Abstract
Gastric cancer (GC) is the second most common cancer cause of cancer-related mortality worldwide. Recent studies have demonstrated the function of microRNAs (miRNAs) in the pathogenesis of GC. miR-876-5p demonstrated an antitumor role in hepatocellular carcinoma and lung cancer; however, the function of miR-876-5p has not yet been fully identified in GC. Thus, the present study aimed to investigate the role of miR-876-5p in GC. The results of the present study demonstrated low expression levels of miR-876-5p in GC tumor tissues. Furthermore, overexpression of miR-876-5p inhibited GC cell proliferation and promoted apoptosis, whilst miR-876-5p knockdown promoted GC cell proliferation and decreased cisplatin sensitivity of GC cells. Transforming growth factor β-receptor 1 was demonstrated to be a potential target gene of miR-876-5p. Overall, the results of the present study suggest that miR-876-5p plays an antitumor role in GC.
Keywords: gastric cancer, miRNA-876-5p, cellular proliferation, apoptosis
Introduction
Gastric cancer (GC) is the second most common cause of cancer-associated mortality worldwide (1). In East Asia, based on GLOBOCAN 2018 data, the average incidence of gastric cancer is 32.1 per 100,000 among men and 13.2 among women (2). For the majority of cases, patients with GC are diagnosed at the advanced stage of disease and thus, successful therapeutic strategies are limited, and the prognosis is poor (3–5). Metastasis is a common trait of advanced GC progression (6); thus, in order to improve the survival rate of patients with GC, investigations that focus on the molecular mechanisms underlying pathogenesis of GC are required.
MicroRNAs (miRNAs) are small conserved non-coding RNAs, typically 20–25 nucleotides in length (7). miRNAs bind to complementary sequences that are frequently present in the 3′-untranslated region (UTR) of the target mRNA, thereby suppressing the translation of the target mRNAs (7). A number of studies have demonstrated that miRNAs play multiple roles in the pathogenesis of GC (8–14).
The function of miR-876-5p has been investigated in multiple types of cancer, including hepatocellular carcinoma (HCC) (15,16) and lung cancer (17). miR-876-5p has been demonstrated to inhibit cell proliferation of HCC and metastasis by targeting the DNMT3A gene (16), and also inhibit epithelial-to-mesenchymal transition (EMT) and metastasis by targeting the BCL6 corepressor-like 1 gene (15). With regard to lung cancer, miR-876-5p has been demonstrated to suppress EMT by targeting bone morphogenetic protein 4 (17). However, the role of miR-876-5p in GC is not yet fully understood. Thus, the present study investigated the functions of miR-876-5p in GC, with the aim of identifying potential, novel therapeutic targets for further development in the future.
Materials and methods
Patients and tissue samples
A total of 16 GC tissues and corresponding adjacent normal tissues were collected from 16 patients with GC between June 2015 and September 2016 who underwent surgery at the Department of Gastroenterology, West China Hospital (Chengdu, China). The age and sex distributions of the 16 patients with GC were listed as follows: Age, 46–72 years; mean age, 56; male/female, 11:5. The inclusion criteria were: i) Age ≥18 years; ii) the absence of radiotherapy, chemotherapy or any adjuvant therapy before hospitalization; iii) the absence of other malignant tumors; and iv) the absence of a family history of genetic diseases. Exclusion criteria were: i) The presence of antibiotics taking within three months before blood collection; ii) the presence of liver insufficiency and, iii) the presence of autoimmune system deficiency. The distance between the tumor tissues and adjacent non-tumor tissues was >5 cm. The histopathological diagnoses of all patients were confirmed by a senior pathologist at West China Hospital. The present study was approved by the Ethics Committee of Sichuan University (Chengdu, China) and all patients provided written informed consent.
