Abstract
Effect of CXCL8 gene silencing-mediated JAK-STAT signaling pathway on epithelial-mesenchymal transition (EMT) of human cutaneous melanoma cells was explored. Eighty patients with cutaneous melanoma were enrolled in the study. Cells were transfected accordingly and divided into five groups: The blank group (human cutaneous melanoma cells), NC group (human cutaneous melanoma cells + blank vector plasmid transfection), CXCL8 siRNA group (human cutaneous melanoma cells + CXCL8 silent expression vector plasmid transfection), AG490 group (human cutaneous melanoma cells + JAK-STAT signal pathway inhibitor transfection), CXCL8 siRNA + AG490 group (human cutaneous melanoma cells + JAK-STAT signaling pathway inhibitor + CXCL8 silent expression vector plasmid transfection). The expression levels of CXCL8, JAK2, STAT3, epithelial cadherin (E-cadherin), neurotrophic cadherin (N-cadherin) and vimentin in tissues and cells were detected by RT-qPCR and western blot analysis. CCK-8 and flow cytometry were used to detect cell proliferation and apoptosis. Compared with adjacent normal tissues, the expression of E-cadherin in human cutaneous melanoma tissues was significantly decreased, whereas the expression of CXCL8, JAK2, STAT3, vimentin and N-cadherin was significantly increased (P<0.05). Compared with the blank group, CXCL8 siRNA group and CXCL8 siRNA + AG490 group had significantly lower expression of CXCL8 (P<0.05). Compared with the blank group, the expression levels of JAK2, STAT3, vimentin and N-cadherin in CXCL8 siRNA group, AG490 group and CXCL8 siRNA + AG490 group were decreased, the expression of E-cadherin was increased, the cell proliferation ability was decreased and apoptosis was increased (P<0.05). Compared with CXCL8 siRNA group, the expression of JAK2, STAT3, vimentin and N-cadherin in CXCL8 siRNA + AG490 group were significantly decreased, the expression of E-cadherin was significantly increased, cell proliferation ability was decreased and apoptosis was increased (P<0.05). In conclusion, CXCL8 gene expression silencing may inhibit EMT and cell proliferation while promoting cell apoptosis of human cutaneous melanoma cells by inhibiting the activation of JAK-STAT signaling pathway.
Keywords: CXCL8 gene, JAK-STAT signaling pathway, cutaneous melanoma, proliferation, apoptosis, epithelial-mesenchymal transition
Introduction
Melanoma originates from neural crest of epiblast and is a malignant melanoma originating from the pigmented areas of the skin (cutaneous melanoma elaborated in this study), mucosa, eye and the central nervous system (1,2). The pathogenesis of melanoma is still unclear. Several studies have reported that melanoma is closely related to factors, such as race, large amount of ultraviolet radiation, abuse of estrogen drugs and immunodeficiency (3–5). Due to various carcinogenic factors, melanoma is one of the most invasive and migratory malignant solid tumors that spreads rapidly and may lead to the death of a great number of patients after several months of diagnosis (6,7). Currently, surgery is still an important treatment for melanoma in patients insensitive to radiotherapy or chemotherapy (8,9). Recent studies have suggested that gene therapy can provide a new direction for the treatment of melanoma, and therefore gene therapy has become a promising research field worldwide (10–12). The pathogenesis of melanoma is a complex multi-step process involving numerous genes (13). The malignant transformation process of normal cells into tumor cells involves various gene changes (14,15). In malignant tumors, the expression of tumor suppressor genes is decreased or the function is absent (16). It is worth noting that the threat of cancer to human life is mainly attributed to malignant growth, due to invasion and metastasis, rather than the growth in the primary site. Melanoma is one of the most invasive and migratory malignant solid tumors (17). Degradation of extracellular matrix and basement membrane is a key step in the complex process of melanoma leaving the primary site, invading the surrounding tissues and passing through the basement membrane to enter the lymph or blood circulation and transfer to newly diffused tissues (18,19).
Epithelial-mesenchymal transition (EMT) was proposed by Greenburg and Hay in 1982 (20). EMT refers to the phenomenon of epithelial cells transforming into mesenchymal cells under specific physiological and pathological conditions (21,22). Cells may lose their connection and polarity, change in morphology, and their migration ability may be enhanced, thus gaining invasion and metastasis abilities (23,24). It has been reported that although melanocytes are not traditional epithelial components, there is EMT in the process of melanoma invading dermal stroma (25). EMT has been reported in previous studies to be involved in the invasion and metastasis of melanoma (26–28). During EMT, there are obvious differences in the expression of EMT-related molecules (29), whose comparison in melanocytes and different melanoma cell lines (30) has led to the discovery of abnormal expression of epithelial cadherin (E-cadherin), neurotrophic cadherin (N-cadherin) and vimentin in melanocytes (31,32).
JAK/STAT signaling pathway is a widely used signal transduction pathway of hematopoietic growth factor, which mediates a number of physiological processes of cells (33). JAK/STAT signaling pathway may play critical roles in ontogenesis, hematopoietic regulation and immune response (34). Persistent activation of JAK/STAT signaling pathway is related to the occurrence of various human tumors (35,36). Current studies have confirmed that external stimulators are closely associated with the regulation of various cell behaviors and the activation of signal transduction pathways in cells (33,37,38). JAK/STAT signal transduction pathway is one of the most commonly studied pathways and plays an important role in the process of cell signal transduction. JAK is a class of intracytoplasmic non-receptor soluble tyrosine protein kinases, including the four subfamilies of JAK1, JAK2, JAK3 and TYK2 (39). STAT is a cytoplasmic protein that can bind to the DNA of the target gene regulatory region, which is the downstream substrate of JAK (40). STAT family includes seven members, i.e., STAT1-4, STAT5A, STAT5B and STAT6 (41). After tyrosine phosphorylation of JAK family, STATs in cytoplasm are recruited and phosphorylated to form dimers into the nucleus (42). In the nucleus, STATs bind to the promoter of target genes, and then activate gene expression, so as to regulate physiological and pathological reactions of cells, and function significantly in the process of cell growth, activation, differentiation and apoptosis (43). Moreover, AG490 is a selective inhibitor of JAK2 tyrosine phosphorylation (44) which can effectively block the downstream signal transduction and STAT3 activation, and then inhibit corresponding physiological and pathological effects.
The role of JAK/STAT signaling pathway has been recognized in multiple human malignancies. However, there are few studies on the aforementioned targets in cutaneous melanoma. Furthermore, chemokin CXCL8, also known as interleukin-8, is a pro-inflammatory molecule that functions significantly in tumor microenvironment and has been screened to be associated with the development of various tumors, considering its roles in directing neutrophils and oligodendrocytes to combat infection and tumor cells in the progression of metastasis. The present study was carried out to investigate the effect of CXCL8 silencing-mediated activation of JAK-STAT signaling pathway on EMT of human cutaneous melanoma cells. This study is expected to provide potential reference for target gene screening during genetic therapy and updated therapeutic approaches for inhibiting the development of cutaneous melanoma.
