA, Niu N, Tang X, Thompson KJ, Wang L, Kocher JP, Subramanian S, Kalari KR. Circular RNAs and their associations with breast cancer subtypes. Oncotarget. 2016;7:80967–80979. doi: 10.18632/oncotarget.13134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qiu M, Xia W, Chen R, Wang S, Xu Y, Ma Z, Xu W, Zhang E, Wang J, Fang T, et al. The circular RNA circPRKCI promotes tumor growth in lung adenocarcinoma. Cancer Res. 2018;78:2839–2851. doi: 10.1158/0008-5472.CAN-17-2808. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting the conclusions of this article are available in the Gene Expression Omnibus repository (GSE124396; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE1243 96).