Cell culture
The human GC cell line AGS-1 cell line and human gastric epithelial mucosa cell line GES-1 were purchased from the China Infrastructure of Cell Line Resources, Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences. The two cell lines were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) at 37°C and 5% CO2. The cells were cultured until they reached 80% confluence. Cisplatin was purchased from Hansoh Pharma. A final concentration of 25 µM of cisplatin was added to the cultures as previously described (18).
miR-876-5p mimics and oligonucleotide transfection
miR-876-5p mimics, miR-876-5p antisense oligonucleotides (ASO) and negative controls (miR-NC mimics and miR-NC ASO) were constructed by Sangon Biotech Co., Ltd. The primer sequences were as follows: miR-876-5p mimics, sense 5′-UGGAUUUCUUUGUGAAUCACCA-3′ and antisense 5′-GUGAUUCACAAAGAAAUCCAUU-3′; mimic control, sense 5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense 5′-ACGUGACACGUUCGGAGAATT-3′; miR-876-5p inhibitors 5′-UGGUGAUUCACAAAGAAAUCCA-3′; and inhibitor control 5′-CAGUACUUUUGUGUAGUACAA-3. A total of 2×105 AGS-1 cells were seeded into 6-well plates overnight and transfected with 50 nM miR-876-5p mimics or miR-NC mimics using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Similarly, AGS-1 cells were seeded and transfected with miR-876-5p ASO (50 nM); miR-NC ASO (50 nM) was used as control. The cell growth and apoptosis assays were performed 24 h later.
Cell proliferation assay
Cell proliferation was assessed using an MTT assay. The AGS-1 cells were seeded into 96-well plates at a density of 5×105 cells/plate. MTT was added to the medium at a final concentration of 0.1 mg/ml. The purple formazan crystals were dissolved using 100 µl of dimethyl sulfoxide and optical density was measured using a microplate reader (Multiskan Sky Thermo Fisher Scientific, Inc.) at a wavelength of 570 nm (19–21).
Reverse transcription-quantitative (RT-q)PCR
Total RNA was extracted from GC tissues and AGS-1 cell lines using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. The RNA was quantified by NanoDrop™ (ThermoFisher Scientific Inc.). Total RNA were reverse transcribed into cDNA using the All-in-One™ miRNA First-Strand cDNA Synthesis kit (Invitrogen, Thermo Fisher Scientific, Inc.). The PCR was performed by the SuperScript™ III Platinum™ SYBR™ Green One-Step qPCR kit (ThermoFisher Scientific Inc.). The primers were synthesized by Sangon Biotech Co., Ltd. The RNA sequences for the primers mentioned above were as follows: miR-876-5p sense 5′-AGGACUUCUCCCUCCUCCCAG-3′ and anti-sense, 5′-UCCUCUUCUCCCUCCAGGGAG-3′; U6 snRNA sense 5′-CTCGCTTCGGCAGCACATATACT-3′; and anti-sense 5′-ACGCTTCACGAATTTGCGTGTC-3′. miR-876-5p expression was normalized to the internal reference gene U6 (18,22). PCR conditions were as follows: Denaturation for 10 min at 95°C, followed by 40 cycles of annealing for 1 min at 95°C and extension for 30 sec at 60°C. The dissolution procedure was designed as follows: 15 sec at 95°C, 30 sec at 60°C and 15 sec at 95°C. Quantification was performed using the 2−∆∆Cq method (23).
Mutated site and target gene prediction
The mutated sites were generated by Gene Site-directed Mutagenesis System (Thermo Fisher Scientific, Inc.). The potential targets of miR-876-5p were predicted by TargetScan software (24–29).
Apoptosis analysis
The apoptotic ability of AGS-1 cells was assessed via flow cytometry using Annexin V-propidium iodide (PI) staining. AGS-1 cells were suspended in Annexin V-fluorescein isothiocyanate (FITC) (1 µg/ml) binding buffer (Abcam) at a density of 5×105 cells/ml and incubated at room temperature for 20 min. Subsequently, PI (0.1 µg/ml) (Abcam) was added to the samples, incubated for 5 min at room temperature and analyzed using a BD FACSVerse™ flow cytometer (BD Biosciences) using the 488 nm excitation line (argon ion or solid-state laser), and emission was detected at 530 nm (green for Annexin V-FITC) and 575–610 nm (orange for PI). The data were analyzed using the BD FACSuite™ version 1.01 (BD Biosciences).