Patients and methods
Reagents and equipment
Laboratory reagents: 95% medical ethanol and absolute ethanol (Tianjin Fuyu Fine Chemical Co., Ltd.); xylene (Shanghai YuanMu Biological Technology Co., Ltd.); polyformaldehyde (Sigma-Aldrich; Merck KGaA); hematoxylin and eosin (H&E; Beijing Solarbio Science & Technology Co., Ltd.); protein denaturant (Shanghai Sibas Biotechnology Development Co., Ltd.), primary polyclonal antibody CXCL8 (Abcam), goat anti-rabbit IgG antibody (Wuhan Boster Biological Technology, Ltd.), ECL kit (Amersham; GE Healthcare). DMEM (HyClone; GE Healthcare), PBS and trypsin (both from Beijing Solarbio Science & Technology Co., Ltd.), BCA kit (23250; Thermo Fisher Scientific, Inc.), Annexin V-FITC/PI apoptotic detection kit (Nanjing KeyGen Biotech Co., Ltd.).
Main experimental equipment: Electronic analytical balance (Shanghai Ping Xuan Scientific Instruments, Ltd.); RM2126 paraffin slicing machine (Shanghai Leica Instruments Ltd.); Olympus fluorescent microscope and inverted fluorescent microscope (both from Olympus Corporation); gel imaging system (ChemiDoc MP; Bio-Rad Laboratories, Inc.); pipette (0.5-10 µl, 20-200 µl, Research® plus 100-1,000 µl; Beijing Gilson Science & Technology Co., Ltd.).
Research subjects
Melanoma tissues were collected from 80 patients with cutaneous melanoma admitted to the General Hospital of Ningxia Medical University (Yinchuan, China) from January 2016 to January 2018. The collected melanoma tissues served as the experimental group and the adjacent normal tissues as the control group. All cases were confirmed by histopathology. The experimental group consisted of 53 males and 27 females, with an average age of 61.55±9.04 years, and the age at onset ranged from 32 to 80 years. Among the 80 cases, there were 50 cases of superficial diffusion and 30 cases of nodular metastasis, 41 cases of non-lymph node metastasis and 39 cases of lymph node metastasis.
Inclusion criteria of eligible cases: i) patients who were diagnosed with cutaneous melanoma by histopathology; ii) patients with complete medical records; iii) patients with no previous history of radiotherapy and chemotherapy. Exclusion criteria: i) Patients who were not treated in the aforementioned hospital after diagnosis; ii) patients who had a history of treatment in other hospitals; iii) patients with other malignant tumors. Tumor tissues in the center of the lesion and non-tumor tissues adjacent to the cancer (5 cm) were collected and frozen in cryopreservation tubes. Next, the tumor tissues were quickly frozen in liquid nitrogen tanks (−196°C). The samples were stored in a refrigerator at −80°C. The study was approved by the Ethics Committee of the General Hospital of Ningxia Medical University. Patients who participated in this research had complete clinical data. Signed informed consents were obtained from the patients or their guardians.
H&E staining
During H&E staining, human cutaneous melanoma tissues and adjacent normal tissues were divided into melanoma group and normal control group, respectively. The main reagent formulations were as follows: i) Hematoxylin staining: Hematoxylin (1 g), ethanol (10 ml), distilled water (200 ml), potassium alum (20 g), HgO (0.5 g); ii) eosin staining: Eosin (2.5-5 g), distilled water (500 ml); iii) hydrochloric acid alcohol differentiation solution: 0.5 ml hydrochloric acid and 100 ml 75% alcohol to mix. a) Tissue sample dewaxing: The tissue sample slices were placed in xylene, soaked fully for 10 min, then replaced in xylene and continuously soaked for 10 min. b) Hydration of tissue samples: The tissue samples soaked in xylene were first immersed in absolute ethanol for 5 min, and then in 95, 85 and 70% ethanol for 5 min for complete hydration. c) Hematoxylin staining, differentiation, and bluing: The dewaxed tissue samples were placed in hematoxylin solution, stained for 5 min, and washed with tap water for 5 min; the stained tissue samples were placed in alcohol hydrochloride differentiation solution for several seconds; the slice was washed for 10 min with tap water, and the nuclear staining was observed under the microscope. d) Eosin staining: Tissue samples were placed in 0.5% eosin solution and stained for 5 min. v) Dehydration: After H&E staining, the tissue samples were put into 95% ethanol I, 95% ethanol II, 100% ethanol I and 100% ethanol II for 2 min, respectively. e) Tissue sample slice air-drying and sealing: The dehydrated tissue sample slice was soaked in xylene twice for 4 min each time, and then the tissue sample slice was dried and sealed with neutral gum. f) Finally, the slices were observed and photographed under a microscope. With the morphological image analysis system, different groups were analyzed (magnification, ×400), and the images were collected randomly. The experiment was repeated 3 times.
RT-qPCR
Some melanoma tissues and adjacent normal tissues were ground into fine powder by adding liquid nitrogen. Total RNA was extracted using TRIzol® reagent (Sangon Biotech Co., Ltd.), and RNA concentration and purity were determined. Total RNA was reverse transcribed into cDNA (total volume, 20 µl) using the PrimeScript™ RT reagent kit (Takara Bio, Inc.), according to the manufacturer's protocol. The reaction conditions of the reverse transcription of RNA into cDNA were: Incubation at 25°C for 10 min, additional incubation at 42°C for 50 min, and heating at 95°C for 5 min. cDNA was diluted with DEPC water and blended. Fluorescent RT-qPCR was performed following the manufacturer's protocol. During RT-qPCR double-labeled fluorescent probes (5′-end of the fluorescent reporting group, 3′-end of the fluorescent quenching group) were used, in which the 5′-end was 6-carboxy-fluorescene-labeled fluorescence group and the 3′-end was 6-carboxytetramethylrhodamine-labeled fluorescence group (both from Applied Biosystems; Thermo Fisher Scientific, Inc.). The primers of CXCL8, JAK2, STAT3, E-cadherin, N-cadherin, vimentin and β-actin were designed and synthesized by Wuhan Boster Biological Technology, Ltd.: CXCL8 forward (5′-3′), ATGGCT GCTGAACCAGTAGA and reverse (5′-3′), CTAGTCTTC GTTTTGAACAG; JAK2 forward (5′-3′), GGGTGTTCGCGT CGCCACTT and reverse (5′-3′), CAGATCGGGCGACCA GAGCG; STAT3 forward (5′-3′), GCGGCAGTTTCTGGC CCCTT and reverse (5′-3′), CGGGCCACAATCCGGGCAAT; E-cadherin forward primer (5′-3′), ATGCTGATGCCCCCA ATACC and reverse (5′-3′), ATCTTGCCAGGTCCTTTGCT; N-cadherin forward (5′-3′), CCGGAGAACAGTCTCCAACTC and reverse (5′-3′), CCCACAAACAGCACCAGTC; vimentin forward (5′-3′), GGTGCAATCGTGATCTGGGA and reverse (5′-3′), GTCTTTGCTCGAATGTGCGG; and β-actin forward primer (5′-3′), CCCAGCACAATGAAGATCAAGATCAT and reverse (5′-3′), ATCTGCTGGAAGGTGGACAGCGA. The reaction system volume was 25 µl and was consisted of cDNA (1 µl), 10X PCR buffer (2.5 µl), 10 mmol/l dNTPs (2 µl), PCR upstream primers (1 µl), PCR downstream primers (1 µl), Taq DNA polymerase (1 µl), and deionized water (16.5 µl). Fluorescent RT-qPCR was performed in ABI PRISM® 7300 system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The reaction conditions were as follows: Pre-denaturation at 95°C for 5 min, denaturation at 94°C for 1 min, annealing at 54°C for 45 sec, extension at 72°C for 1 min, in a total of 30 cycles, followed by extension at 72°C for 10 min. β-actin was used as internal reference. All primers were designed and synthesized by Wuhan Boster Biological Technology, Ltd. The 2−∆∆Cq method (45) was used to express the multiplier relationship between the target gene expression in the experimental group and the control group. The experiment was repeated 3 times independently.
Western blot analysis
Some melanoma and adjacent normal tissues were ground with liquid nitrogen into a homogeneous fine powder and 1 ml of modified RIPA lysis buffer (Beyotime Institute of Biotechnology) was added for protein extraction. Melanoma and adjacent normal tissues were ground into homogenate in an ice bath. Protein lysate was added to the melanoma tissues at 4°C for 30 min and shaken every 10 min for the extraction of total protein. According to the manufacturer's instructions of the BCA kit, the content of total protein was determined and frozen at −80°C. Protein (50 µg) was collected and mixed with denaturant, boiled for 10 min for denaturation, and then separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). After electrophoresis, the protein was transferred from the SDS-PAGE gel to a nitrocellulose membranes using the electrical transfer method. Nitrate cellulose membranes were blocked overnight at 4°C in PBST (PBS + Tween-20) containing 10% skimmed milk powder. The membranes were rinsed with PBST (5 min for 3 times) and incubated overnight with the primary antibodies: rabbit polyclonal CXCL8 antibody (1:1,000; ab84995), rabbit monoclonal JAK2 antibody (1:1,000; ab108596), rabbit anti-STAT3 antibody (1:100, ab76315), rabbit monoclonal E-cadherin antibody (1:1,000; ab194982), rabbit monoclonal vimentin antibody (1:1,000; ab92547) and rabbit polyclonal N-cadherin antibody (1:1,000; ab76057) (all from Abcam), respectively, followed by PBS washing 3 times at room temperature, 5 min each. Next, the membranes were incubated with goat anti-mouse IgG antibody labeled with horseradish peroxidase (1:2,000; ab6789; Wuhan Boster Biological Technology, Ltd.) for 120 min at room temperature. The reference gene was mouse monoclonal β-actin antibody (1:500; ab8226; Abcam). The membranes were washed 3 times with TBST, and ECL kit was used for luminescence reaction. Protein bands were visualized using Bio-Rad Image Reader imaging analyzer (Bio-Rad Laboratories, Inc.). Quantity One software 4.6.6 software (Bio-Rad Laboratories, Inc.) was used for the analysis of the gray value of the bands. The experiment was repeated three times.
Screening of cell lines, cell culture and transfection
Human melanoma cell lines A375 (cat. no. AC148), SK-MEL-28 (cat. no. AC339802) and SK-MEL-1 (cat. no. AC100491) (Shanghai Institute of Cell Research, Chinese Academy of Sciences) were selected for subculture and were inoculated in culture medium. The cells were cultured in a thermostat at 37°C and 5% CO2. The medium consisted of 10% fetal bovine serum, RPMI-1640 medium and penicillin-streptomycin. Every 24-48 h, the medium was replaced, digested and passaged with 0.25% trypsin, and the cells in logarithmic phase were selected for the experiment. The expression of CXCL8 in A375, SK-MEL-28 and SK-MEL-1 cells was detected by fluorescent RT-qPCR, and the optimal cell line was screened out for subsequent experiments.
Cells were divided into five groups: Blank group (human cutaneous melanoma cells), NC group (human cutaneous melanoma cells + blank vector plasmid transfection), CXCL8 siRNA group (human cutaneous melanoma cells + CXCL8 silent expression vector plasmid transfection), AG490 group (human cutaneous melanoma cells + JAK-STAT signal pathway inhibitor transfection), CXCL8 siRNA + AG490 group (human cutaneous melanoma cells + JAK-STAT signaling pathway inhibitor + CXCL8 silent expression vector plasmid transfection). Transfection plasmids were purchased from Thermo Fisher Scientific, Inc., and JAK-STAT signaling pathway inhibitor was purchased from Beijing Baioleibo Technology Co., Ltd. Cells were inoculated into 6-well plates 24 h before transfection. When the cell confluency reached ~50-60%, the cells were transfected into a human cutaneous melanoma cell line by Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). The cells were transfected for 6 h and then cultured for 48 h, before collected for subsequent experiments.
CCK-8 assay for the detection of cell proliferation after transfection
When the cell growth density reached ~80%, all the cells in logarithmic growth phase were washed twice with PBS solution and digested by 0.25% trypsin to form a single cell suspension. After counting, the cells with adjusted density of 3×103 were inoculated into 96-well plates with a volume of 200 µl/well, with 6 replicates made for each sample. Cells were evenly dispersed by gentle shaking. Subsequently, the 96-well plates were incubated at 37°C and 5% CO2 saturated humidity. The plates were taken out at 24, 48, 72 and 96 h, respectively, followed by the addition of 10 µl CCK-8 (Sigma-Aldrich; Merck KGaA) per well for continuous culture for 3 h. The absorbance values (A values) of each well were read at 450 nm by a microplate reader. Each experiment was repeated 3 times. Cell viability curves were plotted with time as abscissa and A value as ordinate.
Flow cytometry
For the apoptosis detection by flow cytometry, human cutaneous melanoma cell lines were selected. The experimental groups were the Blank group, NC group, CXCL8 siRNA group, AG490 group and CXCL8 siRNA + AG490 group. i) Cell collection: Adherent cells were digested and directly collected into 5 ml centrifugal tubes. The number of cells per sample was 1 to 5×106/ml. After centrifugation at 800 × g for 5 min, the culture medium was abandoned. ii) Preparation of cell suspension: Cells were washed with PBS and centrifuged at 800 × g for 5 min, and the supernatant was discarded. iii) Annexin V-FITC/PI double labeled staining by using a flow cytometer (Thermo Fisher Scientific, Inc.), with the apoptotic rate analyzed by ModFit LT 3.0 software (Verity Software House, Inc): 500 µl cells were suspended by a binding buffer, 5 µl PI and 5 µl Annexin V-FITC were added to each tube, and the cells were incubated at room temperature in the dark for 15 min. iv) Annexin V-FITC and PI fluorescence were detected by flow cytometry, and apoptosis was detected.
Statistical analysis
Statistical data were processed and analyzed by the SPSS 21.0 statistical software (IBM Corp.). Normal distribution and homogeneous tests of variance were performed for all data. The parameters of the experimental measurement data conforming to a normal distribution were expressed as the mean ± standard deviation. Data were analyzed between two groups using the independent samples t-test. One-way analysis of variance (ANOVA) was used for comparisons among groups, followed by the Bonferroni post hoc. P<0.05 was considered to indicate a statistically significant difference.
Results
H&E staining in human cutaneous melanoma
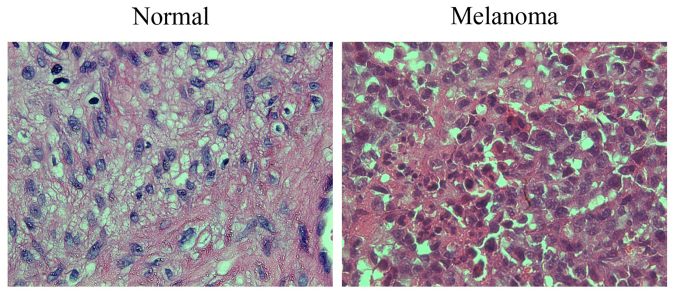
H&E staining revealed that compared with the adjacent normal tissues, the cells in the melanoma tissues presented obvious atypia, had oval appearance with different shapes and sizes in most cases, indicating compact arrangement, obvious nucleoli, mitotic figures, large cytoplasm, diffuse distribution, and increased production of melanin granules between and within cells (Fig. 1).