Western blot analysis
Total protein was extracted from AGS-1 cells using lysis buffer (150 mM NaCl, 50 mM Tris-HCI, 1% Triton X-100 and 0.1% SDS) with Protease Inhibitor Cocktail and Phosphatase Inhibitor Cocktail (Sigma-Aldrich; Merck KGaA). Proteins (10 mg) were separated via SDS-PAGE on a 10% gel and subsequently transferred onto a polyvinylidene difluoride membrane. The membranes were blocked in 5% bovine serum albumin (Sigma-Aldrich; Merck KGaA) for 1 h at room temperature, prior to transforming growth factor β-receptor 1 (TGFBR1) analysis. The membranes were incubated with anti-TGFBR1 (1:1,000; cat. no. ab31013; Abcam) and β-actin (1:1,000; cat. no. ab8226; Abcam) overnight at 4°C. Membranes were washed three times with PBS and subsequently incubated with a horseradish peroxidase-conjugated rabbit IgG antibody (1:2,000; cat. no. ab218695; Abcam) for 2 h at room temperature. Protein bands were visualized using the ECL Western Blotting Detection reagents (GE Healthcare) and images were analyzed using ImageJ software (National Institutes of Health).
Luciferase reporter plasmid transfection
AGS-1 cells were seeded at 1×105 per well and were serum-starved for 6 h pre-transfection. The 3′ untranslated region (UTR) of TGFBR1 was cloned into the reporter plasmid (500 ng) and the pGL3-control (100 ng; Promega Corporation). miR-876-5p (30 nM) were transfected into AGS-1 cells containing the wild-type or mutant 3′-UTR plasmids using Lipofectamine® 2000 (Invitrogen, Thermo Fisher Scientific Inc.) for 5 min at room temperature. Mutants of the 3′ UTR of TGFBR1 were generated using the Site-directed Mutagenesis kit (cat. no. A13282, Thermo Fisher Scientific Inc.). Cells were harvested 24 h later, and the luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega Corporation). The firefly luciferase enzyme activity was normalized to the Renilla luciferase enzyme activity.
Statistical analysis
All data were analyzed using SPSS software version 16.0 (SPSS, Inc.). All experiments were performed in triplicate and data are presented as the mean ± standard deviation. Two-tailed Student's t-test was used in order to analyze the mean values between two groups; one-way ANOVA was used in order to test the mean values among three groups or more, followed by Student-Newman-Keuls post hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-876-5p levels in GC tissues
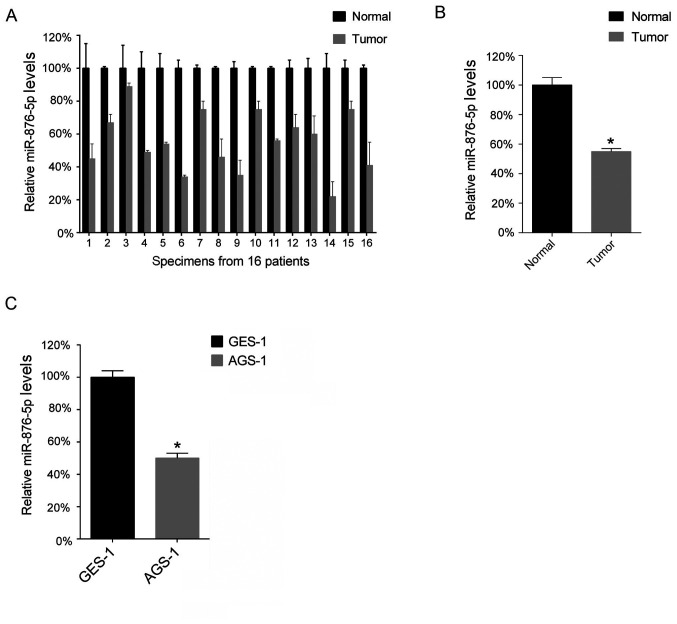
The present study performed a RT-qPCR analysis in order to determine miR-876-5p levels in the 16 GC tissues and 16 corresponding normal adjacent tissues. The results of the present study demonstrated that miR-876-5p levels are downregulated in GC tissues compared with normal tissues (Fig. 1A), as the mean levels of miR-876-5p in the 16 GC tissues was lower compared with that in the 16 normal adjacent tissues (Fig. 1B). Furthermore, the present study analyzed miR-876-5p levels in the GC cell lines and demonstrated that the AGS-1 cell line expressed lower levels of miR-876-5p compared with the GES-1 cell line (Fig. 1C).
Figure 1.
miR-876-5p is downregulated in GC tissue samples. (A) The miR-876-5p levels in 16 GC tissues and 16 corresponding matched normal adjacent tissues were analyzed via RT-qPCR. The miR-876-5p levels in the normal adjacent tissues were arbitrarily defined as 100%. (B) The mean values of miR-876-5p levels in the 16 GC tissues and 16 normal adjacent tissues were calculated and presented. (C) The miR-876-5p levels in AGS-1 cells were analyzed using RT-qPCR. *P<0.05; normal vs. tumor tissues and GES-1 vs. AGS-1 microRNA-876-5p. GC, gastric cancer; RT-qPCR, reverse transcription-quantitative PCR.