Figure 1.
H&E staining for the detection of histopathological changes in tumor and adjacent normal tissues of cutaneous melanoma (×400). Scale bar, 25 µm. H&E, hematoxylin and eosin.
mRNA and protein expression of related genes
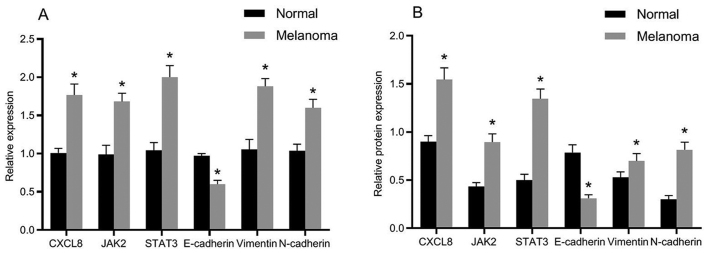
The mRNA and protein expression levels of related genes in normal and melanoma tissues are presented in Fig. 2. Compared with the adjacent normal tissues, the mRNA and protein expression levels of E-cadherin were significantly decreased in melanoma tissues, whereas the mRNA and protein expression levels of CXCL8, JAK2, STAT3, vimentin and N-cadherin were obviously increased. The differences were statistically significant (P<0.05).
Figure 2.
Expression of related genes and proteins in tissues of cutaneous melanoma. (A) Histogram of the mRNA expression of related genes in tissues. (B) Histogram of the protein expression of related proteins in tissues; *P<0.05, compared with adjacent normal tissues.
Screening of cell lines
The expression of CXCL8 gene in A375 human cutaneous melanoma cells was the highest compared with that in SK-MEL-28 and SK-MEL-1 human cutaneous melanoma cells, with statistically significant differences (P<0.05) (Fig. 3). Thus, A375 human cutaneous melanoma cells were screened out for subsequent experiments.
Figure 3.
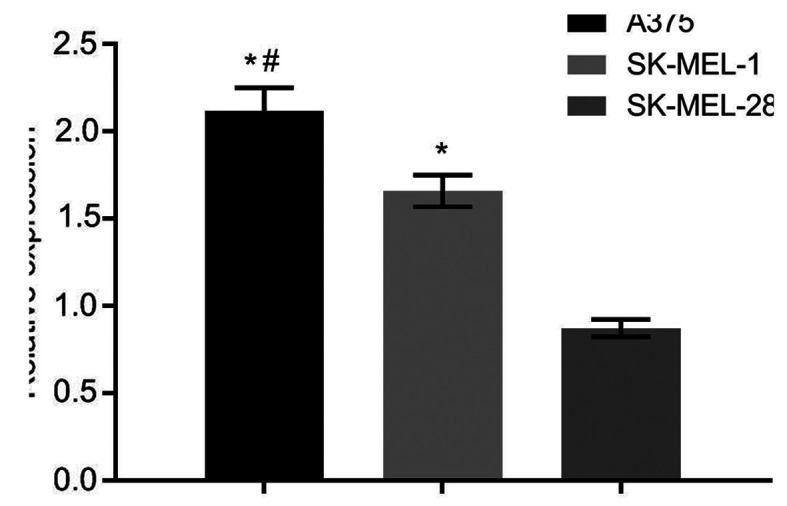
CXCL8 gene expression in different cell lines by fluorescent quantitative PCR. *P<0.05, compared with SK-MEL-28 cell line; #P<0.05, compared with SK-MEL-1 cell line.
mRNA and protein expression of related genes after cell transfection
The mRNA and protein expression levels of related genes after transfection are presented in Fig. 4. The CXCL8 siRNA group and CXCL8 siRNA + AG490 group showed significantly decreased CXCL8 expression compared with the blank group (P<0.05), whereas there was no significant difference in the CXCL8 expression compared with the NC and AG490 groups (P>0.05). Compared with the blank group, the expression levels of JAK2, STAT3, vimentin and N-cadherin in the CXCL8 siRNA group, AG490 group and CXCL8 siRNA + AG490 group were significantly decreased, whereas the expression level of E-cadherin was increased (P<0.05). There was no obvious difference in the expression levels of the related genes compared with those in the CXCL8 siRNA group and AG490 group (P>0.05). Compared with CXCL8 siRNA group, the expression levels of JAK2, STAT3, vimentin and N-cadherin in CXCL8 siRNA + AG490 group were obviously decreased, whereas the E-cadherin expression was significantly increased (P<0.05).
Figure 4.
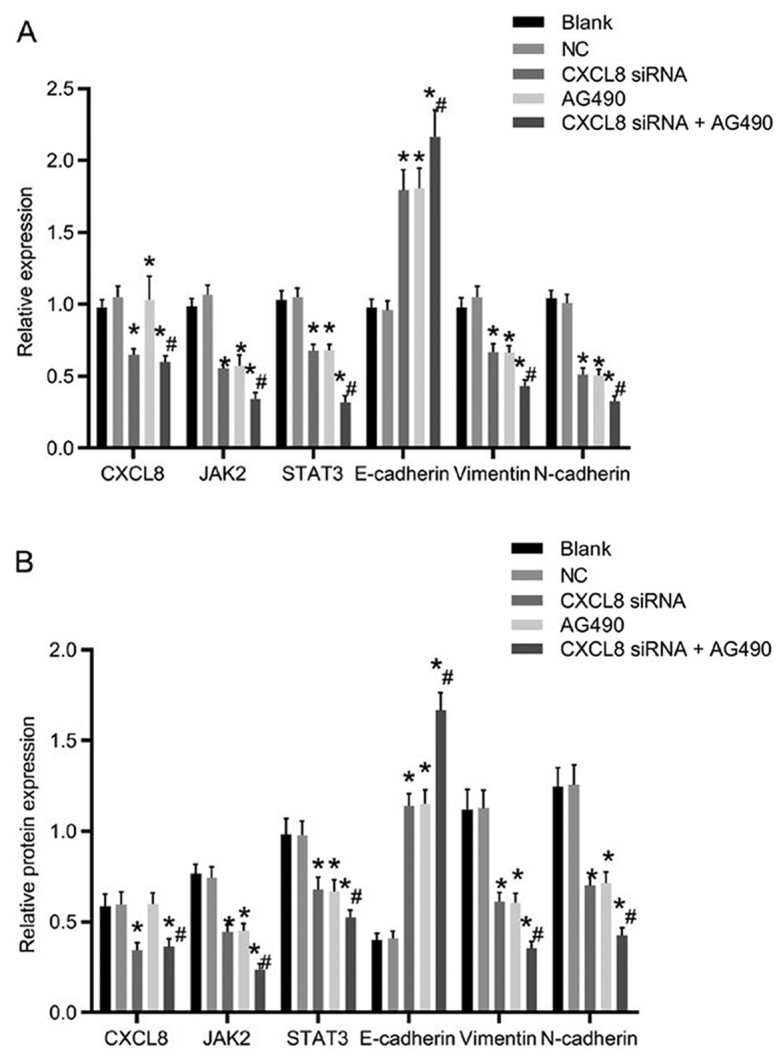
Comparison of the mRNA and protein expression of related genes after transfection in different groups. (A) Histogram of the mRNA expression of related genes in cells. (B) Histogram of the protein expression of related proteins in cells; *P<0.05 compared with the Blank group; #P<0.05 compared with the CXCL8 siRNA group.
Changes of cell proliferation ability after cell transfection
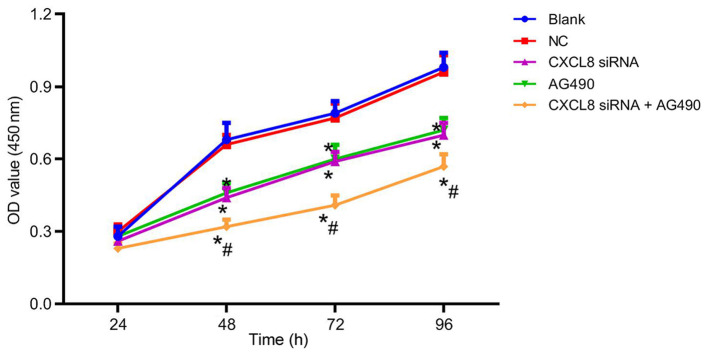
After cell transfection, the cell proliferation ability of each group is presented in Fig. 5. There was no obvious difference in cell proliferation among groups at 24 h (P>0.05). At 48, 72 and 96 h, the cell proliferation ability in the CXCL8 siRNA group, AG490 group and CXCL8 siRNA + AG490 group was decreased compared with that in the blank group (P<0.05). There was no significant difference in the cell proliferation of the NC group at the different time-points (P>0.05); and no significant difference was presented between the cell proliferation ability of the CXCL8 siRNA group and AG490 group at the different time-points (P>0.05). Compared with CXCL8 siRNA group, CXCL8 siRNA + AG490 group showed a significant decrease in cell proliferation (P<0.05).
Figure 5.
Comparison of cell proliferation in each group after cell transfection. *P<0.05, compared with the blank group, and #P<0.05, compared with the CXCL8 siRNA group.
Changes of cell apoptosis ability after cell transfection
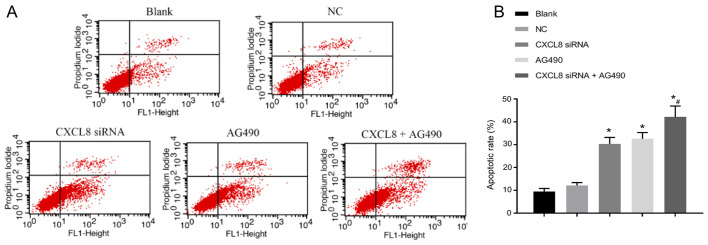
The apoptotic status is shown in Fig. 6. Compared with the blank group, the apoptotic rate of cells in the CXCL8 siRNA group, AG490 group and CXCL8 + AG490 group was increased (P<0.05). There was no significant difference in cell apoptosis when compared with NC group (P>0.05). In addition, no significant difference was detected in cell apoptosis between CXCL8 siRNA group and AG490 group (P>0.05). Compared with CXCL8 siRNA group, CXCL8 siRNA + AG490 group presented obvious increase in cell apoptosis (P<0.05).
Figure 6.
Comparison of cell apoptosis in each group after cell transfection. (A) Flow cytometry results in each group. (B) Comparison histogram of apoptotic rate in each group; *P<0.05, compared with the blank group, and #P<0.05, compared with the CXCL8 siRNA group.
Discussion
Invasion and migration of malignant tumor cells is a complex multi-step and multi-factor process (46,47). Melanoma is highly malignant and prone to hematogenous and lymphatic metastasis with poor prognosis (1). Early detection and diagnosis are particularly important for the prediction of melanoma prognosis (4). The preferred treatment for malignant melanoma is early surgical excision (8). Chemotherapy and radiotherapy have been used to prevent the recurrence of lesions, remove small foci and metastases, and prevent metastasis after surgical resection (9). However, melanoma is insensitive to current radiotherapy and chemotherapy, and most of the anti-neoplastic drugs currently used in clinic have strong toxicity and side-effects, which seriously affect the quality of life of cancer patients (9). Therefore, searching and screening for safe, effective, less toxic side-effects of anticancer strategies are highly valued.
JAK-STAT signal transduction pathway is a classic signal transduction pathway, which participates in cell proliferation, survival, transformation, migration and other processes (33,34). Among the six family members of STATs, STAT1, STAT3 and STAT5 are the most common (41). It has been reported for numerous tumors that blocking the functions of STAT3 and STAT5 alone can inhibit the growth of tumor cells and induce apoptosis (48). Previous studies have shown that STAT3 is expressed at high levels and is activated in cervical and endometrial cancer tissues (49,50). Blocking the persistent signal pathway of STAT3 is considered to be able to inhibit the growth of cancer cells, which may become a new therapeutic target (51). Pan et al (52) have reported that high mRNA expression levels of STAT3 in colon cancer, breast cancer and glioblastoma are related significantly to shorter survival times. In addition, cell proliferation and apoptosis maintain homeostasis under normal conditions, and the mechanism regulating the balance maintains the normal physiological function of the body (53). Once this balance is destroyed, it leads to numerous serious pathological changes, such as tumors, degenerative diseases and autoimmune diseases. Development of tumors are not only related to abnormal proliferation and differentiation of tumor cells, but also to the changes of cell death (54). It has been proven that the reduction of apoptosis can cause tumorigenesis and promote malignant transformation and evolution of cancer cells by escaping (55). Therefore, the strategy of inducing apoptosis of cancer cells has become the focus of cancer treatment in recent decades.
In the present study, H&E staining showed that compared with the adjacent normal tissues, melanoma cells in tissues present obvious atypia, oval appearance with different shapes and sizes in most cases, compact arrangement, obvious nucleoli, mitotic figures, large cytoplasm, diffuse distribution, and increased production of melanin granules between and within cells, showing the biological features of melanoma cells. Subsequent gene expression detection showed that the expression of E-cadherin in melanoma tissues was significantly decreased compared with that of the adjacent normal tissues, whereas the expression levels of CXCL8, JAK2, STAT3, vimentin, and N-cadherin were significantly increased. These results suggest that there may be high CXCL8 expression and activation of JAK-STAT signaling pathway, and presence of EMT in cutaneous melanoma. In the malignant evolution of 90% of epithelial malignant tumors, there may be active downregulation of epithelial cells, decreased polarity of inter-cellular homogeneous adhesion system, disengagement from constraint of inherent organizational structure, acquisition of phenotypes and related gene changes, resulting in cell invasion and migration, with EMT (56,57). During EMT, strong phenotypic characteristics of epithelial cells, such as intercellular adhesion and polar distribution, are replaced by mesenchymal phenotype (decreased adhesion, fibroblast phenotype, increased mobility), which is an important link in malignant evolution and metastasis of cancer cells (58,59). It has been reported that human cancer cells can secrete CXCL8 through autocrine and/or paracrine pathways and interact with chemokine receptors to promote the proliferation and migration of tumor cells (60). Furthermore, important markers of EMT are loss of homogeneous adhesion between epithelial cells (decrease in E-cadherin expression, increase in N-cadherin and vimentin expression) (61,62), resulting in cell adhesion complex disintegration, cell adhesion and cell proliferation disorder, morphological and structural disorders of epithelial cells, loss of cell polarity and contact inhibition, cell growth disorder and infiltration into surrounding tissues (63).