Overexpression of miR-876-5p inhibits cellular proliferation and promotes apoptosis
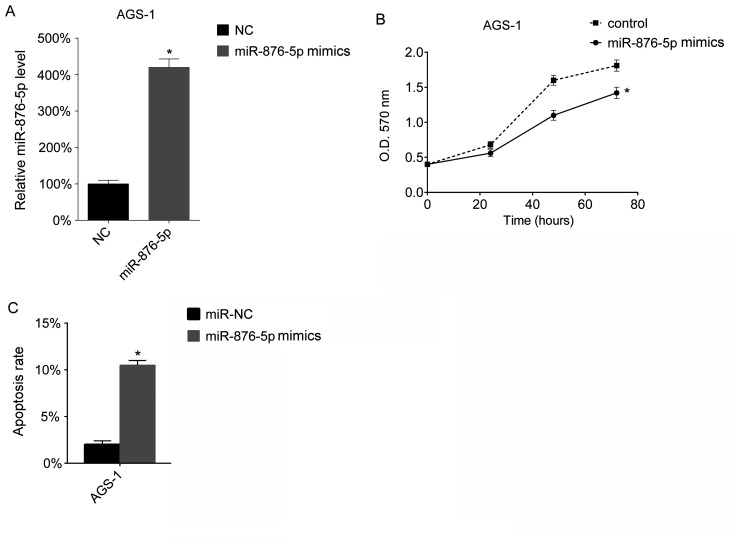
The present study assessed the function of miR-876-5p in vitro. miR-876-5p levels in AGS-1 cells were overexpressed using miR-876-5p mimics. The results of the present study demonstrated that transfection with miR-876-5p mimics increased the miR-876-5p levels in AGS-1 cells (Fig. 2A). Subsequently, cell proliferation was assessed using an MTT assay. The results of the present study demonstrated that overexpression of miR-876-5p inhibits proliferation of AGS-1 cells (Fig. 2B). The present study assessed the apoptotic ability of the transfected cells and demonstrated that miR-876-5p increased the apoptotic rate of AGS-1 cells (Fig. 2C).
Figure 2.
Overexpression of miR-876-5p inhibits proliferation and promotes apoptosis of AGS-1 cells. AGS-1 cells were seeded into 24-well plates at a density of 1×106 cells/well overnight. miR-876-5p mimics were transfected into AGS-1 cells. (A) After 24 h, miR-876-5p levels in AGS-1 cells were analyzed via reverse transcription-quantitative PCR. The miR-876-5p levels in miR-NC mimic-transfected cells. The miR-876-5p levels in the control group were arbitrarily defined as 100%. (B) Cellular proliferation was assessed using an MTT assay 24 h after transfection, at 24, 48 and 72 h. (C) Cell apoptosis rates were assessed by FACS 24 h after transfection with miR-876-5p mimics. *P<0.05, control vs. transfection group. miR-876-5p, microRNA-876-5p; miR-NC, microRNA-control; FACS, fluorescence-activated cell sorting; NC, control; OD, optical density.