Cell line experiment was carried out, and the highest expression level of CXCL8 gene was found in A375 human cutaneous melanoma cells, when compared with that in SK-MEL-28 and SK-MEL-1 human cutaneous melanoma cell lines. Thus, A375 cells were screened out for subsequent experiments. The results for the mRNA and protein expression levels of related genes after transfection were: Compared with the blank group, CXCL8 siRNA group and CXCL8 siRNA + AG490 group showed evidently decreased CXCL8 expression. The expression levels of JAK2, STAT3, N-cadherin and vimentin in CXCL8 siRNA group, AG490 group and CXCL8 siRNA + AG490 group were decreased, whereas the expression of E-cadherin was increased. The aforementioned trends are especially significant in CXCL8 siRNA + AG490 group. In subsequent research of biological characteristics, cell proliferation test results showed that at 48, 72 and 96 h, CXCL8 siRNA group, AG490 group and CXCL8 siRNA + AG490 group had reduced cell proliferation, with more evident decrease of cell proliferation in CXCL8 siRNA + AG490 group. The results indicated that the silenced expression of CXCL8 can inhibit the proliferation of cancer cells, while JAK-STAT signaling pathway activation may promote the proliferation of cancer cells. Apoptotic status of each group further revealed that compared with the blank group, the apoptotic rate in the CXCL8 siRNA group, AG490 group and CXCL8 + AG490 group was increased, and it was in particular significantly increased in the CXCL8 siRNA + AG490 group. These results suggest that suppressed expression of CXCL8 can promote apoptosis of cancer cells, whereas activation of JAK-STAT signaling pathway can inhibit apoptosis of cancer cells.
In conclusion, silencing of CXCL8 gene expression may inhibit EMT and cell proliferation while promoting cell apoptosis of human cutaneous melanoma cells by inhibiting JAK-STAT signaling pathway activation. In clinical practice, the study of the association of CXCL8 and JAK-STAT signaling pathway with EMT and apoptosis of cutaneous melanoma will have essential impact on the diagnosis, treatment and drug screening of cutaneous melanoma, especially with the discovery of more signal transduction substrates, activators and inhibitors.
Acknowledgements
We would like to thank our team members for their work and our colleagues for their valuable scientific advice and helpful suggestions.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
XH wrote the manuscript, analyzed and interpreted the patients' data. LY and TM performed H&E staining, PCR, western blot analysis, CCK-8 assay and flow cytometry, and were responsible for the statistical analysis. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The study was approved by the Ethics Committee of the General Hospital of Ningxia Medical University (Yinchuan, China). Patients who participated in this research had complete clinical data. Signed informed consents were obtained from the patients or their guardians.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Aggarwal R, Dhawan S, Chopra P. Primary gastric melanoma: A diagnostic challenge. J Gastrointest Cancer. 2014;45(Suppl 1):33–35. doi: 10.1007/s12029-013-9530-6. [DOI] [PubMed] [Google Scholar]
- 2.Varamo C, Occelli M, Vivenza D, Merlano M, Lo Nigro C. MicroRNAs role as potential biomarkers and key regulators in melanoma. Genes Chromosomes Cancer. 2017;56:3–10. doi: 10.1002/gcc.22402. [DOI] [PubMed] [Google Scholar]
- 3.Liu-Smith F, Ziogas A. An age-dependent interaction between sex and geographical UV index in melanoma risk. J Am Acad Dermatol. 2017 Dec 2; doi: 10.1016/j.jaad.2017.11.049. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nissen LHC, Pierik M, Derikx LAAP, de Jong E, Kievit W, van den Heuvel TRA, van Rosendael AR, Plasmeijer EI, Dewint P, Verhoeven RHA, et al. Risk factors and clinical outcomes in patients with IBD with melanoma. Inflamm Bowel Dis. 2017;23:2018–2026. doi: 10.1097/MIB.0000000000001191. [DOI] [PubMed] [Google Scholar]
- 5.Hübner J, Waldmann A, Eisemann N, Noftz M, Geller AC, Weinstock MA, Volkmer B, Greinert R, Breitbart EW, Katalinic A. Association between risk factors and detection of cutaneous melanoma in the setting of a population-based skin cancer screening. Eur J Cancer Prev. 2018;27:563–569. doi: 10.1097/CEJ.0000000000000392. [DOI] [PubMed] [Google Scholar]
- 6.Fratangelo F, Camerlingo R, Carriero MV, Pirozzi G, Palmieri G, Gentilcore G, Ragone C, Minopoli M, Ascierto PA, Motti ML. Effect of ABT-888 on the apoptosis, motility and invasiveness of BRAFi-resistant melanoma cells. Int J Oncol. 2018;53:1149–1159. doi: 10.3892/ijo.2018.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramgolam K, Lauriol J, Lalou C, Lauden L, Michel L, de la Grange P, Khatib AM, Aoudjit F, Charron D, Alcaide- Loridan C, et al. Melanoma spheroids grown under neural crest cell conditions are highly plastic migratory/invasive tumor cells endowed with immunomodulator function. PLoS One. 2011;6:e18784. doi: 10.1371/journal.pone.0018784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tchernev G. One step melanoma surgery for patient with thick primary melanomas: ‘To break the rules, you must first master them!’. Open Access Maced J Med Sci. 2018;6:367–371. doi: 10.3889/oamjms.2018.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marinova L, Yordanov K, Sapundgiev N. Primary mucosal sinonasal melanoma - case report and review of the literature. The role of complex treatment-surgery and adjuvant radiotherapy. Rep Pract Oncol Radiother. 2010;16:40–43. doi: 10.1016/j.rpor.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tesic N, Kamensek U, Sersa G, Kranjc S, Stimac M, Lampreht U, Preat V, Vandermeulen G, Butinar M, Turk B, et al. Endoglin (CD105) silencing mediated by shRNA under the control of endothelin-1 promoter for targeted gene therapy of melanoma. Mol Ther Nucleic Acids. 2015;4:e239. doi: 10.1038/mtna.2015.12. [DOI] [PubMed] [Google Scholar]
- 11.Menezes ME, Talukdar S, Wechman SL, Das SK, Emdad L, Sarkar D, Fisher PB. Prospects of gene therapy to treat melanoma. Adv Cancer Res. 2018;138:213–237. doi: 10.1016/bs.acr.2018.02.007. [DOI] [PubMed] [Google Scholar]
- 12.Mohammed-Saeid W, Chitanda J, Al-Dulaymi M, Verrall R, Badea I. Design and evaluation of RGD-modified gemini surfactant-based lipoplexes for targeted gene therapy in melanoma model. Pharm Res. 2017;34:1886–1896. doi: 10.1007/s11095-017-2197-0. [DOI] [PubMed] [Google Scholar]
- 13.Piccinin S, Doglioni C, Maestro R, Vukosavljevic T, Gasparotto D, D'Orazi C, Boiocchi M. p16/CDKN2 and CDK4 gene mutations in sporadic melanoma development and progression. Int J Cancer. 1997;74:26–30. doi: 10.1002/(SICI)1097-0215(19970220)74:1<26::AID-IJC5>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 14.Singer M, Wang C, Cong L, Marjanovic ND, Kowalczyk MS, Zhang H, Nyman J, Sakuishi K, Kurtulus S, Gennert D, et al. A distinct gene module for dysfunction uncoupled from activation in tumor-infiltrating T cells. Cell. 2016;166:1500–1511.e9. doi: 10.1016/j.cell.2016.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y, Han N, Zhou S, Zhou R, Yuan X, Xu H, Zhang C, Yin T, Wu K. The DACH/EYA/SIX gene network and its role in tumor initiation and progression. Int J Cancer. 2016;138:1067–1075. doi: 10.1002/ijc.29560. [DOI] [PubMed] [Google Scholar]
- 16.Volodko N, Gordon M, Salla M, Ghazaleh HA, Baksh S. RASSF tumor suppressor gene family: Biological functions and regulation. FEBS Lett. 2014;588:2671–2684. doi: 10.1016/j.febslet.2014.02.041. [DOI] [PubMed] [Google Scholar]
- 17.Patel M, Boo H, Kandasamy S, Patel D, Iorio A. The advantages of dermoscopy in the diagnosis of acral melanoma from other podiatric lesions: A literature review. J Am Podiatr Med Assoc. 2016;106:3. doi: 10.7547/8750-7315-2016.1.Iorio. [DOI] [Google Scholar]
- 18.Benton G, Kleinman HK, George J, Arnaoutova I. Multiple uses of basement membrane-like matrix (BME/Matrigel) in vitro and in vivo with cancer cells. Int J Cancer. 2011;128:1751–1757. doi: 10.1002/ijc.25781. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y, Sun X, Nan N, Cao KX, Ma C, Yang GW, Yu MW, Yang L, Li JP, Wang XM, et al. Elemene inhibits the migration and invasion of 4T1 murine breast cancer cells via heparanase. Mol Med Rep. 2017;16:794–800. doi: 10.3892/mmr.2017.6638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greenburg G, Hay ED. Cytoskeleton and thyroglobulin expression change during transformation of thyroid epithelium to mesenchyme-like cells. Development. 1988;102:605–622. doi: 10.1242/dev.102.3.605. [DOI] [PubMed] [Google Scholar]
- 21.Han ZH, Wang F, Wang FL, Liu Q, Zhou J. Regulation of transforming growth factor β-mediated epithelial-mesenchymal transition of lens epithelial cells by c-Src kinase under high glucose conditions. Exp Ther Med. 2018;16:1520–1528. doi: 10.3892/etm.2018.6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ning X, Zhang H, Wang C, Song X. Exosomes released by gastric cancer cells induce transition of pericytes into cancer-associated fibroblasts. Med Sci Monit. 2018;24:2350–2359. doi: 10.12659/MSM.906641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dettman RW, Simon HG. Rebooting the collagen gel: artificial hydrogels for the study of epithelial mesenchymal transformation. Dev Dyn. 2018;247:332–339. doi: 10.1002/dvdy.24560. [DOI] [PubMed] [Google Scholar]
- 24.Zhang P, Dai H, Peng L. AGEs induce epithelial to mesenchymal transformation of human peritoneal mesothelial cells via upregulation of STAT3. Glycoconj J. 2019;36:155–163. doi: 10.1007/s10719-019-09861-7. [DOI] [PubMed] [Google Scholar]
- 25.Zhou L, Yang K, Randall Wickett R, Zhang Y. Dermal fibroblasts induce cell cycle arrest and block epithelial-mesenchymal transition to inhibit the early stage melanoma development. Cancer Med. 2016;5:1566–1579. doi: 10.1002/cam4.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li CY, Wang Q, Shen S, Wei XL, Li GX. Oridonin inhibits migration, invasion, adhesion and TGF-β1-induced epithelial-mesenchymal transition of melanoma cells by inhibiting the activity of PI3K/Akt/GSK-3β signaling pathway. Oncol Lett. 2018;15:1362–1372. doi: 10.3892/ol.2017.7421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tai KF, Wang CH. Using adenovirus armed short hairpin RNA targeting transforming growth factor β1 inhibits melanoma growth and metastasis in an ex vivo animal model. Ann Plast Surg. 2013;71(Suppl 1):S75–S81. doi: 10.1097/SAP.0000000000000041. [DOI] [PubMed] [Google Scholar]
- 28.Sinnberg T, Levesque MP, Krochmann J, Cheng PF, Ikenberg K, Meraz-Torres F, Niessner H, Garbe C, Busch C. Wnt-signaling enhances neural crest migration of melanoma cells and induces an invasive phenotype. Mol Cancer. 2018;17:59. doi: 10.1186/s12943-018-0773-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tu Z, Xie S, Xiong M, Liu Y, Yang X, Tembo KM, Huang J, Hu W, Huang X, Pan S, et al. CXCR4 is involved in CD133-induced EMT in non-small cell lung cancer. Int J Oncol. 2017;50:505–514. doi: 10.3892/ijo.2016.3812. [DOI] [PubMed] [Google Scholar]
- 30.Crosson WP, Berlinberg E, Nazarian R. Identification of targetable EMT markers in cancer stem-like cells derived from therapeutic resistant melanoma. Cancer Res. 2017;77 Abst 3882. [Google Scholar]
- 31.Ding Y, Li X, Hong D, Jiang L, He Y, Fang H. Silence of MACC1 decreases cell migration and invasion in human malignant melanoma through inhibiting the EMT. Biosci Trends. 2016;10:258–264. doi: 10.5582/bst.2016.01091. [DOI] [PubMed] [Google Scholar]
- 32.Wels C, Joshi S, Koefinger P, Bergler H, Schaider H. Transcriptional activation of ZEB1 by Slug leads to cooperative regulation of the epithelial-mesenchymal transition-like phenotype in melanoma. J Invest Dermatol. 2011;131:1877–1885. doi: 10.1038/jid.2011.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang JS, Guh JY, Chen HC, Hung WC, Lai YH, Chuang LY. Role of receptor for advanced glycation end-product (RAGE) and the JAK/STAT-signaling pathway in AGE-induced collagen production in NRK-49F cells. J Cell Biochem. 2001;81:102–113. doi: 10.1002/1097-4644(20010401)81:1<102::AID-JCB1027>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 34.Wang R, Huang S, Fu X, Huang G, Yan X, Yue Z, Chen S, Li Y, Xu A. The conserved ancient role of chordate PIAS as a multilevel repressor of the NF-κB pathway. Sci Rep. 2017;7:17063. doi: 10.1038/s41598-017-16624-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Erdman VV, Nasibullin TR, Tuktarova IA, Somova RS, Mustafina OE. Association analysis of polymorphic gene variants in the JAK/STAT signaling pathway with aging and longevity. Russian Journal of Genetics. Russ J Genet. 2019;55:728–737. doi: 10.1134/S1022795419050077. [DOI] [Google Scholar]
- 36.Mullen M, Gonzalez-Perez RR. Leptin-induced JAK/STAT signaling and cancer growth. Vaccines (Basel) 2016;4:26. doi: 10.3390/vaccines4030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mi C, Ma J, Wang KS, Wang Z, Li MY, Li JB, Li X, Piao LX, Xu GH, Jin X. Amorfrutin A inhibits TNF-α induced JAK/STAT signaling, cell survival and proliferation of human cancer cells. Immunopharmacol Immunotoxicol. 2017;39:338–347. doi: 10.1080/08923973.2017.1371187. [DOI] [PubMed] [Google Scholar]