Suppression of miR-876-5p promotes cellular proliferation and inhibits apoptosis
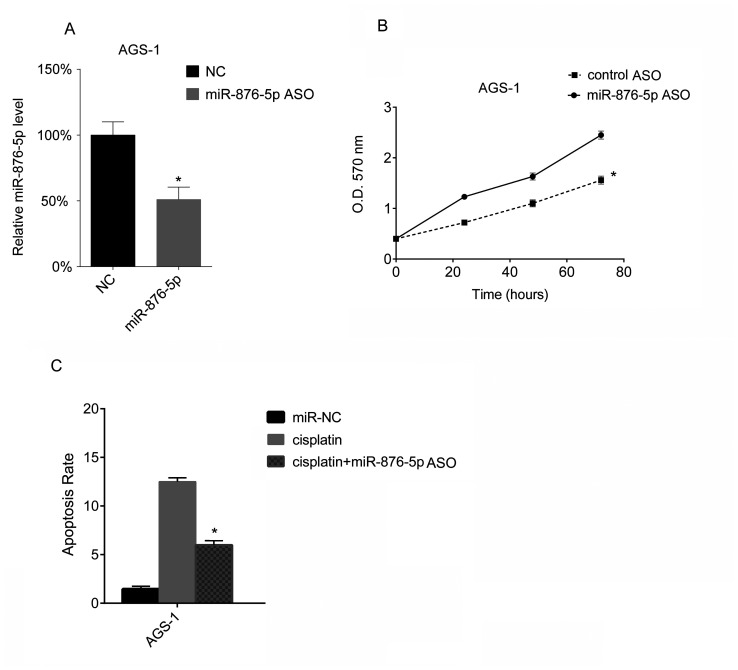
miR-876-5p knockdown in AGS-1 cells was performed in the present study, via miR-876-5p ASO transfection. The miR-876-5p levels of AGS-1 cells were analyzed via RT-qPCR, and the results of the present study demonstrated that miR-876-5p ASO transfection downregulates miR-876-5p levels in AGS-1 (Fig. 3A). Furthermore, the results of the MTT assay in the present study demonstrated that miR-876-5p ASO transfection promotes the proliferation of AGS-1 cells (Fig. 3B). Subsequently, AGS-1 cells were treated with cisplatin following miR-876-5p ASO transfection. The results of the present study demonstrated that cisplatin increased the apoptotic rate of AGS-1 cells, whereas the downregulation of miR-876-5p decreased the apoptotic rate induced by cisplatin (Fig. 3C).
Figure 3.
miR-876-5p knockdown promotes proliferation and inhibits apoptosis of AGS-1 cells. AGS-1 cells were seeded into 24-well plates at a density of 1×106 cells/well overnight. miR-876-5p mimics were transfected into AGS-1 cells. (A) After 24 h, miR-876-5p levels in AGS-1 cells were analyzed using reverse transcription-quantitative PCR. The miR-876-5p levels in miR-NC mimic-transfected cells. The miR-876-5p levels in the control group were arbitrarily defined as 100%. (B) Cellular proliferation was assessed using an MTT assay 24 h after transfection, at 24, 48 and 72 h. (C) Cell apoptosis rates were assessed by FACS 24 h after transfection with miR-876-5p ASO. *P<0.05, cisplatin vs. cisplatin + miR-876-5p ASO. miR-876-5p, microRNA-876-5p; miR-NC, microRNA-control; FACS, fluorescence-activated cell sorting; NC, control; miR-876-5p ASO, microRNA-876-5p antisense oligonucleotides; OD, optical density.
miR-876-5p targets TGFBR1
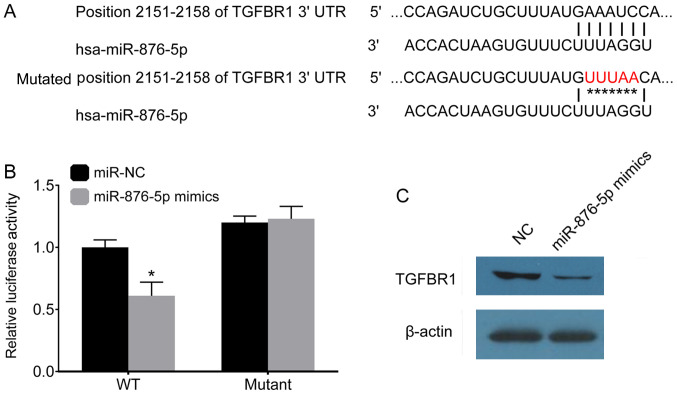
Physiologically, miRNAs play a role in targeting genes. The present study used the TargetScan software in order to predict the potential target genes of miR-876-5p. The results of the present study demonstrated that TGFBR1 may be targeted by miR-876-5p. The present study mutated the 2,151-2,158 position of TGFBR1 (Fig. 4A), in order to investigate the association between miR-876-5p and TGFBR1. The 3′-UTR TGFBR1 mutant was generated and subsequently transfected into a luciferase reporter plasmid. miR-876-5p and the 3′-UTR TGFBR1 mutant were co-transfected into AGS-1 cells. At 24 h post-transfection, miR-876-5p was demonstrated to decrease the luciferase activity of the 3′-UTR of wild-type TGFBR1, but not that of the 3′-UTR of mutated TGFBR1 (Fig. 4B). After 48 h, TGFBR1 protein levels were determined using western blot analysis, and the results of the present study demonstrated that miR-876-5p mimics inhibited the expression of TGFBR1 in AGS-1 cells (Fig. 4D).
Figure 4.
miR-876-5p targets TGFBR1. (A) The binding sites between miR-876-5p and TGFBR1 and the location of the mutant (highlighted in red). (B) miR-876-5p mimics and a plasmid containing either WT or the mutated 3′-UTR TGFBR1 sequence were transfected into AGS-1 cells and luciferase activity was measured after 48 h. (C) The amount of TGFBR1 protein in AGS-1 cells was determined post-transfection via western blot analysis. *P<0.05, WT miR-NC vs. miR-876-5p mimics. miR-876-5p, microRNA-876-5p; TGFBR1, transforming growth factor β-receptor 1; WT, wild-type; 3′-UTR, 3′-untranslated region; miR-NC, microRNA control; NC, control.
Discussion
The results of the present study demonstrated that miR-876-5p levels were downregulated in GC tumor tissues compared with normal adjacent tissues. Furthermore, overexpression of miR-876-5p inhibited cellular proliferation and promoted apoptosis, whereas miR-876-5p knockdown promoted cellular proliferation and inhibited apoptosis. The results of the present study suggest that TGFBR1 is targeted by miR-876-5p. To the best of our knowledge, the results of the present study are the first to report miR-876-5p involvement in human GC. As miR-876-5p may be associated with the survival time of patients with GC, future studies are required with larger patient sample sizes, in order to accurately assess survival time analysis in mice models. Furthermore, future studies are critical in order to assess the association between miR-876-5p levels and the function in drug-treatment pressure.
A previous study demonstrated the involvement of miR-876-5p in viral infections. Wang et al (30) demonstrated that severe human enterovirus led to elevated expression of circulating miR-876-5p. Furthermore, miR-876-5p has been reported to participate in the miRNA-based antiviral defense mechanism against influenza virus in humans, by targeting the matrix protein of human influenza A H1N1 virus (31).
Previous studies have demonstrated that miR-876-5p inhibits cell proliferation, metastasis and EMT in hepatocellular carcinoma and lung cancer (15–17). The results of the present study further confirmed the inhibitory role of miR-876-5p and proposed TGFBR1 as a target gene. TGFBR1 is the receptor of TGF-β, which is known to induce EMT (32). Thus, miR-876-5p may have the potential to regulate EMT through TGFBR1 targeting.
TGF-β has a dual role in cancer development, acting as a tumor suppressor and a tumor promoter; it promotes tumorigenesis indirectly by acting on the tumor microenvironment (33). Thus, there is potential that TGFBR1 may play a similar dual role in the pathogenesis of GC. It is predicted that the inhibition of TGFBR1 by miR-876-5p may result in inactivation of the TGF-β signaling pathway. The results of the present study demonstrated that miR-876-5p targeted TGFBR1, but the role of TGFBR1 is not yet fully understood. Furthermore, the results of the present study suggest that miR-876-5p may be involved in the complex TGF-β signaling pathway by targeting TGFBR1. Another study demonstrated that variants exist in the whole TGFBR1 gene, represented by the single nucleotide polymorphisms, and that genetic variants in TGFBR1 may play a role in the development of GC (34). The results of the present study indicate that miR-876-5p could regulate TGFBR1 by binding to its 3′-UTR. Whether the variants of TGFBR1 have an effect on the regulation function of miR-876-5p warrants further investigations. Overall, the results of the present study confirm the inhibitory role of miR-876-5p in GC.
Acknowledgements
The authors would like to thank Dr Ke Lei (Department of Oncology, West China Hospital, Sichuan University) for discussions regarding the present study.