- 38.Yeh CM, Chang LY, Lin SH, Chou JL, Hsieh HY, Zeng LH, Chuang SY, Wang HW, Dittner C, Lin CY, et al. Epigenetic silencing of the NR4A3 tumor suppressor, by aberrant JAK/STAT signaling, predicts prognosis in gastric cancer. Sci Rep. 2016;6:31690. doi: 10.1038/srep31690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abramovich C, Yakobson B, Chebath J, Revel M. A protein-arginine methyltransferase binds to the intracytoplasmic domain of the IFNAR1 chain in the type I interferon receptor. EMBO J. 1997;16:260–266. doi: 10.1093/emboj/16.2.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khanna P, Chua PJ, Wong BSE, Yin C, Thike AA, Wan WK, Tan PH, Baeg GH. GRAM domain-containing protein 1B (GRAMD1B), a novel component of the JAK/STAT signaling pathway, functions in gastric carcinogenesis. Oncotarget. 2017;8:115370–115383. doi: 10.18632/oncotarget.23265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Slattery ML, Lundgreen A, Kadlubar SA, Bondurant KL, Wolff RK. JAK/STAT/SOCS-signaling pathway and colon and rectal cancer. Mol Carcinog. 2013;52:155–166. doi: 10.1002/mc.21841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma JF, Sanchez BJ, Hall DT, Tremblay AK, Di Marco S, Gallouzi IE. STAT3 promotes IFNγ/TNFα-induced muscle wasting in an NF-κB-dependent and IL-6-independent manner. EMBO Mol Med. 2017;9:622–637. doi: 10.15252/emmm.201607052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nosaka T, Kawashima T, Misawa K, Ikuta K, Mui AL, Kitamura T. STAT5 as a molecular regulator of proliferation, differentiation and apoptosis in hematopoietic cells. EMBO J. 1999;18:4754–4765. doi: 10.1093/emboj/18.17.4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gurbuz V, Konac E, Varol N, Yilmaz A, Gurocak S, Menevse S, Sozen S. Effects of AG490 and S3I-201 on regulation of the JAK/STAT3 signaling pathway in relation to angiogenesis in TRAIL-resistant prostate cancer cells in vitro. Oncol Lett. 2014;7:755–763. doi: 10.3892/ol.2014.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hwang TL, Changchien TT, Wang CC, Wu CM. Claudin-4 expression in gastric cancer cells enhances the invasion and is associated with the increased level of matrix metalloproteinase-2 and −9 expression. Oncol Lett. 2014;8:1367–1371. doi: 10.3892/ol.2014.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 47.Porther N, Barbieri MA. The role of endocytic Rab GTPases in regulation of growth factor signaling and the migration and invasion of tumor cells. Small GTPases. 2015;6:135–144. doi: 10.1080/21541248.2015.1050152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dong B, Liang Z, Chen Z, Li B, Zheng L, Yang J, Zhou H, Qu L. Cryptotanshinone suppresses key onco-proliferative and drug-resistant pathways of chronic myeloid leukemia by targeting STAT5 and STAT3 phosphorylation. Sci China Life Sci. 2018;61:999–1009. doi: 10.1007/s11427-018-9324-y. [DOI] [PubMed] [Google Scholar]
- 49.Chen W, He W, Cai H, Hu B, Zheng C, Ke X, Xie L, Zheng Z, Wu X, Wang H. A-to-I RNA editing of BLCAP lost the inhibition to STAT3 activation in cervical cancer. Oncotarget. 2017;8:39417–39429. doi: 10.18632/oncotarget.17034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wallbillich JJ, Josyula S, Saini U, Zingarelli RA, Dorayappan KD, Riley MK, Wanner RA, Cohn DE, Selvendiran K. High glucose-mediated STAT3 activation in endometrial cancer is inhibited by metformin: Therapeutic implications for endometrial cancer. PLoS One. 2017;12:e0170318. doi: 10.1371/journal.pone.0170318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bao X, Ren T, Huang Y, Sun K, Wang S, Liu K, Zheng B, Guo W. Knockdown of long non-coding RNA HOTAIR increases miR-454-3p by targeting Stat3 and Atg12 to inhibit chondrosarcoma growth. Cell Death Dis. 2017;8:e2605. doi: 10.1038/cddis.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pan Y, Wang S, Su B, Zhou F, Zhang R, Xu T, Zhang R, Leventaki V, Drakos E, Liu W, et al. Stat3 contributes to cancer progression by regulating Jab1/Csn5 expression. Oncogene. 2017;36:1069–1079. doi: 10.1038/onc.2016.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jullien N, Roche C, Brue T, Figarella-Branger D, Graillon T, Barlier A, Herman JP. Dose-dependent dual role of PIT-1 (POU1F1) in somatolactotroph cell proliferation and apoptosis. PLoS One. 2015;10:e0120010. doi: 10.1371/journal.pone.0120010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Horton BL, Williams JB, Cabanov A, Spranger S, Gajewski TF. Intratumoral CD8+ T-cell apoptosis is a major component of T-cell dysfunction and impedes antitumor immunity. Cancer Immunol Res. 2018;6:14–24. doi: 10.1158/2326-6066.CIR-17-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jiang Y, Sheng H, Meng L, Yue H, Li B, Zhang A, Dong Y, Liu Y. RBM5 inhibits tumorigenesis of gliomas through inhibition of Wnt/β-catenin signaling and induction of apoptosis. World J Surg Oncol. 2017;15:9. doi: 10.1186/s12957-016-1084-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Feng X, Zhao L, Shen H, Liu X, Yang Y, Lv S, Niu Y. Expression of EMT markers and mode of surgery are prognostic in phyllodes tumors of the breast. Oncotarget. 2017;8:33365–33374. doi: 10.18632/oncotarget.16497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun B, Zhang D, Zhao N, Zhao X. Epithelial-to-endothelial transition and cancer stem cells: Two cornerstones of vasculogenic mimicry in malignant tumors. Oncotarget. 2017;8:30502–30510. doi: 10.18632/oncotarget.8461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bo H, Zhang S, Gao L, Chen Y, Zhang J, Chang X, Zhu M. Upregulation of Wnt5a promotes epithelial-to-mesenchymal transition and metastasis of pancreatic cancer cells. BMC Cancer. 2013;13:496. doi: 10.1186/1471-2407-13-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Q, Qu C, Xie F, Chen L, Liu L, Liang X, Wu X, Wang P, Meng Z. Curcumin suppresses epithelial-to-mesenchymal transition and metastasis of pancreatic cancer cells by inhibiting cancer-associated fibroblasts. Am J Cancer Res. 2017;7:125–133. [PMC free article] [PubMed] [Google Scholar]
- 60.Liu Q, Li A, Tian Y, Wu JD, Liu Y, Li T, Chen Y, Han X, Wu K. The CXCL8-CXCR1/2 pathways in cancer. Cytokine Growth Factor Rev. 2016;31:61–71. doi: 10.1016/j.cytogfr.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kai Y, Chen QW, Sun YF, Lin JA, Xu JH. Loss of BMI-1 dampens migration and EMT of colorectal cancer in inflammatory microenvironment through TLR4/MD-2/MyD88-mediated NF-κB signaling. J Cell Biochem. 2018;119:1922–1930. doi: 10.1002/jcb.26353. [DOI] [PubMed] [Google Scholar]
- 62.Yang W, Wang JG, Xu J, Zhou D, Ren K, Hou C, Chen L, Liu X. HCRP1 inhibits TGF-β induced epithelial-mesenchymal transition in hepatocellular carcinoma. Int J Oncol. 2017;50:1233–1240. doi: 10.3892/ijo.2017.3903. [DOI] [PubMed] [Google Scholar]
- 63.Xiong S, Xiao GW. Reverting doxorubicin resistance in colon cancer by targeting a key signaling protein, steroid receptor coactivator. Exp Ther Med. 2018;15:3751–3758. doi: 10.3892/etm.2018.5912. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.