Funding
This study was supported by a grant from the Science Supporting Funding of Sichuan University (grant no.20180110).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
HZ, YZ, JY and JX collected patient data and performed cell experiments. JX, SY and LX performed PCR, western blotting and other molecular experiments. YY and XS contributed to the study design and manuscript writing. ZJ and CZ contributed to funding support, data analysis and revising the manuscript for important intellectual content. All the authors have read and approved the manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Sichuan University (Chengdu, China) and the patients provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Sitarz R, Skierucha M, Mielko J, Offerhaus GJA, Maciejewski R, Polkowski WP. Gastric cancer: Epidemiology, prevention, classification, and treatment. Cancer Manag Res. 2018;10:239–248. doi: 10.2147/CMAR.S149619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rawla P, Barsouk A. Epidemiology of gastric cancer: Global trends, risk factors and prevention. Prz Gastroenterol. 2019;14:26–38. doi: 10.5114/pg.2018.80001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mocellin S, Verdi D, Pooley KA, Nitti D. Genetic variation and gastric cancer risk: A field synopsis and meta-analysis. Gut. 2015;64:1209–1219. doi: 10.1136/gutjnl-2015-309168. [DOI] [PubMed] [Google Scholar]
- 4.Wadhwa R, Song S, Lee JS, Yao Y, Wei Q, Ajani JA. Gastric cancer molecular and clinical dimensions. Nat Rev Clin Oncol. 2013;10:643–655. doi: 10.1038/nrclinonc.2013.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riquelme I, Saavedra K, Espinoza JA, Weber H, García P, Nervi B, Garrido M, Corvalán AH, Roa JC, Bizama C. Molecular classification of gastric cancer: Towards a pathway-driven targeted therapy. Oncotarget. 2015;6:24750–24779. doi: 10.18632/oncotarget.4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coccolini F, Montori G, Ceresoli M, Cima S, Valli MC, Nita GE, Heyer A, Catena F, Ansaloni L. Advanced gastric cancer: What we know and what we still have to learn. World J Gastroenterol. 2016;22:1139. doi: 10.3748/wjg.v22.i3.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giordano S, Columbano A. MicroRNAs: New tools for diagnosis, prognosis, and therapy in hepatocellular carcinoma? Hepatology. 2013;57:840–847. doi: 10.1002/hep.26095. [DOI] [PubMed] [Google Scholar]
- 8.Gao F, Zhang C, Zhou C, Sun W, Liu X, Zhang P, Han J, Xian L, Bai D, Liu H, et al. A critical role of toll-like receptor 2 (TLR2) and its' in vivo ligands in radio-resistance. Sci Rep. 2015;5:13004. doi: 10.1038/srep13004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng B, Liang L, Wang C, Huang S, Cao X, Zha R, Liu L, Jia D, Tian Q, Wu J, et al. MicroRNA-148a suppresses tumor cell invasion and metastasis by downregulating ROCK1 in gastric cancer. Clin Cancer Res. 2011;17:7574–7583. doi: 10.1158/1078-0432.CCR-11-1714. [DOI] [PubMed] [Google Scholar]
- 10.Li J, Dong G, Wang B, Gao W, Yang Q. miR-543 promotes gastric cancer cell proliferation by targeting SIRT1. Biochem Biophys Res Commun. 2016;469:15–21. doi: 10.1016/j.bbrc.2015.11.062. [DOI] [PubMed] [Google Scholar]
- 11.Zhou X, Zhu W, Li H, Wen W, Cheng W, Wang F, Wu Y, Qi L, Fan Y, Chen Y, et al. Diagnostic value of a plasma microRNA signature in gastric cancer: A microRNA expression analysis. Sci Rep. 2015;5:11251. doi: 10.1038/srep11251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ibarrola-Villava M, Llorca-Cardeñosa MJ, Tarazona N, Mongort C, Fleitas T, Perez-Fidalgo JA, Roselló S, Navarro S, Ribas G, Cervantes A. Deregulation of ARID1A, CDH1, cMET and PIK3CA and target-related microRNA expression in gastric cancer. Oncotarget. 2015;6:26935. doi: 10.18632/oncotarget.4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ge X, Liu X, Lin F, Li P, Liu K, Geng R, Dai C, Lin Y, Tang W, Wu Z, et al. MicroRNA-421 regulated by HIF-1α promotes metastasis, inhibits apoptosis, and induces cisplatin resistance by targeting E-cadherin and caspase-3 in gastric cancer. Oncotarget. 2016;26:24466–24682. doi: 10.18632/oncotarget.8228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du Y, Wang L, Wu H, Zhang Y, Wang K, Wu D. MicroRNA-141 inhibits migration of gastric cancer by targeting zinc finger E-box-binding homeobox 2. Mol Med Rep. 2015;12:3416–3422. doi: 10.3892/mmr.2015.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu Q, Zhu Q, Zhou Z, Wang Y, Liu X, Yin G, Tong X, Tu K. MicroRNA-876-5p inhibits epithelial-mesenchymal transition and metastasis of hepatocellular carcinoma by targeting BCL6 corepressor like 1. Biomed Pharmacother. 2018;103:645–652. doi: 10.1016/j.biopha.2018.04.037. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y, Xie Y, Li X, Lin J, Zhang S, Li Z, Huo L, Gong R. miR-876-5p acts as an inhibitor in hepatocellular carcinoma progression by targeting DNMT3A. Pathol Res Pract. 2018;214:1024–1030. doi: 10.1016/j.prp.2018.04.012. [DOI] [PubMed] [Google Scholar]
- 17.Bao L, Lv L, Feng J, Chen Y, Wang X, Han S, Zhao H. MiR-876-5p suppresses epithelial-mesenchymal transition of lung cancer by directly down-regulating bone morphogenetic protein 4. J Biosci. 2017;42:671–681. doi: 10.1007/s12038-017-9722-5. [DOI] [PubMed] [Google Scholar]
- 18.Song B, Ji W, Guo S, Liu A, Jing W, Shao C, Li G, Jin G. miR-545 inhibited pancreatic ductal adenocarcinoma growth by targeting RIG-I. FEBS Lett. 2014;588:4375–4381. doi: 10.1016/j.febslet.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 19.Hansen MB, Nielsen SE, Berg K. Re-examination and further development of a precise and rapid dye method for measuring cell growth/cell kill. J Immunol Methods. 1989;119:203–210. doi: 10.1016/0022-1759(89)90397-9. [DOI] [PubMed] [Google Scholar]
- 20.Freimoser FM, Jakob CA, Aebi M, Tuor U. The MTT [3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide] assay is a fast and reliable method for colorimetric determination of fungal cell densities. Appl Environ Microbiol. 1999;65:3727–3729. doi: 10.1128/AEM.65.8.3727-3729.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meletiadis J, Meis JF, Mouton JW, Donnelly JP, Verweij PE. Comparison of NCCLS and 3-(4, 5-dimethyl-2-thiazyl)-2, 5-diphenyl-2H-tetrazolium bromide (MTT) methods of in vitro susceptibility testing of filamentous fungi and development of a new simplified method. J Clin Microbiol. 2000;38:2949–2954. doi: 10.1128/JCM.38.8.2949-2954.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song B, Zhang C, Li G, Jin G, Liu C. MiR-940 inhibited pancreatic ductal adenocarcinoma growth by targeting MyD88. Cell Physiol Biochem. 2015;35:1167–1177. doi: 10.1159/000373995. [DOI] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 25.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garcia DM, Baek D, Shin C, Bell GW, Grimson A, Bartel DP. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat Struct Mol Biol. 2011;18:1139–1146. doi: 10.1038/nsmb.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Friedman RC, Farh KK-H, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome research. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee S, Paulson KG, Murchison EP, Afanasiev OK, Alkan C, Leonard JH, Byrd DR, Hannon GJ, Nghiem P. Identification and validation of a novel mature microRNA encoded by the Merkel cell polyomavirus in human Merkel cell carcinomas. J Clin Virol. 2011;52:272–275. doi: 10.1016/j.jcv.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang RY, Weng KF, Huang YC, Chen CJ. Elevated expression of circulating miR876-5p is a specific response to severe EV71 infections. Sci Rep. 2016;6:24149. doi: 10.1038/srep24149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang H, Li Z, Li Y, Liu J, Li X, Shen T, Duan Y, Hu M, Xu D. A computational method for predicting regulation of human microRNAs on the influenza virus genome. BMC Syst Biol. 2013;7(Suppl 2):S3. doi: 10.1186/1752-0509-7-S2-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu J, Lamouille S, Derynck R. TGF-beta-induced epithelial to mesenchymal transition. Cell Res. 2009;19:156–172. doi: 10.1038/cr.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katz LH, Li Y, Chen JS, Muñoz NM, Majumdar A, Chen J, Mishra L. Targeting TGF-β signaling in cancer. Exp Opin Therapeutic Targets. 2013;17:743–760. doi: 10.1517/14728222.2013.782287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen J, Miao L, Jin G, Ren C, Ke Q, Qian Y, Dong M, Li H, Zhang Q, Ding Y, et al. TGFBR1 tagging SNPs and gastric cancer susceptibility: A two-stage case-control study in Chinese population. Mol Carcinog. 2014;53:109–116. doi: 10.1002/mc.21954. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.